Key Points
Question
Do rare pathogenic variants in genes beyond previously established prostate cancer risk genes contribute to risk of aggressive prostate cancer?
Findings
In this exome-sequencing genetic association study of 17 546 men with aggressive and nonaggressive prostate cancer, an association between known genes BRCA2, ATM, and NBN with aggressive prostate cancer was found. Nominal association evidence was observed for MSH2, XRCC2, and MRE11A.
Meaning
The findings of this study suggest that DNA repair and cancer susceptibility genes can inform disease management in men with nonaggressive prostate cancer, as men carrying deleterious variants in these genes are likely to develop advanced disease.
Abstract
Importance
Germline gene panel testing is recommended for men with advanced prostate cancer (PCa) or a family history of cancer. While evidence is limited for some genes currently included in panel testing, gene panels are also likely to be incomplete and missing genes that influence PCa risk and aggressive disease.
Objective
To identify genes associated with aggressive PCa.
Design, Setting, and Participants
A 2-stage exome sequencing case-only genetic association study was conducted including men of European ancestry from 18 international studies. Data analysis was performed from January 2021 to March 2023. Participants were 9185 men with aggressive PCa (including 6033 who died of PCa and 2397 with confirmed metastasis) and 8361 men with nonaggressive PCa.
Exposure
Sequencing data were evaluated exome-wide and in a focused investigation of 29 DNA repair pathway and cancer susceptibility genes, many of which are included on gene panels.
Main Outcomes and Measures
The primary study outcomes were aggressive (category T4 or both T3 and Gleason score ≥8 tumors, metastatic PCa, or PCa death) vs nonaggressive PCa (category T1 or T2 and Gleason score ≤6 tumors without known recurrence), and metastatic vs nonaggressive PCa.
Results
A total of 17 546 men of European ancestry were included in the analyses; mean (SD) age at diagnosis was 65.1 (9.2) years in patients with aggressive PCa and 63.7 (8.0) years in those with nonaggressive disease. The strongest evidence of association with aggressive or metastatic PCa was noted for rare deleterious variants in known PCa risk genes BRCA2 and ATM (P ≤ 1.9 × 10−6), followed by NBN (P = 1.7 × 10−4). This study found nominal evidence (P < .05) of association with rare deleterious variants in MSH2, XRCC2, and MRE11A. Five other genes had evidence of greater risk (OR≥2) but carrier frequency differences between aggressive and nonaggressive PCa were not statistically significant: TP53, RAD51D, BARD1, GEN1, and SLX4. Deleterious variants in these 11 candidate genes were carried by 2.3% of patients with nonaggressive, 5.6% with aggressive, and 7.0% with metastatic PCa.
Conclusions and Relevance
The findings of this study provide further support for DNA repair and cancer susceptibility genes to better inform disease management in men with PCa and for extending testing to men with nonaggressive disease, as men carrying deleterious alleles in these genes are likely to develop more advanced disease.
This genetic association study examines the association between variants of established prostate cancer risk genes and the risk of aggressive prostate cancer.
Introduction
Germline gene panel testing for prostate cancer (PCa) is recommended for men with advanced PCa to guide treatment and disease management decisions1,2 and for unaffected men with a cancer family history to inform screening decisions and whether to undergo active surveillance once diagnosed.3 Commercially available PCa gene panels for clinical risk assessment include a small number of DNA repair and cancer-related genes (eg, BRCA2, BRCA1, ATM, CHEK2, and HOXB13), with pathogenic variants more common in men with metastatic than low-stage cancer.4,5,6 Additional genes have been suggested for expanded gene panels, although most lack statistical evidence of an association with aggressive disease, as previous PCa sequencing studies have been underpowered due to small sample sizes and the rarity of pathogenic variants.4,5,6,7,8,9 The importance of other forms of coding variation in panel genes, including nonsynonymous variants of uncertain significance (VUS) have also not been extensively examined in PCa. Consequently, our understanding of the genes and spectrum of genetic variation contributing to aggressive PCa susceptibility remains limited. Large-scale genome and candidate panel gene investigations are required to more accurately define aggressive PCa genes and inform guidelines for genetic testing and genetically driven approaches to surveillance and disease management.
Our objective in this study was to investigate the association between rare deleterious variants and VUS across the genome and in candidate genes, particularly DNA repair genes, with clinically significant PCa risk. This investigation was conducted within a large, multistage exome sequencing study of 8361 nonaggressive and 9185 aggressive PCa cases among men of European ancestry, including 2397 cases with distant metastases. Analyses were limited to men of European ancestry due to the availability of data.
Methods
Participants
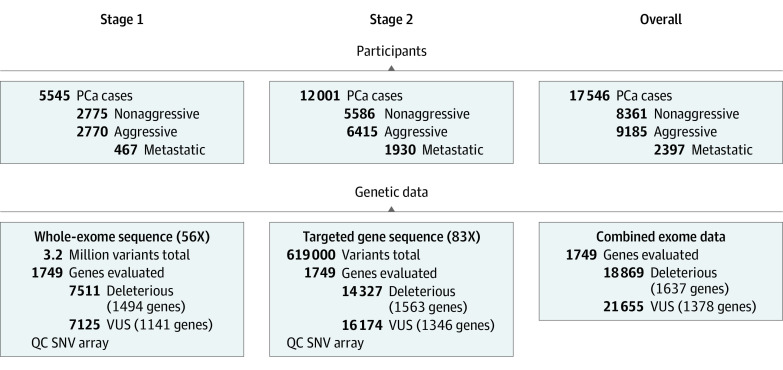
This multistage study included men with aggressive and nonaggressive PCa from 18 epidemiologic studies in Australia, the US, the UK, Finland, Sweden, and other European countries4 (Figure 1; eTable 1 in Supplement 1); details regarding study design, recruitment, and data sources for each study are provided in the eMethods in Supplement 2. Data analysis was performed from January 2021 to March 2023. Aggressive cases involved men who either had category T4 disease or both category T3 and Gleason score greater than or equal to 8 tumors at initial diagnosis, had de novo or metachronous metastatic PCa, or died from PCa, similar to the definition of aggressive PCa suggested by Hurwitz et al.10 Nonaggressive cases involved men with localized disease (category T1/T2) and Gleason score less than or equal to 6 tumors (46.8% of patients with nonaggressive disease had ≥10 years of follow-up, indicating that they were alive and without recurrence). Staging was pathologic when available and clinical otherwise (eMethods in Supplement 2). Grading was based on radical prostatectomy for men who underwent surgery and biopsy otherwise. All participants provided informed consent, and study protocols were approved by respective institutional review boards.
Figure 1. Participants With Prostate Cancer (PCa) and Germline Genetic Sequencing Data in Stage 1 and Stage 2.
Additional sequencing details can be found in the eMethods in Supplement 2 and eTable 2 in Supplement 1. QC indicates quality control; SNV, single-nucleotide variant; VUS, variants of uncertain significance.
Germline Sequencing and Bioinformatic Analysis
Germline DNA sequencing was conducted in 2 stages (Figure 1). In stage 1, whole-exome sequencing data were generated in 5545 patients (Agilent SureSelect Human All Exon v6+UTRs capture kit and the Illumina HiSeq 2500 sequencing platform, with 56X mean targeted exon coverage).4 In stage 2, targeted sequencing of 1749 genes was conducted in 12 001 additional patients (custom probe design and the NovaSeq 6000 sequencing platform, with 83X mean targeted exon coverage) (eMethods in Supplement 2). Genes selected in stage 2 had prior association evidence with either PCa or cancer in general or association evidence with aggressive PCa in stage 1 (eTable 2 in Supplement 1). Quality control and variant calling procedures are described in the eMethods in Supplement 2 and previously.4
Rare variants (minor allele frequency [MAF]<0.01) with a Variant Effect Predictor (VEP) Impact score of high,11 representing variants with a high likelihood of having deleterious (eg, protein truncating or splice altering) functional consequences, or identified as pathogenic or likely pathogenic in the ClinVar database12 were classified as deleterious (eTable 3 in Supplement 1). Rare (MAF<0.01) missense variants, excluding those classified as deleterious, that change 1 or more bases in the amino acid sequence but preserve length11 and had a Rare Exome Variant Ensemble Learner (REVEL) score greater than 0.6, indicating a high likelihood of pathogenicity,13 were classified as VUS.
Statistical Analysis
Single-variant and gene-based analyses were performed for aggressive vs nonaggressive PCa and metastatic vs nonaggressive PCa using logistic regression. As a secondary outcome, we evaluated death due to PCa vs nonaggressive PCa. Gene-based analyses were performed by aggregating variants across a gene to compare carrier frequencies between case groups. Analyses included covariates for study, age at PCa diagnosis, and 10 principal components of ancestry (eMethods in Supplement 2). Stage 1 and 2 analyses were performed separately and meta-analyzed in fixed-effect inverse variance–weighted models. Exome-wide single-variant analyses used a P < 5 × 10−7 significance threshold, and exome-wide gene-based analyses used Bonferroni-adjusted P value thresholds (P < .05/number of genes tested) for each outcome separately to account for multiple testing (P values in the Results section are unadjusted).
We also evaluated 2 focused candidate PCa gene sets: 29 DNA repair and cancer susceptibility genes included in PCa-specific gene panels as previously described3 or with previous association evidence with PCa or disease aggressiveness3,4,5,14,15,16,17,18,19 and an extended set of an additional 167 DNA repair genes20,21,22,23 (eTable 4 in Supplement 1), given the strong evidence of the involvement of DNA repair genes in PCa risk.4,5,6,14,16,21 In candidate gene-based analyses, genes were considered to have nominal association evidence with an unadjusted P < .05 for either primary outcome (aggressive or metastatic PCa). Genes not meeting this criterion but with an odds ratio (OR) greater than or equal to 2 for either outcome and more than 10 carriers were considered to have large effects. With our sample size, we had approximately 80% power to detect associations for genes with OR greater than or equal to 2 and carrier frequencies greater than or equal to 0.60%. Tests of statistical significance were 2-sided. Findings were further investigated for associations with age at PCa diagnosis (using linear regression models) and first-degree PCa family history (available for 69.6% [n = 12 208] of participants), with additional analyses stratified by country (eMethods in Supplement 2). Analyses were performed for deleterious variants and VUS and aggregated across candidate genes. Sensitivity analyses were performed excluding nonaggressive cases with a prostate-specific antigen (PSA) level greater than or equal to 20 ng/mL (to convert to micrograms per liter, multiply by 1) at diagnosis (PSA available for 82.7% [n = 6917] of men with nonaggressive PCa) to examine whether this impacted the results.
Results
Participants
Analyses included 17 546 men of European ancestry (Table 1; eTable 1 in Supplement 1). Of 9185 men with aggressive PCa, 65.7% (n = 6033) died of PCa, 26.1% (n = 2397) had confirmed metastatic disease, 50.7% (n = 4656) had Gleason score greater than or equal to 8 tumors, and 59.0% (n = 5415) had category T3 or T4 disease (Table 1 indicates missing information for each variable). Of patients who died of PCa, 21.6% (n = 1306) had category T1 or T2 disease and Gleason score less than 8 tumors at diagnosis. Most patients with aggressive PCa were older at diagnosis than those with nonaggressive PCa (mean [SD] age, 65.1 [9.2] vs 63.7 [8.0] years).
Table 1. Clinical Characteristics of Study Participants by Study Stage.
| Characteristic | No. (%) | ||
|---|---|---|---|
| Stage 1 | Stage 2 | Stage 1 and 2 | |
| Total patients | 5545 | 12 001 | 17 546 |
| Aggressive PCa | |||
| Patients | 2770 (50.0) | 6415 (53.5) | 9185 (52.3) |
| Age at diagnosis, mean (SD), y | 66.1 (8.8) | 64.7 (9.3) | 65.1 (9.2) |
| First-degree PCa family history | |||
| Yes | 331 (11.9) | 925 (14.4) | 1256 (13.7) |
| No | 1776 (64.1) | 3080 (48.0) | 4856 (52.9) |
| Missing | 663 (23.9) | 2410 (37.6) | 3073 (33.4) |
| Death due to PCa | |||
| Yes | 2052 (74.1) | 3981 (62.1) | 6033 (65.7) |
| No | 311 (11.2) | 369 (5.8) | 680 (7.4) |
| Missing | 407 (14.7) | 2065 (32.2) | 2472 (26.9) |
| Metastatic disease at diagnosis or follow-up | |||
| Yes | 467 (16.9) | 1930 (30.1) | 2397 (26.1) |
| No | 663 (23.9) | 1849 (28.8) | 2512 (27.3) |
| Missing | 1640 (59.2) | 2636 (41.1) | 4276 (46.6) |
| Category | |||
| T1 | 410 (14.8) | 1045 (16.3) | 1455 (15.8) |
| T2 | 367 (13.2) | 946 (14.7) | 1313 (14.3) |
| T3 | 1277 (46.1) | 2035 (31.7) | 3312 (36.1) |
| T4 | 654 (23.6) | 1449 (22.6) | 2103 (22.9) |
| Missing | 62 (2.2) | 940 (14.7) | 1002 (10.9) |
| Gleason score at diagnosis | |||
| ≤6 | 197 (7.1) | 923 (14.4) | 1120 (12.2) |
| 7 | 490 (17.7) | 1311 (20.4) | 1801 (19.6) |
| 8-10 | 1862 (67.2) | 2794 (43.6) | 4656 (50.7) |
| Missing | 221 (8.0) | 1387 (21.6) | 1608 (17.5) |
| PSA at diagnosis, median (IQR), ng/mL | 30.9 (10.0-105.0) | 18.0 (7.5-66.0) | 20.4 (8.0-77.0) |
| Missing | 661 (23.9) | 1184 (18.5) | 1845 (20.1) |
| Nonaggressive PCa | |||
| Patients | 2775 (50.0) | 5586 (46.5) | 8361 (47.7) |
| Age at diagnosis, mean (SD), y | 67.5 (7.0) | 61.8 (7.7) | 63.7 (8.0) |
| First-degree PCa family history | |||
| Yes | 467 (16.8) | 1003 (18.0) | 1470 (17.6) |
| No | 1816 (65.4) | 2810 (50.3) | 4626 (55.3) |
| Missing | 492 (17.7) | 1773 (31.7) | 2265 (27.1) |
| Category | |||
| T1 | 2383 (85.9) | 4110 (73.6) | 6493 (77.7) |
| T2 | 392 (14.1) | 1476 (26.4) | 1868 (22.3) |
| Gleason Score ≤6 at diagnosis | 2773 (99.9) | 5586 (100) | 8359 (99.9) |
| Missing | 2 (0.07) | 0 (0) | 2 (0.02) |
| PSA at diagnosis, median (IQR), ng/mL | 7.1 (5.0-10.4) | 5.9 (4.3-8.4) | 6.2 (4.4-9.1) |
| Missing | 659 (23.7) | 785 (14.1) | 1444 (17.3) |
| Follow-up years, median (IQR) | 11.4 (10.0-14.0) | 8.5 (4.2-12.0) | 10.0 (6.0-13.0) |
| Missing | 260 (9.4) | 84 (1.5) | 344 (4.1) |
Abbreviations: PCa, prostate cancer; PSA, prostate-specific antigen.
Exome-Wide Analysis
Individual Variants
Of 72 025 variants identified in 1749 genes tested either within or meta-analyzed across stages 1 and 2, only missense variant Ile179Thr in KLK3 on chromosome 19q13.3, a previously reported PSA locus,24 was exome-wide significant (rs17632542, c.536T>C; aggressive MAF, 7.8%; nonaggressive MAF, 4.7%; OR, 1.72; 95% CI, 1.56-1.89; P = 1.0 × 10−28) (eTable 5 and eTable 6 in Supplement 1, eFigure 1, eFigure 2, and eFigure 3 in Supplement 2).
Gene-Based
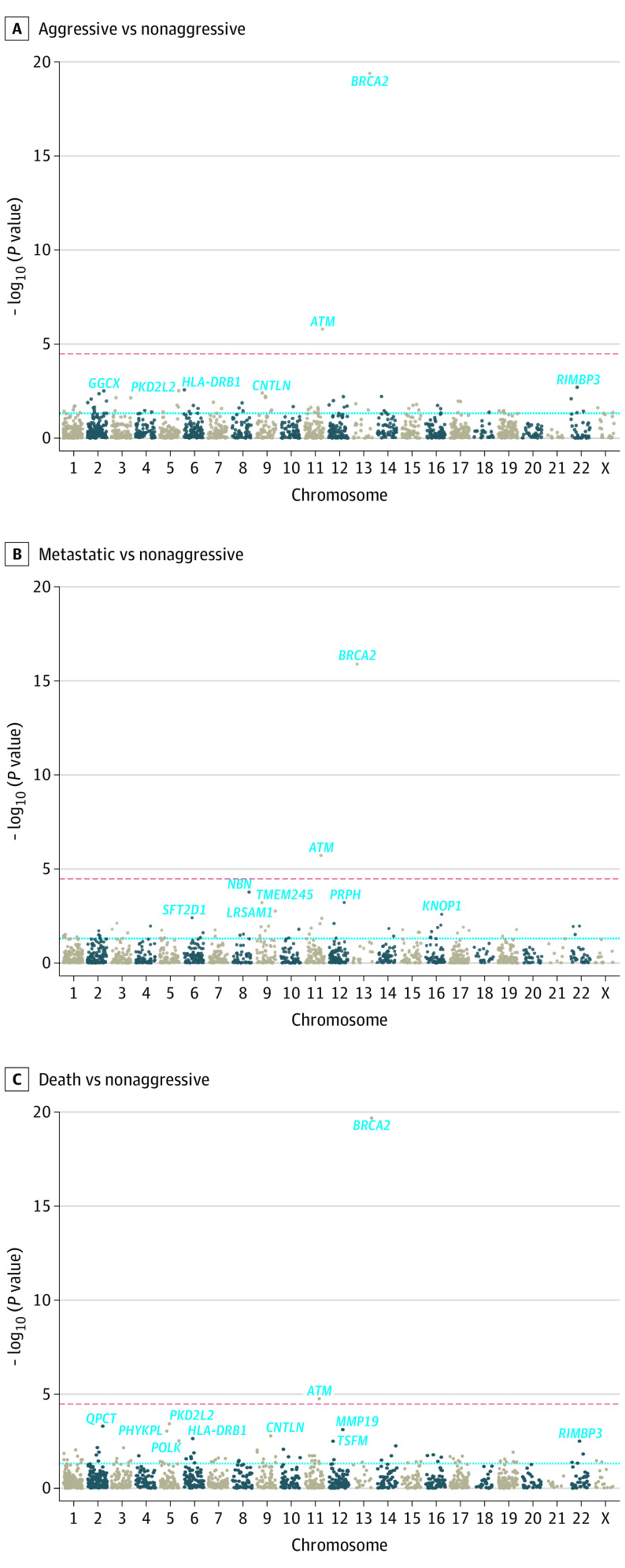
In gene-based analyses of deleterious variants, BRCA2 and ATM were exome-wide significant (Table 2, Figure 2; eTable 7, eTable 8, and eTable 9 in Supplement 1). The carrier frequency of deleterious BRCA2 alleles was significantly higher in aggressive cases (2.4%; OR, 4.29; 95% CI, 3.15-5.86; P = 4.0 × 10−20) and metastatic cases (3.0%; OR, 5.61; 95% CI, 3.73-8.44; P = 1.3 × 10−16) compared with nonaggressive cases (0.7%). Significantly higher carrier frequencies were also observed for deleterious ATM alleles in aggressive cases (1.6%; OR, 2.17; 95% CI, 1.58-2.99; P = 1.6 × 10−6) and metastatic cases (1.9%; OR, 2.80; 95% CI, 1.83-4.29; P = 1.9 × 10−6) compared with nonaggressive cases (0.7%). Although not exome-wide significant, higher carrier frequencies were also observed for deleterious NBN alleles in metastatic cases (0.5%; OR, 5.16; 95% CI, 2.19-12.12; P = 1.7 × 10−4) compared with nonaggressive cases (0.2%). The effect of NBN was similar when excluding the Slavic founder variant c.657del5 (rs587776650)25 (OR, 5.93; 95% CI, 1.82-19.33; P = .003) (eTable 10 in Supplement 1). Results for men who died of PCa were similar to results for those with aggressive PCa (Figure 2; eTable 8 in Supplement 1). Results were largely unchanged when excluding patients with nonaggressive PCa with a PSA level greater than 20 ng/mL at diagnosis in sensitivity analyses (eFigure 4 in Supplement 2).
Table 2. Risk of Aggressive PCa Associated With Rare Deleterious Variants in Candidate PCa Genes.
| Chromosome | Gene | Men carrying deleterious variants, No. | Aggressive vs nonaggressive | Metastatic vs nonaggressive | ||||
|---|---|---|---|---|---|---|---|---|
| Nonaggressive (n = 8361) | Aggressive (n = 9185) | Metastatic (n = 2397) | OR (95% CI) | P value | OR (95% CI) | P value | ||
| PCa panel genes (n = 16) | ||||||||
| 13 | BRCA2 | 55 | 222 | 72 | 4.29 (3.15-5.86) | 4.0 × 10−20a | 5.61 (3.73-8.44) | 1.3 × 10−16a |
| 11 | ATM | 60 | 142 | 45 | 2.17 (1.58-2.99) | 1.6 × 10−06a | 2.80 (1.83-4.29) | 1.9 × 10−06a |
| 2 | MSH2 | 7 | 20 | 4 | 3.27 (1.29-8.31) | .01a | 2.45 (0.64-9.44) | .19 |
| 22 | CHEK2 | 107 | 147 | 38 | 1.30 (1.00-1.69) | .05 | 1.35 (0.90-2.02) | .15 |
| 8 | NBN | 13 | 29 | 13 | 1.99 (0.98-4.05) | .06 | 5.16 (2.19-12.12) | 1.7 × 10−04a |
| 3 | MLH1 | 28 | 37 | 8 | 1.47 (0.88-2.48) | .14 | 1.83 (0.77-4.31) | .17 |
| 16 | PALB2 | 15 | 35 | 10 | 1.57 (0.80-3.06) | .19 | 1.82 (0.74-4.47) | .19 |
| 17 | HOXB13 | 173 | 145 | 30 | 0.87 (0.69-1.10) | .24 | 0.81 (0.53-1.22) | .31 |
| 2 | EPCAM | 6 | 4 | 0 | 0.49 (0.13-1.90) | .30 | NA | .93 |
| 17 | TP53 | 9 | 21 | 6 | 1.47 (0.57-3.81) | .43 | 3.35 (0.70-16.02) | .13 |
| 17 | BRCA1 | 25 | 28 | 10 | 1.24 (0.70-2.22) | .46 | 1.65 (0.75-3.62) | .21 |
| 17 | RAD51D | 3 | 8 | 2 | 1.67 (0.41-6.78) | .47 | 2.06 (0.31-13.86) | .46 |
| 17 | BRIP1 | 18 | 14 | 5 | 0.76 (0.35-1.65) | .49 | 1.33 (0.47-3.80) | .59 |
| 2 | MSH6 | 46 | 51 | 13 | 1.06 (0.70-1.62) | .77 | 1.04 (0.54-2.01) | .91 |
| 17 | RAD51C | 6 | 8 | 1 | 1.09 (0.32-3.69) | .89 | 0.46 (0.04-4.78) | .52 |
| 7 | PMS2 | 14 | 17 | 5 | 0.96 (0.45-2.06) | .92 | 1.06 (0.35-3.22) | .92 |
| PCa candidate DNA repair genes (n = 13) | ||||||||
| 7 | XRCC2 | 2 | 8 | 2 | 6.24 (1.24-31.4) | .03a | 8.05 (0.69-94.34) | .10 |
| 2 | BARD1 | 10 | 18 | 5 | 1.57 (0.70-3.53) | .28 | 3.03 (0.90-10.21) | .07 |
| 16 | SLX4 | 12 | 20 | 7 | 1.39 (0.65-2.96) | .40 | 2.40 (0.82-6.98) | .11 |
| 2 | GEN1 | 17 | 14 | 5 | 0.72 (0.30-1.70) | .45 | 2.10 (0.58-7.68) | .26 |
| 8 | WRN | 43 | 54 | 14 | 1.17 (0.77-1.80) | .46 | 1.04 (0.53-2.03) | .91 |
| 19 | ERCC2 | 28 | 37 | 9 | 1.14 (0.68-1.91) | .62 | 1.38 (0.61-3.11) | .44 |
| 3 | XPC | 16 | 17 | 6 | 1.17 (0.57-2.42) | .67 | 1.32 (0.48-3.67) | .59 |
| 1 | MUTYH | 141 | 164 | 50 | 1.04 (0.82-1.33) | .74 | 1.17 (0.82-1.67) | .37 |
| 11 | MRE11A | 11 | 12 | 6 | 0.87 (0.37-2.06) | .75 | 4.58 (1.36-15.39) | .01a |
| 4 | FAM175A | 9 | 11 | 2 | 0.96 (0.37-2.45) | .93 | 1.74 (0.30-10.24) | .54 |
| 3 | BAP1 | 0 | 1 | 0 | NA | .95 | NA | NA |
| 3 | ATR | 53 | 56 | 15 | 0.99 (0.66-1.49) | .96 | 1.58 (0.76-3.28) | .22 |
| 2 | FANCL | 87 | 93 | 26 | 1.01 (0.74-1.38) | .97 | 1.01 (0.63-1.63) | .97 |
| Potentially novel PCa DNA repair genes (n = 8) | ||||||||
| 5 | POLK | 48 | 85 | 20 | 1.58 (1.08-2.30) | .02a | 1.77 (1.00-3.15) | .05 |
| 6 | POLH | 1 | 11 | 3 | 12.12 (1.50-98.18) | .02a | 5.88 (0.32-107.77) | .23 |
| 15 | NEIL1 | 66 | 49 | 9 | 0.65 (0.44-0.96) | .03a | 0.42 (0.20-0.89) | .02a |
| 13 | LIG4 | 7 | 15 | 6 | 1.89 (0.73-4.89) | .19 | 2.34 (0.70-7.77) | .17 |
| 19 | POLD1 | 6 | 10 | 2 | 2.08 (0.69-6.26) | .19 | 1.50 (0.22-10.49) | .68 |
| 8 | NEIL2 | 3 | 8 | 4 | 2.43 (0.63-9.40) | .20 | 3.81 (0.75-19.28) | .11 |
| 8 | RRM2B | 3 | 10 | 3 | 2.06 (0.55-7.76) | .29 | 2.52 (0.42-15.01) | .31 |
| 6 | MSH5 | 12 | 19 | 7 | 1.39 (0.65-3.00) | .40 | 3.11 (1.16-8.33) | .02a |
Abbreviations: NA, not applicable; OR, odds ratio; PCa, prostate cancer.
P < .05.
Figure 2. Exome-Wide Gene-Based Results Meta-Analyzed Across Stages 1 and 2 by Prostate Cancer (PCa) Outcome.
A, With 87 of 1460 (6.0%) tested genes having P < .05. B, With 62 of 1314 (4.7%) tested genes having P < .05. C, With 93 of 1422 (6.5%) tested genes having P < .05. The red dashed line indicates a Bonferroni-adjusted significance threshold; the blue dashed line indicates P = .05.
Exome-wide significant findings were further evaluated by considering functional domains within BRCA2 (transactivation, RAD51-binding including 8 BRC repeats, MEILB2-binding, DNA-binding, helical domain, and OB1-2)26 and ATM (FAT and PI3K)27 (eFigure 5 in Supplement 2). Men carrying deleterious BRCA2 helical domain alleles had a 7.5-fold (95% CI, 1.69-33.4; P = .008) increased risk of aggressive compared with nonaggressive PCa, while men carrying deleterious BRCA2 MEILB2-binding domain alleles had a 9.3-fold (95% CI, 1.18-73.7; P = .03) increased risk of aggressive compared with nonaggressive PCa. Men carrying deleterious ATM FAT and PI3K alleles had similar or lower risks of aggressive disease compared with overall ATM gene-based findings. The limited number of men carrying deleterious alleles within a given domain makes it difficult to draw conclusions from this analysis pending follow-up investigations in larger studies. We did not consider NBN domains given the small number of men carrying deleterious NBN alleles (n = 26).
PCa Panel and Candidate DNA Repair Genes
PCa Aggressiveness
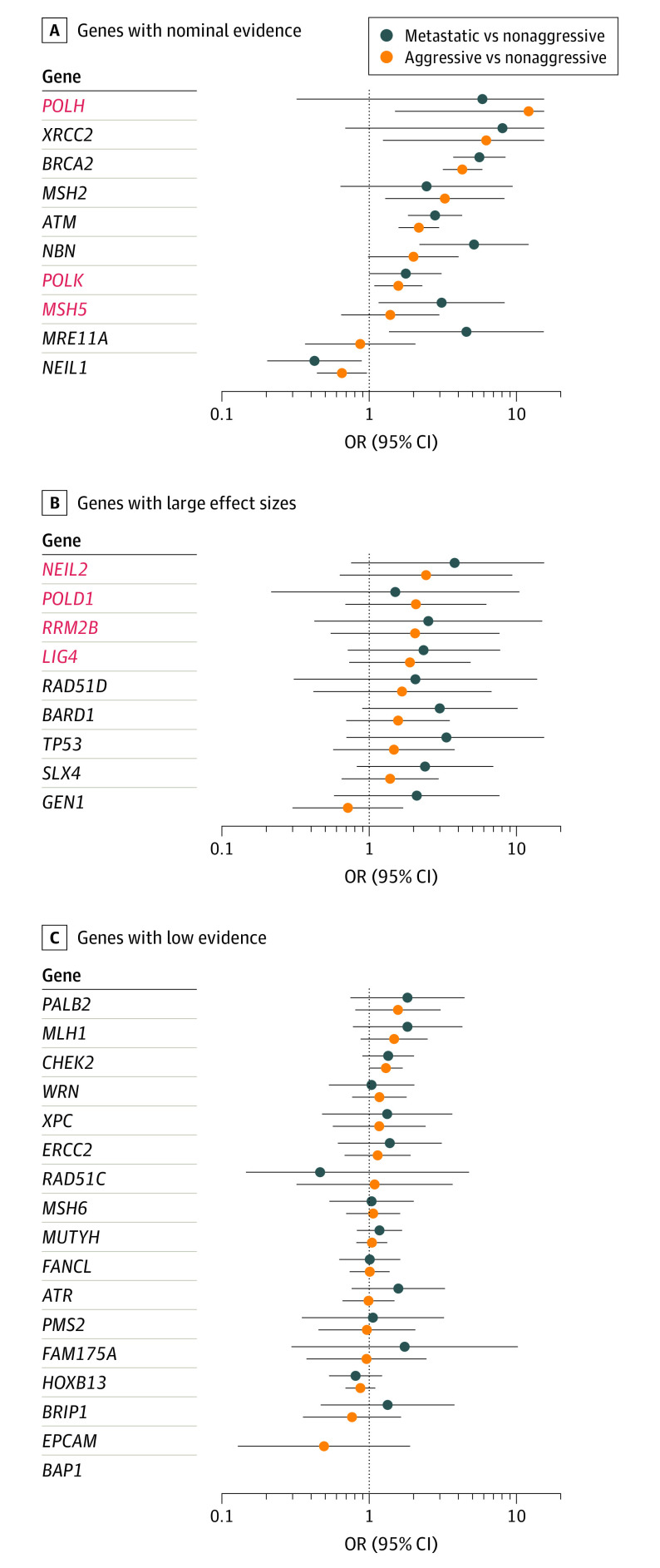
Examining rare deleterious variants within the 29 candidate genes (eMethods, eTable 4 in Supplement 1), in addition to BRCA2, ATM, and NBN, we found nominal association evidence (P < .05) for MSH2 (aggressive OR, 3.27; 95% CI, 1.29-8.31; P = .01), XRCC2 (aggressive OR, 6.24; 95% CI, 1.24-31.4; P = .03), and MRE11A (metastatic OR, 4.70; 95% CI, 1.39-15.90; P = .01) (Figure 3). Five other genes had evidence of large effects (OR≥2), but differences were not statistically significant: TP53, RAD51D, BARD1, GEN1, and SLX4 (Figure 3). Deleterious variants in these 11 genes were carried by 5.6% of patients with aggressive PCa and 7.0% of those with metastatic PCa compared with 2.3% of those with nonaggressive PCa, which decreased to 1.3% upon excluding BRCA2, 1.5% upon excluding ATM, and 0.8% upon excluding NBN (eTable 11 in Supplement 1). While the remaining 26 genes had low association evidence, we found that PALB2, MLH1, BRCA1, FAM175A, and ATR had ORs greater than 1.5, while CHEK2 had a P value close to the nominal threshold (OR, 1.30; P = .05) for its association with aggressive PCa (Figure 3).
Figure 3. Risk of Aggressive Prostate Cancer (PCa) Associated With Rare Deleterious Variants in Candidate PCa Genes.
A, Genes with nominal P values (P < .05). B, Genes with large effect sizes (odds ratio [OR] ≥2) but nonsignificant P values (P > .05). C, Genes with low evidence (P > .05 and OR <2). Red font represents potentially novel candidate PCa DNA repair genes with positive associations. Error bars indicate 95% CIs, which are reported in Table 2.
To assess whether our findings have implications for disease progression, we compared carrier frequencies between 686 men initially diagnosed with nonaggressive PCa (category T1/T2 and Gleason score ≤6 tumors) whose PCa later progressed to metastatic disease or died from PCa and 4141 men initially diagnosed with nonaggressive disease who had at least 10 years of follow-up to confirm that they did not progress to metastatic disease or die from PCa. Men carrying rare deleterious alleles in any of the 11 candidate genes had a 2-fold increased risk of progressing to lethal disease (OR, 2.00; 95% CI, 1.13-3.53; P = .02), which was similar, although not significant upon excluding BRCA2, ATM, and NBN (OR, 2.09; 95% CI, 0.65-6.67; P = .21) (eTable 12 in Supplement 1).
Examining an extended set of 167 additional DNA repair genes (eTable 4 in Supplement 1), we detected nominal positive association evidence for 3 genes: POLK (aggressive OR, 1.58; 95% CI, 1.08-2.30; P = .02), POLH (aggressive OR, 12.12; 95% CI, 1.50-98.2; P = .02), and MSH5 (metastatic OR, 3.11; 95% CI, 1.16-8.33; P = .02), with a nominal protective effect observed for NEIL1 (aggressive metastatic OR, 0.42; 95% CI, 0.20-0.89; P = .02) and large nonsignificant effect sizes (OR≥2) observed for 4 other genes: LIG4, POLD1, NEIL2, and RRM2B (Figure 2, Table 2; eFigure 6 in Supplement 2 and eTable 10 in Supplement 1).
Country Heterogeneity, Age at PCa Diagnosis, and Family History
Of the 19 associated genes described above (11 candidate and 8 additional DNA repair genes), there was effect heterogeneity by country for risk of aggressive PCa associated with BRCA2 (heterogeneity P = .02), with a null association observed for Finnish men, as previously reported,4 and POLK (P = .02) (eTable 13 in Supplement 1). Men carrying deleterious BRCA2 alleles had a 1.02-year younger age at nonaggressive PCa diagnosis (95% CI, −2.70 to 0.65; P = .23) and a 4.34-year younger age at metastatic PCa diagnosis (95% CI, −6.30 to −2.39; P = 1.3 × 10−5; heterogeneity vs nonaggressive, P = .01) (eTable 14 in Supplement 1). Men carrying deleterious NEIL2 alleles had a 4.67-year older age at nonaggressive PCa diagnosis (95% CI, −2.22 to 11.57; P = .18) and a 6.81-year younger age at metastatic PCa diagnosis (95% CI, −14.15 to 0.53; P = .07; heterogeneity vs nonaggressive P = .03). No other genes had associations with age at diagnosis that significantly differed by disease aggressiveness. Carrying deleterious ATM alleles was associated with a 1.83-year younger age at diagnosis of any PCa (95% CI, −2.88 to −0.79; P = 5.5 × 10−4) (eTable 14 in Supplement 1), while carrying rare deleterious alleles across the 11 candidate genes was associated with a 1.86-year younger age at diagnosis of any PCa (95% CI, −2.24 to −1.29; P = 1.1 × 10−10) (eTable 15 in Supplement 1). However, this association became null upon excluding BRCA2, ATM, and NBN (β = 0.24; 95% CI, −0.84 to 1.31; P = .66) (eTable 15 in Supplement 1).
Higher carrier frequencies in men with vs without a first-degree PCa family history were observed for ATM (1.8% vs 1.1%; P = .003) and PALB2 (0.6% vs 0.2%; P = .003) (eTable 16 in Supplement 1); however, these differences were not statistically significant after adjusting for multiple testing. We observed carrier frequency differences by family history status when aggregating across the 11 genes (4.8% in family history–positive and 3.9% in family history–negative men; P = .03); however, this difference was not significant after excluding BRCA2, ATM, and NBN (P = .57) (eTable 17 in Supplement 1).
Rare VUS
Considering rare missense VUS across the 11 genes with significant or nominal evidence or large effect sizes based on deleterious variants, we found nominal association evidence (P < .05) with metastatic PCa for WRN (OR, 6.35; 95% CI, 1.22-33.17; P = .028) and MRE11A (OR, 2.25; 95% CI, 1.03-4.94; P = .043), with large nonsignificant effects (OR≥2) observed for NBN, TP53, RAD51D, and XRCC2 (Figure 2; eFigure 6 and eFigure 7 in Supplement 2 and eTable 10 in Supplement 1). Aggregating VUS across the 11 genes, we observed a null association with PCa aggressiveness (eTable 18 and eTable 19 in Supplement 1). No individual genes were associated with age at diagnosis (eTable 20 in Supplement 1); however, carrying VUS in any of the 11 genes was weakly associated with a 1.88-year younger age at metastatic diagnosis (95% CI, −3.65 to −0.11; P = .04) (eTable 21 in Supplement 1). Only ATM VUS had carrier frequencies that differed by family history status, with 3.7% of men with metastatic PCa with a family history carrying ATM VUS compared with 0.8% of those with metastatic PCa without a family history (P = 9.9 × 10−4) (eTable 22 and eTable 23 in Supplement 1).
Discussion
In this large exome sequencing study of PCa, we found rare deleterious variants in BRCA2, ATM, and NBN were strongly associated with susceptibility to aggressive and/or metastatic PCa, while rare deleterious variants in MSH2, XRCC2, and MRE11A and rare VUS in WRN and MRE11A were nominally (P < .05) associated with disease aggressiveness. Nominal positive associations were also observed for rare deleterious variants in potentially novel DNA repair genes POLK, POLH, and MSH5. Based on estimated effect sizes alone (OR≥2), we also report deleterious variants and/or VUS in 9 other genes with large nonsignificant findings. Only missense variant Ile179Thr in KLK3 on chromosome 19q13.3 was associated with aggressive disease in exome-wide analyses. This variant is strongly associated with constitutive PSA levels,24 which may result in overdetection of less-aggressive tumors and contribute to the observed association with aggressive PCa also reported in previous studies, although evidence suggests that this variant may also independently influence disease progression.28,29
We found evidence of an association for deleterious variants in several previously implicated aggressive PCa genes (BRCA2, ATM, MSH2, NBN, and TP53).4,5,8,14,15,16,17,30,31,32,33,34,35 We also provide support for positive germline associations with deleterious variants and/or VUS in 13 additional genes with limited or no previous evidence of germline associations with aggressive PCa risk (nominal: MRE11A, XRCC2, POLK, POLH, and MSH5; and large effect sizes [OR≥2]: RAD51D, SLX4, BARD1, GEN1, LIG4, POLD1, NEIL2, and RRM2B).5,14 However, several of these genes have been implicated in PCa onset and progression. Expression of LIG4 in PCa tumors has been linked to advanced Gleason scores, positive nodal involvement, and early biochemical recurrence.36 Loss of BARD1 has been shown to increase sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors in PCa cell lines.37 The gene MRE11A is involved in DNA double-strand break repair and interacts with ATM, NBN, and BRCA1, and MRE11A expression has been positively associated with PCa progression and poor survival.38 SLX4 is a phosphorylation target for ATM and ATR protein kinases,39 and biallelic SLX4 variants have been associated with a subtype of Fanconi anemia FA-P.40 Men carrying Lynch syndrome variants in mismatch repair genes have been shown to have a 2-fold increased risk of PCa,41 which these data support with 2 genes (MSH2 and MSH5) associated with aggressive PCa.16,21,42 The DNA polymerase genes POLK, POLH, and POLD1 play important roles in DNA replication, and polymerase dysregulation often occurs in cancers.43 Germline POLD1 variants have been associated with colorectal and endometrial cancer risk.44
Of the genes with association evidence for deleterious variants, MRE11A, NBN, XRCC2, TP53, and RAD51D also had VUS association evidence, with similar effect sizes observed for all but RAD51D, supporting the role of protein coding alterations in these genes in aggressive PCa susceptibility. However, it is important to note limitations in studying VUS, including the difficulty of differentiating pathogenic from nonpathogenic VUS. As such, functional testing and other methods to improve the classification of VUS are needed to study this form of coding variation, particularly for genes with evidence of deleterious variants associated with aggressive PCa. Furthermore, large-scale replication sequencing studies in non-European ancestry populations in whom VUS frequencies may vary will also inform whether future genetic testing guidelines may benefit from integrating VUS. Such studies will be important to determine whether our findings based on men of European ancestry are generalizable across diverse populations.
Germline gene panel testing in PCa is currently limited to a few genes recommended for men with a cancer family history or high risk or advanced disease.2,3 Of genes included in the panels, we found significant or nominal evidence for BRCA2, ATM, MSH2, and NBN. We observed nominal positive evidence for 5 candidate genes not currently included in PCa gene panels (XRCC2, MRE11A, POLK, POLH, and MSH5). Our results support the exclusion of genes that have not been informative of PCa risk in previous studies or the current study, such as PMS2, RAD51C, EPCAM, and BRIP1. However, the involvement of candidate genes in PCa aggressiveness needs to be confirmed in larger studies and in diverse populations.
For men with aggressive PCa, genetic testing could inform treatment decisions and disease management, particularly for men carrying deleterious alleles in DNA repair genes. For example, PARP inhibitors are approved treatments for women with breast and ovarian cancer carrying pathogenic BRCA1 and BRCA2 alleles.45,46,47,48 PARP inhibitors were approved by the US Food and Drug Administration and included in the NCCN guidelines for treatment of metastatic castration-resistant PCa in men with pathogenic germline or somatic BRCA2 and BRCA1 alleles.1,2,49 As such, it may be important to extend testing guidelines, as this information would help identify which patients are at risk of progression to lethal forms of PCa at a time when curative treatment may be possible. It will be important for large prospective and multiancestry studies to further investigate whether men with nonaggressive PCa carrying deleterious variants are more likely to advance to lethal disease. Furthermore, consistent with previous studies, we observed a significantly younger age at PCa diagnosis in men carrying deleterious variants in BRCA24,50 and ATM27 and a 1.86-year younger age at diagnosis, on average, in men carrying deleterious variants in any of the 11 PCa risk genes, which is important information that could guide screening decisions. While this investigation used extreme definitions of disease aggressiveness to identify and characterize genetic contributions to aggressive PCa, future work aimed at understanding genetic risk of other clinically relevant tumor subgroups (eg, Gleason score 4 + 3 vs 3 + 4 tumors) will be important.
Limitations
This study has limitations. While this investigation represents what is, to our knowledge, the largest PCa exome sequencing study to date, our study had limited power (approximately 80%) to detect exome-wide significant associations with single variants or genes with OR less than 2.0 and carrier frequencies less than 0.6%. Many genes with large estimated effect sizes (OR≥2) had P values <.20, further emphasizing the need for large samples size to achieve even nominal (unadjusted P < .05) statistical significance in gene-based tests of rare variants with true effect sizes of similar magnitude. Previous rare coding variation investigations have focused on aggregating across genes given the lack of statistical power to evaluate genes individually. With our relatively large sample size, we integrated statistical evidence and carrier frequency to evaluate candidate genes individually and provide insight into which genes are important to aggressive PCa risk that may guide decisions regarding expanding gene panels. Detecting novel genes with weaker effects and lower carrier frequencies at exome-wide significance will require sample sizes ranging from tens to hundreds of thousands of men (eTable 24 in Supplement 1). Furthermore, although 46.8% of patients with nonaggressive PCa had 10 or more years of follow-up confirming they were alive and without recurrence, another limitation of this study is the potential for misclassification of nonaggressive cases. Our investigation included studies spanning many years, and Gleason score grading has changed over time along with treatment recommendations, which were part of our criteria used to define aggressive and nonaggressive disease (eg, Gleason score and category). Furthermore, treatment information was not available, which could further define disease progression. However, such misclassification would bias results toward the null rather than lead to false-positive findings. In addition, as study participants were limited to men of European ancestry, sequencing investigations in diverse populations will be crucial to determine whether our findings are generalizable across populations.
Conclusions
The findings of this genetic association study implicate genes to be considered for gene panel testing in PCa that would be beneficial to a broad group of men, including those diagnosed at any stage of the disease. Germline genetic testing holds potential to inform risk of disease onset and aggressiveness, screening for family members, and tailored disease management to improve outcomes of men diagnosed with PCa. Additional large-scale sequencing studies, particularly in diverse populations, are needed to confirm the involvement or lack thereof of the genes lacking strong statistical evidence reported here in PCa aggressiveness.
eTable 1. Clinical Characteristics of Participants by Study
eTable 2. Genes Included in Stage 2 Targeted Exome Sequencing
eTable 3. Description of Rare Deleterious Variants and VUS Among 1,749 Genes
eTable 4. Candidate Prostate Cancer Gene Sets
eTable 5. Association Results for Variants Significantly Associated With Aggressive Versus Non-Aggressive Prostate Cancer
eTable 6. Association Between Individual Rare Deleterious Variants and Variants Of Uncertain Significance (VUS) and Prostate Cancer Risk
eTable 7. Rare Deleterious Variant Gene-Based Association Results for Aggressive Versus Non-Aggressive Prostate Cancer Risk
eTable 8. Rare Deleterious Variant Gene-Based Association Results for Death Due to Prostate Cancer Versus Non-Aggressive Prostate Cancer Risk
eTable 9. Rare Deleterious Variant Gene-Based Association Results for Metastatic Versus Non-Aggressive Prostate Cancer Risk
eTable 10. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants and VUS in Candidate Prostate Cancer Genes
eTable 11. Risk of Progressing Aggressive Prostate Cancer Associated With Deleterious Variants and VUS in Candidate Prostate Cancer Genes Aggregated
eTable 12. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants Aggregated Across Candidate Prostate Cancer Genes Stratified By Country
eTable 13. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants In Candidate Prostate Cancer Genes Stratified By Country
eTable 14. Association Results for Deleterious Variants in Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 15. Association Results for Deleterious Variants Aggregated Across Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 16. Association Between Deleterious Variants in Candidate Prostate Cancer Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 17. Association Between the Aggregate of Deleterious Variants Across Candidate Prostate Cancer Genes and Family History Of Prostate Cancer Stratified by Disease Aggressiveness
eTable 18. Risk of Aggressive Prostate Cancer Associated With the Aggregate of VUS Across Candidate Prostate Cancer Genes Stratified By Country
eTable 19. Risk of Aggressive Prostate Cancer Associated With VUS in Candidate Prostate Cancer Genes Stratified by Country
eTable 20. Association Results for VUS in Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 21. Association Results for the Aggregate of VUS Across Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 22. Association Between VUS in Candidate Prostate Cancer Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 23. Association Between the Aggregate of VUS Across Candidate PCa Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 24. Sample Sizes Needed to Detect Exome-Wide Significant Associations With Aggressive Prostate Cancer
eMethods. Detailed Methods
eReferences
eFigure 1. Manhattan Plot of Association Results Between Genetic Variants and Risk of Aggressive Versus Non-Aggressive Prostate Cancer
eFigure 2. Manhattan Plot of Association Results Between Genetic Variants and Risk of PC Death Versus Non-Aggressive Prostate Cancer
eFigure 3. Manhattan Plot of Association Results Between Genetic Variants and Risk of Metastatic Versus Non-Aggressive Prostate Cancer
eFigure 4. Sensitivity Analyses Comparing Gene-Based Results for Aggressive Versus Non-Aggressive Disease Including and Excluding Non-Aggressive Cases With PSA>20
eFigure 5. Carrier Frequencies and Effects for Functional Domains Within BRCA2 and ATM
eFigure 6. Carrier Frequencies of Rare Deleterious Variants and VUS in PCa Panel Genes and Candidate and Novel PCa DNA Repair Genes by PCa Aggressiveness
eFigure 7. Comparison of OR Between Rare VUS and Rare Deleterious Variant Gene-Based Analyses
eAppendix. Supplementary Note
Data Sharing Statement
References
- 1.Schaeffer EM, Srinivas S, Adra N, et al. NCCN guidelines insights: prostate cancer, version 1.2023. J Natl Compr Canc Netw. 2022;20(12):1288-1298. doi: 10.6004/jnccn.2022.0063 [DOI] [PubMed] [Google Scholar]
- 2.Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate cancer, version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(5):479-505. doi: 10.6004/jnccn.2019.0023 [DOI] [PubMed] [Google Scholar]
- 3.Giri VN, Knudsen KE, Kelly WK, et al. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol. 2020;38(24):2798-2811. doi: 10.1200/JCO.20.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darst BF, Dadaev T, Saunders E, et al. Germline sequencing DNA repair genes in 5545 men with aggressive and nonaggressive prostate cancer. J Natl Cancer Inst. 2021;113(5):616-625. doi: 10.1093/jnci/djaa132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. doi: 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plym A, Dióssy M, Szallasi Z, et al. DNA repair pathways and their association with lethal prostate cancer in African American and European American men. J Natl Cancer Inst Cancer Spectr. 2021;6(1):pkab097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei J, Yang W, Shi Z, et al. Observed evidence for guideline-recommended genes in predicting prostate cancer risk from a large population-based cohort. Prostate. 2021;81(13):1002-1008. doi: 10.1002/pros.24195 [DOI] [PubMed] [Google Scholar]
- 8.Matejcic M, Patel Y, Lilyquist J, et al. Pathogenic variants in cancer predisposition genes and prostate cancer risk in men of African ancestry. JCO Precis Oncol. 2020;4:32-43. doi: 10.1200/PO.19.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledet EM, Burgess EF, Sokolova AO, et al. Comparison of germline mutations in African American and Caucasian men with metastatic prostate cancer. Prostate. 2021;81(7):433-439. doi: 10.1002/pros.24123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurwitz LM, Agalliu I, Albanes D, et al. ; Prostate Cancer Cohort Consortium (PC3) Working Group . Recommended definitions of aggressive prostate cancer for etiologic epidemiologic research. J Natl Cancer Inst. 2021;113(6):727-734. doi: 10.1093/jnci/djaa154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren W, Gil L, Hunt SE, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(database issue):D980-D985. doi: 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877-885. doi: 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mijuskovic M, Saunders EJ, Leongamornlert DA, et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br J Cancer. 2018;119(1):96-104. doi: 10.1038/s41416-018-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leongamornlert D, Saunders E, Dadaev T, et al. ; UKGPCS Collaborators . Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110(6):1663-1672. doi: 10.1038/bjc.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leongamornlert DA, Saunders EJ, Wakerell S, et al. Germline DNA repair gene mutations in young-onset prostate cancer cases in the UK: evidence for a more extensive genetic panel. Eur Urol. 2019;76(3):329-337. doi: 10.1016/j.eururo.2019.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na R, Zheng SL, Han M, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71(5):740-747. doi: 10.1016/j.eururo.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart SN, Ellingson MS, Schahl K, et al. Determining the frequency of pathogenic germline variants from exome sequencing in patients with castrate-resistant prostate cancer. BMJ Open. 2016;6(4):e010332. doi: 10.1136/bmjopen-2015-010332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697-1708. doi: 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284-1289. doi: 10.1126/science.1056154 [DOI] [PubMed] [Google Scholar]
- 21.Saunders EJ, Dadaev T, Leongamornlert DA, et al. ; UK Genetic Prostate Cancer Study Collaborators; UK ProtecT Study Collaborators; PRACTICAL Consortium . Gene and pathway level analyses of germline DNA-repair gene variants and prostate cancer susceptibility using the iCOGS-genotyping array. Br J Cancer. 2016;114(8):945-952. doi: 10.1038/bjc.2016.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst. 2012;104(9):670-681. doi: 10.1093/jnci/djs177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ming M, He YY. PTEN in DNA damage repair. Cancer Lett. 2012;319(2):125-129. doi: 10.1016/j.canlet.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann TJ, Passarelli MN, Graff RE, et al. Genome-wide association study of prostate-specific antigen levels identifies novel loci independent of prostate cancer. Nat Commun. 2017;8:14248. doi: 10.1038/ncomms14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varon R, Vissinga C, Platzer M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93(3):467-476. doi: 10.1016/S0092-8674(00)81174-5 [DOI] [PubMed] [Google Scholar]
- 26.Ikegami M, Kohsaka S, Ueno T, et al. High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat Commun. 2020;11(1):2573. doi: 10.1038/s41467-020-16141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson Q, Brook MN, Dadaev T, et al. ; PRACTICAL Consortium . Rare germline variants in ATM predispose to prostate cancer: a PRACTICAL consortium study. Eur Urol Oncol. 2021;4(4):570-579. doi: 10.1016/j.euo.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Shi Z, Wei J, et al. KLK3 germline mutation I179T complements DNA repair genes for predicting prostate cancer progression. Prostate Cancer Prostatic Dis. 2022;25(4):749-754. doi: 10.1038/s41391-021-00466-6 [DOI] [PubMed] [Google Scholar]
- 29.Helfand BT, Roehl KA, Cooper PR, et al. Associations of prostate cancer risk variants with disease aggressiveness: results of the NCI-SPORE Genetics Working Group analysis of 18,343 cases. Hum Genet. 2015;134(4):439-450. doi: 10.1007/s00439-015-1534-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateo J, Cheng HH, Beltran H, et al. Clinical outcome of prostate cancer patients with germline DNA repair mutations: retrospective analysis from an international study. Eur Urol. 2018;73(5):687-693. doi: 10.1016/j.eururo.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Yu H, Li S, et al. Rare germline pathogenic mutations of DNA repair genes are most strongly associated with grade group 5 prostate cancer. Eur Urol Oncol. 2020;3(2):224-230. doi: 10.1016/j.euo.2019.12.003 [DOI] [PubMed] [Google Scholar]
- 32.Shi Z, Lu L, Resurreccion WK, et al. Association of germline rare pathogenic mutations in guideline-recommended genes with prostate cancer progression: a meta-analysis. Prostate. 2022;82(1):107-119. doi: 10.1002/pros.24252 [DOI] [PubMed] [Google Scholar]
- 33.Rusak B, Kluźniak W, Wokołorczykv D, et al. Inherited NBN mutations and prostate cancer risk and survival. Cancer Res Treat. 2019;51(3):1180-1187. doi: 10.4143/crt.2018.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell KN, Cheng HH, Powers J, et al. Inherited TP53 variants and risk of prostate cancer. Eur Urol. 2022;81(3):243-250. doi: 10.1016/j.eururo.2021.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bancroft EK, Page EC, Brook MN, et al. ; IMPACT Study Collaborators . A prospective prostate cancer screening programme for men with pathogenic variants in mismatch repair genes (IMPACT): initial results from an international prospective study. Lancet Oncol. 2021;22(11):1618-1631. doi: 10.1016/S1470-2045(21)00522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grupp K, Roettger L, Kluth M, et al. Expression of DNA ligase IV is linked to poor prognosis and characterizes a subset of prostate cancers harboring TMPRSS2:ERG fusion and PTEN deletion. Oncol Rep. 2015;34(3):1211-1220. doi: 10.3892/or.2015.4080 [DOI] [PubMed] [Google Scholar]
- 37.Dillon KM, Bekele RT, Sztupinszki Z, et al. PALB2 or BARD1 loss confers homologous recombination deficiency and PARP inhibitor sensitivity in prostate cancer. NPJ Precis Oncol. 2022;6(1):49. doi: 10.1038/s41698-022-00291-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Xu WH, Wei Y, et al. Elevated MRE11 expression associated with progression and poor outcome in prostate cancer. J Cancer. 2019;10(18):4333-4340. doi: 10.7150/jca.31454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svendsen JM, Smogorzewska A, Sowa ME, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138(1):63-77. doi: 10.1016/j.cell.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43(2):142-146. doi: 10.1038/ng.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymond VM, Mukherjee B, Wang F, et al. Elevated risk of prostate cancer among men with Lynch syndrome. J Clin Oncol. 2013;31(14):1713-1718. doi: 10.1200/JCO.2012.44.1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burmester JK, Suarez BK, Lin JH, et al. Analysis of candidate genes for prostate cancer. Hum Hered. 2004;57(4):172-178. doi: 10.1159/000081443 [DOI] [PubMed] [Google Scholar]
- 43.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96-110. doi: 10.1038/nrc2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palles C, Cazier JB, Howarth KM, et al. ; CORGI Consortium; WGS500 Consortium . Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136-144. doi: 10.1038/ng.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tutt ANJ, Garber JE, Kaufman B, et al. ; OlympiA Clinical Trial Steering Committee and Investigators . Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med. 2021;384(25):2394-2405. doi: 10.1056/NEJMoa2105215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523-533. doi: 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 47.Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737):245-251. doi: 10.1016/S0140-6736(10)60893-8 [DOI] [PubMed] [Google Scholar]
- 48.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33(3):244-250. doi: 10.1200/JCO.2014.56.2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091-2102. doi: 10.1056/NEJMoa1911440 [DOI] [PubMed] [Google Scholar]
- 50.Kote-Jarai Z, Leongamornlert D, Saunders E, et al. ; UKGPCS Collaborators . BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105(8):1230-1234. doi: 10.1038/bjc.2011.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Clinical Characteristics of Participants by Study
eTable 2. Genes Included in Stage 2 Targeted Exome Sequencing
eTable 3. Description of Rare Deleterious Variants and VUS Among 1,749 Genes
eTable 4. Candidate Prostate Cancer Gene Sets
eTable 5. Association Results for Variants Significantly Associated With Aggressive Versus Non-Aggressive Prostate Cancer
eTable 6. Association Between Individual Rare Deleterious Variants and Variants Of Uncertain Significance (VUS) and Prostate Cancer Risk
eTable 7. Rare Deleterious Variant Gene-Based Association Results for Aggressive Versus Non-Aggressive Prostate Cancer Risk
eTable 8. Rare Deleterious Variant Gene-Based Association Results for Death Due to Prostate Cancer Versus Non-Aggressive Prostate Cancer Risk
eTable 9. Rare Deleterious Variant Gene-Based Association Results for Metastatic Versus Non-Aggressive Prostate Cancer Risk
eTable 10. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants and VUS in Candidate Prostate Cancer Genes
eTable 11. Risk of Progressing Aggressive Prostate Cancer Associated With Deleterious Variants and VUS in Candidate Prostate Cancer Genes Aggregated
eTable 12. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants Aggregated Across Candidate Prostate Cancer Genes Stratified By Country
eTable 13. Risk of Aggressive Prostate Cancer Associated With Deleterious Variants In Candidate Prostate Cancer Genes Stratified By Country
eTable 14. Association Results for Deleterious Variants in Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 15. Association Results for Deleterious Variants Aggregated Across Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 16. Association Between Deleterious Variants in Candidate Prostate Cancer Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 17. Association Between the Aggregate of Deleterious Variants Across Candidate Prostate Cancer Genes and Family History Of Prostate Cancer Stratified by Disease Aggressiveness
eTable 18. Risk of Aggressive Prostate Cancer Associated With the Aggregate of VUS Across Candidate Prostate Cancer Genes Stratified By Country
eTable 19. Risk of Aggressive Prostate Cancer Associated With VUS in Candidate Prostate Cancer Genes Stratified by Country
eTable 20. Association Results for VUS in Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 21. Association Results for the Aggregate of VUS Across Candidate Prostate Cancer Genes and Age at Diagnosis Stratified by Disease Aggressiveness
eTable 22. Association Between VUS in Candidate Prostate Cancer Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 23. Association Between the Aggregate of VUS Across Candidate PCa Genes and Family History of Prostate Cancer Stratified by Disease Aggressiveness
eTable 24. Sample Sizes Needed to Detect Exome-Wide Significant Associations With Aggressive Prostate Cancer
eMethods. Detailed Methods
eReferences
eFigure 1. Manhattan Plot of Association Results Between Genetic Variants and Risk of Aggressive Versus Non-Aggressive Prostate Cancer
eFigure 2. Manhattan Plot of Association Results Between Genetic Variants and Risk of PC Death Versus Non-Aggressive Prostate Cancer
eFigure 3. Manhattan Plot of Association Results Between Genetic Variants and Risk of Metastatic Versus Non-Aggressive Prostate Cancer
eFigure 4. Sensitivity Analyses Comparing Gene-Based Results for Aggressive Versus Non-Aggressive Disease Including and Excluding Non-Aggressive Cases With PSA>20
eFigure 5. Carrier Frequencies and Effects for Functional Domains Within BRCA2 and ATM
eFigure 6. Carrier Frequencies of Rare Deleterious Variants and VUS in PCa Panel Genes and Candidate and Novel PCa DNA Repair Genes by PCa Aggressiveness
eFigure 7. Comparison of OR Between Rare VUS and Rare Deleterious Variant Gene-Based Analyses
eAppendix. Supplementary Note
Data Sharing Statement