Abstract
Basal forebrain cholinergic neurons (BFCN) participate in processes of learning, memory, and attention. Little is known about the genes expressed by BFCN and the extracellular signals that control their expression. Previous studies showed that bone morphogenetic protein (BMP) 9 induces and maintains the cholinergic phenotype of embryonic BFCN. We measured gene expression patterns in septal cultures of embryonic day 14 mice and rats grown in the presence or absence of BMP9 by using species-specific microarrays and validated the RNA expression data of selected genes by immunoblot and immunocytochemistry analysis of their protein products. BMP9 enhanced the expression of multiple genes in a time-dependent and, in most cases, reversible manner. The set of BMP9-responsive genes was concordant between mouse and rat and included genes encoding cell-cycle/growth control proteins, transcription factors, signal transduction molecules, extracellular matrix, and adhesion molecules, enzymes, transporters, and chaperonins. BMP9 induced the p75 neurotrophin receptor (NGFR), a marker of BFCN, and Cntf and Serpinf1, two trophic factors for cholinergic neurons, suggesting that BMP9 creates a trophic environment for BFCN. To determine whether the genes induced by BMP9 in culture were constituents of the BFCN transcriptome, we purified BFCN from embryonic day 18 mouse septum by using fluorescence-activated cell sorting of NGFR+ cells and profiled mRNA expression of these and NGFR– cells. Approximately 30% of genes induced by BMP9 in vitro were overexpressed in purified BFCN, indicating that they belong to the BFCN transcriptome in situ and suggesting that BMP signaling contributes to maturation of BFCN in vivo.
Keywords: nerve growth factor receptor, neuronal development, septum, microarray, fluorescence-activated cell sorting
Bone morphogenetic proteins (BMPs) play critical roles in the development of the nervous system, and there is growing evidence that BMPs regulate the expression of neurotransmitter phenotype, including the cholinergic phenotype (1–4) of basal forebrain cholinergic neurons (BFCN) that project to the neocortex and hippocampus and are important in the processes of attention, learning, and memory (5). Previous studies identified multiple BMP-regulated target genes in a variety of cells, including nervous tissue. However, although BMP target genes have been characterized in the specification of catecholaminergic and serotonergic neurons (6, 7), little is known about BMP target genes implicated in BFCN determination. BFCN are defined by their neuroanatomical location and the neurotransmitter that they synthesize and release, i.e., acetylcholine. The latter process requires a concerted expression of three genes, encoding choline acetyltransferase (Chat), the vesicular acetylcholine transporter (Vacht), and the choline transporter 1 (Cht1). Several additional features of these cells include expression of acetylcholinesterase, the neurotrophin receptors NGFR and TRKA, the expression of certain neurotransmitter receptors (e.g., GABAA), and estrogen receptors. However, beyond these attributes, not much is known about the genes expressed by BFCN. In the current study, we performed microarray gene expression profiling in mouse and rat primary septal neurons treated with BMP9 (growth/differentiation factor 2). BMP9 up-regulated the expression of numerous genes, and this effect was reversible in most cases. Moreover, multiple BMP9-induced genes are highly expressed in purified BFCN suggesting that, in addition to providing a differentiating signal for their neurotansmitter phenotype, BMP9 may also act to induce other phenotypic features of these neurons.
Materials and Methods
Cell Culture. Dissociated septal cells from embryonic day (E) 14 mice or rats were plated on polyL-lysine/laminin-coated tissue culture dishes, and grown in DMEM containing 10% heat inactivated FBS and FGF2 (20 ng/ml). BMP9 (10 ng/ml; Wyeth) or vehicle was added immediately after plating and again every 24 h (5). All experiments with animals were approved by the Boston University Institutional Animal Care and Use Committee.
Purification of BFCN by FACS. Dissociated cells from E18 mice were suspended in 2% FBS in PBS (FACS buffer) at a concentration of 5 × 107 cells per ml and immunolabeled with affinity-purified rabbit anti-p75 neurotrophin receptor (NGFR) antibody (Advanced Targeting Systems, San Diego; 1:100 dilution), for 45 min at 4°C. After two washes, cells were centrifuged through a cushion of 4% BSA, resuspended in FACS buffer, and incubated with a goat anti-rabbit Alexa-fluor conjugated secondary antibody (20 μl/ml; Invitrogen) at 4°C for 30 min. The cells were washed, pelleted through a BSA cushion, and resuspended in 1% FBS in DMEM. The appropriate controls (unlabeled cells and cells incubated only with the secondary antibody) were performed in parallel. NGFR+ and NGFR– cells were sorted and purified by using FACS (MoFlo, Cytomation, Ft. Collins, CO). The background was measured from cells exposed only to the secondary antibody, and dead and false-positive cells were excluded by using the summit for moflo acquisition and sort control software (Cytomation).
Oligonucleotide Microarrays. RNA was extracted by using the guanidinium thiocyanate-phenol/chloroform method. Amplification of cRNA, RNeasy spin column purification (Qiagen, Valencia, CA), and cRNA fragmentation were performed as described in ref. 8. A spiked standard curve was used for normalization and conversion to RNA frequencies as described in refs. 8 and 9. Reaction mixtures were hybridized to Affymetrix Mu11KsubA and Mu11KsubB (mouse cultures), RG_U34A (rat cultures), or Mouse Genome 430 2.0 (purified BFCN) arrays. The arrays were stained with Streptavidin R-phycoerythrin (Molecular Probes) by using the GeneChip Fluidics Station 400 and scanned with a Hewlett–Packard GeneArray Scanner according to the manufacturer's instructions. Data were collected and analyzed by microarray suite 4.0 software.
RT-PCR. RNA samples were also used for RT-PCR by using the Superscript One-Step RT-PCR with Platinum Taq (Invitrogen Life Technologies). First-strand cDNA synthesis was performed with 25 ng of total RNA, oligo dT primer, and reverse transcriptase at 48°C (45 min). Primers used included Gapd (BD Biosciences, Franklin Lakes, NJ); Ngfr (forward: CACCACCTCCAGAGCGAGACCTCATAG; reverse: GAACATCAGCGGTCGGAATG); Chat (forward: CGGGATCCTGCCTCATCTCTGGTGT; reverse: GGCGGAATTCAATCACAACAT); Vacht (forward: AGCGGGCCTTTCATTGATCG; reverse: GGCGCACGTCCACCAGGAAGG); and Cht1 (forward: CGGGGAACCATTGAATTCGTTGAAGTCTAC; reverse: GGGGCAAGCTTCCACTTTCAAATAGATACT). PCR was performed by using Platinum TaqDNA polymerase with a denaturing step for 2 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 58°C (except for Chat at 57°C), and 2 min at 72°C, and terminated by an elongation step at 72°C for 7 min. PCR products were size-fractionated on a 10% polyacrylamide gel and stained with ethidium bromide.
Immunoblots. Septal cultures from E14 mice were treated for 3 days with BMP9 (10 ng/ml) or vehicle, cells were harvested, and equal protein amounts of cell lysates were processed for SDS/PAGE. Proteins were electroblotted onto poly(vinylidene difluoride) membranes and blocked. Membranes were probed with antibodies to the Na+/K+ ATPase α-1 subunit (Upstate Biotechnology, Lake Placid, NY; 1:10,000); noggin (Regeneron Pharmaceuticals, Tarrytown, NY; RP57-16, 50 ng/ml); caveolin (Transduction Laboratories, Lexington, KY; C37120, 1:1,000); NGFR (affinity-purified goat polyclonal antibody, Santa Cruz Biotechnology; 1:500); and to cyclin-dependent kinase 5 (Santa Cruz Biotechnology; 1:100). Bands were visualized by using an enhanced chemiluminescence reagent (DuPont/NEN).
Immunocytochemistry. E14 septal cells were plated on cover-glass coated with polyD-lysine and laminin and fixed 3 days after in methanol for 5 min at –10°C, or fixed in 3% paraformaldehyde/0.02% glutaraldehyde for 10 min and incubated at room temperature with 0.1% Triton X-100. The cells were washed with PBS, blocked for 60 min with 1% BSA, and incubated overnight with primary antibodies in 1% BSA and for 40 min with the appropriate fluorescent Alexa-conjugated secondary antibodies (Molecular Probes). Slides were mounted by using Prolong Antifade (Molecular Probes). Double immunofluorescence staining with anti-Na+/K+ ATPase α-1 monoclonal antibody (2 μg/ml) and anti-VACHT goat affinity-purified polyclonal antibody (1:500; Chemicon), was done in parallel with negative and positive controls. Immunofluorescence staining with anti-NGFR (Santa Cruz Biotechnology) and anti-noggin (3 μg/ml) antibodies was also performed.
Results
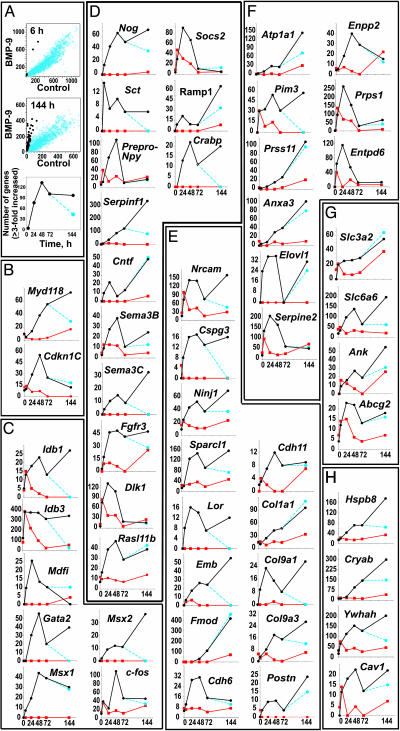
Pattern of Gene Expression After BMP9 Treatment. We treated cells from E14 mouse septum with BMP9 or vehicle (control cells), harvested their RNA at 20 min and 6, 24, 48, 72, and 144 h, and analyzed mRNA expression by using the Mu11K microarrays. BMP9 activated the expression of multiple genes in a time-dependent fashion. Fig. 1A shows genes whose mRNA abundance was increased 3-fold or more by BMP9 as compared with the control at any one of the six time points examined. By 6 h of treatment, only 5 genes fulfilled this criterion, but by 24 h, 77 genes were induced. At 48 h, there were 134 induced genes, and this number stabilized at 102 and 98 by 72 and 144 h, respectively. When BMP9 treatment was terminated after the initial 72 h, the number of genes that fulfilled our 3-fold-over-control criterion dropped to 44 3 days later (i.e., in the absence of BMP9 for another 72 h). These data show that the continuous presence of BMP9 is required for many of its differentiating activities. Similar qualitative and quantitative patterns of change in gene expression evoked by BMP9 were observed in rat primary septal cultures, examined with the RG-U34A array (Table 2, which is published as supporting information on the PNAS web site).
Fig. 1.
BMP9 induction of gene expression in cultured cells from E14 mouse septum. Cells were grown for varying periods of time in the presence or absence of BMP9 (10 ng/ml). Microarray analysis was performed on the RNA purified from the cultures. (A) The abundance of particular mRNA species, in parts per million (ppm) based on internal standard curve calibrations, in BMP9-treated cells versus their abundance in control cells, depicting the trends at the beginning and the end of the time course (i.e., at 6 and 144 h, respectively). Genes whose expression is up-regulated at least 3-fold by BMP9 are indicated by black circles. Time course (Bottom) of the total number of affected genes (black circles), including those whose expression remained elevated 72 h after BMP9 was removed from the cultures (blue circles). (B–H) Graphs represent those genes whose expression was up-regulated at least 3-fold by BMP9 (black circle) over controls (red circles) at any two of the six time points examined. Some cells were treated for 3 days with BMP9, washed on the third day, and were incubated for an additional 3 days in the absence of BMP9 (blue circles). The data are expressed in ppm on the ordinates and time (hours) on the abscissas. (B) Cell cycle-associated genes. (C) Transcription factors. (D) Signaling molecules. (E) Extracellular matrix and adhesion molecules. (F) Enzymes and enzyme inhibitors. (G) Transporters. (H) Chaperonins.
Genes whose expression was induced by BMP9 fall into several categories and encode proteins that can be classified as regulators of cell-cycle/growth control, transcription factors, signal transduction molecules (receptor ligands, receptors, and modulators of signaling), extracellular matrix and adhesion molecules, enzymes, transporters, and chaperonins. For the final data analysis, we elected to focus only on those genes whose transcripts were induced by BMP9 to levels at least 3-fold over controls at any two of the six time points examined. Using this rather stringent criterion, we arrived at a total number of 52 BMP9 responsive murine genes. This set represents the most responsive genes, and multiple additional genes are induced by BMP9 to a lesser extent.
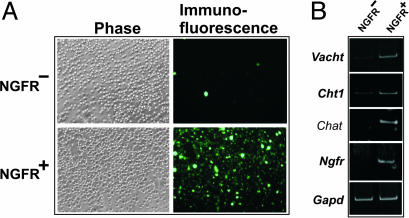
Multiple BMP9-Induced Genes in Septal Cultures Belong to the BFCN Transcriptome. BFCN neurons express a number of additional markers other than cholinergic, e.g., NGFR. In the rat, our data show that BMP9 induced the expression of the latter gene (Table 2), suggesting that BMP9 may induce other genes characteristic of BFCN. To test this hypothesis, and recognizing the limitations inherent in the interpretation of data obtained from heterogeneous cellular models, we profiled the transcriptome of purified septal cholinergic neurons. These cells are amenable to purification by FACS because >90% of them express NGFR and, similarly, >90% of NGFR-expressing cells are cholinergic (10, 11). Moreover, the adult-like pattern of septal NGFR-expressing cells is established by E17 in the rat (12). Thus, we used an anti-NGFR antibody to label BFCN in a suspension of mouse E18 septal cells and used FACS to obtain the NGFR+ (i.e., BFCN) and NGFR– (non-BFCN) cells. As expected, the FACS-purified BFCN were highly enriched in the NGFR protein and mRNA as well as in the mRNA encoding three markers of cholinergic neurons, Chat, Vacht, and Cht1 (Fig. 2) as compared with the NGFR– cells. The purified BFCN expressed multiple mRNA species in excess of those found in the NGFR– fraction. The latter fraction, which was presumably heterogeneous, was enriched in GABAergic markers, e.g., glutamic acid decarboxylase (data not shown). Remarkably, the BFCN overexpressed ≈30% of the genes induced by BMP9 in primary septal cells (Table 1), indicating that multiple BMP9 responsive genes in the primary neurons belong to the gene expression repertoire of differentiated septal cholinergic neurons.
Fig. 2.
Analysis of FACS-purified BFCN. (A) Visualization of NGFR positive and negative septal cells after FACS. After immunostaining of dissociated cells from E18 mouse septa with anti-NGFR polyclonal antibody and Alexa-fluor conjugated secondary antibody, the cells were sorted and purified by FACS (see Materials and Methods), and aliquots of the positive and negative fractions were analyzed by phase and fluorescence microscopy. (B) RT-PCR of BFCN markers for Vacht (814 bp), Cht1 (840 bp), Chat (267 bp), Ngfr (657 bp), and Gapd (983 bp; as control) of RNAs obtained from NGFR positive and negative cell fractions.
Table 1. Multiple genes induced by BMP9 in primary septal cell culture are enriched in septal NGFR+ neurons.
| Gene symbol | Gene name | Enrichment, fold |
|---|---|---|
| Cell cycle/growth control | ||
| Cdkn1c | Cyclin-dependent inhibitor 1C Transcription factors | 2 |
| Idb1 | Inhibitor of DNA binding 1 | 3 |
| Idb3 | Inhibitor of DNA binding 3 | 5 |
| Fos | FBJ osteosarcoma oncogene (c-fos) Signal transduction | 5 |
| Bmpr1a | Bone morphogenetic protein receptor type 1A | 2 |
| Ramp1 | Receptor (calcitonin) activity modifying protein 1 | 2 |
| Rbp1 | Retinol binding protein 1, cellular | 2 |
| Sema3c | Semaphorin 3C | 2 |
| Fgfr3 | Fibroblast growth factor receptor 3 Extracellular matrix/adhesion | 4 |
| Col9a1 | Procollagen type 9 α 1 | 2 |
| Cspg3 | Chondroitin sulfate proteoglycan 3 (neurocan) | 2 |
| Fath | Fat tumor suppressor homolog | 3 |
| Sparcl1 | SPARC-like 1 (mast9, hevin) Enzymes/inhibitors | 6 |
| Elovl1 | Elongation of very long chain fatty acids | 2 |
| Plod2 | Procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | 2 |
| Atp1a1 | ATPase, Na+/K+ transporting, α 1 polypeptide | 3 |
| Cyp7b1 | Cytochrome P450, family 7, subfamily b, polypeptide 1 | 3 |
| Serpine2 | Serine (or cysteine) proteinase inhibitor, clade E, member 2 (nexin-1) | 3 |
| Prss11 | Protease, serine, 11 (lgf binding) Chaperonins | 5 |
| Serpinh1 | Serine (or cysteine) proteinase inhibitor, clade H, member 1 (HSP47) | 4 |
Cell Cycle and Growth Control. Treatment of primary septal neurons with BMP9 induced the expression of two growth inhibitory proteins, the cyclin-dependent kinase inhibitor 1C (Cdkn1c/p57Kip2) and Myd118/Gadd45β (Fig. 1B). Whereas the induction of Cdkn1c was transient, the induction of Myd118 waned only upon BMP9 withdrawal. Interestingly, Cdkn1c expression was also higher in purified BFCN than in NGFR– cells (Table 1).
Transcription Factors. BMP9 increased the expression of eight transcription factors (Fig. 1C). Among these transcription factors, we found genes encoding the helix-loop-helix Idb1 and Idb3 transcriptional regulators. These two transcripts, initially expressed in control cultures, dropped their levels after 6 h. However, in the presence of BMP9, their levels remained elevated at all times. High Idb1 and Idb3 expression was also a feature of purified BFCN. BMP9 induced four genes with little or no expression in control cultures, namely Mdfi (I-mf; inhibitor of MyoD family), Gata2, Msx1, and Msx2. BMP9 also increased the expression of the ubiquitous transcription factor c-fos, with peak levels at 48 h. In rat septal cultures, BMP9 induced the expression of Roaz (also termed Oaz; Table 2).
Signal Transduction. BMP9 induced the expression of seven signal transduction-related genes that were absent or had low expression levels in control cultures. Three of them, namely those encoding secretin (Sct), semaphorin E (Sema3C), and cellular retinoic acid binding protein (Crabp), were undetectable once BMP9 was withdrawn, whereas those encoding noggin (Nog), pigment epithelium derived factor (Serpinf1, Pedf/Sdf3), ciliary neurotrophic factor (Cntf), and receptor activity modifying protein 1 (Ramp1; an accessory protein for G protein-coupled receptors for calcitonin and related peptides) showed a decline in their expression levels or remained unchanged upon BMP9 withdrawal (Fig. 1D). The expression of most of the genes that had detectable basal mRNA levels in control cultures peaked between 24 and 48 h in the presence of BMP9. Among them we found the precursor of neuropeptide Y (prepro-Npy), the extracellular secreted protein semaphorin A (Sema3B), and two genes that are important during neuronal differentiation, δ-like 1 (Dlk1; a ligand for the Notch signaling pathway) and suppressor of cytokine signaling 2 (Socs2; expressed highly in neurons). BMP9 also induced the expression of Rasl11b, an immediate early gene that down-regulates the actions of TGF-β (Fig. 1D) (13). In addition, BMP9 induced the expression of three receptors: fibroblast growth factor receptor 3 (Fgfr3), Ngfr, a marker of BFCN, and BMP receptor type 1A (Bmpr1a). The latter two were detected only in the rat cultures, because the murine microarray does not contain probes for these genes (Table 2). Also, in the rat, BMP9 induced the expression of glypican-3 (Gpc3), olfactomedin-1 (Olfm1), and cytosolic retinol-binding protein (Rbp1; Table 2). Moreover, the abundance of transcripts encoding Sema3C, Ramp1, Fgfr3, Bmpr1a, and Rbp1 was significantly higher in purified BFCN than in NGFR– cells (Table 1).
Extracellular Matrix and Adhesion. BMP9 up-regulated the expression of several adhesion proteins strongly expressed in the nervous system, including Nrcam/Bravo, neurocan (Cspg3), ninjurin (Ninj1), sparc-like protein 1 (Sparcl1), loricrin (Lor), and embigin (Emb). Although many of the BMP9-induced genes encode proteins mostly studied in the context of bone and cartilage formation, including fibromodulin (Fmod), cadherin-6 (Cdh6), cadherin-11 (Cdh11), type I collagen (Col1a1), and type IX collagen (Col9a1), there is growing evidence that several of them function in the adult CNS and are important for cell sorting and aggregation during CNS development. In addition, BMP9 up-regulated the expression of osteoblast-specific factor-2 (Postn), a putative adhesion protein related to neuronal fasciclin 1. In the rat, two other genes were also up-regulated by BMP9, Fath (a member of the cadherin superfamily of proteins not represented in the murine microarray) and fibronectin (Fn1; which did not meet the criteria for inclusion in Fig. 1E). Col9a1, Cspg3, Fath, and Sparcl1 were also overexpressed in purified BFCN as compared with NGFR– cells (Table 1).
Enzymes and Enzyme Inhibitors. BMP9 induced the expression of several enzymes and enzyme inhibitors (Fig. 1F). A sustained induction was observed for (Na/K)ATPase α1 subunit (Atp1a1); Pim3, a member of the Pim family of serine/threonine kinases, also induced by synaptic activity and during embryonic development of the CNS; insulin-like growth factor-binding protein 5 protease (Prss11); annexin A3 (Anxa3), a phospholipid-binding protein endowed with the activity of inositol 1,2-cyclic phosphate phosphodiesterase, which also acts as a phospholipase A2 inhibitor; and a protein termed elongation of very long chain fatty acids (Fen1/Elo2, Sur4/Elo3, yeast-like 1, Elovl1), involved in the biosynthesis of very long chain fatty acids and sphingolipids. In contrast, the induction of the following genes was transient: protease nexin-1 (Serpine2), a thrombin and urokinase inhibitor with an amino acid sequence identical to glial-derived neurite promoting factor; autotaxin/lysophospholipase D (Enpp2), which catalyzes the hydrolysis of lysophosphatidylcholine into choline and lysophosphatidic acid; phosphoribosylpyrophosphate synthetase 1 (Prps1), required for the de novo and salvage pathways of purine and pyrimidine biosynthesis; and ectonucleoside triphosphate diphosphohydrolase 6 precursor (Entpd6 or Cd39l2), an enzyme that hydrolyzes extracellular nucleoside tri- and/or diphosphates. In the rat (Table 2), four other genes were also up-regulated by BMP9: palmitoyl-protein thioesterase (Ppt2) and procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (Plod2), both not represented on the murine microarray, and lipoprotein lipase and cytochrome P450 oxysterol 7-α-hydroxylase (Cyp7b1; which were not up-regulated by BMP9 in the mouse). Among these BMP9-responsive enzyme-encoding genes Atp1a1, Cyp7b1, Elovl1, Plod2, Prss11, and Serpine2 were also overexpressed in purified BFCN as compared with the NGFR– cells (Table 1).
Transporters. Four genes classified as transporters emerged as targets for BMP9 (Fig. 1G). Primary cultures treated with BMP9 maintained high expression levels of members 2 (Slc3a2/Cd98/4f2hc) and 6 (Slc6a6/Taut) of the solute carrier families 3 and 6, respectively. Slc3a2 encodes a protein belonging to the activators of the dibasic and neutral amino acid transport, and Slc6a6 encodes a sodium-dependent taurine transporter. In addition, BMP9 up-regulated the expression of progressive ankylosis protein (Ank), a membrane protein that regulates intra- and extracellular levels of inorganic pyrophosphate, and member 2 (Abcg2) of the superfamily of ATP-binding cassette transporters, best known as one of several proteins conferring a multidrug resistance phenotype to cancer cells.
Chaperonins. Three genes coding for chaperonins were induced by BMP9 in a sustained fashion (Fig. 1H), including heat shock 27-kDa protein 8 (Hspb8), the B chain of α-crystallin (Cryab), and 14-3-3 η (Ywhah). Also, mRNA levels for caveolin (Cav1) rose within the first 24 h and were maintained for the remaining time in the presence of BMP9. In the rat (Table 2), BMP9 induced the expression of neurofilament medium (Nfm) and heat shock protein 47 (Serpinh1). The latter transcript was also significantly enriched in purified BFCN as compared with the NGFR– cells (Table 1).
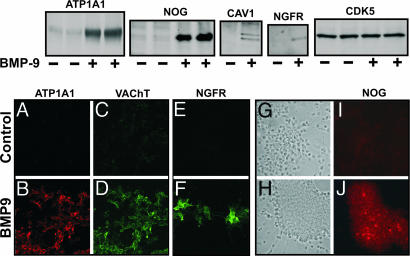
Detection of BMP9-Induced Increases in Gene Expression by Western Blot and Immunocytochemistry. To verify the data obtained by microarray analysis, we chose four of the genes induced by BMP9 and performed Western blot and immunocytochemistry of the corresponding proteins in samples from E14 mouse septal cultures that were treated with BMP9 for 72 h. In all cases, our results confirmed the data obtained with the microarray assays. Two of the chosen genes, encoding (Na+/K+)-ATPase α1 subunit and noggin, were strongly induced by BMP9 in both the mouse and rat (Fig. 1 and Table 2), and their protein products were also increased (Fig. 3 Upper). Double immunofluorescence staining with an antibody to (Na+/K+)-ATPase α1 and an antibody against VACHT revealed overlapping labeling of cells that had been treated with BMP9 (Fig. 3 A–D). Immunofluorescence staining with an antibody to human noggin recognized some cells within the cell clusters that typically develop after treatment of embryonic septal cultures with BMP9 (Fig. 3 I and J; phase-contrast microphotography, Fig. 3 G and H). A third gene, coding for caveolin-1, was chosen for being strongly induced in the mouse (Fig. 1H) but only at 144 h in the rat. Caveolin-1 protein was indeed induced by BMP9 (Fig. 3 Upper). The fourth protein investigated, NGFR, was selected because it is a marker of BFCN, and its mRNA was strongly induced by BMP9 in the rat microarray analysis (Table 2). The immunoblot (Fig. 3 Upper) revealed a band corresponding to this protein that was confirmed by immunofluorescence, with strong labeling of cells of neuronal morphology in cultures treated with BMP9 (Fig. 3 E and F). Finally, as a negative control, we analyzed a protein, cyclin-dependent kinase 5 (CDK5), which showed no change in its mRNA expression nor protein levels when cells were treated with BMP9 (Fig. 3 Upper).
Fig. 3.
Analysis of BMP9-induced proteins. (Upper) Western blots of selected proteins from control and BMP9-treated neuronal cultures. Septal cultures from E14 mice were treated for 3 days with BMP9 (10 ng/ml) or vehicle. Cells were harvested and processed for SDS/PAGE as described in Materials and Methods. (Lower) Immunocytochemistry (A–J) of cultures are treated as in Upper.(A–D) Double immunofluorescence staining with anti-Na+/K+ ATPase α-1 and anti-VACHT antibodies, done in parallel with negative and positive controls. (E and F) Immunofluorescence staining with anti-NGFR antibody. Phase-contrast (G and H) and immunofluorescence (I and J) staining with anti-Noggin antibody of the same field. All pictures were obtained with a ×20 objective.
Discussion
The transcriptome profiling performed on purified BFCN and cultured septal cells treated with BMP9 revealed that a set of genes that is normally highly expressed in differentiated BFCN is induced by BMP9 in vitro. These data indicate that BMP-mediated signaling is important for the maturation of BFCN in a way that extends beyond their neurotransmitter phenotype.
Some of the responsive genes may contain BMP response elements, whereas others may be up-regulated as a consequence of the activity of autocrine molecules or transcription factors induced by BMP9. An example of the former may be the helix-loop-helix transcription regulator Idb1. Both the murine and rat septal cultures required BMP9 to maintain Idb1 expression, and its mRNA was also found to be expressed in purified BFCN. IDB1 acts by binding basic helix-loop-helix transcriptional activators, thus preventing the latter from interacting with the DNA. Idb1 expression is regulated by BMPs through a BMP response element located in its promoter (14). Idb1 and Idb3 (a related gene also responsive to BMP9 and expressed in purified BFCN) are functional in the developing and adult brain (15). Our data show that BMP9 also induced the expression of the zinc-finger transcription factor Gata2. Previous studies in several systems showed that Gata2 expression is induced by other BMPs (16–19). Mice with a targeted deletion of Gata2 die in utero and have a number of neurological abnormalities (20). Finally, the induced expression of c-fos by BMP9 agrees with previous studies, which show that BMPs regulate the expression of this gene (21) and that the expression of Chat and Vacht may be regulated by c-fos (22). c-fos was also overexpressed in purified BFCN.
In rat septal cultures, BMP9 induced the expression of two transcription factors, Roaz and Cbfb (Pebp2b). These data are of particular interest because ROAZ binds SMAD1, and the resulting complex binds to BMP response elements in the promoter regions of BMP-responsive genes (23, 24). This finding demonstrates that the abundance of Roaz mRNA is increased by BMP treatment, a result that suggests that the expression of Roaz itself is regulated by BMPs. Our data are also consistent with previous reports indicating that in the CNS, BMPs up-regulate the expression of the homeobox genes Msx1 (25) and Msx2 (26). However, whereas Msx1 was expressed in NGFR+, it was absent in NGFR–, suggesting that Msx1 may be a requirement for the maintenance of the BFCN phenotype at this developmental stage. Two previously uncharacterized BMP9 targets encoding signal transduction proteins that may participate in cholinergic differentiation were also found, Cntf and Serpinf1. The products of these target genes, CNTF and PEDF (SERPINF1), are known to promote neuronal survival and cholinergic differentiation. Although adult cholinergic septal neurons do not respond to CNTF by increasing CHAT activity (as spinal cord motorneurons do), they do respond to CNTF after axotomy (e.g., by fimbria-fornix transection) by maintaining the expression of Ngfr (27). PEDF can induce neuronal differentiation (28) and is neuroprotective for cholinergic motorneurons (29). Moreover, in cultured cerebellar granule cells, PEDF induced the expression of nerve growth factor and its receptors, Ngfr and Trka (30). In our previous studies, the induction of the cholinergic phenotype by BMP9 was highly potentiated by FGF2, which by itself had no activity (2). Here, we show that BMP9 induces Fgfr3 and purified BFCN also express this receptor.
Other BMP9-induced signal transduction-related genes are implicated in neuronal differentiation. The Dlk1 gene belongs to the EGF-like homeotic protein family, which includes the Notch receptor and its ligands, such as Delta and Serrate (31). SOCS2 belongs to the suppressor of cytokines signaling family of proteins that inhibit Janus kinases and signal transducer and activator of transcription proteins signaling (32). In addition, Socs2 is expressed in the developing nervous system, at ages consistent with a role in neural differentiation, in neural progenitor cells and neurons, but not in astrocytes (33). Moreover, its expression is dramatically increased by CNTF (33), whose gene is responsive to BMP9. Finally, BMP9 induced the expression of two retinoid-binding proteins, cellular retinoic acid binding protein I (Crabp1 in the mouse) and cellular retinol binding protein (Rbp1 in the rat). These proteins appear to play a role in neuronal differentiation (34–36). Rbp1 expression was also higher in purified BFCN than in the NGFR– cells.
In addition to our data showing BMP9 induced expression of genes implicated in neuronal and cholinergic differentiation, our results also show the up-regulation of genes involved in cell–cell interactions, neuronal plasticity, cell-cycle control, and apoptosis. BMP9 induced the expression of two cadherins, Cdh6 and Cdh11, which are relevant in the establishment of motoneuron differentiation (37) and specific motoneuron sorting (38), and participate in the establishment of axoaxonal, axoglial, and glio-glial contacts (39). The cell adhesion molecule NRCAM has been implicated in the development of axon tracts (40). The induction of Cspg3, Ninj1, Emb, and Sparcl1 have been associated with neuronal remodeling and repair. This fact is noteworthy, because there are reports suggesting that BMPs may be involved in neuronal plasticity after traumatic or hypoxic brain injury (41–48). Sparcl1, which was also overexpressed in purified BFCN, is highly expressed in the developing and adult CNS and contains one follistatin domain (49); thus, like noggin, it may interact with BMPs. The rise in Sparcl1 expression upon treatment with BMP9, preceding those of Col1a1 and Hspb8, may not be surprising because SPARC (secreted protein, acidic, cysteine-rich) can bind to collagen type I and regulate its production (50). In this regard, it should be pointed out that BMP9 induced the concerted expression of genes associated with the laying down of extracellular collagen matrix. Three of these genes encode collagens, including the Col9a1, Col9a3, and Col1a1. The mRNA for Col9a1 was also enriched in purified BFCN as compared with NGFR– cells. Moreover, BMP9 induced the expression of Serpinh1, a chaperonin that is necessary for the proper processing of procollagens in the endoplasmic reticulum (51), and Plod2, important in collagen crosslinking. Significantly, Plod2 and Serpinh1 transcripts were also overexpressed in purified BFCN, suggesting that the synthesis and processing of collagens constitutes a BMP-regulated and heretofore unknown property of these cells. Lastly, within the group classified as chaperonins, BMP9 induced the expression of Ywhah, initially considered to be brain-specific and now recognized as being involved in many biological processes, such as cell-cycle control, signal transduction, apoptosis, and long-term potentiation (52).
BMP9 induced the expression of several genes encoding proteins that modulate BMP signaling, including Bmpr1a, Nog, Gpc3, Roaz, and Rasl11b, suggesting that BMP9 may adjust its own activity through a feedback mechanism. Neural precursor cells express BMPR1A, and its activation in these cells induces the expression of Ngfr (53). Our data showing the induction of Ngfr by BMP9 are consistent with the possibility that BMPR1A mediates this effect. Bmpr1a mRNA expression was also higher in purified BFCN as compared with NGFR– cells, suggesting that BMP signaling is functional in vivo. Moreover, purified BFCN expressed other genes necessary for BMP signal transduction including Bmpr2, Smad1, Samd5, and Smad4 (data not shown). As noted above, BMP9 induces the expression of Nog, and noggin is a BMP antagonist capable of binding multiple BMPs. Nog was also expressed by purified BFCN and its expression was 40% higher in these cells than in NGFR– cells (data not shown).
In summary, our analysis shows that BMP9 induces the expression of multiple genes in cultured basal forebrain cells. A large fraction of these genes belongs to the BFCN transcriptome, indicating that BMP signaling participates in the maturation of these neurons. Moreover, several BMP9-induced genes encode proteins with trophic activities for BFCN, suggesting that BMP signaling participates in the generation of a favorable milieu for these cells. Among the BMP9-responsive genes, however, only a few have known actions in the specification of neuronal and/or cholinergic phenotype; others are previously uncharacterized targets, and their functions in BFCN remain to be determined.
Supplementary Material
Acknowledgments
We thank Dr. Alan Ho for his expertise and help in the FACS analysis. This research was supported by Alzheimer's Association Grant IIRG-00-207 (to I.L.-C.) and National Institutes of Health Grants AG09525 (to J.K.B.), NS042793 (to J.K.B.), NS044238 (to B.B.), MH059775 (to B.E.S.), and NS30791 (to B.E.S.).
Author contributions: I.L.-C., M.T.F., and J.K.B. designed research; I.L.-C., M.T.F., T.J.M., V.P.K., B.E.S., V.D., B.B., and J.K.B. performed research; R.S.T. contributed new reagents/analytic tools; I.L.-C., M.T.F., and J.K.B. analyzed data; and I.L.-C. and J.K.B. wrote the paper.
Abbreviations: BFCN, basal forebrain cholinergic neurons; BMP, bone morphogenetic protein; CNTF, ciliary neurotrophic factor; En, embryonic day n; NGFR, p75 neurotrophin receptor; VACHT, vesicular acetylcholine transporter.
References
- 1.Fann, M.-J. & Patterson, P. H. (1994) J. Neurochem. 63, 2074–2079. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Coviella, I., Berse, B., Krauss, R., Thies, R. S. & Blusztajn, J. K. (2000) Science 289, 313–316. [DOI] [PubMed] [Google Scholar]
- 3.Nonner, D., Barrett, E. F., Kaplan, P. & Barrett, J. N. (2001) J. Neurochem. 77, 691–699. [DOI] [PubMed] [Google Scholar]
- 4.Nonner, D., Panickar, K., Barrett, E. F. & Barrett, J. N. (2004) J. Neurochem. 91, 77–87. [DOI] [PubMed] [Google Scholar]
- 5.Bartus, R. T. (2000) Exp. Neurol. 163, 495–529. [DOI] [PubMed] [Google Scholar]
- 6.Goridis, C. & Rohrer, H. (2002) Nat. Rev. Neurosci. 3, 531–541. [DOI] [PubMed] [Google Scholar]
- 7.Galter, D., Bottner, M., Krieglstein, K., Schomig, E. & Unsicker, K. (1999) Eur. J. Neurosci. 11, 2444–2452. [DOI] [PubMed] [Google Scholar]
- 8.Byrne, M. C., Whitley, M. Z. & Follettie, M. T. (2000) in Current Protocols in Molecular Biology, ed. Ausubel, F. M. (Wiley, New York), 4th Ed., pp. 22.2.1–22.2.13.
- 9.Hill, A. A., Brown, E. L., Whitley, M. Z., Tucker-Kellogg, G., Hunter, C. P. & Slonim, D. K. (2001) Genome Biol. 2, 0055.1–0055.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawbarn, D., Allen, S. J. & Semenenko, F. M. (1988) Neurosci. Lett. 94, 138–144. [DOI] [PubMed] [Google Scholar]
- 11.Woolf, N. J., Gould, E. & Butcher, L. L. (1989) Neuroscience 30, 143–152. [DOI] [PubMed] [Google Scholar]
- 12.Koh, S. & Loy, R. (1989) J. Neurosci. 9, 2999–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piek, E., Van Dinther, M., Parks, W. T., Sallee, J. M., Bottinger, E. P., Roberts, A. B. & Ten Dijke, P. (2004) Biochem. J. 383, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Rovira, T., Chalaux, E., Massague, J., Rosa, J. L. & Ventura, F. (2002) J. Biol. Chem. 277, 3176–3185. [DOI] [PubMed] [Google Scholar]
- 15.Lyden, D., Young, A. Z., Zagzag, D., Yan, W., Gerald, W., O'Reilly, R., Bader, B. L., Hynes, R. O., Zhuang, Y., Manova, K., et al. (1999) Nature 401, 670–677. [DOI] [PubMed] [Google Scholar]
- 16.Bilodeau, M. L., Boulineau, T., Greulich, J. D., Hullinger, R. L. & Andrisani, O. M. (2001) In Vitro Cell Dev. Biol. Anim. 37, 185–192. [DOI] [PubMed] [Google Scholar]
- 17.Xu, R. H., Ault, K. T., Kim, J., Park, M. J., Hwang, Y. S., Peng, Y., Sredni, D. & Kung, H. f. (1999) Dev. Biol. 208, 352–361. [DOI] [PubMed] [Google Scholar]
- 18.Maeno, M., Mead, P. E., Kelley, C., Xu, R. H., Kung, H.-f., Suzuki, A., Ueno, N. & Zon, L. I. (1996) Blood 88, 1965–1972. [PubMed] [Google Scholar]
- 19.Tsarovina, K., Pattyn, A., Stubbusch, J., Muller, F., van der, W. J., Schneider, C., Brunet, J. F. & Rohrer, H. (2004) Development (Cambridge, U.K.) 131, 4775–4786. [DOI] [PubMed] [Google Scholar]
- 20.Nardelli, J., Thiesson, D., Fujiwara, Y., Tsai, F. Y. & Orkin, S. H. (1999) Dev. Biol. 210, 305–321. [DOI] [PubMed] [Google Scholar]
- 21.Ohta, S., Hiraki, Y., Shigeno, C., Suzuki, F., Kasai, R., Ikeda, T., Kohno, H., Lee, K., Kikuchi, H., Konishi, J., et al. (1992) FEBS Lett. 314, 356–360. [DOI] [PubMed] [Google Scholar]
- 22.Kaufer, D., Friedman, A., Seidman, S. & Soreq, H. (1998) Nature 393, 373–377. [DOI] [PubMed] [Google Scholar]
- 23.Hata, A., Seoane, J., Lagna, G., Montalvo, E., Hemmati-Brivanlou, A. & Massague, J. (2000) Cell 100, 229–240. [DOI] [PubMed] [Google Scholar]
- 24.Shim, S., Bae, N. & Han, J. K. (2002) Nucleic Acids Res. 30, 3107–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuta, Y., Piston, D. W. & Hogan, B. L. (1997) Development (Cambridge, U.K.) 124, 2203–2212. [DOI] [PubMed] [Google Scholar]
- 26.Graham, A., Francis-West, P., Brickell, P. & Lumsden, A. (1994) Nature 372, 684–686. [DOI] [PubMed] [Google Scholar]
- 27.Panni, M. K., Atkinson, J. & Sofroniew, M. V. (1999) Neuroscience 89, 1113–1121. [DOI] [PubMed] [Google Scholar]
- 28.Steele, F. R., Chader, G. J., Johnson, L. V. & Tombran-Tink, J. (1993) Proc. Natl. Acad. Sci. USA 90, 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bilak, M. M., Becerra, S. P., Vincent, A. M., Moss, B. H., Aymerich, M. S. & Kuncl, R. W. (2002) J. Neurosci. 22, 9378–9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yabe, T., Herbert, J. T., Takanohashi, A. & Schwartz, J. P. (2004) J. Neurosci. Res. 77, 642–652. [DOI] [PubMed] [Google Scholar]
- 31.Fleming, R. J. (1998) Semin. Cell Dev. Biol. 9, 599–607. [DOI] [PubMed] [Google Scholar]
- 32.Hilton, D. J., Richardson, R. T., Alexander, W. S., Viney, E. M., Willson, T. A., Sprigg, N. S., Starr, R., Nicholson, S. E., Metcalf, D. & Nicola, N. A. (1998) Proc. Natl. Acad. Sci. USA 95, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnley, A. M., Faux, C. H., Rietze, R. L., Coonan, J. R. & Bartlett, P. F. (2002) Nat. Neurosci. 5, 1155–1162. [DOI] [PubMed] [Google Scholar]
- 34.Maden, M., Horton, C., Graham, A., Leonard, L., Pizzey, J., Siegenthaler, G., Lumsden, A. & Eriksson, U. (1992) Mech. Dev. 37, 13–23. [DOI] [PubMed] [Google Scholar]
- 35.Liu, R. Z., Denovan-Wright, E. M., Degrave, A., Thisse, C., Thisse, B. & Wright, J. M. (2004) Eur. J. Biochem. 271, 339–348. [DOI] [PubMed] [Google Scholar]
- 36.Bhasin, N., Maynard, T. M., Gallagher, P. A. & LaMantia, A. S. (2003) Dev. Biol. 261, 82–98. [DOI] [PubMed] [Google Scholar]
- 37.Marthiens, V., Padilla, F., Lambert, M. & Mege, R. M. (2002) Mol. Cell Neurosci. 20, 458–475. [DOI] [PubMed] [Google Scholar]
- 38.Price, S. R., Marco Garcia, N. V., Ranscht, B. & Jessell, T. M. (2002) Cell 109, 205–216. [DOI] [PubMed] [Google Scholar]
- 39.Takai, Y., Shimizu, K. & Ohtsuka, T. (2003) Curr. Opin. Neurobiol. 13, 520–526. [DOI] [PubMed] [Google Scholar]
- 40.Brummendorf, T., Kenwrick, S. & Rathjen, F. G. (1998) Curr. Opin. Neurobiol. 8, 87–97. [DOI] [PubMed] [Google Scholar]
- 41.Lewen, A., Soderstrom, S., Hillered, L. & Ebendal, T. (1997) NeuroReport 8, 475–479. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., Chang, C. F., Morales, M., Chou, J., Chen, H. L., Chiang, Y. H., Lin, S. Z., Cadet, J. L., Deng, X., Wang, J. Y., et al. (2001) Stroke 32, 2170–2178. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima, K., Takizawa, T., Ochiai, W., Yanagisawa, M., Hisatsune, T., Nakafuku, M., Miyazono, K., Kishimoto, T., Kageyama, R. & Taga, T. (2001) Proc. Natl. Acad. Sci. USA 98, 5868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakashima, K. & Taga, T. (2002) Mol. Neurobiol. 25, 233–244. [DOI] [PubMed] [Google Scholar]
- 45.Martinez, G., Carnazza, M. L., Di Giacomo, C., Sorrenti, V. & Vanella, A. (2001) Brain Res. 894, 1–11. [DOI] [PubMed] [Google Scholar]
- 46.Helm, G. A., Alden, T. D., Sheehan, J. P. & Kallmes, D. (2000) Neurosurgery 46, 1213–1222. [DOI] [PubMed] [Google Scholar]
- 47.Chang, C. F., Morales, M., Chou, J., Chen, H. L., Hoffer, B. & Wang, Y. (2002) Neuropharmacology 43, 418–426. [DOI] [PubMed] [Google Scholar]
- 48.Araki, T. & Milbrandt, J. (2000) J. Neurosci. 20, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendis, D. B., Ivy, G. O. & Brown, I. R. (1996) Brain Res. 730, 95–106. [DOI] [PubMed] [Google Scholar]
- 50.Hambrock, H. O., Nitsche, D. P., Hansen, U., Bruckner, P., Paulsson, M., Maurer, P. & Hartmann, U. (2003) J. Biol. Chem. 278, 11351–11358. [DOI] [PubMed] [Google Scholar]
- 51.Nagai, N., Hosokawa, M., Itohara, S., Adachi, E., Matsushita, T., Hosokawa, N. & Nagata, K. (2000) J. Cell Biol. 150, 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferl, R. J., Manak, M. S. & Reyes, M. F. (2002) Genome Biol. 3, REVIEWS3010.1–3010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panchision, D. M., Pickel, J. M., Studer, L., Lee, S. H., Turner, P. A., Hazel, T. G. & McKay, R. D. (2001) Genes Dev. 15, 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.