Abstract
The search for understanding immunotherapy response has sparked interest in diverse areas of oncology, with artificial intelligence (AI) and radiomics emerging as promising tools, capable of gathering large amounts of information to identify suitable patients for treatment. The application of AI in radiology has grown, driven by the hypothesis that radiology images capture tumor phenotypes and thus could provide valuable insights into immunotherapy response likelihood. However, despite the rapid growth of studies, no algorithms in the field have reached clinical implementation, mainly due to the lack of standardized methods, hampering study comparisons and reproducibility across different datasets. In this review, we performed a comprehensive assessment of published data to identify sources of variability in radiomics study design that hinder the comparison of the different model performance and, therefore, clinical implementation. Subsequently, we conducted a use-case meta-analysis using homogenous studies to assess the overall performance of radiomics in estimating programmed death-ligand 1 (PD-L1) expression. Our findings indicate that, despite numerous attempts to predict immunotherapy response, only a limited number of studies share comparable methodologies and report sufficient data about cohorts and methods to be suitable for meta-analysis. Nevertheless, although only a few studies meet these criteria, their promising results underscore the importance of ongoing standardization and benchmarking efforts. This review highlights the importance of uniformity in study design and reporting. Such standardization is crucial to enable meaningful comparisons and demonstrate the validity of biomarkers across diverse populations, facilitating their implementation into the immunotherapy patient selection process.
Subject terms: Cancer imaging, Predictive markers
Introduction
Cancer immunotherapy, particularly immune checkpoint inhibitors (ICI), has emerged as the gold standard for treating various cancers, including lung, renal, and melanoma1–4. The remarkable success achieved with ICI has generated optimism for its potential application in treating numerous other types of cancer. However, the variability in patient responses makes it necessary to identify biomarkers capable of predicting individual responses to ICI. This crucial step is instrumental in enhancing patient stratification, maximizing treatment efficacy, detecting treatment resistance and thus minimizing potential harm for those who may not benefit. Various tissue-based predictive biomarkers have been proposed, such as microsatellite instability (MSI)5,6, tumor mutational burden (TMB)7, programmed death-ligand 1 (PD-L1) expression8, and tumor-infiltrating lymphocyte (TIL) count9. However, these biomarkers often require invasive procedures to obtain tumor tissue for analysis, and their accuracy in identifying suitable candidates for immunotherapy remains suboptimal10. Radiomics analysis, in combination with machine learning (ML) methods, efficiently extracts meaningful information from medical images, enabling three-dimensional evaluation of tumors throughout the entire body, and repeated assessments over the course of cancer treatment11. In particular, extracting radiomics features from standard-of-care CT images, a widely used imaging technique for cancer staging and follow-up, offers a valuable tool with potential for developing predictive biomarkers in the context of immunotherapy12–15. This is especially pertinent in cancer immunotherapy, where treatment may occur after the initial diagnosis, in pretreated patients with evolving tumors and non-reachable lesions16,17. The non-invasive nature of radiomics applications thus becomes highly valuable.
In fact, the emergence of encouraging radiomics signatures for predicting response to immunotherapy has caused a boom in research endeavors in this field. Nevertheless, the absence of standardized protocols and benchmarking studies of biological validation of such signatures poses a significant challenge for the application of these signatures in clinical practice. Despite numerous radiomics studies predicting response across various tumor types, inconsistencies persist in data selection, model construction, and outcome definition. To assess the reliability of predictive radiomics studies, standardization research criteria such as the Radiomics Quality Score (RQS)11 and the CLEAR checklist18 have been introduced19. However, low RQS have been reported in most published radiomics studies, indicating poor documentation practices and limited reproducibility20. Efforts are emerging to develop PRISMA-AI guidelines21 that will define standardized frameworks, comprehensive method descriptions, and data-sharing practices in radiomics-based studies, as well as, allow study comparison, validation, and meta-analysis efforts in this domain.
In this review, we provide an overview of the current state of radiomics-based biomarkers to guide the use of immunotherapy through a comprehensive examination. It encompasses the potential biases and variations in the currently developed radiomics pipelines that challenge the comparison of studies through meta-analysis. Additionally, we present a short case study featuring a meta-analysis of studies predicting PD-L1 status from CT imaging, comparing radiomics ML and deep learning (DL) models. By examining the existing literature and conducting a meta-analysis, we aim to offer valuable insights and perspectives on the efficacy and reliability of radiomics as immunotherapy biomarkers.
Results
Uncovering potential sources of variability in radiomics study design
We conducted a systematic review encompassing all studies utilizing ML or DL techniques in CT imaging for predicting either direct response to immunotherapy or any surrogate biomarker of response. Our findings highlight the significant diversity in study design among publications aiming to create similar predictive models (Fig. 1). This variability in methodology presents a challenge when attempting to compare the performance of these models through meta-analysis. In this section, we aim to summarize all these studies and the differences among them.
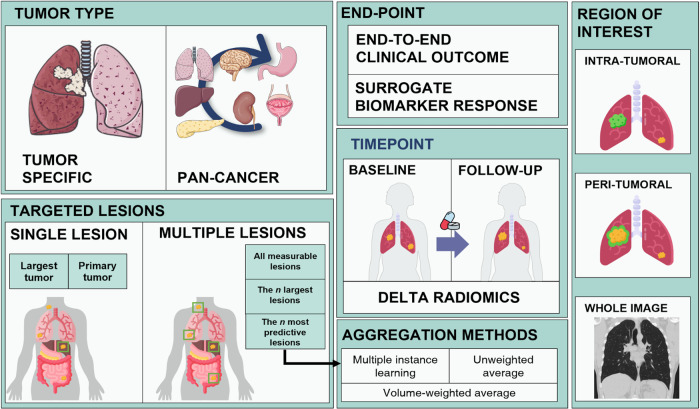
Fig. 1.
Potential sources of variability in radiomics study design including features related to the cohort setting (specific signature for a single tumor type or pan-cancer), end-point (for clinical outcome such as response yes/no or for predicting molecular surrogate biomarkers such as programmed death-ligand 1 [PD-L1] expression), number of lesions, imaging timepoints and region of interest. n: number of lesions.
Cohort setting
The characteristics that define the tumor phenotype and make it more responsive to immunotherapy can encompass tumor-specific aspects or those that can be expressed and captured across multiple tumor types. Consequently, researchers have pursued two approaches in the development and validation of radiomics-based biomarkers. One approach being tumor specific, aiming to create validated biomarkers within each type. While the other approach addresses this challenge by incorporating multiple tumor types and considering the location of metastatic disease when feeding the models.
To date, the majority of radiomics studies on immunotherapy response prediction have focused on non-small cell lung cancer (NSCLC), benefiting from the availability of larger datasets and a higher degree of treatment responses in this tumor type. In fact, most of the studies exploring radiomics to predict direct response or surrogates of response to immunotherapy (i.e., biological and molecular markers used as predictors of a patient’s response such as PD-L1) have been done in NSCLC populations. Other tumor types including melanoma, gastric, head and neck, bladder and kidney cancers have been investigated to a lesser extent and only few studies have developed predictive radiomics models in pan-cancer settings12,14. Despite the increasing number of lung cancer and melanoma patients receiving immunotherapy as part of standard care, it is noteworthy that only around 30% of the articles included cohorts larger than 200 patients, and merely 22% reported the utilization of external validation cohorts (Supplementary Table 1).
The development of tumor type-specific radiomics signatures allows finding radiomics features unique to that population; however, it reduces the generalization of the methods to other tumor types that are less common or rarely treated with immunotherapy. On the other hand, pan-cancer approaches require the use of larger cohorts for the model to comprehend the inherent heterogeneity of the population, thereby reducing the bias towards the response probability of each tumor type.
Outcome evaluation
Studies focusing on predictive radiomics signatures and immunotherapy can be categorized into two types: those aiming to directly predict clinical outcome and those focused on predicting known surrogate biomarkers. However, the lack of standardization regarding outcome definition poses a significant challenge, making it difficult to compare and assess the predictive capabilities of the resulting radiomics signatures.
One major challenge is the wide range of clinical endpoints used to assess treatment response. The most relevant measure for evaluating the benefit of immunotherapy treatment in patients is overall survival (OS). While certain radiomics studies have considered OS as the clinical endpoint13,15,22–33, most studies rely on tumor size changes by the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1)16. From the RECIST assessment, multiple measurements can be computed and used as endpoints, including progression-free survival (PFS)29–32,34–38, disease control (which gathers complete response (CR), partial response (PR), and stable disease (SD))14,22,28,34,35,39–46 or objective response rate (ORR)23,47–50. However, it is important to note that these response evaluations are considered surrogate endpoints for OS, and their reliability is hindered by their inherent subjectivity and variability, challenging the development of reproducible models51,52. Furthermore, the wide range of response evaluation criteria derived from RECIST53,54 (e.g., PFS, ORR, disease control) also limits the direct comparison of radiomics signatures across studies.
Similarly, when predicting molecular surrogate biomarkers (such as PD-L1 expression), many studies tend to discretize the target variable and transform it into a classification problem. However, these biomarker cutoffs are subjected to the primary tumor biology or the type of treatment. Therefore, the lack of standardized cutoff values further complicates the comparison of radiomics signatures for predicting surrogate biomarkers in immunotherapy. In addition to the heterogeneity in endpoint definitions, it is important to consider that the performance of radiomics signatures predicting surrogate biomarkers will be inherently limited by the predictive capacity of the surrogate biomarker itself. This implies that the effectiveness of the radiomics signatures in predicting treatment response will be constrained by the predictive capabilities of the surrogate biomarker being used.
Study design regarding number of lesions, region of interest and time-points
Another relevant point in the study design for immunotherapy radiomics signatures is the selection of target lesions for analysis. Many radiomics studies rely on delineating and extracting features from a single selected tumor (~63% of the studies found in the review), often the primary or largest lesion, arguing that the single chosen lesion can represent the whole disease. However, in patients with metastases at multiple sites, heterogeneous immunophenotypes can drive different immune responses55,56. Therefore, analyzing only one lesion per patient may not fully capture the tumor heterogeneity and limit the predictive capacity of the model. To partially overcome this limitation, feature aggregation methods such as average, volume-weighted average, or attention-based multiple instance learning (MIL) are commonly used14,41,49,57. Additionally, the analysis of inter- and intra-lesion heterogeneity through radiomics studies to capture the whole metastatic disease has also been considered as a potential indicator of immunotherapy response58.
Moreover, with the aim of providing the model with all the potential relevant data and knowing the effect of surrounding tumor microenvironment for immunotherapy response, certain studies have also explored the value of incorporating peritumoral area information into predictive models for predicting response to immunotherapy22,50. Nevertheless, the models obtained more relevant information from the intratumoral features. Some studies have also shown that intratumoral 3D radiomics features provide more informative insights compared to using only 2D radiomics features28.
Finally, regarding the imaging time-points, the majority of studies in radiomics research have focused on the development of predictive biomarkers using baseline scans, which refer to the scans obtained just before initiating treatment. This approach facilitates improved patient selection for treatment decision-making. However, some studies have demonstrated enhanced outcomes by analyzing changes in the radiomics tumor phenotype between baseline and early follow-up time points, commonly known as early readouts or delta radiomics signatures13,15,22,29,43,47,59. Such approaches enable the capture of response or progression patterns that may go unnoticed by radiologists, thereby potentially preventing patients to stay longer under ineffective treatment. Some of these studies have shown that tracking these changes in CT scans provides better prognostic value compared to the current standard of care, RECIST13,60. It is important to note, however, that these early readouts do not represent true predictive biomarkers per se, but rather serve as indicators of early response, and should be thought of as alternative response criteria themselves, rather than predictive biomarkers. This is because at the time these early readouts are assessed, treatment decisions have already been made, and the patient is already receiving immunotherapy.
Radiomics feature selection and model implementation
Fifty percent of the pipelines implemented for hand-crafted radiomics analysis correspond to Least Absolute Shrinkage and Selection Operator (LASSO) for feature selection (implemented in 40% of the studies), followed by a logistic regression for classification (implemented in 25% of the studies). Multiple studies have highlighted the benefits of utilizing LASSO as the feature selection method due to its efficacy in high-dimensional data regression, thereby mitigating the risk of overfitting30,49. In terms of classification method, several studies have explored the performance of different classification algorithms for predicting response (such as support vector machine (SVM), Random Forest (RF), decision tree and k-nearest neighbor) (Supplementary Figure 1). All of them showed that logistic regression had similar or slightly better performance than other more complex classifiers28,38,50,61.
Only a few studies have used more advanced DL methods to predict response to immunotherapy13,32,45,46. These methods are data-hungry and need large cohorts of patients, as well as reliable and objective annotations, to achieve good performance. However, gathering this amount of data regarding immunotherapy treatment response is still challenging. For that reason, most of the CT-based DL models currently developed are focused on predicting surrogates of response such as PD-L1 status32,62–64.
Case-study: meta-analysis for predicting PD-L1 status from CT imaging comparing DL vs classical ML
In order to get a better understanding of the overall performance of the radiomics signatures as predicting biomarkers for immunotherapy, we conducted a meta-analysis of all the studies that implemented CT-based radiomics with classical ML or DL to predict PD-L1 status. Figure 2 shows a flow chart illustrating the systematic review conducted in PubMed, outlining the predefined inclusion and exclusion criteria.
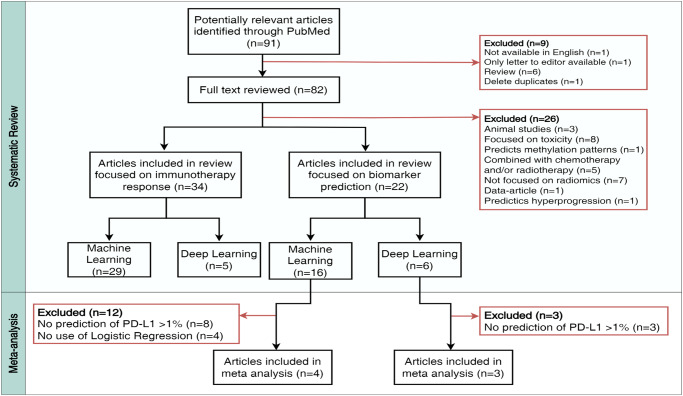
Fig. 2.
Referred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram for study illustrating the number of records screened in the review and articles included and excluded, outlining the predefined inclusion and exclusion criteria. In total, 56 articles were included in the review and 35 articles were excluded, reasons for exclusion were reported. Seven studies exploring CT-based radiomics models for predicting programmed death-ligand 1 (PD-L1) expression were included in the meta-analysis.
We identified a total of 56 articles developing CT-based predictive signatures in patients treated with immunotherapy; 34 for predicting direct response and 22 predicting surrogate molecular biomarkers (Supplementary. Detailed results systematic review). In Supplementary Table 1, all included papers are listed. We reviewed the CLEAR guidelines for all these studies (Supplementary Table 2). However, we could not include harmonized image preprocessing techniques or feature selection methods. Accounting for the previously described variability in the methods of radiomics signatures and with the aim of investigating the most standardized models, we found seven comparable studies to perform the meta-analysis. All of them predicted PD-L1 expression assessed as tumor proportion score (TPS) ≥ 1%, using the area under the curve (AUC) as the evaluation metric and implementing either logistic regression (n = 4)61,64–66 or DL (n = 3)62,64,67 as the predictive model. External validation performance was also explored in three studies applying logistic regression methods and one DL modeling.
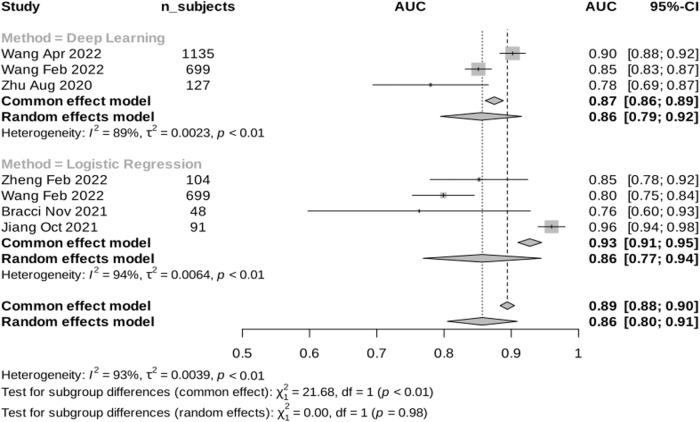
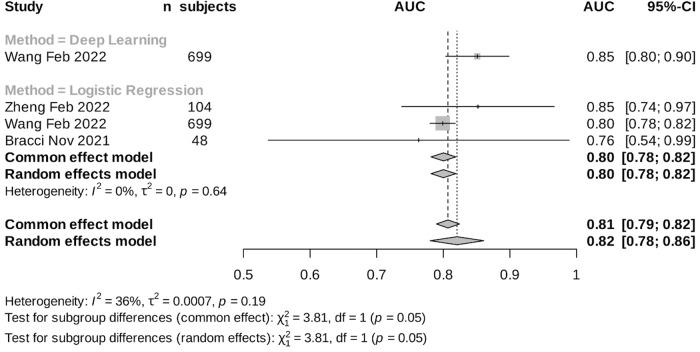
The included papers showed varying performance in predicting PD-L1 expression, with AUROCs ranging from 0.76 to 0.96 in both logistic regression and DL methods. In the internal validation, the logistic regression models showed a pooled AUC of 0.86 (95%CI 0.77–0.94, i²= 94%) while the DL method exhibited a pooled AUROC of 0.86 (95%CI 0.79–0.92, i²= 89%), using random effects model (Fig. 3). Interestingly, our findings revealed that the performance across different studies for logistic regression remained comparable in the external validation set, yielding an estimated AUROC of 0.80 (95%CI 0.78–0.82, i²= 0%) (Fig. 4). These findings indicate low heterogeneity between studies in the external validation performance in contrast to the higher heterogeneity in the internal set. There was not enough data from DL studies to evaluate the heterogeneity in the external set. Notably, studies utilizing logistic regression and DL methods demonstrated similar results in the internal set, with a combined estimated AUROC of 0.86 (95% CI 0.80–0.91), despite DL models having access to a larger dataset compared to logistic regression studies.
Fig. 3.
Meta-analysis results: Internal validation performance of the reported studies that implemented CT-based radiomics with classical machine learning (ML) or deep learning (DL) for predicting programmed death-ligand 1 (PD-L1) expression.
Fig. 4.
Meta-analysis results: External validation performance of the reported studies that implemented CT-based radiomics with machine learning (ML) or deep learning (DL) for predicting programmed death-ligand 1 (PD-L1) expression.
Discussion
The application of artificial intelligence (AI) to improve patient stratification towards better treatment selection is of growing interest in both radiology and oncology fields. Numerous studies have focused on uncovering radiological features of tumors that could predict response patterns to immunotherapy. However, the lack of standardized and homogeneous frameworks employed in these studies, as well as scarce data sharing, present challenges when comparing results and validating radiomics models, ultimately, hindering their integration into clinical practice. In this review, we aimed to shed light on the factors that contribute to the variability in radiomics studies, rendering the available models incomparable. Additionally, we conducted a comprehensive literature review specifically targeting studies that investigated CT-based radiomics signatures for predicting response to immune checkpoint inhibitors (ICI) and surrogate biomarkers of response to ICI.
Remarkably, our findings revealed that despite the abundance of studies predicting direct response to immunotherapy, only a limited number of these studies employed similar methods, making them unsuitable for meta-analysis. This significant variability in methodology poses challenges in terms of study comparisons and reproducibility across different datasets. Similar challenges have arisen in the development of other potential predictive biomarkers based on biological samples such as PD-L1 expression or TMB, highlighted in debates surrounding the heterogeneous distribution of these markers in tumor samples, variations in staining techniques, and establishment of appropriate thresholds, among other issues68–70. Addressing these challenges requires benchmarking studies that facilitate the comparison of established methods with novel techniques across diverse cohorts, thereby promoting advancement and standardization in the field.
Nonetheless, some studies investigating the development of radiomics signatures for predicting programmed death-ligand 1 (PD-L1) expression in tumors met the necessary criteria for meaningful pooling and meta-analysis. Consequently, our meta-analysis exclusively focused on studies examining CT-based radiomics for predicting PD-L1 status. This case study highlighted the promising performance of the reported models in predicting PD-L1 expression, with area under the receiver operating characteristic curve (AUROC) values ranging from 0.7 to 0.9. While these models have demonstrated positive outcomes and exhibited limited heterogeneity in accuracy during external validation, questions persist regarding the lack of widespread adoption in clinical practice. One potential contributing factor could be the absence of a reported correlation between PD-L1 prediction and treatment response. Furthermore, it is important to acknowledge the potential influence of publication bias, which may result in a prevalence of positive results, possibly overshadowing scientifically crucial findings from studies that may not achieve high accuracy despite employing sound methodologies.
Developing multi-center studies is essential to demonstrate the applicability of these methods across large and heterogeneous datasets, ensuring reliability and fairness by encompassing diverse populations and machines from various institutions. Concerns regarding data privacy and patient data monetization have slowed down the development of large-scale multi-center models. Nevertheless, efforts have been made in this field to provide more secure methods of data sharing and decentralized model training, such as federated learning71,72, where models can be trained on multi-institutional data without leaving the respective institutions, thus safeguarding data privacy. Moreover, some studies have highlighted potential improvements of predictive models through multimodal approaches that combine radiomics with histopathology or genomics73,74. Still, this requires representative heterogeneous data ideally from multiple centers, including all sources of information, which has been a notable limitation thus far. Finally, integration of radiomics-based biomarkers into clinical practice hinges on the critical aspects of explainability and trustability, ensuring that healthcare professionals can comprehend and rely on these complex data-driven insights to make informed patient care decisions.
Moreover, the path to integrating radiomics into clinical practice, even when all the previous limitations are considered, still relies on biological translation of the predictive models. Certain studies have made substantial progress in this direction by correlating radiomics predictions with biological and molecular markers like PD-L130, cellular pathways39 or cytotoxic immunophenotype14. Other studies have focused on developing models that aim to predict directly the molecular properties of the tumor from surgical resections or biopsies75. However, ongoing investigation in this direction is needed to enhance the reliability and applicability of these models for seamless integration into routine clinical practice.
In conclusion, the journey towards establishing radiomics-based biomarkers is challenging, requiring technical development of imaging assays and computational methods, validation encompassing sensitivity, specificity, and reproducibility evaluations, biological validation, as well as proving clinical relevance ideally through embedding them in prospective clinical trials. Despite the considerable interest and expectations from the scientific community, as well as the abundance of papers exploring imaging phenotypes derived from radiomics as potential biomarkers of response to immunotherapy, these tools have yet to be implemented in clinical practice. To make a substantial impact on clinical trials and medical practice, larger prospective studies with appropriate external validation datasets, focusing on the clinical applicability of these signatures, are crucial.
Fortunately, changes are underway in the field that should facilitate the exploration of these novel biomarkers and their potential applicability in the clinic. The imaging scientific community, through collaborative efforts and consortia supported by the EU commissioner, is working to bridge the gap between research and real-world application. Among the most significant initiatives is the EUCAIM project, which is dedicated to establish an infrastructure for over 60 million cancer images from over 100,000 cancer patients with the goal to develop and benchmark trustworthy AI tools. Together, we strive to pave the way for the true integration of radiomics-based biomarkers into clinical decision-making, ultimately improving the care of cancer patients.
Methods
Detailed description of the of the systematic review methodology
Search strategy
A search was conducted in the PubMed electronic database for potential articles published at date October 1st, 2022. The search strategy used was (((“Radiomics” OR “CT based biomarker” OR “imaging based biomarker” OR “imaging marker” OR “imaging biomarker”) AND (“Immunotherapy”[Mesh] OR “ipilimumab” OR “tremelimumab” OR “CTLA-4” OR pembrolizumab” OR nivolumab” OR “Immuno Checkpoint Inhibitors”[Mesh] OR “cemiplimab” OR “atezolizumab” OR “immune checkpoint blockade” OR “avelumab” OR “durvalumab” OR “PD-L1” OR “PD-1”)) AND (((“Tomography, X-Ray Computed” [Mesh] OR “Computed Tomography” OR “CT”) NOT “Positron Emission Tomography”) NOT “PET”). Our search terms did not include specific cancer types or outcome types. Finally, we also considered any articles referred to us by experts, identified during the prior scoping search, or found in the references section of the full-text articles we evaluated.
Instead of only assessing studies based on hand-crafted radiomics applied to classical machine learning (ML) models, studies that employed deep learning (DL) techniques were also examined. Articles were evaluated systematically on title and full-text level, and reasons for exclusion were noted. All studies which were potentially relevant for the paper were included in a data extraction table.
Study selection and eligibility criteria
According to the inclusion criteria, we focused exclusively on systemic treatments involving immune checkpoint inhibitors (ICI) alone. Articles were included if they were (i) primary studies that investigated (ii) response to ICIs alone by using (iii) classical ML or DL on (iv) human tumor lesions and (v) written in the English language.
We excluded studies of ICI in combination with other therapies. If the study included patients who received immunotherapy, chemotherapy and/or radiotherapy, we only included them in case the results for immunotherapy were assessed separately. Other forms of immunotherapy, such as monoclonal antibodies, vaccines, immune system modulators, or T-cell transfer therapy, were beyond the scope of our review. Predicting hyperprogression, toxicity and methylation patterns were also considered outside the scope of this review.
The included articles were divided in two different categories, based on the type of predicted outcome; prediction of end-to-end ICI response or biomarkers for response. Then, for every outcome category, we divided the studies based on the applied methods: conventional ML and DL approaches. From each article, we reported the used methods, main results and the reported conclusions and limitations. Regarding the methods, we collected the feature aggregation and selection, and the implemented ML algorithm. We filtered the results from some studies with additional experiments regarding other endpoints, as defined in the exclusion criteria.
Statistical analysis
To obtain an overall estimation, the area under the curve (AUC) with 95% confidence interval (CI) was calculated for each study. No p-values were reported for pooled AUCs. Heterogeneity estimation was assessed and reported in all analyses using means of I2 and a statistical test to evaluate the similarity of results across studies (homogeneity test). Both fixed and random effects models were applied regardless of the homogeneity test outcome. When the p-value was greater than 0.05 (indicating no significant heterogeneity), the fixed effects model was used, assuming a common effect size. Conversely, the random effects model, employing the DerSimonian-Laird method, was utilized to account for heterogeneity. Due to limited statistical power in detecting heterogeneity, the random effects model was employed for subgroup analysis.
Internal validation results, accounting for cross-validation and internal split, were used for the meta-analysis. External validation was also analyzed when applicable in an additional experiment. All the analyses were implemented using R v(4.2.2) and package metafor.
Supplementary information
Acknowledgements
This research has been funded by the Comprehensive Program of Cancer Immunotherapy & Immunology II (CAIMI-II) supported by the BBVA Foundation (grant 53/2021). RPL is supported by LaCaixa Foundation, a CRIS Foundation Talent Award (TALENT19-05), the FERO Foundation, the Instituto de Salud Carlos III-Investigacion en Salud (PI18/01395 and PI21/01019), the Prostate Cancer Foundation (18YOUN19) and the Asociacion Española Contra el Cancer (AECC) (PRYCO211023SERR). ML is supported by the PERIS PIF-Salut Grant. OP is supported by a La Caixa INPhINIT Fellowship.
Author contributions
M.L. and R.P.L. designed the study. M.L. and B.G. contributed to data collection and assembly. M.L., B.G., O.P., R.P.L. interpreted and analyzed the data. V.N., R.D. and P.C. performed statistical revision. M.L., B.G., V.N., P.C., O.P., R.D., P.N., S.T., R.B.T., E.S. and R.P.L. wrote and reviewed the report and approved the final version for submission. M.L. and B.G. contributed equally to this work and manuscript preparation and should be considered co-first authors.
Competing interests
ES, has received speakers fees from GE healthcare and is a co-founder and shareholder of Lucida Medical Ltd. EG declares research funding from Novartis, Roche, Thermo Fisher, AstraZeneca, Taiho, BeiGene, Janssen. EG also reports consultant or advisor role for Roche, Ellipses Pharma, Boehringer Ingelheim, Janssen Global Services, Seattle Genetics, Thermo Fisher, MabDiscovery, Anaveon, F-Star Therapeutics, Hengrui, Sanofi, Incyte, Medscape and speaker bureau from Merck Sharp & Dohme, Roche, Thermo Fisher, Lilly, Novartis, SeaGen. RD declares advisory role for Roche, Foundation Medicine, received a speaker’s fee from Roche, Ipsen, Amgen, Servier, Sanofi, Libbs, Merck Sharp & Dohme, Lilly, AstraZeneca, Janssen, Takeda, Bristol Myers Squibb, GlaxoSmithKline, Gilead and research grants from Merck, Novartis, Daiichi-Sankyo, GlaxoSmithKline and AstraZeneca. PN declares advisory or consultat role for MSD ONCOLOGY, BAYER and speaker’s fee from Novartis. No other competing interests are disclosed by any author.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marta Ligero, Bente Gielen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00534-9.
References
- 1.Long GV, et al. Nivolumab for patients with advanced melanoma treated beyond progression: analysis of 2 phase 3 clinical trials. JAMA Oncol. 2017;3:1511–1519. doi: 10.1001/jamaoncol.2017.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postow MA, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer J, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le DT, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J. Clin. Oncol. 2020;38:11–19. doi: 10.1200/JCO.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marabelle A, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J. Clin. Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan TA, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daud AI, et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J. Clin. Oncol. 2016;34:4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 2019;5:1614–1618. doi: 10.1001/jamaoncol.2019.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilard C, et al. Cancer immunotherapy: it’s time to better predict patients’ response. Br. J. Cancer. 2021;125:927–938. doi: 10.1038/s41416-021-01413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 12.Sun R, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 13.Trebeschi S, et al. Prognostic value of deep learning-mediated treatment monitoring in lung cancer patients receiving immunotherapy. Front Oncol. 2021;11:609054. doi: 10.3389/fonc.2021.609054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ligero M, et al. A CT-based radiomics signature is associated with response to immune checkpoint inhibitors in advanced solid tumors. Radiology. 2021;299:109–119. doi: 10.1148/radiol.2021200928. [DOI] [PubMed] [Google Scholar]
- 15.Dercle L, et al. Early readout on overall survival of patients with melanoma treated with immunotherapy using a novel imaging analysis. JAMA Oncol. 2022;8:385–392. doi: 10.1001/jamaoncol.2021.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez-Sánchez A, et al. Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat. Genet. 2020;52:582–593. doi: 10.1038/s41588-020-0630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen PHD, et al. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nat. Commun. 2021;12:227. doi: 10.1038/s41467-020-20171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kocak B, et al. CheckList for EvaluAtion of Radiomics research (CLEAR): a step-by-step reporting guideline for authors and reviewers endorsed by ESR and EuSoMII. Insights Imaging. 2023;14:75. doi: 10.1186/s13244-023-01415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. 2020;11:1–16. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramlee S. et al. Radiomic signatures associated with CD8+ tumour-infiltrating lymphocytes: a systematic review and quality assessment study. Cancers. 14. 10.3390/cancers14153656 (2022). [DOI] [PMC free article] [PubMed]
- 21.Cacciamani GE, et al. PRISMA AI reporting guidelines for systematic reviews and meta-analyses on AI in healthcare. Nat. Med. 2023;29:14–15. doi: 10.1038/s41591-022-02139-w. [DOI] [PubMed] [Google Scholar]
- 22.Khorrami M, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol. Res. 2020;8:108–119. doi: 10.1158/2326-6066.CIR-19-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peisen, F. et al. Combination of whole-body baseline CT radiomics and clinical parameters to predict response and survival in a stage-IV melanoma cohort undergoing immunotherapy. Cancers. 14. 10.3390/cancers14122992 (2022). [DOI] [PMC free article] [PubMed]
- 24.Tunali, I. et al. Hypoxia-related radiomics and immunotherapy response: a multicohort study of non-small cell lung cancer. JNCI Cancer Spectr. 5. 10.1093/jncics/pkab048 (2015). [DOI] [PMC free article] [PubMed]
- 25.Schraag A, et al. Baseline clinical and imaging predictors of treatment response and overall survival of patients with metastatic melanoma undergoing immunotherapy. Eur. J. Radio. 2019;121:108688. doi: 10.1016/j.ejrad.2019.108688. [DOI] [PubMed] [Google Scholar]
- 26.Corino, V. D. A. et al. A CT-based radiomic signature can be prognostic for 10-months overall survival in metastatic tumors treated with nivolumab: an exploratory study. Diagnostics (Basel).11. 10.3390/diagnostics11060979 (2021). [DOI] [PMC free article] [PubMed]
- 27.Zerunian M, et al. CT based radiomic approach on first line pembrolizumab in lung cancer. Sci. Rep. 2021;11:6633. doi: 10.1038/s41598-021-86113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ungan G, et al. Metastatic melanoma treated by immunotherapy: discovering prognostic markers from radiomics analysis of pretreatment CT with feature selection and classification. Int J. Comput Assist Radio. Surg. 2022;17:1867–1877. doi: 10.1007/s11548-022-02662-8. [DOI] [PubMed] [Google Scholar]
- 29.Guerrisi A, et al. Exploring CT texture parameters as predictive and response imaging biomarkers of survival in patients with metastatic melanoma treated with PD-1 inhibitor nivolumab: a pilot study using a delta-radiomics approach. Front Oncol. 2021;11:704607. doi: 10.3389/fonc.2021.704607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jazieh, K. et al. Novel imaging biomarkers predict outcomes in stage III unresectable non-small cell lung cancer treated with chemoradiation and durvalumab. J. Immunother. Cancer. 10. 10.1136/jitc-2021-003778 (2022). [DOI] [PMC free article] [PubMed]
- 31.Nardone V, et al. Radiomics predicts survival of patients with advanced non-small cell lung cancer undergoing PD-1 blockade using Nivolumab. Oncol. Lett. 2020;19:1559–1566. doi: 10.3892/ol.2019.11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He B-X, et al. Deep learning for predicting immunotherapeutic efficacy in advanced non-small cell lung cancer patients: a retrospective study combining progression-free survival risk and overall survival risk. Transl. Lung Cancer Res. 2022;11:670–685. doi: 10.21037/tlcr-22-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazzaschi G, et al. Integrated CT imaging and tissue immune features disclose a radio-immune signature with high prognostic impact on surgically resected NSCLC. Lung Cancer. 2020;144:30–39. doi: 10.1016/j.lungcan.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, et al. A multi-omics-based serial deep learning approach to predict clinical outcomes of single-agent anti-PD-1/PD-L1 immunotherapy in advanced stage non-small-cell lung cancer. Am. J. Transl. Res. 2021;13:743–756. [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B, et al. Combination of computed tomography imaging-based radiomics and clinicopathological characteristics for predicting the clinical benefits of immune checkpoint inhibitors in lung cancer. Respir. Res. 2021;22:189. doi: 10.1186/s12931-021-01780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dercle L, et al. Identification of non–small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin. Cancer Res. 2020;26:2151–2162. doi: 10.1158/1078-0432.CCR-19-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladwa R, et al. Computed tomography texture analysis of response to second-line nivolumab in metastatic non-small cell lung cancer. Lung Cancer Manag. 2020;9:LMT38. doi: 10.2217/lmt-2020-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, et al. A CT-based radiomics approach to predict nivolumab response in advanced non-small-cell lung cancer. Front Oncol. 2021;11:544339. doi: 10.3389/fonc.2021.544339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trebeschi S, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019;30:998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Imaging biomarkers to predict and evaluate the effectiveness of immunotherapy in advanced non-small-cell lung cancer. Front Oncol. 2021;11:657615. doi: 10.3389/fonc.2021.657615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M, et al. A combined-radiomics approach of CT images to predict response to anti-PD-1 immunotherapy in NSCLC: a retrospective multicenter study. Front Oncol. 2021;11:688679. doi: 10.3389/fonc.2021.688679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji Z, et al. Use of radiomics to predict response to immunotherapy of malignant tumors of the digestive system. Med Sci. Monit. 2020;26:e924671. doi: 10.12659/MSM.924671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z-L, et al. Pilot study of CT-based radiomics model for early evaluation of response to immunotherapy in patients with metastatic melanoma. Front Oncol. 2020;10:1524. doi: 10.3389/fonc.2020.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malone ER, et al. Predictive radiomics signature for treatment response to nivolumab in patients with advanced renal cell carcinoma. Can. Urol. Assoc. J. 2022;16:E94–E101. doi: 10.5489/cuaj.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren, Q. et al. Assessing the robustness of radiomics/deep learning approach in the identification of efficacy of anti–PD-1 treatment in advanced or metastatic non-small cell lung carcinoma patients. Front Oncol. 12. 10.3389/fonc.2022.952749 (2022). [DOI] [PMC free article] [PubMed]
- 46.Rundo F, et al. Three-dimensional deep noninvasive radiomics for the prediction of disease control in patients with metastatic urothelial carcinoma treated with immunotherapy. Clin. Genitourin. Cancer. 2021;19:396–404. doi: 10.1016/j.clgc.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Gong J, et al. A short-term follow-up CT based radiomics approach to predict response to immunotherapy in advanced non-small-cell lung cancer. Oncoimmunology. 2022;11:2028962. doi: 10.1080/2162402X.2022.2028962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Z, et al. A radiomics model predicts the response of patients with advanced gastric cancer to PD-1 inhibitor treatment. Aging. 2022;14:907–922. doi: 10.18632/aging.203850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park KJ, et al. Radiomics-based prediction model for outcomes of PD-1/PD-L1 immunotherapy in metastatic urothelial carcinoma. Eur. Radio. 2020;30:5392–5403. doi: 10.1007/s00330-020-06847-0. [DOI] [PubMed] [Google Scholar]
- 50.Yuan G, et al. Development and validation of a contrast-enhanced CT-based radiomics nomogram for prediction of therapeutic efficacy of anti-PD-1 antibodies in advanced HCC patients. Front. Immunol. 2020;11:613946. doi: 10.3389/fimmu.2020.613946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhl CK, et al. Validity of RECIST Version 1.1 for response assessment in metastatic cancer: a prospective, multireader study. Radiology. 2019;290:349–356. doi: 10.1148/radiol.2018180648. [DOI] [PubMed] [Google Scholar]
- 52.Garralda E, Laurie SA, Seymour L, de Vries EGE. Towards evidence-based response criteria for cancer immunotherapy. Nat. Commun. 2023;14:3001. doi: 10.1038/s41467-023-38837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seymour L, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–e152. doi: 10.1016/S1470-2045(17)30074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodi FS, et al. Immune-modified response evaluation criteria in solid tumors (imRECIST): refining guidelines to assess the clinical benefit of cancer immunotherapy. J. Clin. Oncol. 2018;36:850–858. doi: 10.1200/JCO.2017.75.1644. [DOI] [PubMed] [Google Scholar]
- 55.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun R. et al. Imaging approaches and radiomics: toward a new era of ultraprecision radioimmunotherapy? J. Immunother. Cancer. 1010.1136/jitc-2022-004848 (2022). [DOI] [PMC free article] [PubMed]
- 58.Himoto Y. et al. Computed tomography-derived radiomic metrics can identify responders to immunotherapy in ovarian cancer. JCO Precis. Oncol. 3. 10.1200/PO.19.00038 (2019). [DOI] [PMC free article] [PubMed]
- 59.Tunali I, et al. Novel clinical and radiomic predictors of rapid disease progression phenotypes among lung cancer patients treated with immunotherapy: an early report. Lung Cancer. 2019;129:75–79. doi: 10.1016/j.lungcan.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trebeschi S, et al. Development of a prognostic AI-monitor for metastatic urothelial cancer patients receiving immunotherapy. Front Oncol. 2021;11:637804. doi: 10.3389/fonc.2021.637804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Z, et al. CT-based hand-crafted radiomic signatures can predict PD-L1 expression levels in non-small cell lung cancer: a two-center study. J. Digit Imag. 2021;34:1073–1085. doi: 10.1007/s10278-021-00484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C, et al. Non-invasive measurement using deep learning algorithm based on multi-source features fusion to predict PD-L1 expression and survival in NSCLC. Front. Immunol. 2022;13:828560. doi: 10.3389/fimmu.2022.828560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C, et al. Deep learning to predict EGFR mutation and PD-L1 expression status in non-small-cell lung cancer on computed tomography images. J. Oncol. 2021;2021:5499385. doi: 10.1155/2021/5499385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C, et al. Predicting EGFR and PD-L1 status in NSCLC patients using multitask AI system based on CT images. Front Immunol. 2022;13:813072. doi: 10.3389/fimmu.2022.813072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Y-M, et al. A CT-based radiomics signature for preoperative discrimination between high and low expression of programmed death ligand 1 in head and neck squamous cell carcinoma. Eur. J. Radio. 2022;146:110093. doi: 10.1016/j.ejrad.2021.110093. [DOI] [PubMed] [Google Scholar]
- 66.Bracci S, et al. Quantitative CT texture analysis in predicting PD-L1 expression in locally advanced or metastatic NSCLC patients. Radio. Med. 2021;126:1425–1433. doi: 10.1007/s11547-021-01399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y, et al. A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas. Ann. Transl. Med. 2020;8:930. doi: 10.21037/atm-19-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39:154–173. doi: 10.1016/j.ccell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M, Wang S, Trapani JA, Neeson PJ. Challenges of PD-L1 testing in non-small cell lung cancer and beyond. J. Thorac. Dis. 2020;12:4541–4548. doi: 10.21037/jtd-2019-itm-010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duvivier H. L. et al. Pembrolizumab in patients with tumors with high tumor mutational burden: results from the targeted agent and profiling utilization registry study. J. Clin. Oncol. JCO2300702 (2023). [DOI] [PubMed]
- 71.Lu MY, et al. Federated learning for computational pathology on gigapixel whole slide images. Med. Image Anal. 2022;76:102298. doi: 10.1016/j.media.2021.102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rieke N, et al. The future of digital health with federated learning. NPJ Digit. Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vanguri RS, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat. Cancer. 2022 doi: 10.1038/s43018-022-00416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boehm KM, et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat. Cancer. 2022;3:723–733. doi: 10.1038/s43018-022-00388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang Y, et al. Biology-guided deep learning predicts prognosis and cancer immunotherapy response. Nat. Commun. 2023;14:5135. doi: 10.1038/s41467-023-40890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.