Abstract
The estrogen receptor (ER) modulates transcription by forming complexes with other proteins and then binding to the estrogen response element (ERE). We have identified a novel interaction of this receptor with the POU transcription factors Brn-3a and Brn-3b which was independent of ligand binding. By pull-down assays and the yeast two-hybrid system, the POU domain of Brn-3a and Brn-3b was shown to interact with the DNA-binding domain of the ER. Brn-3–ER interactions also affect transcriptional activity of an ERE-containing promoter, such that in estradiol-stimulated cells, Brn-3b strongly activated the promoter via the ERE, while Brn-3a had a mild inhibitory effect. The POU domain of Brn-3b which interacts with the ER was sufficient to confer this activation potential, and the change of a single amino acid in the first helix of the POU homeodomain of Brn-3a to its equivalent in Brn-3b can change the mild repressive effect of Brn-3a to a stimulatory Brn-3b-like effect. These observations and their implications for transcriptional regulation by the ER are discussed.
Transcriptional regulation by the complex interaction of different classes of transcription factors allows a limited number of proteins to elicit diverse effects on gene expression, depending on the expression of other proteins, such as tissue-specific factors and signals which may influence their interactions (reviewed in references 39, 40, 59, and 68 and references therein). We were interested in looking at proteins which interact with the transcription factors Brn-3a and Brn-3b and modulate the regulation of gene expression by these proteins. These two proteins belong to the POU (Pit-Oct-Unc) family of transcription factors (21, 25, 26, 42, 66, 70, 73, 76). Members of this class of transcription factors are defined on the basis of the common POU domain, which consists of two highly conserved regions, the POU-specific domain and the POU homeodomain, which are separated by a poorly conserved linker region. The POU domain acts as the DNA-binding domain which recognizes and binds specific DNA sequences present in target gene promoters but is also involved in protein-protein interactions (3, 72, 73). There are three known members of the Brn-3 family of transcription factors, namely, Brn-3a (also known as Brn-3.0) (21, 42, 65), Brn-3b (also called Brn-3.2) (42, 65, 70), and Brn-3c (also known as Brn-3.1) (21, 52), which are encoded by different genes (65, 77). Furthermore, different isoforms of Brn-3a and Brn-3b which result from alternative splicing of the genes encoding these two proteins have been identified (21, 43, 65, 70).
The Brn-3 proteins show restricted homology outside the conserved carboxyl-terminal POU domain and the amino-terminal POU IV box (21, 65, 70). Since the studies reported here were carried out with Brn-3a and Brn-3b, references to Brn-3 proteins will pertain to observations with Brn-3a or Brn-3b and not Brn-3c. Sequence differences between Brn-3a and Brn-3b proteins are paralleled by different effects on promoters which contain binding sites recognized by both proteins. For instance, cellular promoters of genes encoding α-internexin (7), SNAP 25 (33), and pro-opiomelanocortin (POMC) (21), which contain the Brn-3 DNA recognition site, were activated by Brn-3a, while Brn-3b repressed α-internexin gene promoter activity but had little effect on the SNAP-25 promoter. In addition, Brn-3a was found to be an activator of a reporter construct containing its binding site, while Brn-3b inhibited basal activity of this promoter (6, 48). Both proteins appear to recognize and bind to the same DNA sequence element in the double-stranded conformation (6, 8, 48) but were also capable of binding to single-stranded DNA. This was demonstrated by the preferential binding of both Brn-3a and Brn-3b to the antisense strand of the DNA binding site identified in the α-internexin promoter (8). The requirement for the same binding site but with different effects on gene expression may form the basis for the regulation of expression of genes whose promoters contain the Brn-3 DNA binding site in tissues which coexpress the different transcription factors.
Both Brn-3a mRNA and Brn-3b mRNA were detected in regions of the brain as well as in sensory neurons (21, 25, 70). However, Brn-3a and Brn-3b mRNAs were also detected in tissues of the reproductive tract (9). A number of other POU domain transcription factors, such as Tst-1 (25, 47, 76), Sperm-1 (1), Brn-5 (2), and Oct-6 (64), have been detected in the testis, while Oct-3/4 has also been identified in the ovary (61). The precise roles of these POU domain transcription factors in the reproductive tract are still not clear. In studies to identify a role for Brn-3 transcription factors in these tissues, we have examined the observed interaction of Brn-3a and Brn-3b proteins with the estrogen receptor (ER).
The ER proteins are members of the nuclear hormone receptor family which are highly expressed in tissues of the reproductive tract (see references 14 and 56 and references therein as well as references 30 and 30a) but have also been detected in many other tissues, including specific regions of the brain and sensory neurons (see references 13, 29, 62, and 62a and references therein). While the mechanism by which the ER regulates the activity of target genes is not clear, it has been shown that, classically, the receptor mediates its effect by binding as a complex with other proteins to specific DNA sequences, the estrogen response elements (EREs), which are found in the promoters of the estrogen-responsive genes (for reviews, see references 54 and 55), although other possible mechanisms involving protein-protein interactions have recently been identified (57, 63, 71, 75, 78). Like other steroid receptors, conversion of the native ER into a functional complex following ligand binding involves changes in the proteins which are associated with the receptor (31, 74). Thus interactions of the ER with other proteins play a critical role in the function of the receptor. A number of accessory proteins complexed with the ER in either the inactive or active states have been identified. Some of these include heat shock protein 90 (hsp 90) (10, 23), hsp 70 (35), p56 (35), ERAP160 (24), RIP 141 (11), SRC-1 (53), and p45 and p48 (35). It appears that the interaction of some of these proteins with the ER may directly activate transcription, while others are critical for the receptor binding to ERE with subsequent activation of the ER complex.
Mukherjee and Chambon (49) demonstrated that the ER binds the ERE only in the presence of accessory proteins, since the purified receptor protein failed to bind the ERE in vitro but readily formed a complex in the presence of crude extracts from HeLa or yeast (Saccharomyces cerevisiae) cells. They identified a 45-kDa yeast single-stranded DNA-binding protein, DBSF (DNA binding stimulatory factor), which did not bind to the ERE itself but facilitated binding of the purified ER to the ERE. In addition, Landel et al. (34) isolated two proteins, p45 and p48, which were required for efficient and stable binding of the ER to the ERE in vitro, in a manner which was independent of ligand binding. Studies by Lannigan et al. (35–37) indicated that the ER bound with higher affinity to an ERE-containing DNA sequence when it was in the single-stranded conformation compared with a weaker association observed with double-stranded ERE. It was suggested that the ERE is structurally labile and could form a unique tertiary structure which is required for binding of the receptor (35–38). Binding of the ER and its associated proteins may result in dissociation of the double-stranded DNA sequence to a single-stranded conformation with a subsequent increase in the receptor-DNA complex. Thus it is possible that the proteins interacting with the active complex may help to stabilize this association and increase the ER binding to the ERE.
Here we demonstrate a novel interaction of the 46-kDa single-stranded DNA-binding Brn-3a protein and 35-kDa Brn-3b protein with the ER protein in vivo and in vitro and show that this association can regulate ER binding to the ERE. Furthermore, these interactions appear to differentially regulate the transcriptional activity of an ERE-containing promoter.
MATERIALS AND METHODS
Plasmid DNAs.
The vit-tk CAT reporter construct containing a functional ERE was derived from the vitellogenin (vit) gene with region −331 to −87 cloned upstream of the thymidine kinase promoter (tk) which drives the expression of the chloramphenicol acetyltransferase (CAT) gene. The ERE luciferase vector contains the ERE sequence (consensus sequence underlined) 5′-CTAGAAAGTCAGGTCACAGTGACCTGATCAAT-3′ cloned into the pGL2 vector (Promega). The coding sequences of Brn-3a and Brn-3b were cloned into Bluescript and used for in vitro translation (67). The coding sequences of Brn-3a or Brn-3b and the POU domains were cloned under the control of the Moloney murine leukemia virus promoter into the pLTR expression vector, which was modified by a cryptic splice site in the simian virus 40 3′ untranslated region (65). The full-length and POU domain Brn-3 mutants were made as described by Dawson et al. (15). ER antibodies were obtained either from NovaCastra (Vector Laboratories, Peterborough, United Kingdom) (mouse anti-human) or from Stress Gen Biotech. Corp. (via Bioquote, Ltd., Yorkshire, United Kingdom) for anti-ER antibodies (SRA1010) raised against residues 582 to 595 of the receptor. Antibodies against Brn-3a or Brn-3b proteins were obtained from BAbCo (Berkeley, Calif.), and anti-Bad monoclonal antibodies were obtained from Transduction Laboratories.
Oligonucleotides used for the electrophoretic mobility shift assays (EMSAs) were the ERE sequence 5′ TCAGGTCACAGTGACCTG 3′ and CRE as the nonspecific competitor (5′ GCATAAATAAT 3′).
Generation of proteins.
In vitro translation was carried out with the single-step transcription-translation system with the TNT-coupled reticulocyte lysate (Promega) according to the manufacturer’s protocol. One microgram of plasmid (containing cDNA encoding either Brn-3a or Brn-3b, wild-type or truncated ER, or the control luciferase protein) and 40 μCi of [35S]methionine were used in a 50-μl reaction mixture. The labelled translated proteins were assessed by polyacrylamide gel electrophoresis (PAGE) analysis of 5 μl of the products. Glutathione S-transferase (GST) fusion proteins were generated by cloning the appropriate DNA fragment into the bacterial expression vector pGEX-2T, which allows expression under isopropyl-1-thio-β-d-galactopyranoside (IPTG) induction. The fusion proteins were isolated by affinity chromatography with glutathione-coupled Sepharose beads according to the manufacturer’s protocol (Pharmacia Biotech, Inc.). The bands of the protein products were observed by Coomassie staining after sodium dodecyl sulfate (SDS)-PAGE analysis.
Protein-protein interaction.
The protein-protein interaction assay was performed according to the method described by Baniahmad et al. (4). Briefly, Brn-3a or Brn-3b–GST fusion proteins linked to the glutathione-Sepharose beads were prepared and stored in NENT buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris [pH 8], 0.5% Nonidet P-40, 0.5% milk powder). Prior to use, approximately 1 to 2 μg of the fusion proteins was incubated in 20% milk powder in NENT buffer for 15 min at room temperature. The beads were washed in 1 ml of NENT buffer and once in 1 ml of transcription wash buffer (20 mM HEPES [pH 7.9], 60 mM NaCl, 1 mM dithiotreitol, 6 mM MgCl2, 8.2% glycerine, 0.1 mM EDTA). Following SDS-PAGE analysis and densitometry, the volumes of in vitro-translated proteins were adjusted so that relatively equal amounts of each protein were used. The in vitro-translated ER proteins or the equivalent amounts of luciferase control proteins were then incubated with the beads in 100 μl of transcription buffer for 1 h at room temperature. The beads were washed (five times with 1 ml of NENT buffer), and the proteins were solubilized in SDS loading buffer, heated to 100°C for 5 min, and resolved on an SDS–12% polyacrylamide gel. Following electrophoresis, the gel was dried and exposed to radiographic film or a PhosphorImager screen. The amounts of protein retained following the interaction studies were assessed by comparing the intensity of the bands resulting after the protocol with that resulting when equivalent amounts of proteins (input) were run on a similar gel.
To confirm that the interaction was not dependent on contaminating DNA, the interaction studies were carried out by a modified method as described by Lai and Herr (32a). In brief, the reaction was carried out as before, but one sample of in vitro-translated protein was incubated with 50 μg of ethidium bromide (EtBr) for 15 min prior to incubation with the GST fusion proteins. This amount of ethidium bromide was also maintained during washes.
Yeast two-hybrid studies.
Yeast two-hybrid studies were carried out with the HybriZAP GAL4 DNA-binding domain vector (pBDGAL4) obtained from Stratagene and the pGAD424 GAL4 activation domain vector (Clontech). The manufacturer’s recommended protocol was used to confirm in vivo interactions. Briefly, DNAs encoding either the POU domains of Brn-3a (Brn-3a POUpBD) and Brn-3b (Brn-3b POUpBD) or the amino-terminal domain of Brn-3a (Brn-3aN pBD) were cloned into the GAL4 DNA-binding domain vector so that they were in frame with the GAL4 DNA-binding domain sequence. The DNA encoding the wild-type ER was cloned into the GAL4 activation domain vector (ER-pGAD). These vectors were cotransformed into competent HF7c yeast, a modified yeast (Saccharomyces cerevisiae) strain which expressed no endogenous GAL4 and which carries the auxotrophic markers leucine (leu2) and tryptophan (trp1) to allow for selection of cells carrying pGAD424 and pBDGAL4, respectively, and histidine (his3) for selection of cells transformed with interacting proteins. In addition, a second reporter gene, lacZ, could be used to assay for interacting proteins. Following cotransformation with the two vectors, the cells were plated out onto synthetic dropout media agar plates which lacked leucine, tryptophan, and histidine. Colonies were reselected, grown in the dropout media (no leucine, tryptophan, or histidine), and then tested for lacZ promoter activity as described in the manufacturer’s protocol. The Brn-3 and the ER-pGAD vectors were also grown on plates containing no tryptophan or leucine, respectively, to show expression of the selection marker.
Immunoprecipitation.
Immunoprecipitation was carried out to assess the interaction between Brn-3 proteins and the ER in vivo. Protein extracts were made either from rat tissues, such as brain, ovary, and kidney (negative control). Tissues were homogenized in extraction buffer containing 50 mM Tris-HCl (pH 8.0), 170 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, and 10 μg of the protease inhibitors leupeptin, aprotinin, and pepstatin per ml plus 1 mM phenylmethylsulfonyl fluoride. The tissue homogenate was centrifuged at 14,000 × g for 10 min to pellet debris. The supernatant was precleared by incubation with 25 μl of protein A-protein G-agarose slurry for 30 to 60 min at 4°C. After centrifugation, the supernatant was incubated overnight at 4°C with either 10 μl of the anti-ER antibody SRA1010, 10 μl of antibody to the Bad protein (which does not interact with the Brn-3 proteins), or no antibody. The immunocomplexes were then collected by incubation with 30 μl of the protein A-protein G-agarose slurry for 30 min. The agarose beads were washed five times with buffer containing 10 mM NaCl, 1 mM EDTA, 20 mM Tris (pH 8), and 0.5% Nonidet P-40 and then boiled in 1× SDS sample buffer (2% SDS, 10% glycerol, 62 mM Tris-HCl [pH 6.8], 1% β-mercaptoethanol) and loaded on an SDS–12% polyacrylamide gel. A Western blot was produced, and this was probed with the antibodies to the Brn-3 proteins (1:2,000 dilution).
EMSA.
The EMSA was carried out as described by Theil et al. (67). Briefly, 3 μl of the in vitro-translated ER, Brn-3 proteins, or luciferase (control) protein was added as indicated to 2 μl of 10× EMSA buffer (10 mM HEPES [pH 7.9], 60 mM KCl, 4% Ficoll, 1 mM dithiothreitol, 1 mM EDTA) containing 2 μg of poly(dI-dC) to prevent nonspecific interactions, specific or nonspecific competitor oligonucleotides, or antibodies (where specified) and kept at room temperature for 5 min. One nanogram of 5′-end-labelled oligonucleotide probe (labelled with T4 kinase and purified on a Sephadex G-25 column) was then added, and this combination was mixed briefly, spun in a microcentrifuge for 5 s, and then incubated on ice for 45 min to 1 h. The DNA-protein complexes were resolved from free DNA by gel electrophoresis on a 7% polyacrylamide gel run in 0.5× Tris-borate-EDTA for 2 to 2.5 h at 4°C. The gel was dried and exposed to a double layer of film if 35S-labelled proteins were used, since the first layer eliminated the 35S activity.
Cell culture, DNA transfection, and assays.
MCF7 cells were routinely grown in Dulbecco’s modified Eagle’s medium, which contained l-glutamate and phenol red and was supplemented with 10% fetal calf serum and 10 ng of insulin per ml. When grown in medium which contains no phenol red and dextran-coated charcoal-stripped serum, these cells become strongly responsive to the ligand, estradiol (5). Therefore, before the experiments were carried out, subconfluent cells were maintained in phenol red-free Dulbecco’s modified Eagle’s medium containing 10% dextran-coated charcoal-stripped fetal calf serum (prepared according to the method described by Migliaccio et al. [46]) and 10 ng of insulin per ml for 72 h. The medium was replaced by 5 ml of fresh medium 12 h prior to transfection. Transfection of plasmid DNA was carried out according to the method of Gorman (22). Routinely, 5 μg of reporter DNA and 5 μg of each expression vector were transfected into 5 × 105 cells. After transfection, the medium on the cells was replaced with fresh medium and 10−8 M 17β-estradiol was added to the cells. Cells were harvested after 72 h. The amount of DNA taken up by the cells in each case was measured by the slot blotting of 15 μl of the extract and hybridization with a probe derived from the ampicillin resistance gene in the plasmid vector. Differences in the intensity of the bands were measured by densitometry and used to equalize the volumes of extract used for subsequent CAT or luciferase assays.
CAT assays were carried out as described previously (22), and luciferase assays were done as described by the manufacturer’s (Promega) protocol, with results measured on a Turner 20-e luminometer.
RESULTS
Brn-3a interaction with the DNA-binding domain of the ER.
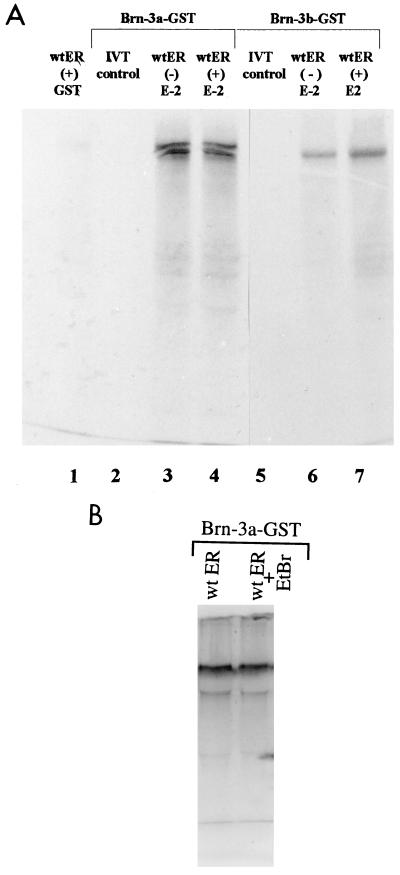
To identify factors which interacted with Brn-3 proteins, we used the matrix-bound fusion protein of GST combined with Brn-3a (Brn-3a–GST) or Brn-3b (Brn-3b–GST) for in vitro protein-protein interaction studies. In this study, the Brn-3 fusion proteins were incubated with 35S-labelled in vitro-translated wild-type ER under the conditions described in Materials and Methods. The intensities of the bands (Fig. 1A) represent the relative amounts of the labelled ER protein retained by the Brn-3–GST fusion proteins and indicated that the full-length receptor interacted with Brn-3a (lane 3) and Brn-3b (lane 6). This ER–Brn-3 protein association was a specific interaction, since the in vitro-translated wild-type ER did not interact with the GST moiety of the fusion protein (lane 1) and the Brn-3–GST fusion proteins did not retain any of the 35S-labelled in vitro-translated control containing the luciferase protein (lanes 2 and 5). Furthermore, the interaction between wild-type ER protein and Brn-3a or Brn-3b was not modified by the presence of the ligand 17β-estradiol (lanes 4 and 7, respectively). Thus, the interactions between this receptor and Brn-3a or Brn-3b can occur in the absence or presence of 17β-estradiol in vitro.
FIG. 1.
Brn-3a and Brn-3b proteins interact with the wild-type (wt) ER protein in vitro. (A) SDS-PAGE analysis of products retained by the Brn-3a–GST or Brn-3b–GST fusion proteins, which were incubated with either 35S-labelled in vitro-translated (IVT) wild-type ER protein in the absence (lanes 3 and 6, respectively) or in the presence (lanes 4 and 7) of 10−8 M 17-β-estradiol or with an in vitro-translated control containing the luciferase protein (lanes 2 and 5). The reaction was carried out as described in Materials and Methods, and the gel was dried and exposed to film overnight. To show that the GST moiety of the fusion protein did not interact with the in vitro-translated proteins, the protein was incubated with ER under similar conditions (lane 1). (B) This protein interaction is independent of the presence of contaminating DNA. This was shown by carrying out the pull-down assay in either the absence (lane 1) or the presence (lane 2) of 50 μg of EtBr prior to and during the protein-protein interaction protocol described above.
To confirm that this protein-protein interaction was not dependent on the presence of contaminating DNA, these studies were repeated in the presence of EtBr, which should prevent any association of proteins that is dependent on contaminating DNA (32a). Fifty micrograms of EtBr was added to one sample of the in vitro-translated wild-type ER approximately 30 min prior to incubation with the fusion protein. The samples with or without EtBr were then mixed with the Brn-3–GST fusion protein and incubated as before. EtBr was maintained in the test sample during the incubation period and in the subsequent washes. As seen in Fig. 1B, the intensities of the bands representing the ER protein retained by association with Brn-3a were the same. This result indicates that the EtBr had no effect on the interaction between Brn-3a and the ER, and so this association was not dependent on the presence of contaminating DNA.
Domain C of ER interacts with the POU domain of Brn-3.
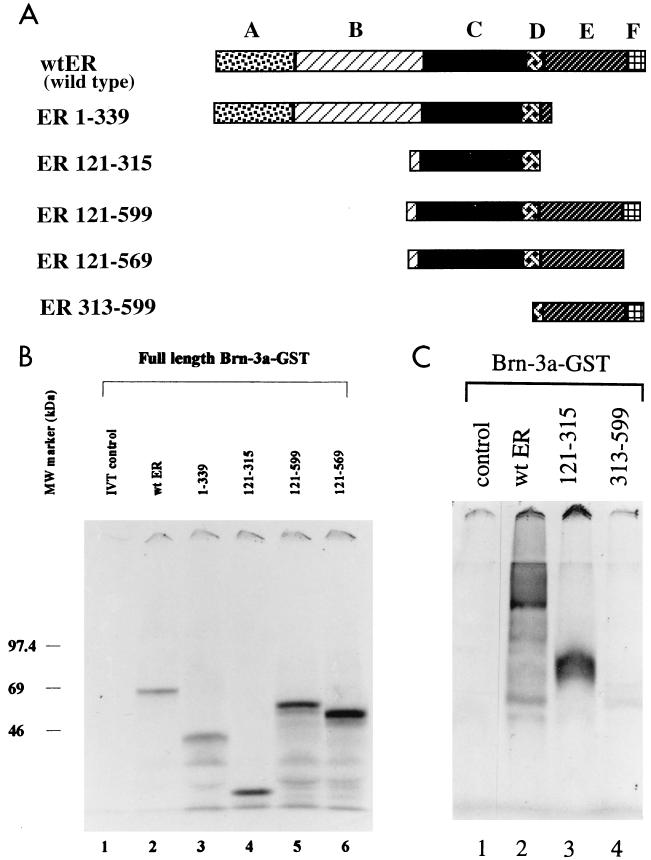
To identify the domain or domains of the ER which interacted with the Brn-3 proteins, we used truncated ER proteins in which different domains were deleted (Fig. 2A) in similar pull-down experiments. The volumes of in vitro-translated proteins used for the interaction studies were adjusted after densitometric scanning of a test gel so that similar amounts of each protein were used. The ER constructs did not bind to the GST moiety of the fusion protein (not shown), and the Brn-3a–GST fusion protein did not retain any proteins from the labelled in vitro control reaction (lane 1). As expected, the wild-type ER bound to Brn-3a–GST protein with approximately 7% of the input proteins retained (Fig. 2B, lane 2). Construct ER 1–339 (nucleotide positions 1 to 339), containing domains A, B, and C but not the carboxyl-terminal domains of the ER, was retained by the Brn-3a protein in a manner similar to that observed with wild-type ER (lane 3). The DNA-binding C domain (ER 121–339) on its own interacted with Brn-3a protein (lane 4), and approximately 37% of the input protein was retained. Similar amounts of the truncated proteins containing domains C to F (ER 121–599) (lane 5) or with domains C to E (ER 121–169) (lane 6) were also retained. This indicated that the constructs containing the DNA-binding C domain can interact with Brn-3 proteins with the ER proteins lacking the A and B domains being retained more readily in these studies.
FIG. 2.
(A) Schematic diagram showing the structure of the truncated ER proteins used to study the domains which contribute to the interaction of the receptor with Brn-3 proteins. The numbers indicate the positions of the amino acids in the wild-type receptor. (B) Brn-3 proteins can interact with ER proteins containing the DNA-binding C domain. Pull-down assays were carried out with the full-length Brn-3a–GST fusion protein and wild-type or truncated ER containing the C domain. The proteins retained following the interaction studies were resolved by SDS-PAGE analysis on a 12% polyacrylamide denaturing gel. Approximately 7% of the input protein of either wild-type ER (lane 2) or ER 1–339 (lane 3) was retained by Brn-3a compared with 37% of the input proteins of the ER 121–315 (lane 4) construct containing the C domain only or ER 121–599 (lane 5) and ER 121–569 (lane 6), which also contain other domains in the carboxyl terminus of the ER. Lane 1 shows the labelled in vitro-translated (IVT) luciferase protein incubated under the same conditions with Brn-3a. (C) The C domain (ER 121–315) was critical for the interactions with Brn-3 proteins. Protein interaction studies were carried out with the receptor clone ER 313–599, which lacked domain C but contained carboxyl-terminal domains D to F (lane 4), and the results were compared with those for either full-length ER (lane 2) or ER 121–315 (lane 3). Incubation with the control protein is shown in lane 1.
To check that this interaction with Brn-3 required the C domain of ER, these studies were repeated with a construct containing amino acids 313 to 599 which lacked the C domain. As expected, interactions with the Brn-3 fusion proteins were observed with the wild-type ER (Fig. 2c, lane 2) and C domain (lane 3) but not with ER 313–599 (lane 4). This suggests that the DNA-binding C domain of the receptor was critical for mediating the interaction with the Brn-3 proteins, since only ER proteins containing this domain resulted in binding, and the strongest interaction was observed with the isolated C domain. Another POU domain protein, Pit-1, which exhibited weak protein-protein interactions with the ER, was shown to interact with the DNA-binding C domain of the ER via its own DNA-binding POU domain (27).
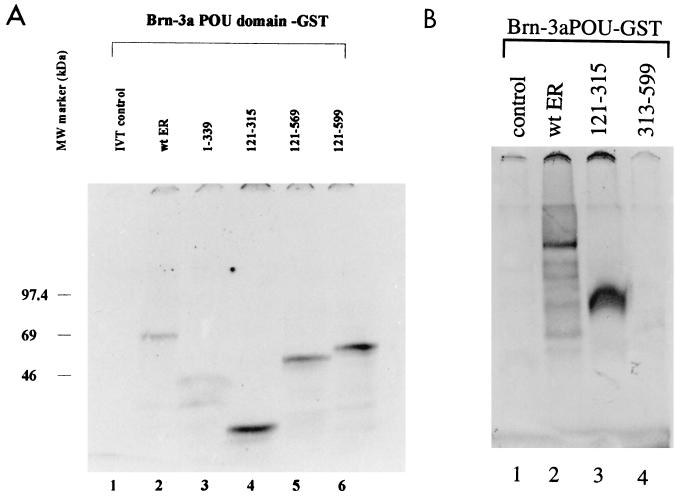
To establish whether the POU domains of Brn-3a and Brn-3b were sufficient for this interaction with the ER protein, fusion proteins containing the POU domains of Brn-3a and Brn-3b were used in similar interaction studies with the wild-type and truncated ER proteins. Figure 3A shows the results of interaction studies with the Brn-3a POU domain and indicates that the interactions were similar to that observed with the full-length protein. Thus, the associations between the Brn-3a POU protein and either wild-type ER (lane 2) or ER 1–339 (lane 3) were similar, whereas the C domain of the ER showed much stronger interaction with the POU domain (lane 3). In addition, the constructs which contain domains C to F but lack the amino-terminal A and B domains also appeared to associate more readily with the Brn-3a POU domain (lanes 4 to 6). However, ER 313–599, which lacks the C domain but contains domains D to F, did not bind the Brn-3a POU domain (Fig. 3B, lane 4) compared with the observed interaction with the wild-type ER (lane 2) and the C domain only (lane 3). Similar results were obtained with Brn-3b (data not shown).
FIG. 3.
(A) ER proteins can interact with the POU domain of Brn-3a. Interaction studies of wild-type (wt) or truncated ER with Brn-3a POU–GST fusion protein were carried out as described before. The Brn-3a POU protein was incubated with either labelled in vitro-translated (IVT) luciferase protein (lane 1), wild-type ER (lane 2), ER 1–339 (lane 3), ER 121–315 (lane 4), ER 121–569 (lane 5), or ER 121–599 (lane 6). The retained proteins were resolved and analyzed by SDS-PAGE and autoradiography. MW, molecular mass. (B) The C domain (ER 121–315) also mediated the interactions with the Brn-3 POU domain. ER 313–599, which lacked domain C but contained carboxyl-terminal domains D to F, displayed no binding (lane 4) compared with either full-length ER (lane 2) or ER 121–315 (lane 3). Incubation with the control protein is shown in lane 1.
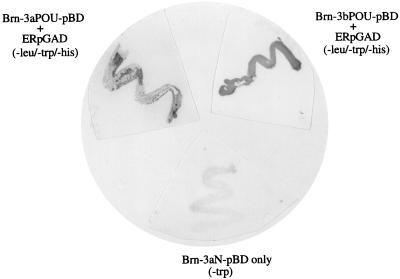
The interaction between the wild-type ER and the POU domain of the Brn-3 proteins was confirmed by the yeast two-hybrid system. The POU domains of Brn-3a (Brn-3a POUpBD) and Brn-3b (Brn-3b POUpBD) and the amino-terminal region of Brn-3a (Brn-3aN pBD) were cloned into the yeast two-hybrid vector containing the GAL4 DNA-binding domain and were cotransformed with the GAL4 activation domain vector containing the cDNA sequence encoding the wild-type ER (ER-pGAD). As expected, when the individual plasmids were transformed into yeast cells, colonies were observed when cells containing the Brn-3 pBD constructs were grown on medium lacking tryptophan and ER-pGAD cells were grown on plates lacking leucine, but not when they were grown in selection media lacking leucine, tryptophan, and histidine. However, cotransformation of Brn-3a pBD or Brn-3b pBD cells with ER-pGAD resulted in a number of colonies when the cells were plated on agar which lacked leucine, tryptophan, and histidine, indicating a functional interaction between the POU domains and the ER. No colonies were obtained when ER-pGAD cells were cotransformed with Brn-3aN pBD and grown in this selection medium, suggesting that the amino-terminal domain of Brn-3a does not interact with the ER.
Clones resulting from the interaction of the Brn-3a or Brn-3b POU domain and ER were reselected by growth on fresh plates lacking leucine, tryptophan, and histidine, and these colonies were assayed for activity of the second reporter gene, lacZ. As seen in Fig. 4, colonies resulting from either Brn-3a or Brn-3b POUpBD interacting with ER showed strong β-galactosidase activity. The cells transformed with the individual plasmids such as Brn-3aN pBD (Fig. 4, lower panel) gave rise to clones in medium lacking tryptophan but containing histidine and leucine, but when assayed for β-galactosidase activity, all colonies remained white, indicating no promoter activity. This was also observed with colonies obtained when plasmids containing Brn-3 POU domain or ER coding sequences were transformed alone and grown in media lacking only tryptophan or leucine, respectively (not shown). These results therefore indicate that the Brn-3a or Brn-3b POU domains must interact with the ER protein to activate histidine and lacZ promoter activity. These results therefore confirm that the interaction with the ER occurs via the isolated POU domain of the Brn-3 proteins and also occurs in vivo.
FIG. 4.
Interaction between the POU domains of Brn-3a and Brn-3b with the ER as demonstrated by the yeast two-hybrid system. The POU domains of Brn-3a (Brn-3a POUpBD) and Brn-3b (Brn-3b POUpBD) and the amino terminus of Brn-3a (Brn-3aN pBD) were cloned into the GAL4 DNA-binding domain vector. The full-length cDNA encoding the wild-type ER was cloned into the GAL4 activation domain vector (ER-pGAD). The different Brn-3-containing vectors either were transformed individually or were transformed together with ER-pGAD into competent HF7c yeast and plated out onto agar plates lacking tryptophan only (−trp) or tryptophan, leucine, and histidine (−leu/−trp/−his). Clones which resulted from interaction between the POU domains and ER when grown on agar plates lacking leucine, tryptophan, and histidine were reselected and assayed for activity of the second reporter gene coding for β-galactosidase (top panels). Brn-3aN pBD did not give rise to any clones when cotransformed with ER-pGAD and plated on media lacking leucine, tryptophan, and histidine, but there was growth on agar plates lacking tryptophan. These colonies remained white after the assay for lacZ promoter activity (lower panel).
Brn-3 proteins can be immunoprecipitated with the ER.
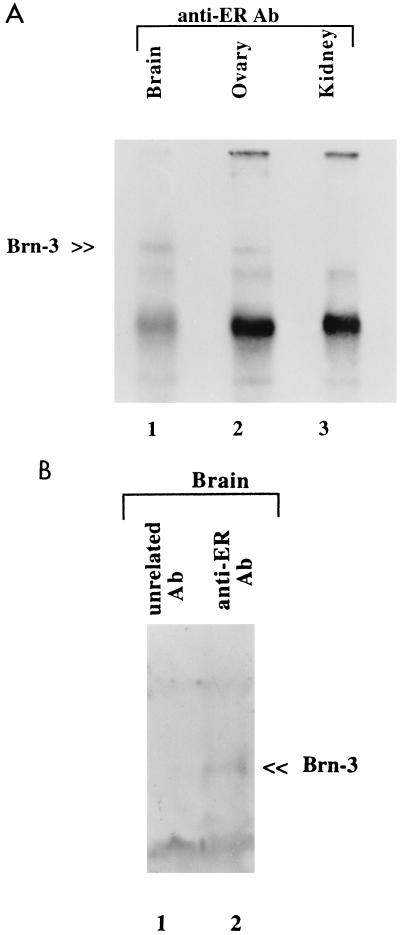
To confirm that this interaction can occur under physiological conditions, immunoprecipitation studies were performed with homogenate from tissues known to express these factors. Proteins were extracted from rat brain and ovary, both of which express Brn-3a, Brn-3b, and the ER protein, as well as from kidney, which has no detectable Brn-3 proteins. Immunoprecipitation was carried out with precleared lysates and the ER antibody SRA1010. The specificity of the interactions was assessed by incubation of the extracts with antibody to a protein known not to interact with the Brn-3 proteins or with no antibody. Following collection of the immunocomplexes, the proteins were resolved by SDS-PAGE analysis on a 12% polyacrylamide denaturing gel. This was transferred to a membrane filter, which was probed with anti-Brn-3a or anti-Brn-3b antibodies. Figure 5 shows the results following immunoblotting with Brn-3b antibodies, which were similar to those for Brn-3a (not shown). The specific bands representing Brn-3b (indicated) were detected when proteins extracted from brain (Fig. 5A, lane 1) and ovary (lane 2) but not kidney (lane 3) were immunoprecipitated with the ER antibody. A band was not present when the control anti-Bad antibody was incubated with protein extracts (Fig. 5B, lane 1) or with no antibody in the first step (not shown).
FIG. 5.
(A) Brn-3 proteins can be immunoprecipitated with the ER from protein extracts of tissues which coexpress these two factors. Protein extracts from either rat brain (lane 1), ovary (lane 2), or kidney (which lacks Brn-3) (lane 3) were precleared and then incubated with anti-ER antibodies (Ab). Immunocomplexes were collected, washed, and resolved by SDS-PAGE on a 12% polyacrylamide gel and then detected after Western blotting and probing with the anti-Brn-3 antibodies. The position of the Brn-3b protein is indicated. This Brn-3 band was not observed if no antibody was added after the preclearing step (not shown). The other bands are nonspecific or represent the immunoglobulin G complex resulting from use of the protein A-protein G-agarose slurry. (B) Brn-3 protein is specifically retained by the ER, since addition of antibody specific for the unrelated Bad protein (lane 1) to protein extracted from brain does not immunoprecipitate Brn-3b, as observed with the ER antibody (lane 2).
Thus, the ER association with the Brn-3 proteins can occur both in vivo and in vitro. In addition, this interaction was observed in the absence of DNA, is independent of binding of the ligand estradiol, and appears to require the DNA-binding C domain of the ER and the POU domain of the Brn-3 proteins.
Brn-3 increases the affinity of ER binding to the ERE.
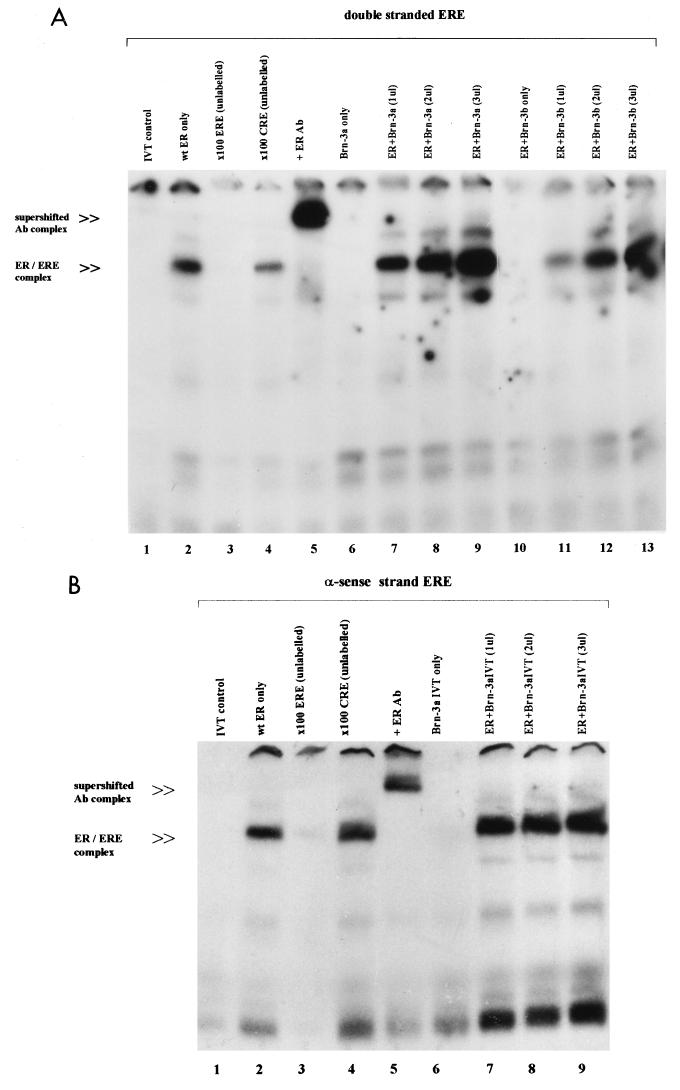
Since the DNA-binding C domain of the ER is involved in the interaction with the Brn-3 proteins, we wanted to find out if this association affected the binding of the receptor to its DNA recognition site, the ERE, and so modulated transcriptional regulation by the receptor complex. We therefore used the EMSA to analyze the effect of addition of increasing amounts of Brn-3a on the ER complex bound to an ERE. Figure 6A shows the interaction of the ER with labelled double-stranded ERE derived from the vitellogenin gene promoter. As expected, there was specific binding of the ER to the ERE, which gave rise to an ER-ERE complex (lane 2) which was not present when the in vitro-translated control containing the luciferase protein was incubated with the labelled probe (lane 1). This complex was specifically competed away upon addition of unlabelled ERE oligonucleotide (lane 3), but not by a nonspecific DNA sequence (lane 4). Furthermore, it was supershifted when antibodies to the ER were added to the reaction mixture (lane 5), indicating the presence of the receptor in this complex. While in vitro-translated Brn-3a or Brn-3b did not appear to interact with the labelled double-stranded ERE oligonucleotide in the absence of the ER (lanes 6 and 10), addition of increasing amounts (1 to 3 μl) of either Brn-3a (lanes 7 to 9) or Brn-3b (lanes 11 to 13) in the presence of the receptor resulted in much stronger intensity of the ER-ERE complex. This complex of increasing intensity can be supershifted by the ER antibodies (data not shown), indicating the presence of increasing amounts of the receptor protein in the complex. Therefore, both Brn-3a and Brn-3b specifically enhance the ER complex-DNA interaction, since addition of a related POU protein, Oct-2, had no effect on ER-ERE complex formation (data not shown).
FIG. 6.
(A) Brn-3a and Brn-3b enhance the binding of the ER complex on double-stranded ERE. EMSA was performed with 32P-labelled double-stranded ERE and in vitro-translated wild-type (wt) ER and Brn-3 proteins under the conditions described in Materials and Methods. The resulting ER-ERE complex was resolved from free labelled probe on a 7% nondenaturing polyacrylamide gel, which was dried and exposed to film. The labelled double-stranded ERE probe was incubated with the ER proteins and no competitor (lane 2) or with a 100-fold molar excess of either unlabelled ERE oligonucleotide (lane 3) or unlabelled nonspecific (CRE) competitor oligonucleotide (lane 4). One microliter of anti-ER (human) antibodies (Ab) was added to the reaction mixture in lane 5, resulting in the supershifted ER-ERE complex indicated. Brn-3a and Brn-3b were incubated with the labelled ERE on their own (lanes 6 and 10, respectively) or in the presence of the ER and increasing amounts (1 to 3 μl) of Brn-3a (lanes 7 to 9) or Brn-3b (lanes 11 to 13). The in vitro-translated (IVT) control luciferase protein was incubated with the labelled ERE and is shown in lane 1. (B) Effect of Brn-3 protein on the ER complex binding to the single-stranded antisense (α-sense) ERE DNA. EMSA showing the effect of addition of increasing amounts of Brn-3 to the single-stranded ERE DNA. The position of the main ER-ERE complexes is indicated. Lane 2 shows the ER protein incubated with labelled antisense ERE and no competitor, and lanes 3 and 4 show a 100-fold molar excess of either unlabelled ERE oligonucleotide (lane 3) or unlabelled nonspecific (CRE) competitor oligonucleotide (lane 4). Anti-ER (human) antibody (Ab) was added to the reaction mixture in lane 5. Lanes 6 to 9 show the effect of incubation of Brn-3a with the labelled ERE on its own (lane 6) or in the presence of the ER and increasing amounts (1 to 3 μl) of Brn-3a in vitro-translated protein (lanes 7 to 9). The in vitro-translated control containing the luciferase protein was incubated with the labelled ERE (lane 1).
Studies by others have shown that the ER bound to ERE more readily if the DNA was in the single-stranded conformation, and this requires a single-stranded DNA-binding protein for efficient binding, although this protein did not appear to bind to the ERE DNA itself (50–53). Since the Brn-3 proteins have been shown to bind single-stranded DNA, it is possible that interaction of the ER with Brn-3 somehow enhanced binding of the ER complex to the double-stranded DNA by facilitating changes in the ERE conformation. Therefore we wanted to see if increasing amounts of the Brn-3 proteins could significantly increase binding of the ER complex to single-stranded ERE in the same manner as that observed on double-stranded ERE. The experiments were therefore repeated with labelled single-stranded ERE (sense and antisense strands). We observed that while the ER did not bind to the sense strand (data not shown), it bound strongly and specifically to the antisense strand of the ERE (Fig. 6b). Thus, in the presence of the labelled antisense ERE, the ER protein formed an ER-ERE complex (lane 2) which was not present when the control protein was incubated under the same conditions (lane 1). This band was specifically competed by addition of cold ERE oligonucleotide (lane 3), but not the nonspecific competitor (lane 4), and was supershifted in the presence of the ER antibody (lane 5). Brn-3a protein did not appear to stably bind to the ERE (lane 6), but addition of 1 μl of Brn-3a protein resulted in a small increase in the protein-DNA complex in the presence of ER (lane 7). However when increasing amounts (2 and 3 μl) of Brn-3a were added (lanes 7 to 9), there was no significant increase in the intensity of this complex which was observed on double-stranded ERE.
The ER therefore associates more readily with the ERE in the presence of Brn-3a or Brn-3b proteins, and this effect is more significant on double-stranded ERE than the changes observed on the single-stranded antisense ERE to which the ER binds.
Brn-3a and Brn-3b can modulate the activity of a promoter containing an ERE.
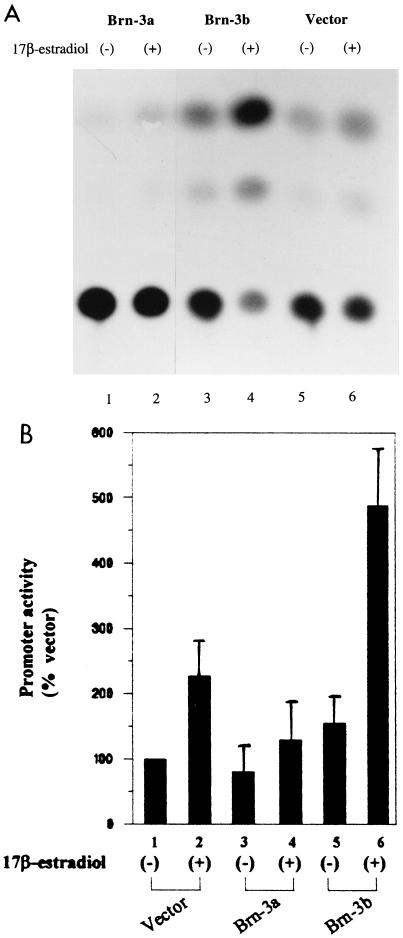
To study the functional relevance of the interaction between the Brn-3 proteins and the ER, cotransfection experiments were carried out in which the vit-tk CAT reporter construct containing a functional ERE (derived from region −331 to −87 of the vitellogenin gene [see Materials and Methods]) was cotransfected with pLTR expression vectors, which drive the expression of Brn-3a and Brn-3b (65). Figure 7A shows a representative CAT assay following the cotransfection of the proteins with the treatment indicated, while Fig. 7B shows the results of at least three independent experiments. Results were expressed as a percentage of the promoter activity observed when the empty expression vector was cotransfected with the reporter construct. In the absence of 17β-estradiol, Brn-3a had a slight inhibitory effect on the basal activity of the promoter, while Brn-3b mildly activated it compared with the control. However, upon stimulation with 10−8 M 17β-estradiol ligand, overexpression of Brn-3b resulted in strong activation of the promoter, while Brn-3a was still a weak repressor compared with the empty vector. Thus, while both Brn-3a and Brn-3b can bind to the ER, they appear to mediate different effects on promoter activity. Cotransfection of Brn-3a or Brn-3b with the empty pBLCAT2 reporter vector containing the thymidine kinase promoter had no effect on the promoter activity (6).
FIG. 7.
Brn-3a and Brn-3b modulate the activity of an ERE-containing promoter. The CAT assay results show the effect of cotransfection of a reporter construct containing the ERE sequence derived from the vitellogenin gene promoter with Brn-3 expression vectors. Studies were carried out with MCF7 cells, which were grown in medium containing no phenol red and supplemented with dextran-coated charcoal-stripped serum. Cells were either untreated (−) or were stimulated with 10−8 M 17β-estradiol (+) after transfection. (A) Representative assay showing CAT activity after transfection and the treatments indicated. (B) Results of at least three independent CAT assays showing activity resulting from cotransfection with Brn-3a or Brn-3b with the reporter construct expressed as a percentage of the activity of the empty vector (bar 1). Bar 2 shows the basal activity of the empty vector in stimulated cells, while the effects of overexpression of Brn-3a (bars 3 and 4) or Brn-3b (bars 5 and 6) in untreated or stimulated cells are shown by bars 3 to 6.
Brn-3a and Brn-3b mediate their effect via the ERE.
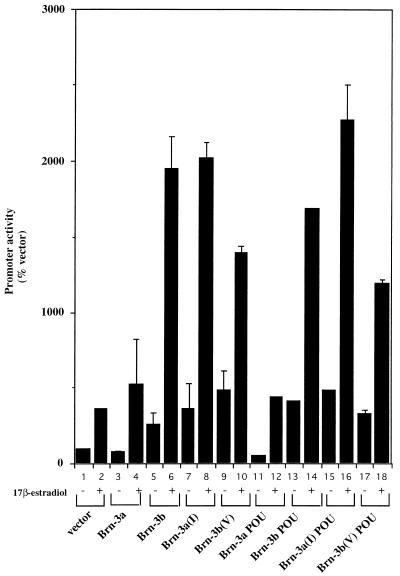
To establish if the effect observed on transcription of this ERE-containing promoter was mediated via the ERE alone, cotransfection experiments were carried out with a reporter construct containing just the ERE sequence cloned upstream of the thymidine kinase promoter and driving the expression of the luciferase gene (see Materials and Methods). The results shown in Fig. 8 are expressed as a percentage of the empty expression vector (bar 1). As before, in the untreated cells, Brn-3a acted as a weak repressor (bar 3) while Brn-3b mildly activated the promoter (bar 5). In the presence of 17β-estradiol, Brn-3a no longer repressed the promoter but had little significant effect on its activity (bar 4) compared with the vector only in stimulated cells (bar 2). However, Brn-3b was a strong activator of the promoter upon stimulation with the ligand (bar 6). Therefore, the opposite effect on the transcriptional activity by Brn-3a and Brn-3b appeared to be mediated via the ERE.
FIG. 8.
Brn-3a and Brn-3b have opposite effects on activity of a heterologous promoter containing the ERE sequence only. Luciferase assays were carried out after cotransfection of Brn-3a or Brn-3b and the ERE-containing reporter construct. Transfections were carried out in MCF7 cells, which were grown in phenol red-less medium supplemented with dextran-coated charcoal-stripped serum. Cells were either untreated (−) or were stimulated with 10−8 M 17β-estradiol (+) after transfection. Results are expressed as a percentage of the activity of the empty vector in untreated cells (bar 1). Bar 2 shows the basal activity of the empty vector in stimulated cells, while the effects of overexpression of wild-type Brn-3a (bars 3 and 4) or Brn-3b (bars 5 and 6) on promoter activity in untreated or stimulated cells are shown by bars 3 to 6. Full-length Brn-3a mutants in which the amino acid valine at position 22 of the POU homeodomain was converted to isoleucine of Brn-3b [Brn-3a(I)] were also used (bars 7 and 8). The effect of the reciprocal substitution in Brn-3b (Brn-3b(V) is shown by bars 9 and 10. The effect of cotransfection of expression vectors containing only the POU domain of the Brn-3 proteins is also shown with Brn-3a POU by bars 11 and 12 and with Brn-3b POU by bars 13 and 14. Expression mutants containing the changed amino acid residue in the POU homeodomain, Brn-3a POU(I) and Brn-3b POU(V), were also used in these experiments (bars 15 and 16 and 17 and 18, respectively). These results were reproduced in at least three independent experiments.
Since the ER can interact with the Brn-3 proteins via their POU domains, we were interested in testing whether this domain was sufficient to confer transcriptional activity to the ERE-containing promoter. Therefore, similar cotransfection experiments were carried out with expression vectors containing the isolated POU domains of Brn-3a and Brn-3b. The results (Fig. 8, bars 11 to 14) indicated that the POU domain was sufficient to confer some of the effect of the full-length proteins to the ERE-containing promoter. Thus the Brn-3a POU domain repressed promoter activity in untreated cells but had little effect on 17β-estradiol-stimulated cells (bars 11 and 12). Similarly, the POU domain of Brn-3b mildly activated the promoter in unstimulated cells, and addition of the ligand 17β-estradiol resulted in a significant but smaller increase in promoter activity (bars 13 and 14) than that of the full-length Brn-3b.
Valine-to-isoleucine conversion in the first helix of the POU domain changes Brn-3a to an activator.
It was previously shown that Brn-3a and Brn-3b share very high sequence homology within the POU domain, with only seven amino acid differences in this region. Six of these changes are in the linker region which joins the POU-specific domain and POU homeodomain and which is poorly conserved between different POU factors. However one significant difference was the conversion of a valine residue in Brn-3a to isoleucine in Brn-3b at position 22 in the first helix of the POU homeodomain, a region which has been associated with protein-protein interactions in other POU proteins (32, 58, 72). We have shown that the POU domain interacts with the ER and was sufficient to mediate the effects on transcription. Therefore, we were interested in checking whether the difference in this critical amino acid in the homeodomain contributed to the differences in the effects of Brn-3a and Brn-3b on promoter activity and whether changing this could modify the transcriptional effect. Therefore, mutants of Brn-3a and Brn-3b were constructed in which the valine residue in the Brn-3a POU domain was converted to isoleucine found at this position in the Brn-3b POU domain [Brn-3a(I)]. The reciprocal isoleucine-to-valine mutation was made in the Brn-3b POU domain [Brn-3b(V)]. These mutations were made in the full-length Brn-3a and Brn-3b cDNA sequences as well as in the isolated POU domains, and then the sequences were cloned into an expression vector and used in cotransfection studies with the ERE reporter.
As seen in Fig. 8, bars 7 and 8, the conversion of valine to isoleucine at this position resulted in Brn-3a behaving in a manner which was comparable to that of Brn-3b. Thus, in the untreated cells, it mildly activated the promoter, while it behaved as a strong activator of transcription in the presence of 17β-estradiol compared with the repressive effect of the wild-type Brn-3a (bars 3 and 4). Moreover, there was a small but significant and reproducible decrease in activation by Brn-3b(V) (bars 9 and 10) compared with that by the wild-type Brn-3b (bars 5 and 6). Therefore, the mutated Brn-3b was still capable of activating transcription, but to a reduced extent. Changing this amino acid in the isolated POU domain expression constructs also gave rise to similar changes on transcriptional activity. Brn-3aPou(I) was a strong activator of gene transcription (bars 15 and 16), comparable to the full-length wild-type Brn-3b and greater than the Brn-3b POU domain on its own in stimulated cells. The Brn-3bPou(V) mutant again resulted in a small but reproducible decrease in promoter activity (bars 17 and 18) compared with the Brn-3b POU domain on its own.
Therefore, the amino acid at this position must play a critical role in the ability of these factors to modulate transcriptional activity under these conditions and on this promoter. Since both Brn-3 proteins bind to the ER, it is unlikely that amino acids in this position would influence the Brn-3–ER associations. However, this region may provide the interface for interactions with other proteins, with the presence of valine or isoleucine facilitating binding of different proteins to Brn-3a or Brn-3b. Therefore, when associated with the ER, Brn-3b may bind to a protein which behaves as an activator in stimulated cells, while Brn-3a may be associated with a different protein which has little significant effect on transcription. This would explain why the amino acid conversion in this position so significantly influences transcriptional activation by the mutants.
DISCUSSION
Nuclear receptors such as the ER are complexed with a number of proteins in both the active and inactive states, and the associated proteins are critical for modifying the state of the complex and hence affecting gene transcription. While a number of proteins interacting with the ER have been identified and studied, the mechanism by which this receptor modulates gene transcription is still unclear. The ER has been shown to be present in a large number of different tissues (13, 14, 29, 56). While some of the factors associated with the receptor are ubiquitously expressed, others may be cell-specific factors which are different in each cell type. The ability of nuclear receptors to interact with POU proteins to modulate transcription of target genes has been demonstrated by the synergistic interaction of the POU transcription factor Pit-1 with the thyroid hormone receptor to activate growth hormone gene transcription (12) and with the ER to modulate expression of the prolactin promoter (16, 27).
We report on the novel interaction of the POU domain transcription factors Brn-3a and Brn-3b with the ER and show that these interactions can modulate the transcriptional activity of promoters containing an ERE site. Isolation of an immunocomplex formed between Brn-3 proteins and the ER from tissue extracts which coexpress these two proteins indicates that this interaction can occur in these cell types under physiological conditions. Furthermore, the association between the Brn-3 proteins and the receptor appears to be independent of either of these proteins binding to DNA or binding of the ligand 17β-estradiol to the ER. Landel et al. (34) reported a similar association of the ER with two uncharacterized proteins with sizes of 45 and 48 kDa (p45 and p48) with the ER which were critical for the association of the ER with the ERE, but whose interaction with the receptor was not affected by estrogen agonists or antagonists. The Brn-3–ER association appears to be mediated via the POU domain of the Brn-3 protein and the DNA-binding C domain of the ER, since these two domains can interact in the absence of other domains of either protein, as shown by in vitro studies as well as in vivo by the yeast two-hybrid system. Furthermore, a construct lacking the C domain did not bind to the Brn-3 proteins. This requirement for the DNA-binding domains of both proteins in this interaction is similar to that observed with Pit-1 and the ER which demonstrated weak protein-protein interaction via their respective DNA-binding domains (16, 27). Different domains of the ER may contribute to the interactions, since the constructs which lack the A and B activation domains but contain the DNA-binding C domain were more readily retained by both Brn-3 proteins.
A number of studies have indicated that there are multiple variants of the ER mRNA in normal or malignant breast tissue (17, 19, 30, 30a, 41, 45, 50, 51) and in endometrial carcinoma (44) which may encode different isoforms of the protein. Some of these variants can alter transcriptional activity by the receptor (17, 18, 28, 29), and it is possible that these changes may be a result of altered interaction with proteins, such as Brn-3, which associates with the ER to form the transcriptionally active complex. Furthermore, isoforms of Brn-3 proteins have also been identified (21, 43, 63, 69), and while the differences in these spliced variants lie outside the POU domain, it is possible that these changes may alter the ER–Brn-3 interactions and/or modify the effects on gene transcription.
We have also shown that both Brn-3a and Brn-3b appear to enhance the binding of the ER complex to the ERE more significantly when the DNA is in a double-stranded conformation compared with the single antisense strand to which the ER complex can bind. Thus addition of increasing amounts of Brn-3 proteins resulted in a complex of increasing intensity which was shown to contain the ER. Lannigan and Notides (35) demonstrated that while the ER bound to double-stranded DNA containing an ERE sequence, it binds preferentially and with higher affinity to the single-stranded antisense sequence (lower strand) of this DNA. Furthermore, they showed that the ERE consensus DNA sequences form tertiary structures with which the ER interacts (36, 37). We also found that the ER complex can bind to the single antisense strand (single-stranded ERE) but not the sense strand of the ERE. However, on the antisense single-stranded ERE, we found that addition of Brn-3 resulted in a small increase in the intensity of the ER-ERE complex, but addition of increasing amounts of Brn-3 did not change this substantially, as seen on double-stranded ERE. Studies of Mukherjee and Chambon (49) showed that a 45-kDa yeast single-stranded DNA-binding protein, DBSF, derived from cell extracts was required for the efficient and stable association of purified ER with the ERE. The mechanism by which the DBSF protein mediated the ER-ERE complex formation is not known, but this protein did not appear to bind to the double-stranded ERE. We have previously shown that both Brn-3a and Brn-3b bound preferentially to the antisense strand of their binding site in the α-internexin promoter (8). It is therefore possible that Brn-3 proteins act in a manner similar to the yeast DBSF protein. Thus, while Brn-3a and Brn-3b may not form a stable complex with the ERE on their own, interaction with the ER may facilitate binding of the complex to the ERE by helping induce changes in the conformation of the labile ERE structure or by helping stabilize a secondary structure which the single-stranded ERE is thought to form to allow the ER complex to bind more readily (35, 37, 38).
The ER–Brn-3 interaction also influenced the ability of the ER complex to modulate gene transcription. Associations with Brn-3a or Brn-3b alter transcriptional activity via an ERE such that Brn-3a coexpressed with the ERE acted as a mild repressor or had little effect, while Brn-3b was a strong activator of gene transcription in ligand-stimulated MCF7 cells. We also found that the POU domain of the Brn-3 proteins, via which these proteins interacted with the ER, was sufficient to mediate their distinct transcriptional effects, as observed with the full-length proteins. Interestingly, Brn-3a and Brn-3b have a high level of homology in the POU domain, with only seven amino acid differences, six of which occur in the poorly conserved linker region. However, there is a critical change from isoleucine in Brn-3a to valine at position 22 in the first helix of the POU homeodomain (42). The change of this single amino acid in the POU homeodomain can significantly modify the effect of both the full-length Brn-3a and Brn-3a POU domain, converting this protein from a repressor to an activator of ERE-containing promoters. The reciprocal change of the isoleucine to valine in wild-type Brn-3b as well as in the isolated POU domain resulted in a small but significant decrease in the ability of this domain to activate transcription.
The region containing this mutation is located on the surface of the POU domain and is thought to be critical for protein-protein interaction. Thus, in the case of the related POU factors Oct-1 and Oct-2, Oct-1 but not Oct-2 interacts with the herpes simplex virus transactivator Vmw65. Substitution of the alanine found at this position in Oct-2 with the glutamic acid residue found in Oct-1 allows the mutant Oct-2 to interact with Vmw65 (32a, 58). This effect appears to depend on the length of the side chain of the amino acid rather than the charge, since replacement of the glutamic acid in Oct-1 with glutamine allows binding to Vmw65, but this does not occur with alanine or aspartic acid present. It has been demonstrated that the amino acid change at this position in Brn-3a and Brn-3b may allow interaction with different proteins, which may contribute to the effect of these two factors on promoter activity (15). Since both Brn-3 proteins can interact with the ER in the presence or the absence of the ligand, this region should not influence the Brn-3–ER association. Rather, it is possible that this region allows interactions with other proteins and that the amino acid difference may result in the valine of Brn-3a associating with a protein that confers little activation potential, while the isoleucine residue of Brn-3b allows recruitment of an activator in ligand-stimulated cells.
It is interesting that on an ERE-containing promoter and in the presence of 17β-estradiol, Brn-3a had a weak inhibitory effect or no effect on promoter activity, while Brn-3b strongly activated this promoter. A number of studies have shown that upon the binding of the Brn-3 proteins to their own DNA recognition sequence, Brn-3a generally behaves as an activator of gene transcription, while Brn-3b represses the activity of a number of these promoters. In some instances, this effect is dependent upon the presence of valine in Brn-3a and isoleucine in Brn-3b at position 22 in the POU homeodomain (15, 48).
Thus, it is possible that upon the binding of Brn-3 proteins to their own site, their conformation is such as to allow interaction with proteins which results in activation of gene transcription by Brn-3a and repression by Brn-3b. On the ERE, Brn-3 factors primarily interact with the ER, and the protein-protein interactions may result in different conformations of these factors compared to their configuration if they are bound to DNA. This may allow interaction with a different set of proteins, thus giving rise to the different effects on promoter activity observed. The ability of transcriptional regulators to mediate such interaction without necessarily binding to DNA has been observed with other proteins, including the ER. For instance, on the brain creatine kinase gene promoter which is activated by the ER, transcriptional activation requires a functional transactivation domain (AF-2) and the DNA-binding C domain of the receptor, but it does not appear to require binding of the ER to the consensus ERE DNA to affect promoter activity (60, 63). On the AP-1 site in the ovalbumin gene promoter, the ER was shown to interact with Jun protein found in Jun-Jun homodimers or Jun-Fos heterodimers to modulate AP-1-directed gene activity (20, 63, 75). These combinatorial interactions with different sets of proteins represent a mechanism by which one set of transcription factors can modulate the expression of a large number of genes.
The interactions between Brn-3 transcription factors and the ER, which has been demonstrated in vitro as well as in vivo, therefore represents a novel interaction involving Brn-3 proteins which does not require binding to DNA. The ER–Brn-3 association can differentially affect gene transcription via an ERE and may therefore be important in activation of the functional ER complex and control of transcription of target genes in tissues which coexpress these factors. While the exact mechanism by which this is achieved remains unclear, it appears that interaction with other as yet unidentified proteins may modulate the effect of the Brn-3 proteins on ER-dependent gene expression. It would be interesting to see if any other proteins which have so far been identified as being associated with the ER may interact directly with the Brn-3 proteins. Identification of other factors which interact with Brn-3 proteins to affect transcription in tissues which coexpress Brn-3 and ER may help our understanding of the complex process by which this receptor mediates its effect on gene transcription.
ACKNOWLEDGMENTS
We thank Pirkko Hentuu and Sue Hoare (Molecular Endocrinology Laboratories, Imperial Cancer Research Fund, London) for provision of and assistance with some of the clones and cell lines used in this study.
This work was supported by the Medical Research Council.
REFERENCES
- 1.Andersen B, Pearse R V, Schleger P N, Cichon Z, Marcus M D, Bardin C W, Rosenfeld M G. Sperm-1: a POU domain gene transiently expressed immediately before meiosis 1 in the male germ cell. Proc Natl Acad Sci USA. 1993;90:11084–11088. doi: 10.1073/pnas.90.23.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen B, Schoneman M D, Pearse R V, Jenne K, Sugarman J, Rosenfeld M G. Brn-5 is a divergent POU domain factor highly expressed in layer IV of the neocortex. J Biol Chem. 1993;268:23390–23398. [PubMed] [Google Scholar]
- 3.Aurora R, Herr W. Segments of POU domain influence one another’s DNA-binding specificity. Mol Cell Biol. 1992;12:455–467. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baniahmad C, Nawaz Z, Baniahmad A, Gleeson M A G, Tsi M, O’Malley B W. Enhancement of human estrogen receptor activity by SPT6: a potential coactivator. Mol Endocrinol. 1995;9:34–43. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- 5.Berthois Y, Katzenellenbogen J A, Katzenellenbogen B S. Phenol red tissue culture media is a weak estrogen: implications concerning the study of estrogen responsive cells in culture. Proc Natl Acad Sci USA. 1986;83:2496–2500. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budhram-Mahadeo V, Theil T, Morris P J, Lillycrop K A, Moroy T, Latchman D S. The DNA target site for the Brn-3 POU family transcription factors can confer responsiveness to cyclic AMP and removal of serum in neuronal cells. Nucleic Acids Res. 1994;22:3092–3098. doi: 10.1093/nar/22.15.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budhram-Mahadeo V, Morris P J, Lakin N D, Theil T, Ching G Y, Lillycrop K A, Moroy T, Liem R K H, Latchman D S. Activation of the α-internexin promoter by the Brn-3a transcription factor is dependent on the N-terminal region of the protein. J Biol Chem. 1995;270:2853–2858. doi: 10.1074/jbc.270.6.2853. [DOI] [PubMed] [Google Scholar]
- 8.Budhram-Mahadeo V, Morris P J, Lakin N D, Dawson S J, Latchman D S. The different activities of the two activation domains of the Brn-3a transcription factor are dependent on the context of the binding site. J Biol Chem. 1996;271:9108–9113. doi: 10.1074/jbc.271.15.9108. [DOI] [PubMed] [Google Scholar]
- 9.Budhram-Mahadeo V. Regulation and function of POU domain transcription factors, Brn-3a and Brn-3b. Ph.D. thesis. London, United Kingdom: University of London; 1995. [Google Scholar]
- 10.Catelli M G, Binart N, Jung-Testas I, Renoir J M, Baulieu E E, Feramisco J R, Welch W J. The common 90-kd protein component of non-transformed 8s steroid receptors is a heat shock protein. EMBO J. 1985;4:3131–3135. doi: 10.1002/j.1460-2075.1985.tb04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavailles V, Dauvios S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang W, Zhou W, Theill L E, Baxter J D, Schaufele F. An activation function in Pit-1 required selectively for synergistic transcription. J Biol Chem. 1996;271:17733–17738. doi: 10.1074/jbc.271.30.17733. [DOI] [PubMed] [Google Scholar]
- 13.Ciocca D R, Vargas Roig L M. Estrogen receptor in human nontarget tissues: biological and clinical implications. Endocr Rev. 1995;16:35–62. doi: 10.1210/edrv-16-1-35. [DOI] [PubMed] [Google Scholar]
- 14.Cunha G R, Cooke P S, Bigsby R, Brody J R. Ontogeny of sex steroid receptors in mammals. In: Parker M G, editor. Nuclear hormone receptors. London, United Kingdom: Academic Press; 1991. pp. 235–268. [Google Scholar]
- 15.Dawson S J, Morris P J, Latchman D S. A single amino acid change converts an inhibitory transcription factor into an activator. J Biol Chem. 1996;271:11631–11633. doi: 10.1074/jbc.271.20.11631. [DOI] [PubMed] [Google Scholar]
- 16.Day R N, Koike S, Sakai M, Muramatsu M, Maurer R A. Both Pit-1 and the estrogen receptor are required for estrogen responsiveness of the rat prolactin gene. Mol Endocrinol. 1990;12:1964–1971. doi: 10.1210/mend-4-12-1964. [DOI] [PubMed] [Google Scholar]
- 17.Fuqua S A, Fitzgerald S D, Chamness G C, Tandon A K, McDonnell D P, Nawaz Z, O’Malley B W, Greene G L, McGuire W L. Variant human breast tumour receptor with constitutive transcriptional activity. Cancer Res. 1991;51:105–109. [PubMed] [Google Scholar]
- 18.Fuqua S A, Fitzgerald S D, Allred D C, Elledge R M, McDonnell D P, Nawaz Z, O’Malley B W, Greene G L, McGuire W L. Inhibition of estrogen receptor action by a naturally occurring variant in human breast tumours. Cancer Res. 1992;52:483–486. [PubMed] [Google Scholar]
- 19.Garcia T, Lehrer S, Bloomer W D, Schachter B. A variant estrogen receptor messenger ribonucleic acid is associated with reduced levels of estrogen binding in human mammary tumours. Mol Endocrinol. 1988;3:687–693. doi: 10.1210/mend-2-9-785. [DOI] [PubMed] [Google Scholar]
- 20.Gaub M-P, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the Fos-Jun complex. Cell. 1990;63:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- 21.Gerrero M R, McEvilly R J, Turner E, Lin C R, O’Connell S, Jenne K J, Hobbs M V, Rosenfeld M G. Brn-3.0, a POU domain protein expressed in the sensory, immune and endocrine systems that functions on elements distinct from known octamer motifs. Proc Natl Acad Sci USA. 1993;90:10841–10845. doi: 10.1073/pnas.90.22.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman C M. High efficiency transfer into mammalian cells. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 23.Green S, Chambon P. The oestrogen receptor: from perspective to mechanism. In: Parker M G, editor. Nuclear hormone receptors. London, United Kingdom: Academic Press; 1991. pp. 15–38. [Google Scholar]
- 24.Halachimi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 25.He X, Treacy M N, Simmons D M, Ingraham H A, Swanson L S, Rosenfeld M G. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–42. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 26.Herr W, Strum R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham P A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. The POU domain: a large conserved region in the mammalian pit-1 Oct-1, Oct-2 and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 27.Holloway J M, Szeto D P, Scully K M, Glass C K, Rosenfeld M G. Pit-1 binding to specific DNA sites as a monomer or dimer determines gene-specific use of a tyrosine dependent synergy domain. Genes Dev. 1995;9:1992–2006. doi: 10.1101/gad.9.16.1992. [DOI] [PubMed] [Google Scholar]
- 28.Ince B A, Zhuang Y, Wrenn C K, Shapiro D J, Katzenellenbogen B S. Powerful dominant negative mutants of the human estrogen receptor. J Biol Chem. 1993;268:14026–14032. [PubMed] [Google Scholar]
- 28a.Ince B A, Schodin D J, Shapiro D J, Katzenellenbogen B S. Repression of endogenous estrogen receptor activity in MCF-7 human breast cancer cells by dominant negative estrogen receptor. Endocrinology. 1993;136:3194–3199. doi: 10.1210/endo.136.8.7628351. [DOI] [PubMed] [Google Scholar]
- 29.Keffer D A, Holdregger C. The ontogeny of the estrogen receptor: the brain and pituitary. Dev Brain Res. 1985;19:1386–1395. doi: 10.1016/0165-3806(85)90190-7. [DOI] [PubMed] [Google Scholar]
- 30.Kuiper G G M J, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J A. Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Kuiper G G M J, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J A. Comparison of the ligand binding specificity and transcript tissue distribution of the estrogen receptor α and β. Endocrinology. 1996;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 31.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 32.Lai J S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeodomain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 32a.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakin N D, Morris P J, Theil T, Sato T N, Moroy T, Wilson M C, Latchman D S. Regulation of neurite outgrowth and SNAP-25 gene expression by the Brn-3a transcription factor. J Biol Chem. 1995;270:15858–15863. doi: 10.1074/jbc.270.26.15858. [DOI] [PubMed] [Google Scholar]
- 34.Landel C C, Kushner P J, Greene F L. The interaction of human estrogen receptor with DNA is modulated by receptor-associated proteins. Mol Endocrinol. 1994;14:1407–1419. doi: 10.1210/mend.8.10.7854357. [DOI] [PubMed] [Google Scholar]
- 35.Lannigan D A, Notides A C. Estrogen receptor selectively binds the coding strand of an estrogen responsive element. Proc Natl Acad Sci USA. 1989;86:863–867. doi: 10.1073/pnas.86.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lannigan D A, Notides A C. A novel mechanism for eukaryotic gene expression. The involvement of DNA tertiary structure in estrogen receptor recognition of its target nucleotide sequence. Biochem Pharmacol. 1990;40:2579–2585. doi: 10.1016/0006-2952(90)90574-5. [DOI] [PubMed] [Google Scholar]
- 37.Lannigan D A, Tomashek J J, Obourn J D, Notides A C. Analysis of estrogen receptor interaction with tertiary-structured estrogen responsive elements. Biochem Pharmacol. 1993;45:1921–1928. doi: 10.1016/0006-2952(93)90452-3. [DOI] [PubMed] [Google Scholar]
- 38.Lannigan D A, Koszewski N J, Notides A C. Estrogen-responsive elements contain non-B DNA. Mol Cell Endocrinol. 1993;94:47–54. doi: 10.1016/0303-7207(93)90050-t. [DOI] [PubMed] [Google Scholar]
- 39.Latchman D S. Eukaryotic transcription factors. 2nd ed. London, United Kingdom: Academic Press; 1995. [Google Scholar]
- 40.Latchman D S. Gene regulation—a eukaryotic perspective. 3rd ed. London, United Kingdom: Chapman; 1998. [Google Scholar]
- 41.Leygue E R, Watson P H, Murphy L C. Estrogen receptor variants in normal human mammary tissue. J Natl Cancer Inst. 1996;88:284–290. doi: 10.1093/jnci/88.5.284. [DOI] [PubMed] [Google Scholar]
- 42.Lillycrop K A, Budhram-Mahadeo V, Lakin N D, Terrenghi G, Wood J N, Polak J M, Latchman D S. A novel POU family transcription factor is closely related to Brn-3 but has a distinct expression pattern in neuronal cells. Nucleic Acids Res. 1992;20:5093–5096. doi: 10.1093/nar/20.19.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y Z, Dawson S J, Latchman D S. Alternative splicing of the Brn-3a and Brn-3b transcription factor mRNA is regulated in neuronal cells. J Mol Neurosci. 1996;7:77–85. doi: 10.1007/BF02736850. [DOI] [PubMed] [Google Scholar]
- 44.Marsigliante S, Muscella A, Ciardo V, Puddlefoot J R, Leo G, Vinson G P, Storelli C. Multiple isoforms of the oestrogen receptor in endometrial cancer. J Mol Endocrinol. 1995;14:365–374. doi: 10.1677/jme.0.0140365. [DOI] [PubMed] [Google Scholar]
- 45.McGuire W L, Chamness G C, Fuqua S A. Abnormal estrogen receptor in clinical breast cancer. J Steroid Biochem Mol Biol. 1992;43:243–247. doi: 10.1016/0960-0760(92)90214-4. [DOI] [PubMed] [Google Scholar]
- 46.Migliaccio A, Pagano M, Auricchio F. Immediate and transient stimulation of protein tyrosine phosphorylation by estradiol in MCF-7 cells. Oncogene. 1993;8:2183–2191. [PubMed] [Google Scholar]
- 47.Monuki E S, Khun R, Weinmaster G, Trapp B D, Lemke G. Expression and activity of POU transcription factor SCIP. Science. 1990;249:1300–1303. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- 48.Morris P J, Theil T, Ring C J A, Lillycrop K A, Moroy T, Latchman D S. The opposite and antagonistic effects of the closely related POU family transcription factors Brn-3a and Brn-3b on the activity of a target promoter are dependent on differences in the POU domain. Mol Cell Biol. 1994;14:6907–6914. doi: 10.1128/mcb.14.10.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee R, Chambon P. A single-stranded DNA-binding protein promotes the binding of the purified oestrogen receptor to its response element. Nucleic Acids Res. 1990;18:5713–5716. doi: 10.1093/nar/18.19.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy L C, Dolzlaw H. Variant estrogen receptor mRNA species detected in human breast cancer biopsy samples. Mol Endocrinol. 1989;3:687–693. doi: 10.1210/mend-3-4-687. [DOI] [PubMed] [Google Scholar]
- 51.Murphy L C, Wang M, Coutt A, Dolzlaw H. Novel mutations in the estrogen receptor messenger RNA in human breast cancers. J Clin Endocrinol Metab. 1996;81:1420–1427. doi: 10.1210/jcem.81.4.8636345. [DOI] [PubMed] [Google Scholar]
- 52.Ninkina N N, Stevens G E M, Wood J N, Richardson W D. A novel Brn-3 like POU transcription factor expressed in subsets of rat sensory and spinal cord neurons. Nucleic Acids Res. 1993;21:3175–3182. doi: 10.1093/nar/21.14.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 54.Parker M G, editor. Nuclear hormone receptors. London, United Kingdom: Academic Press; 1991. [Google Scholar]
- 55.Parker M G. Steroid and related receptors. Curr Opin Cell Biol. 1993;1:512–518. doi: 10.1016/0955-0674(93)90016-j. [DOI] [PubMed] [Google Scholar]
- 56.Pasqualini J R, Sumida C. Ontogeny of steroid receptors in the reproductive system. Int Rev Cytol. 1986;101:275–315. doi: 10.1016/s0074-7696(08)60251-x. [DOI] [PubMed] [Google Scholar]
- 57.Patrone C, Ma Z Q, Pollio G, Agrati P, Parker M G, Maggi A. Cross coupling between insulin and estrogen receptor in human neuroblastoma cell line. Mol Endocrinol. 1994;10:499–507. doi: 10.1210/mend.10.5.8732681. [DOI] [PubMed] [Google Scholar]
- 58.Pomerantz J L, Kristie T M, Sharp P A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;11:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- 59.Pugh B F. Mechanisms of transcription complex assembly. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 60.Ritchie M E, Trask R V, Fontanet H L, Billadello J J. Multiple positive and negative elements regulate human brain creatine kinase gene expression. Nucleic Acids Res. 1991;19:6231–6240. doi: 10.1093/nar/19.22.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scholer H R, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germline specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 62.Scoville S A, Bufton S M, Liuzzi F J. Estrogen regulates neurofilament gene expression in adult rat dorsal root ganglion neurons. Exp Neurol. 1997;146:586–599. doi: 10.1006/exnr.1997.6565. [DOI] [PubMed] [Google Scholar]
- 62a.Shughrue P J, Komme B, Merchenthaler I. The distribution of estrogen receptor beta mRNA in the rat hypothalamus. Steroids. 1996;61:678–681. doi: 10.1016/s0039-128x(96)00222-x. [DOI] [PubMed] [Google Scholar]
- 63.Sukovich D A, Mukherjee R, Benfield P A. A novel cell-type-specific mechanism for estrogen receptor-mediated gene activation in the absence of an estrogen-responsive element. Mol Cell Biol. 1994;14:7134–7143. doi: 10.1128/mcb.14.11.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki N, Rohdewold H, Neuman T, Gruss P, Scholer H R. Oct-6: a POU transcription factor expressed in embryonal stem cells and in developing brain. EMBO J. 1990;9:3723–3732. doi: 10.1002/j.1460-2075.1990.tb07585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theil T, McLean-Hunter S, Zornig M, Moroy T. Mouse Brn-3 family of POU transcription factors: a new amino terminal domain is crucial for the oncogenic activity of Brn-3A. Nucleic Acids Res. 1993;21:5921–5929. doi: 10.1093/nar/21.25.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Theil T, Zechner U, Klett C, Adolph S, Moroy T. Chromosomal localization and cDNA sequences of the murine Brn-3 family of developmental control genes. Cytogenet Cell Genet. 1994;66:267–271. doi: 10.1159/000133709. [DOI] [PubMed] [Google Scholar]
- 67.Theil T, Rodel B, Spiegelhalter F, Moroy T. Short isoform of POU factor Brn-3b can form a heterodimer with Brn-3a that is inactive for octamer motif binding. J Biol Chem. 1995;270:30958–30964. doi: 10.1074/jbc.270.52.30958. [DOI] [PubMed] [Google Scholar]
- 68.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 69.Treacy M N, Rosenfeld M G. Expression of a family of POU-domain protein regulatory genes during development of the central nervous system. Annu Rev Neurosci. 1992;15:139–165. doi: 10.1146/annurev.ne.15.030192.001035. [DOI] [PubMed] [Google Scholar]
- 70.Turner E E, Jenne K J, Rosenfeld M G. Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron. 1994;12:205–218. doi: 10.1016/0896-6273(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 71.Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J Biol Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- 72.Verrijzer C P, van Oosterhout J A W M, van der Vliet P C. The Oct-1 POU domain mediates interactions between Oct-1 and other POU proteins. Mol Cell Biol. 1992;12:542–551. doi: 10.1128/mcb.12.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verrijzer C P, van der Vliet P C. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Peters G A, Zeng X, Tang M, Khan S A. Yeast two hybrid system demonstrates that estrogen receptor dimerization is ligand dependent in vivo. J Biol Chem. 1995;270:23322–23329. doi: 10.1074/jbc.270.40.23322. [DOI] [PubMed] [Google Scholar]
- 75.Webb P, Lopez G N, Uht R M, Kushner P J. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- 76.Wegner M, Drolet D W, Rosenfeld M G. POU-domain proteins: structure and function of developmental regulators. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 77.Xia Y, Andersen B, Mehrabain M, Diep A T, Warden C H, Mohandas T, McEvilly R J, Rosenfeld M G, Lusis A J. Chromosomal organization of mammalian POU domain factors. Genomics. 1993;18:126–130. doi: 10.1006/geno.1993.1435. [DOI] [PubMed] [Google Scholar]
- 78.Yang N N, Venugopalan M, Hardikar S, Glasebrook A. Identification of an estrogen response element activated by metabolites of 17β-estradiol and raloxifene. Science. 1996;273:1222–1225. doi: 10.1126/science.273.5279.1222. [DOI] [PubMed] [Google Scholar]