Abstract
Measurable residual disease (MRD), defined as the population of cancer cells that persist following therapy, serves as the critical reservoir for disease relapse in acute myeloid leukemia and other malignancies. Understanding the biology enabling MRD clones to resist therapy is necessary to guide the development of more effective curative treatments. Discriminating between residual leukemic clones, preleukemic clones, and normal precursors remains a challenge with current MRD tools. Here, we developed a single-cell MRD (scMRD) assay by combining flow cytometric enrichment of the targeted precursor/blast population with integrated single-cell DNA sequencing and immunophenotyping. Our scMRD assay shows high sensitivity of approximately 0.01%, deconvolutes clonal architecture, and provides clone-specific immunophenotypic data. In summary, our scMRD assay enhances MRD detection and simultaneously illuminates the clonal architecture of clonal hematopoiesis/preleukemic and leukemic cells surviving acute myeloid leukemia therapy.
ScMRD assay integrates mutation and immunophenotype at single cell resolution and distinguishes preleukemic vs. leukemic clones.
INTRODUCTION
Acute myeloid leukemia (AML) is a heterogeneous set of hematologic malignancies characterized by expansion of immature myeloid blasts (1). Although most patients with AML show an initial response to therapy (60 to 80%), relapse remains the fundamental challenge to achieving durable cures (1). Measurable residual disease (MRD) represents a critical, therapy-resistant cancer cell reservoir responsible for disease recurrence (2). Accurately identifying relevant MRD clones is necessary to risk stratify patients and to guide further therapy to prevent overt relapse and achieve durable remissions.
MRD is defined by the presence of residual leukemic cells that are not detectable by conventional morphologic assessment. MRD status has notable prognostic value in AML independent of pretreatment genetics/cytogenetics (2), and the presence of MRD is associated with adverse outcomes in a spectrum of AML therapies—including chemotherapy, venetoclax-based therapies, and allogeneic stem cell transplant (3–6). The most common MRD detection methods use multicolor flow cytometry (MFC) analysis of blasts or real-time quantitative polymerase chain reaction (RT-qPCR) of specific mutations or gene fusions detected at diagnosis. MFC MRD testing relies on abnormal immunophenotypic profiles that may not be readily identifiable on all leukemic cells. The current sensitivity of MFC MRD testing is 0.1%, as defined by European Leukemia Net MRD guidelines (2). In addition, flow cytometry is frequently unable to distinguish between phenotypic abnormalities of regenerating precursors, clonal hematopoiesis (CH), and residual leukemic blasts and therefore cannot fully predict the leukemic potential of an immunophenotypically abnormal population. RT-qPCR is highly sensitive and specific; however, the utility of this method is limited as only a small percentage of patients with AML have a detectable, leukemia-specific mutation or gene fusion that is informative for molecular tracking (7). Moreover, it is not feasible to simultaneously detect/quantitate the spectrum of mutations specific to resistant AML clones.
Recently, bulk-tumor next-generation sequencing (NGS) has emerged as a promising technology to expand on the arsenal of MRD monitoring techniques (7). While the sensitivity of bulk NGS (including error-corrected) and MFC-based MRD testing may be further improved, the specificity of these analyses faces both theoretical and empirical barriers due to the clonal complexity of AML (8).
Despite the utility of these assays, there remains a pressing clinical need for discrimination of residual leukemic cells from the mutated CH/preleukemic cells that do not invariably portend relapse (6, 9). Residual subclones may have different leukemic potentials, while ancestral clones may only result in CH rather than frank AML (10), which cannot be accurately delineated by bulk MRD assays. In addition, bulk NGS MRD assays may detect mutations in mature populations lacking leukemic potential, which is often seen in patients receiving differentiation inducing therapies [e.g. isocitrate dehydrogenase (IDH) 1/2 inhibitors] (11). These biological complexities require a nuanced interpretation of MRD assays, ultimately hampering the clinical utility of bulk-sequencing and MFC-based approaches. With 10 to 30% of MRD-negative and 40 to 70% of MRD-positive patients ultimately relapsing (12), there remains a critical need for a sensitive and specific MRD assay to inform clinical intervention.
Single-cell DNA (scDNA) sequencing technology has recently been used to study clonal architecture in AML, distinguish CH/preleukemic versus leukemic clones, and detect mutations in remission samples (8, 13–15). To build upon these pioneer studies and resolve the challenges associated with both bulk NGS and MFC MRD testing, we have developed a novel multiplex single-cell MRD (scMRD) assay by combining flow cytometric enrichment of the targeted precursor/blast population with integrated scDNA sequencing and immunophenotyping. We further developed a single-nucleotide polymorphism (SNP)–based computational algorithm to deconvolute multiplexed data from samples run in parallel.
RESULTS
Limit of mutation detection with the scMRD assay
We hypothesized that combining precursor/blast enrichment and scDNA + protein sequencing technology would increase the sensitivity of MRD detection and delineate MRD clonal architecture. We reasoned that the MRD clones responsible for relapse primarily reside in immature compartments; thus, we used fluorescence-assisted cell sorting (FACS) to enrich viable CD34+ and/or CD117+ progenitors (including CD34+CD117−, CD34+CD117+, and CD34−CD117+) before loading cells onto the Mission Bio Tapestri single-cell sequencing platform (Fig. 1A). As the antibodies (CD45, CD34, CD117, CD4, and CD8) used for flow sorting were also present in the protein panel, we specifically tested and selected the clones that did not block the binding of the same targets recognized by antibodies present in the protein panel (table S1). The custom scDNA panel contains 109 amplicons covering 31 genes known to be involved in haematologic malignancies (8). To increase assay throughput and reduce cost, we multiplexed samples from different patients into each integrated scDNA + protein run. The results of the multiplexed runs were then computationally deconvoluted and used for both single-sample and aggregated analyses.
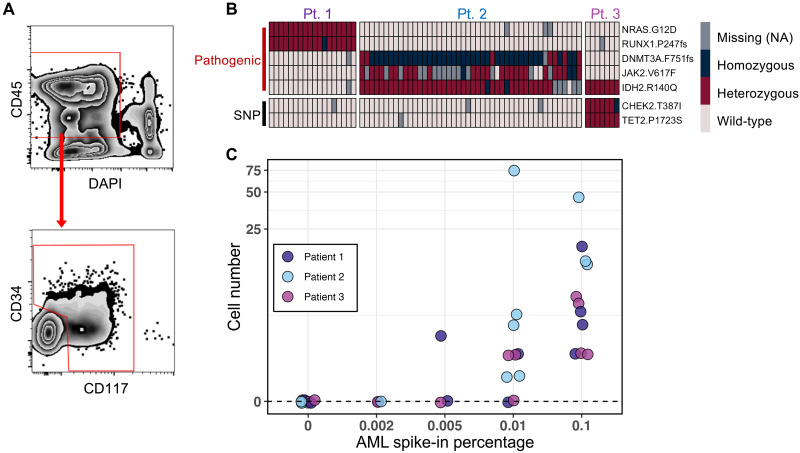
Fig. 1. Limit of mutation detection with the scMRD assay.
(A) Schema of the gating strategy used for flow cytometric enrichment of live CD34+ and/or CD117+ cells after spiking AML cells into normal bone marrow. For clinical MRD samples included in this study, the abnormal blasts were confirmed to be positive for CD34 and/or CD117. (B) Representative heatmap showing mutation calling of spiked-in AML blasts in a limiting dilution experiment testing a sensitivity of 0.1%. Each column represents a single cell. (C) Summary of mutation detection at various sensitivity levels. This plot represents two independent experiments. NA, not applicable.
To evaluate the sensitivity of the scMRD assay, we performed a limiting dilution study in which AML blasts from three genetically distinct AML samples harboring clonal mutations were mixed with 10 million normal bone marrow mononuclear cells before multiplexing to test different sensitivity thresholds (10,000 cells, 0.1%; 1000 cells, 0.01%; 500 cells, 0.005%; 200 cells, 0.002%). Normal bone marrow cells were obtained from patients diagnosed with early-stage mature B or T cell lymphoma that did not have bone marrow involved as assessed by morphology and flow cytometry (table S2). Mutations in AML samples were as follows: patient 1 (AML1), NRAS p.G12D/RUNX1 p.P247fs; patient 2 (AML2), JAK2 p.V617F/IDH2 p.R140Q/DNMT3A p.F751fs; patient 3 (AML3), IDH2 p.R140Q (table S3). FACS-enriched CD34+ and/or CD117+ cells were multiplexed, subjected to the Tapestri microfluidics platform, and sequenced. Expected pathologic mutations were identified in all 11 replicates at a limiting dilution threshold of 0.1% (Fig. 1, B and C, and table S4). Mutations were also identified in 8 of 10 and 1 of 3 replicates at a threshold of 0.01 and 0.005%, respectively, and mutations were not identified in blank controls (0 of 9 replicates) or when present at 0.002% (0 of 3 replicates) (Fig. 1C and table S4). Limiting dilution analysis (16) estimated a sensitivity of 0.0077% [95% confidence interval (CI), 0.004 to 0.0153%] when using a threshold of detecting ≥1 mutated cell (see Methods). Increasing the threshold to detection of ≥3 cells resulted in an estimated sensitivity of 0.0578% (95% CI, 0.03 to 0.111%). These data demonstrate the high sensitivity of mutation detection using the scMRD assay.
Application of the scMRD assay to AML patient samples
We next applied our scMRD assay to 30 cryopreserved postinduction chemotherapy MRD samples obtained from 29 patients with AML (median age, 71 years old; 15 males and 14 females; see table S5). Among these, seven samples (from six patients) were obtained post–allogeneic hematopoietic stem cell transplant (allo-HSCT). The immunophenotype of leukemic blasts at diagnosis and MRD was confirmed to be CD34+ and/or CD117+ for each patient/sample used in our study. MRD was scored as negative in two samples by MFC and in six samples by bulk NGS (table S5). The median percentage of MRD-positive cells by MFC was 0.3% (interquartile range, 0.1 to 0.8%). The median cell number of these samples was 2.6 million (ranging from 0.6 to 14.1 million) with a viability range of 27 to 55%. FACS-enriched CD34+ and/or CD117+ viable cells were multiplexed with up to five unique patient samples per run and processed via the Tapestri platform (50 to 120 thousand cells per run; mean, 77 thousand) (Fig. 2A and table S6).
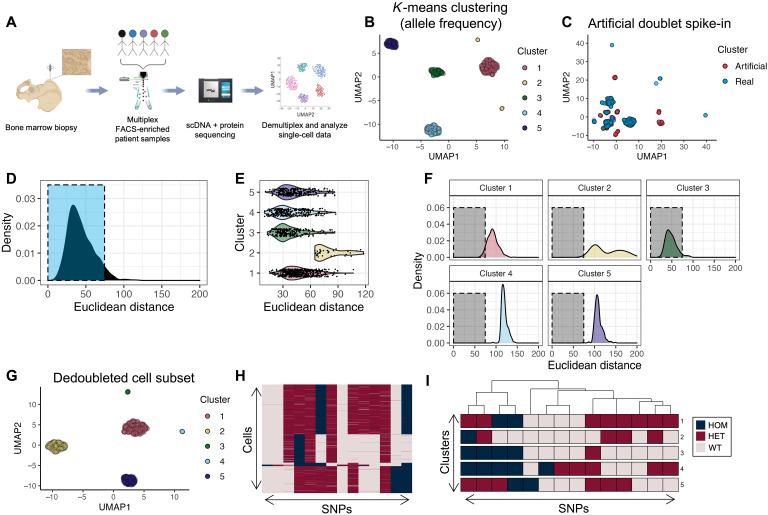
Fig. 2. Workflow and computational demultiplexing of scMRD data.
(A) Schema of scMRD workflow (generated via BioRender). (B) Predoublet exclusion K-means clustering and UMAP analysis of SNP allele frequencies in cells perfectly genotyped for the top 14 SNPs. (C) UMAP plot showing the results of clustering real cells (blue) with artificial doublets (red). (D) Distribution of Euclidean distances from real cells to their respective cluster centers. (E) Violin plot showing by-cluster Euclidean distances of each real cell to the respective cluster center. (F) By-cluster Euclidean distances of each real cell to an artificial doublet cluster center. (G) Postdoublet exclusion K-means clustering and UMAP analysis of SNP allele frequencies. (H) Heatmap showing SNP genotypes in singlet clusters. (I) Heatmap showing the most common SNP profile for each cluster. (B) to (I) show a representative example (MRD5) of the computational pipeline output. UMAP, uniform manifold approximation and projection; HOM, homozygous; HET, heterozygous; WT, wild-type.
Given that unique samples from multiple patients were included in each scMRD run, we developed a computational approach to deconvoluting different individuals in each sequencing run at the single-cell level. Downstream demultiplexing of scMRD sequencing data used germline SNPs covered by the custom scDNA panel (see Methods). We filtered our dataset based on genotyping call rate to include cells with complete genotyping information for the top 10 to 15 SNPs in each run and performed K-means clustering on cells with nonmissing SNP allele frequencies (Fig. 2B). To identify and remove doublets within each scMRD run, we implemented a method to simulate artificial doublet SNP profiles and exclude putative real doublets based on similarity to an artificial cluster center (see Methods). We first randomly sampled our dataset to produce a pool of cells with even representation of each cluster. From this cell pool, we sampled two cells at a time and averaged their SNP allele frequency profiles until we generated 10% doublets relative to the total dataset. After excluding homotypic doublets (i.e., doublets composed of two cells from the same cluster), we reclustered the artificial heterotypic doublets with real cells and applied a Euclidean distance metric to assess the similarity between the SNP profiles of real cells and artificial cluster centers (Fig. 2, C to G). Following doublet detection and exclusion, we then classified additional cells in our dataset according to their germline SNP profile (Fig. 2, H and I). To achieve this, we calculated a Hamming distance between each cell and the most common SNP profile for each cluster. Cells were assigned to clusters based on the available SNP information matching perfectly to one cluster, while being a Hamming distance of ≥3 to every other cluster. For some multiplexed runs, the maximum Hamming distance between clusters was <3, in which case we reduced the classification filter to accommodate this. Cells from each cluster were then queried for any pathogenic mutations identified by bulk NGS at diagnosis, remission, or relapse. This approach enabled us to deconvolute multiplexed scMRD runs and assign sequenced cells to the specific patient from which they were derived without the need to leverage patient-specific somatic mutation information. Using this approach, we classified an average of 2074 sorted cells per run (1215 to 2777 cells, SD = 585.8), reflecting an average of 36.4% of total cells in each run (11.5 to 74.4%; table S6). Demultiplexing enabled assignment of hotspot mutations (i.e., DNMT3A p.R882H) present in multiple samples within the same multiplex (Fig. 3).
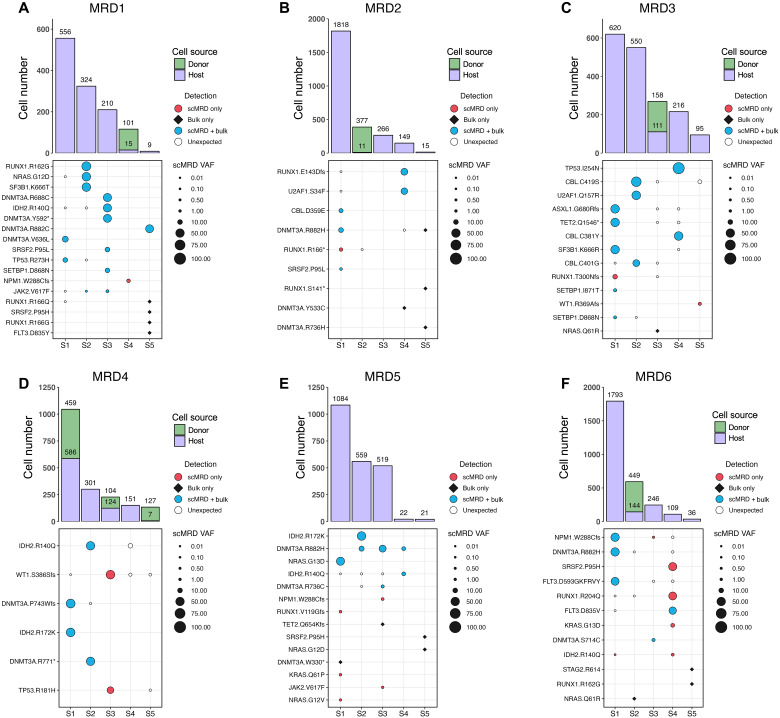
Fig. 3. Deconvolution plots for individual scMRD runs.
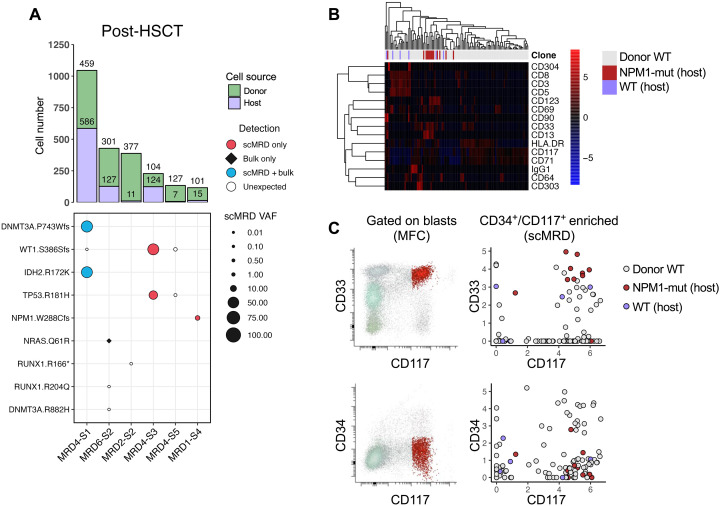
(A to F) Computationally recovered cell number per sample (top) and VAF of mutations detected by scMRD, bulk NGS, or both assays (bottom). Each plot represents a single multiplexed run, and each column corresponds to a unique patient sample. “Unexpected” detection denotes mutations detected by scMRD but not reported at any bulk NGS time point (diagnosis, remission, and relapse). For MRD2, we did not detect mutations in two samples (MRD2-S3 and MRD2-S5) and, thus, could not confirm the identity of these samples. However, these samples had a considerable difference in cell input for sequencing and cell recovery after deconvolution (see table S5). We reasoned that the sample with higher cell number input corresponded to the sample with much higher recovery. MRD1-S4, MRD2-S2, MRD3-S3, MRD4-S1/S3/S5, and MRD6-S2 were post–allo-HSCT. MRD3-S3 and MRD6-S2 were from the same patient but obtained at different time points.
To assess the misclassification rate of our demultiplexing algorithm, we designed a synthetic multiplexing control experiment in which the ground-truth identity of each cell was known (fig. S1). To this end, we sequenced five additional AML patient samples (unsorted and nonmultiplexed) via the Tapestri platform. After determining the clonal architecture for these samples, we isolated a single clone from each sample and merged these barcodes to generate a synthetic dataset composed of 15,542 cells from the five samples. To better represent real scMRD data, we simulated heterotypic doublets and injected them into the dataset at a rate of 7.23% before subjecting the data to our demultiplexing algorithm. Within the synthetic multiplex, we identified nine germline SNPs that were sequenced in each of the five runs and had sufficiently divergent genotypes between the samples. As described for real scMRD data, a subset of cells with complete genotyping information for these nine SNPs (fig. S1A) was subjected to K-means clustering, which partitioned the samples into five clusters (fig. S1, B and C). Demulitplexing enabled resolution of each SNP profile after doublet removal (fig. S1, D and E), and subsequent cell classification based on Hamming distance filters described in the methods (fig. S1F). We sought to assess how the rates of cell classification and misclassification were influenced by missing SNP information in perfect cells (i.e., cells with complete genotyping for all nine SNPs). Our demultiplexing algorithm classified 87.98% of perfect cells (fig. S1G) with a misclassification rate of 0.035% (fig. S1H). We then randomly sampled one to eight SNPs over 50 replicates and calculated the mean percentage of perfect cells classified and misclassified, which revealed that the loss of SNP information results in a lower classification rate with minimal effect on cell misclassification (fig. S1, G and H). Demultiplexing of all possible cells in this control experiment, including those with missing genotypes, resulted in classification of 49.07% of cells with a misclassification rate of 0.235% (fig. S1I).
Overall, the results for MRD status and mutation presence were concordant between bulk NGS and scMRD in 23 of 30 (76.7%) samples and for 44 of 76 (57.9%) mutations, respectively (fig. S2A). Among the mutations detected by both scMRD and bulk NGS, variant allelic frequencies (VAFs) were significantly higher by scMRD (P = 0.011, paired Wilcoxon signed-rank test) (fig. S2B). Among the 33 discordant mutations covered by both scMRD and bulk NGS panels, scMRD identified 17 mutations that were missed/unreported by bulk NGS, including RUNX1 (n = 4), NPM1 (n = 3), KRAS (n = 2), IDH2 (n = 2), WT1 (n = 2), NRAS, JAK2, TP53, and SRSF2 mutations (Fig. 3), 15 (88.2%) of which were associated with and present at relapse. Conversely, there were 16 mutations that were detected by bulk NGS but missed by scMRD, including DNMT3A (n = 4), RUNX1 (n = 4), NRAS (n = 3), SRSF2 (n = 2), STAG2, TET2, and FLT3TKD mutations (Fig. 3). Only 4 of 16 (25% versus 88.2%, P = 0.0004, Fisher’s exact test) were present at relapse. There were four MRD samples with eight mutations (two RUNX1, two DNMT3A, two NRAS, SRSF2, and STAG2) not detected by scMRD. Although these samples had slightly lower numbers of starting viable cells and computationally recovered cells compared to others [starting viable cells per sample, median: 1.5 million versus 2.6 million (P = 0.08); computationally recovered cells per sample, median: 64 versus 413 (P = 0.05); Mann-Whitney test], the cause may be multifactorial including that specific mutations (i.e., NRAS) may reside in mature/differentiated compartments not sampled with our scMRD assay (8).
Resolution of clonal architecture and clone-specific immunophenotype
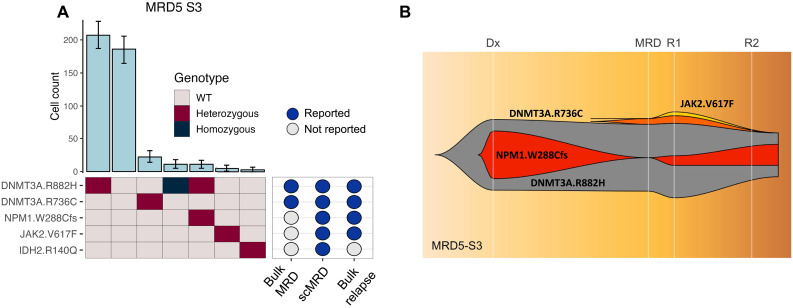
We next assessed the ability of scMRD profiling to differentiate MRD based on clonal architecture, including discrimination between leukemic clones and single-mutant clones that harbor CH/preleukemic mutations. We found that scMRD readily deconvolved CH/preleukemic versus leukemic clonal architecture in demultiplexed samples (Fig. 4A and fig. S3). In sample MRD5-S3, bulk NGS detected DNMT3A p.R882H, DNMT3A p.R736C, and TET2 p.Q654Kfs (not covered by scMRD panel) mutations at the remission time point, while scMRD detected both DNMT3A mutations in distinct clones, with one subclone harboring co-occuring DNMT3A p.R882H/NPM1 p.W288Cfs (NPM1 scVAF = 1.31%) mutations and another with a JAK2 p.V617F (JAK2 scVAF = 0.69%) mutation. Bulk NGS at the time of subsequent relapse (time point R1) revealed the presence of both the NPM1 p.W288Cfs (VAF = 5%) and JAK2 p.V617F (VAF = 2%) mutations (Fig. 4B). Bulk profiling at a second time point during relapse (R2) revealed further expansion of the NPM1 p.W288Cfs mutation (VAF = 11%), while the JAK2 mutation was not detected at this subsequent time point. These data demonstrate that scMRD may enable resolution of residual preleukemic clones (i.e., DNMT3A alone), bystander clones (i.e., JAK2 clone), and leukemic clones (comutant DNMT3A/NPM1) that persist at relapse.
Fig. 4. Clonal analysis of MRD5-S3.
(A) Clonal barplot of a patient (MRD5-S3) illustrating scMRD-specific detection of NPM1 and JAK2 mutations that were present at relapse. (B) Fish plot showing clonal evolution based on bulk NGS at diagnosis, remission (6 month after diagnosis), relapse time point 1 (R1; 7 months after diagnosis), and relapse time point 2 (R2; 11 months after diagnosis).
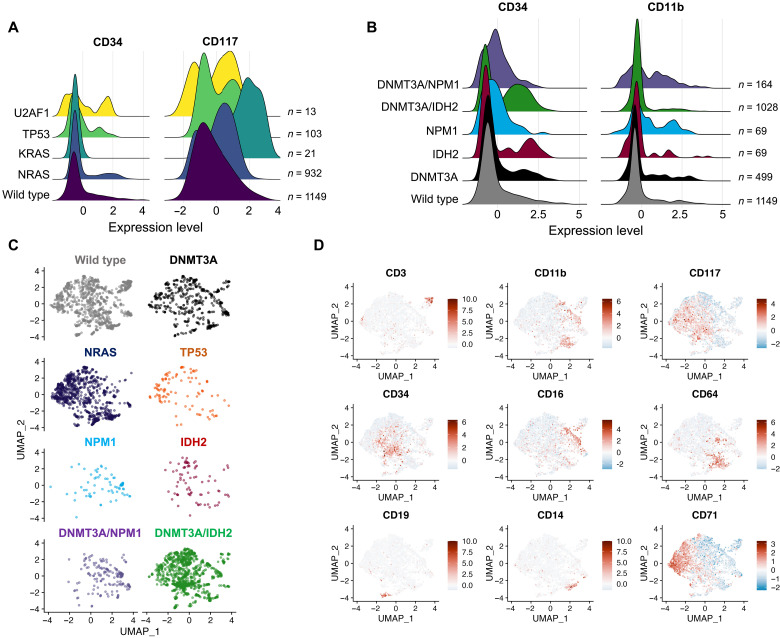
Integration of scMRD immunophenotypic analysis enabled identification of mutation and clone-specific expression of key cell surface proteins (Fig. 5, A and B and fig. S4, A to C). Compared to wild-type clones, NRAS-mutant clones had decreased expression of CD34 [log2 fold change (log2FC) = −0.772, P < 8.2 × 10−16] and CD117 (log2FC = −0.542, P < 3.8 × 10−16), while U2AF1-mutant clones showed increased expression of CD34 (log2FC = 2.03, P < 0.012). In contrast, NPM1 clones displayed higher CD11b (log2FC = 0.381, P < 7.7 × 10−8) and CD13 expression (log2FC = 1.43, P < 1.38 × 10−14) compared to DNMT3A clones. This distinct immunophenotype is well described in patients with NPM1-mutated AML (14). TP53-mutant clones had increased CD71 expression (log2FC = 1.66, P < 4.4 × 10−5) (16). We observed highly similar immunophenotypes between wild-type and DNMT3A-mutant cells, suggesting that CH clones (at least with DNMT3A mutation) may not have overtly aberrant surface protein expression (Fig. 5, C and D). Differential immunophenotypic states were identified when comparing CH/preleukemic and leukemic clones within and between patients (Fig. 5, B to D, and fig. S4, B and C). Comutant DNMT3A/IDH2 or DNMT3A/NPM1 cells showed consistently aberrant immunophenotypes, with DNMT3A/IDH2 cells characterized by increased CD34 (log2FC = 0.461, P < 9.6 × 10−8, versus DNMT3A) and CD117 (log2FC = 1.13, P < 1.7 × 10−29, versus DNMT3A) expression (Fig. 5, C and D, and fig. S4, B and C) and DNMT3A/NPM1-comutated cells showed more highly expressed granulocytic/monocytic markers such as CD11b (log2FC = 0.513, P < 3.2 × 10−21) and CD16 (log2FC = 0.436, P < 1.1 × 10−34) (Fig. 5, C and D). These data highlight that integrated genomic/immunophenotypic analysis at the MRD time point has the potential to distinguish between CH/preleukemic and leukemic clones that portend a substantively higher likelihood of relapse.
Fig. 5. Clone- and mutation-specific immunophenotype.
(A) Clone-specific immunophenotype. (B) Differential surface marker expression between CH/preleukemic versus leukemic clones. (C and D) UMAP analysis of immunophenotypes of single-mutant versus compound-mutant clones. Distribution of cells in UMAP space with cells colored by genotype (C) and UMAP depiction of cell immunophenotype including markers for T cells (CD3), myeloid cells (CD11b and CD33), stem/progenitors (CD117 and CD34), monocytes (CD14 and CD64), granulocytes (CD16), B cells (CD19), and erythroid cells (CD71) (D). Data are centered log ratio–normalized, centered, and scaled on a by-run basis.
Identification of donor and host cell origin in post–allo-HSCT samples
Within our cohort, the seven samples (from six patients) obtained post–allo-HSCT represent an admixture of donor and recipient cells. Germline SNP-based deconvolution identified distinct nonmutant clusters consistent with donor origin, which were confirmed as donor samples by matching the SNP profile of paired pretransplant samples. Donor-host pairs were successfully recovered for all seven samples (Figs. 3, A to F, and 6A). Chimerism was calculated on the basis of recovered host versus donor cells and correlated with the available results by bulk short tandem repeat genotyping on unsorted bone marrow samples. The levels of host cells detected by scMRD assay were higher than those by short tandem repeat testing in four relapsed patients (median of difference, 19.7%; P = 0.1, Wilcoxon matched-pairs signed-rank test; P = 0.02, paired t test), suggesting that host cells show enrichment in immature compartments and represent an early indicator of relapse. Integrated immunophenotypic analysis of post–allo-HSCT samples showed distinct cell surface protein expression between donor and host cells. Analysis of sample MRD1-S4 revealed clear separation of donor and NPM1-mutant host cells, with the former containing a subset of spiked-in CD3+CD8+ T cells and the latter displaying aberrant increased expression of CD33 (log2FC = 2.77, P < 1.64 × 10−2) and CD13 (log2FC = 3.9, P < 1.43 × 10−7) (Fig. 6B). In addition, expression of the T cell activation marker, CD69, was observed not only in a subset of donor and host T cells but also unexpectedly in host leukemic cells, consistent with previous studies showing that CD69 may be expressed in leukemic stem cells and, thus, may represent a surface marker for MRD detection (17). Abnormal immunophenotype of these host leukemic blasts identified by MFC was also detectable by scMRD, with elevated coexpression of CD33 and CD117 on NPM1-mutant cells and characteristically low levels of CD34 (Fig. 6C).
Fig. 6. scDNA + protein analysis enables simultaneous identification of donor cells and MRD.
(A) Aggregated deconvolution plot showing mutations detected and host-donor chimerism of post–allo-HSCT samples from six different patients included in the study. MRD6-S2 and MRD3-S3 were from the same patients; therefore, only the former is shown. MRD2-S2 had no mutations detected by either bulk NGS or scMRD assay. MRD4-S3 had an HDAC1 p.P243L mutation not covered by the scMRD panel. Refer to Fig. 4 for multiplex context for each of these samples. (B) Row-scaled heatmap of differential surface maker expression between donor and host cells in MRD1-S4. (C) Concordance of immunophenotype of MRD cells between MFC and scMRD in MRD1-S4. Number of cells analyzed for (B) and (C): donor WT = 97, Host WT = 6, and Host NPM1-mut = 13.
DISCUSSION
Our study illustrates the feasibility of single-cell genotypic and immunophenotypic profiling at the remission time point to enumerate and delineate MRD through blast enrichment and scDNA + protein technology. Our data demonstrate that scMRD profiling readily resolves clonal architecture and has the potential to distinguish between single-mutant CH/preleukemic versus leukemic clones with multiple co-occurring mutations. As CH/preleukemic clones do not invariably portend relapse (6, 9), this distinction may be critical for clinical decision-making. The integration of mutation and immunophenotypic information further enhances MRD detection by identifying genotype-specific protein expression patterns. This can be potentially used to isolate relevant clones for functional validation and studying MRD biology and therapeutic vulnerabilities. Given the increased use of molecular/cell surface–targeting therapeutic modalities for patients with AML patients (18, 19), assessing expression of surface markers in relevant MRD clones with defined mutational repertoires may provide further guidance for treatment. Future integration of scDNA + protein sequencing with clone-level transcriptional analysis will provide additional insights into the molecular pathways mediating leukemic cell persistence following different AML therapies.
The multiplexing approach implemented in this study is reliable and cost effective; however, more work is needed to optimize this assay. While scMRD detected many mutations missed by bulk NGS, some mutations were also missed by scMRD. There are several explanations: First, the starting cell numbers of these samples with mutations missed by scMRD assay were slightly lower, leading to lower computational cell recovery compared to clinical profiling performed on fresh samples. In clinical practice, fresh samples with higher cell numbers should be used for scMRD testing, which will certainly improve cell recovery and sensitivity. In addition, surface marker expression may change over time during sample storage, which can limit the ability to call specific immunophentypic subsets. Second, our use of FACS for blast enrichment overall can alter mutation allele burden compared to bulk sequencing, with some mutations being enriched in immature hematopoietic stem/progenitor cell (HSPC) compartments and others enriched in differentiated clonal progeny. For example, previous studies have shown that NRAS-mutated clones express high levels of CD11b, a marker often indicative of more differentiated myeloid cells (neutrophils and monocytes) (8). Consistent with this notion, a subset of NRAS mutations were missed by the scMRD assay. Notably, only a small portion of mutations missed by scMRD were associated with relapse. High-purity depletion of mature compartments, as in our flow enrichment, may theoretically decrease the burden of these mutations, including those CH mutations present in mature myeloid progeny with reduced/absent leukemic potential. Enrichment for CD34+ and/or CD117+ cells may not capture MRD cells in a minority of AML patients, particularly those with the highest level of monocytic differentiation (20). In clinical practice, magnetic bead depletion of nonleukemic compartments (CD3+ T cells, CD20+ B cells, and CD16+ granulocytes) followed by scMRD assessment will allow for more accurate detection and delineation of MRD in a more cost-effective manner and can serve as an adjunct to bulk NGS performed before therapy and at relapse.
Future applications of this approach should also seek to further optimize computational demultiplexing to maximize the rate of cell classification while minimizing misclassification. Notably, the synthetic multiplexing control experiment estimated a low misclassification rate of 0.235%. While this is encouraging, in practice, multiplexed single-cell profiling may have higher misclassification rates. Combining our SNP demultiplexing method with an orthogonal technique such as cell hashing may provide the best approach to excluding doublets and to accurately assigning cells to specific patients. Allele dropout remains an additional limitation of scDNA sequencing technology, and, to our knowledge, there is currently no gold standard approach to correcting for it. In the setting of scMRD demultiplexing, allele dropout may affect SNP genotypes and contribute to higher misclassification rates. Inclusion of a larger amplicon panel, and using more than 10 to 15 SNPs to classify cells/patients, may help to minimize the deleterious effects of allele dropout at any given SNP locus. Panel development that accounts for linkage disequilibrium may represent a viable path forward to counter allele dropout (21). As scDNA + protein technology becomes more affordable and widespread, we anticipate rapid improvements in experimental design, application, and algorithmic support.
Another limitation of scMRD profiling is related to the relatively small number of cells recovered for sequencing. Only ~3 to 5% of the cell input into Tapestri was recovered after all processing steps, including computational analysis. This translates to an average recovery of 200 to 500 cells per sample at the MRD time point. Traditionally, a cluster of 20 abnormal cells is deemed adequate for MFC MRD calling. Although cell loss is likely stochastic on the basis of our data, additional studies are needed to demonstrate that the mutations and clonal architecture derived from such a small sample of cells are representative of the bulk tumor. A direct comparison of sensitivity and specificity between MFC, bulk NGS, and scMRD assays is not straightforward as the following: (i) Enriched blasts from banked specimens of suboptimal quality were used for scMRD in our study, and (ii) the amplification methods are different between scMRD (amplicon) and bulk NGS (hybrid capture). Despite our limiting dilution experiments revealing a sensitivity of approximately 0.01% for the scMRD assay, a prospective study is needed to compare the clinical utility of scMRD to other MRD assays such as MFC, droplet digital PCR, and/or error-corrected NGS. As scDNA profiling improves, including with respect to cell recovery for single-cell mutational analysis, we will be able to further enrich our understanding of MRD in AML and other diseases at a single-cell, clonal level.
In conclusion, our scMRD assay not only enables sensitive MRD detection but also achieves sufficient resolution to characterize the clonal architecture of preleukemic/leukemic cells that persist after therapy, which may ultimately increase the specificity of MRD results. Larger studies are needed to comprehensively elucidate the clonal architecture of MRD and study the dynamics of clonal evolution by comparing clonal architecture and immunophenotype at diagnosis and relapse. Our study paves the road for delineating the genetic and phenotypic properties of high-risk MRD clones and better understanding the molecular underpinnings of MRD in AML.
METHODS
Patient samples
Bone marrow aspirates were received in the clinical laboratory at Memorial Sloan Kettering Cancer Center (MSKCC). After 5 days with all necessary clinical tests being completed, the leftover cells were deemed as medical waste, and mononuclear cells were obtained by centrifugation on Ficoll from bone marrow and viably frozen. Corresponding MFC MRD was obtained from diagnostic pathology reports with a reporting sensitivity of 0.1%. Uninvolved (both morphologically and immunophenotypically) bone marrow aspirates from patients with an early-stage mature B or T cell lymphoma were used as normal controls. AML patient samples underwent high-throughput genetic sequencing with an Food and Drug Administration–approved hybrid capture–based targeted deep sequencing assay of 500 genes (IMPACT-heme) or by an NGS platform panel composed of 49 genes that are recurrently mutated in myeloid disorders (RainDance Technologies ThunderBolts Myeloid Panel) with a reporting VAF cutoff at 2%. Informed consent was obtained from patients according to protocols approved by the Institutional Review Boards and in accordance with the Declaration of Helsinki. This study was approved by MSKCC Institutional Review Board (#12-245, #13-037, and #16-1591).
Cell enrichment
Patient samples were thawed, washed with FACS buffer, and quantified using a Countess cell counter. Cells (0.5 × 106 to 4.0 × 106 viable cells) were then resuspended in cell staining buffer (#420201, BioLegend) and incubated with TruStain FcX and 1× Tapestri blocking buffer for 15 min on ice. Cells were incubated with antihuman CD4 (clone: OKT4)–Allophycocyanin (APC)/Cy7 (dilution, 1:30), antihuman CD8 (clone: RPA-T8)–BV711 (dilution, 1:30), antihuman CD34 (clone: Qbend)–APC (dilution, 1:10), antihuman CD117 (clone: A3C6E2)–Phycoerythrin (dilution, 1:75), and antihuman CD45 (clone: Q17A19)–Alexa Flour 488 (dilution, 1:30) for 15 min on ice (table S1). Then, TotalSeq-D Human Heme Oncology Cocktail, V1.0 (# 399906, BioLegend) containing the pool of 45 oligo-conjugated antibodies was added and incubated for an additional 30 min on ice. Cells were then washed three times with cell staining buffer (#420201, BioLegend), followed by resuspension of the cells in 4′,6-diamidino-2-phenylindole (DAPI) containing FACS buffer. DAPI-negative and CD45-positive viable cells were gated. After exclusion of CD4- and CD8-positive lymphocytes, CD34+/CD117−, CD34+/CD117+, and CD34−/CD117+ populations were combined for sorting using a SH800S cell sorter. In MRD1 run, 1000 sorted CD4- versus CD8-positive T cells from two individual samples were spiked in, respectively.
scDNA and protein library preparation and sequencing
Enriched cells were resuspended in Tapestri cell buffer and quantified using a Countess cell counter (Invitrogen). Single cells (1000 to 3000 cells/μl) were encapsulated using a Tapestri microfluidics cartridge and lysed. A forward primer mix (30 μM each) for the antibody tags was added before barcoding. Barcoded samples were then subjected to targeted PCR amplification of a custom 109 amplicons covering 31 genes known to be involved in AML. DNA PCR products were then isolated from individual droplets and purified with Ampure XP beads. The DNA PCR products were then used as a PCR template for library generation as above and repurified using Ampure XP beads. Protein PCR products (supernatant from Ampure XP bead incubation) were incubated with Tapestri pullout oligo (5 μM) at 96°C for 5 min, followed by incubation on ice for 5 min. Protein PCR products were then purified using streptavidin C1 beads (Invitrogen), and beads were used as a PCR template for the incorporation of i5/i7 Illumina indices, followed by purification using Ampure XP beads. All libraries, both DNA and protein, were quantified using an Agilent Bioanalyzer and pooled for sequencing on an Illumina NovaSeq by the MSKCC Integrated Genomics Core.
Data processing and variant filtering
FASTQ files from scDNA + protein samples were processed via the TapestriV2 pipeline as described previously (8). This analytics platform trims adaptor sequences, aligns sequencing reads to the hg19 reference genome, calls cells based on completeness of amplicon sequencing reads for each barcode, and calls variants using GATKv3.7 best practices. After pipeline processing, data for each run were aggregated into H5 files, which were downloaded and read into R using the rhdf5 package. Downstream processing was conducted using custom scripts in R, which is available at https://github.com/RobinsonTroy/scMRD. Low-quality variants and cells were then excluded based on filtering cutoffs for genotype quality score (<30), read depth (<10) alternate allele frequency (<20%), and presence in <0.1% of cells. The details of scMRD computational demultiplexing and scMRD protein analysis are included in the Supplementary Materials.
Limit of detection study analysis
For each multiplexed AML spike-in run, the numerical genotype matrix was extracted from each respective H5 file in R. Each of the three AMLs harbored >1 pathogenic mutation, except for patient 3, which contained a single IDH2 p.R140Q mutation. To increase the confidence in accurate cell calling, two additional heterozygous germline SNPs (CHEK2 p.T387I and TET2 p.P1723S) were identified as private to patient 3. The curated list of known variants included mutations/SNPs present in patient 1 (NRAS p.G12D and RUNX1 p.P247fs), patient 2 (DNMT3A p.F751fs, JAK2 p.V617F, and IDH2 p.R140Q), and patient 3 (IDH2 p.R140Q, CHEK2 p.T387I, and TET2 p.P1723S). After filtering, all cells were queried for variants included in this list. Cells harboring the expected variants were then filtered on the basis of the requirement that real cells must contain at least two pathogenic mutations (patients 1 and 2) or one pathogenic mutation and two SNPs (patient 3). Limiting dilution analysis was conducted using the Extreme Limiting Dilution Analysis software (22), where the AML spike-in cell number was treated as “dose,” and the number of replicates in which the leukemic fraction was detected was treated as “repsonse.” Output of the analysis provided an estimated sensitivity with an associated confidence interval.
scMRD computational demultiplexing
Deconvolution of multiplexed scMRD runs was reliant on the presence of germline SNPs. Suspected SNPs were verified via referencing the Ensembl SNP database through the BioMart R package and were tallied for nonmissing genotyping information within the filtered numerical genotype matrix.
To maximize variability in SNP genotype between patients in each multiplex, the candidate SNP list was filtered to include SNPs with a minor allele frequency of <0.5. An additional filter was applied to only include SNPs that had divergent genotypes (wild-type or heterozygous, wild-type or homozygous, and heterozygous or homozygous) in a set number of cells, which was defined by
This calculation was based on the reasoning that the fraction of cells that must be divergent in genotype for demultiplexing is proportional to the size of the dataset and the number of samples included in the multiplex. On the basis of an estimated false positive rate of 10−3 for the Tapestri platform, the product of these two values is adjusted accordingly. After filtering, the top 20 SNPs with the lowest percentage of missing genotypes were selected. Principal components analysis was then run on cells with complete genotyping information for these 20 SNPs, and the top n SNPs that explained a proportion of variance ≥0.99 were selected for downstream analysis. K-means clustering was performed on a subset of cells with complete SNP genotypes, where the number of clusters for partitioning was set equal to the number of unique patient samples in a given multiplex. Doublet identification and exclusion were conducted by first evenly sampling cells from all clusters to form a pool of cells with equal representation of each cluster. Artificial doublets were then generated via sampling the cell pool two cells at random and averaging the SNP profiles until the proportion of artificial doublets approached 10% of the total number of cells in the dataset. Homotypic doublets (i.e., those created by within-cluster mixing) were excluded, and the resulting dataset of heterotypic doublets was merged with real cells and reclustered to produce real and artificial cluster centers. Euclidean distances were then measured between each real cell and (i) its respective cluster and (ii) the artificial cluster center. The distribution of distances between 95% of cells to their respective clusters was used as a cutoff to exclude cells that were within this distance to the artificial cluster center. This process was repeated 10 times, with random replacement of NA values with allele frequencies of 0, 50, or 100, and cells were excluded if their distance was within the doublet gate in all replicates. After removing doublets and low-quality cells with high similarity to artificial doublets, the most common SNP profile was tallied for each cluster. To classify additional cells, Hamming distance was calculated between all cells and each SNP profile, without penalizing SNPs with missing genotypes. Cells were assigned to clusters based on perfect matching between the available cell SNP genotypes and the SNP profile of one cluster, while being a Hamming distance of ≥3 from every other cluster. For SNP profile comparisons that had a maximum Hamming distance of <3, this filter was reduced accordingly. After cell classification, each cluster was quieried for pathogenic mutations detected by bulk NGS at the diagnosis, remission, and relapse (if applicable) time points, and the cell number per cluster was tallied. For post–allo-HSCT samples, we referenced bulk NGS data (when available) from both donor and host samples used in this study to identify which scMRD populations were of donor origin.
Synthetic multiplexing
As input for the synthetic multiplexing experiment, five AML patient samples (labeled samples A to E) were sequenced via the TapestriV2 scDNA + protein platform independently (unsorted and nonmultiplexed). Each sample was processed separately to assess clonal architecture, and a single-mutant clone from each sample was extracted. Clones from each sample were merged into a dataset with a total of 15,542 cells (16,754 cells with heterotypic doublets added). Computational demultiplexing was performed as described in the “scMRD computational demultiplexing” section on the synthetic dataset containing simulated doublets. Nine germline SNPs with sufficiently divergent genotypes across the five samples were used as input into the algorithm. To assess our algorithm’s performance on classifiying perfect cells, we randomly sampled one to eight SNPs for 50 replicates and calculated the mean classification and misclassification rate for each sampling. After removing duplicate samplings, SDs were calculated for the percent of cells classified or misclassified for each number of available SNPs.
Single-cell protein analysis
For each demultiplexed sample that underwent both scDNA + protein sequencing, single-cell protein data were extracted from H5 files as raw counts. With the exception of the five samples in MRD2, which were not subjected to single-cell protein sequencing, all other samples (n = 25) were used for aggregate protein analysis. Each demultiplexed patient sample was analyzed independently for clonality of mutations and clone-specific immunophenotype. For samples with detected mutations, the protein count matrices were filtered for cells classified into high-confidence clones (≥3 cells) and were used for subsequent aggregate analysis. Protein counts for each run were converted to a Seurat object using the Seurat R package. Count data were then normalized using a centered log ratio transformation to normalize for differences in antibody-derived tag sequencing depth across cells. Normalized protein data from each run were merged, then scaled, and centered on a by-run basis. To minimize technical variability driven by background signal, we called the “vars.to.regress” argument set to the included isotype controls [immunoglobulin G1 (IgG1), IgG2a, and IgG2b] in Seurat’s “ScaleData” function. Clone and mutation information was supplied as metadata and used for downstream aggregate analysis using functions within Seurat. Differential expression of surface markers was assessed using a Wilcoxon rank sum test in Seurat’s “FindMarkers” function. P values denote adjusted P values.
Plotting and graphical representation
All bar plots and scatter plots were generated using the ggplot2 package in R. All heatmaps were generated using the pheatmap R package, except for the heatmap in fig. S1, which was generated using the Superheat R package. The OncoPrint shown in fig. S2 was produced using the Complex Heatmap package in R. The uniform manifold approximation and projection (UMAP) plots, density plots, and violin plots, in Fig. 5 and figs. S4 and S5, were generated using the Seurat R package. The radar plot displayed in fig. 4 was produced with the fmsb package in R. The fish plot in Fig. 4B was generated using the fishplot R package.
Acknowledgments
Funding: This study was funded by the Center for Hematologic Malignancies at MSKCC and in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. T.M.R. is supported by the American Society for Hematology Graduate Hematology Award. R.L.B is supported by a National Cancer Institute grant (R00-CA-248460-03). L.A.M. is supported by a National Cancer Institute grant (K99 CA252002-01A1). S.F.C. is supported by a National Cancer Institute grant (K08 CA241371-01A1). M.R. is supported by an intramural grant from Department of Pathology at MSK. R.L.L. is supported by a Cycle For Survival Innovation Grant and National Cancer Institute R35 CA197594. W.X. is supported by Alex’s Lemonade Stand Foundation and the Runx1 Research Program, a Cycle for Survival’s Equinox Innovation Award in Rare Cancers, MSK Leukemia SPORE (Career Enhancement Program, NIH/NCI P50 CA254838-01), and a National Cancer Institute grant (K08CA267058-01).
Author contributions: W.X. selected, annotated and processed samples, and prepared scDNA + protein libraries. T.M.R. performed computational analysis. R.L.B. and A.S. helped oversee computational analysis. S.P. helped processing the normal bone marrow samples. Y.L. helped annotate bulk NGS sequencing data. Q.G., J.Z., X.S., and C.F. provided clinical samples. R.N., L.A.M., and S.F.C. helped scDNA + protein library preparation. A.L., A.G., and A.D. helped design the limit of detection (LOD) study. M.R., R.L.L., and W.X. designed and supervised the entire study. T.M.R., R.L.L., and W.X. wrote and all authors approved the manuscript.
Competing interests: L.A.M. has received honoraria from Mission Bio and serves on the Mission Bio Speakers Bureau. S.F.C. is a consultant for and holds equity interest in Imago Biosciences, none of which are directly related to the content of this paper. A.S. and A.L. hold employment at Mission Bio. A.G. received research funding from Celularity, ADC Therapeutics, Aprea, AROG, Pfizer, Prelude, and Trillium; received research funding from and served as a consultant for Aptose and Daiichi Sankyo; served as a consultant and member of advisory committees for Astellas, Celgene, and Genentech; received research funding from, served as a consultant for, and was a member of advisory committees for AbbVie; and received honoraria from Dava Oncology. A.D. served as a consultant for Incyte, EUSA Pharma, and Loxo and receives research support from Roche and Takeda. M.R. is on the scientific advisory board in Auron Pharmaceutical for which he received equity support. He receives research funding from Celularity, Roche-Genentech, Beat AML, and NGM and travel fund from BD Biosciences. R.L.L. is on the supervisory board of QIAGEN and is a scientific advisor to Imago, Mission Bio, Syndax. Zentalis, Ajax, Bakx, Auron, Prelude, C4 Therapeutics, and Isoplexis for which he receives equity support. R.L.L. receives research support from Ajax and Abbvie and has consulted for Incyte, Janssen, Morphosys, and Novartis. He has received honoraria from Astra Zeneca and Kura for invited lectures and from Gilead for grant reviews. W.X. has received research support from Stemline Therapeutics. The authors declare that they have no other competing interests.
Data and materials availability: All scDNA + protein sequencing data used in this analysis are available to researchers for purposes of reproducing or extending the analysis via the dbGaP accession number phs003233. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and are included in the dbGAPaccession. The code used for this study have been deposited in a permanent Zenodo repository and are accessible at https://doi.org/10.5281/zenodo.8169057.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.H. Döhner, A. H. Wei, F. R. Appelbaum, C. Craddock, C. D. DiNardo, H. Dombret, B. L. Ebert, P. Fenaux, L. A. Godley, R. P. Hasserjian, R. A. Larson, R. L. Levine, Y. Miyazaki, D. Niederwieser, G. Ossenkoppele, C. Röllig, J. Sierra, E. M. Stein, M. S. Tallman, H.-F. Tien, J. Wang, A. Wierzbowska, B. Löwenberg, Diagnosis and management of AML in adults: 2022 ELN recommendations from an international expert panel. Blood 140, 2022, 1345–1377 [DOI] [PubMed] [Google Scholar]
- 2.M. Heuser, S. D. Freeman, G. J. Ossenkoppele, F. Buccisano, C. S. Hourigan, L. L. Ngai, J. M. Tettero, C. Bachas, C. Baer, M. C. Béné, V. Bücklein, A. Czyz, B. Denys, R. Dillon, M. Feuring-Buske, M. L. Guzman, T. Haferlach, L. Han, J. K. Herzig, J. L. Jorgensen, W. Kern, M. Y. Konopleva, F. Lacombe, M. Libura, A. Majchrzak, L. Maurillo, Y. Ofran, J. Philippe, A. Plesa, C. Preudhomme, F. Ravandi, C. Roumier, M. Subklewe, F. Thol, A. A. van de Loosdrecht, B. A. van der Reijden, A. Venditti, A. Wierzbowska, P. J. M. Valk, B. L. Wood, R. B. Walter, C. Thiede, K. Döhner, G. J. Roboz, J. Cloos, 2021 update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 138, 2753–2767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Terwijn, W. L. J. van Putten, A. Kelder, V. H. J. van der Velden, R. A. Brooimans, T. Pabst, J. Maertens, N. Boeckx, G. E. de Greef, P. J. M. Valk, F. W. M. B. Preijers, P. C. Huijgens, A. M. Dräger, U. Schanz, M. Jongen-Lavrecic, B. J. Biemond, J. R. Passweg, M. van Gelder, P. Wijermans, C. Graux, M. Bargetzi, M. C. Legdeur, J. Kuball, O. de Weerdt, Y. Chalandon, U. Hess, L. F. Verdonck, J. W. Gratama, Y. J. M. Oussoren, W. J. Scholten, J. Slomp, A. N. Snel, M. C. Vekemans, B. Löwenberg, G. J. Ossenkoppele, G. J. Schuurhuis, High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: Data from the HOVON/SAKK AML 42A study. J. Clin. Oncol. 31, 3889–3897 (2013). [DOI] [PubMed] [Google Scholar]
- 4.D. Araki, B. L. Wood, M. Othus, J. P. Radich, A. B. Halpern, Y. Zhou, M. Mielcarek, E. H. Estey, F. R. Appelbaum, R. B. Walter, Allogeneic hematopoietic cell transplantation for acute myeloid leukemia: Time to move toward a minimal residual disease-based definition of complete remission? J. Clin. Oncol. 34, 329–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. S. Hourigan, L. W. Dillon, G. Gui, B. R. Logan, M. Fei, J. Ghannam, Y. Li, A. Licon, E. P. Alyea, A. Bashey, H. J. Deeg, S. M. Devine, H. F. Fernandez, S. Giralt, M. Hamadani, A. Howard, R. T. Maziarz, D. L. Porter, B. L. Scott, E. D. Warlick, M. C. Pasquini, M. E. Horwitz, Impact of conditioning intensity of allogeneic transplantation for acute myeloid leukemia with genomic evidence of residual disease. J. Clin. Oncol. 38, 1273–1283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. W. Pratz, B. A. Jonas, V. Pullarkat, C. Recher, A. C. Schuh, M. J. Thirman, J. S. Garcia, C. D. DiNardo, V. Vorobyev, N. S. Fracchiolla, S. P. Yeh, J. H. Jang, M. Ozcan, K. Yamamoto, A. Illes, Y. Zhou, M. Dail, B. Chyla, J. Potluri, H. Döhner, Measurable residual disease response and prognosis in treatment-naïve acute myeloid leukemia with venetoclax and azacitidine. J. Clin. Oncol. 40, 855–865 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Jongen-Lavrencic, T. Grob, D. Hanekamp, F. G. Kavelaars, A. al Hinai, A. Zeilemaker, C. A. J. Erpelinck-Verschueren, P. L. Gradowska, R. Meijer, J. Cloos, B. J. Biemond, C. Graux, M. van Marwijk Kooy, M. G. Manz, T. Pabst, J. R. Passweg, V. Havelange, G. J. Ossenkoppele, M. A. Sanders, G. J. Schuurhuis, B. Löwenberg, P. J. M. Valk, Molecular minimal residual disease in acute myeloid leukemia. N. Engl. J. Med. 378, 1189–1199 (2018). [DOI] [PubMed] [Google Scholar]
- 8.L. A. Miles, R. L. Bowman, T. R. Merlinsky, I. S. Csete, A. T. Ooi, R. Durruthy-Durruthy, M. Bowman, C. Famulare, M. A. Patel, P. Mendez, C. Ainali, B. Demaree, C. L. Delley, A. R. Abate, M. Manivannan, S. Sahu, A. D. Goldberg, K. L. Bolton, A. Zehir, R. Rampal, M. P. Carroll, S. E. Meyer, A. D. Viny, R. L. Levine, Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587, 477–482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.X. Chen, B. L. Wood, Monitoring minimal residual disease in acute leukemia: Technical challenges and interpretive complexities. Blood Rev. 31, 63–75 (2017). [DOI] [PubMed] [Google Scholar]
- 10.S. Loghavi, C. D. DiNardo, K. Furudate, K. Takahashi, T. Tanaka, N. J. Short, T. Kadia, M. Konopleva, R. Kanagal-Shamanna, N. R. Farnoud, S. Pierce, J. D. Khoury, J. L. Jorgensen, K. P. Patel, N. Daver, M. Yilmaz, L. J. Medeiros, H. Kantarjian, F. Ravandi, S. A. Wang, Flow cytometric immunophenotypic alterations of persistent clonal haematopoiesis in remission bone marrows of patients with NPM1-mutated acute myeloid leukaemia. Br. J. Haematol. 192, 1054–1063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.E. M. Stein, C. D. DiNardo, D. A. Pollyea, A. T. Fathi, G. J. Roboz, J. K. Altman, R. M. Stone, D. J. DeAngelo, R. L. Levine, I. W. Flinn, H. M. Kantarjian, R. Collins, M. R. Patel, A. E. Frankel, A. Stein, M. A. Sekeres, R. T. Swords, B. C. Medeiros, C. Willekens, P. Vyas, A. Tosolini, Q. Xu, R. D. Knight, K. E. Yen, S. Agresta, S. de Botton, M. S. Tallman, Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. S. Hourigan, R. P. Gale, N. J. Gormley, G. J. Ossenkoppele, R. B. Walter, Measurable residual disease testing in acute myeloid leukaemia. Leukemia 31, 1482–1490 (2017). [DOI] [PubMed] [Google Scholar]
- 13.L. W. Dillon, J. Ghannam, C. Nosiri, G. Gui, M. Goswami, K. R. Calvo, K. E. Lindblad, K. A. Oetjen, M. D. Wilkerson, A. R. Soltis, G. Sukumar, C. L. Dalgard, J. Thompson, J. Valdez, C. B. DeStefano, C. Lai, A. Sciambi, R. Durruthy-Durruthy, A. Llanso, S. Gulati, S. Wang, A. Ooi, P. K. Dagur, J. P. McCoy, P. Burr, Y. Li, C. S. Hourigan, Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from non-malignant clonal hematopoiesis. Blood Cancer Discov. 2, 319–325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Ediriwickrema, A. Aleshin, J. G. Reiter, M. R. Corces, T. Köhnke, M. Stafford, M. Liedtke, B. C. Medeiros, R. Majeti, Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 4, 943–952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.K. Morita, F. Wang, K. Jahn, T. Hu, T. Tanaka, Y. Sasaki, J. Kuipers, S. Loghavi, S. A. Wang, Y. Yan, K. Furudate, J. Matthews, L. Little, C. Gumbs, J. Zhang, X. Song, E. Thompson, K. P. Patel, C. E. Bueso-Ramos, C. D. DiNardo, F. Ravandi, E. Jabbour, M. Andreeff, J. Cortes, K. Bhalla, G. Garcia-Manero, H. Kantarjian, M. Konopleva, D. Nakada, N. Navin, N. Beerenwinkel, P. A. Futreal, K. Takahashi, Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat. Commun. 11, 5327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Rodriguez-Meira, R. Norfo, W. X. Wen, A. L. Chédeville, H. Rahman, J. O’Sullivan, G. Wang, E. Louka, W. W. Kretzschmar, A. Paterson, C. Brierley, J.-E. Martin, C. Demeule, M. Bashton, N. Sousos, A. Hamblin, H. Guermouche, F. Pasquier, C. Marzac, F. Girodon, M. Drummond, C. Harrison, I. Plo, S. E. W. Jacobsen, B. Psaila, S. Thongjuea, I. Antony-Debré, A. J. Mead, Deciphering TP53 mutant cancer evolution with single-cell multi-omics. bioRxiv 10.1101/2022.03.28.485984 [Preprint] (2022). 10.1101/2022.03.28.485984. [DOI]
- 17.K. Sachs, A. L. Sarver, K. E. Noble-Orcutt, R. S. LaRue, M. L. Antony, D. Chang, Y. Lee, C. M. Navis, A. L. Hillesheim, I. R. Nykaza, N. A. Ha, C. J. Hansen, F. K. Karadag, R. J. Bergerson, M. R. Verneris, M. M. Meredith, M. L. Schomaker, M. A. Linden, C. L. Myers, D. A. Largaespada, Z. Sachs, Single-cell gene expression analyses reveal distinct self-renewing and proliferating subsets in the leukemia stem cell compartment in acute myeloid leukemia. Cancer Res. 80, 458–470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. B. Leick, H. Silva, I. Scarfò, R. Larson, B. D. Choi, A. A. Bouffard, K. Gallagher, A. Schmidts, S. R. Bailey, M. C. Kann, M. Jan, M. Wehrli, K. Grauwet, N. Horick, M. J. Frigault, M. V. Maus, Non-cleavable hinge enhances avidity and expansion of CAR-T cells for acute myeloid leukemia. Cancer Cell 40, 494–508.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.F. Perna, S. H. Berman, R. K. Soni, J. Mansilla-Soto, J. Eyquem, M. Hamieh, R. C. Hendrickson, C. W. Brennan, M. Sadelain, Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 32, 506–519.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Pei, A. E. Gillen, I. T. Shelton, B. M. Stevens, M. Gasparetto, K. Engel, S. Staggs, Y. Wang, W. Showers, A. Inguva, M. L. Amaya, M. Minhajuddin, A. Winters, S. B. Patel, H. Tolison, A. Krug, T. N. Young, J. Schowinsky, C. M. Mahon, C. A. Smith, D. A. Pollyea, C. T. Jordan, A novel type of monocytic leukemia stem cell revealed by the clinical use of venetoclax-based therapy. bioRxiv 10.1101/2022.12.04.519036 [Preprint] (2022). 10.1101/2022.12.04.519036. [DOI]
- 21.N. Javed, Y. Farjoun, T. J. Fennell, C. B. Epstein, B. E. Bernstein, N. Shoresh, Detecting sample swaps in diverse NGS data types using linkage disequilibrium. Nat. Commun. 11, 3697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Hu, G. K. Smyth, ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S6
Tables S1 to S6