Abstract
The p53 tumor suppressor gene product is known to act as part of a cell cycle checkpoint in G1 following DNA damage. In order to investigate a proposed novel role for p53 as a checkpoint at mitosis following disruption of the mitotic spindle, we have used time-lapse videomicroscopy to show that both p53+/+ and p53−/− murine fibroblasts treated with the spindle drug nocodazole undergo transient arrest at mitosis for the same length of time. Thus, p53 does not participate in checkpoint function at mitosis. However, p53 does play a critical role in nocodazole-treated cells which have exited mitotic arrest without undergoing cytokinesis and have thereby adapted. We have determined that in nocodazole-treated, adapted cells, p53 is required during a specific time window to prevent cells from reentering the cell cycle and initiating another round of DNA synthesis. Despite having 4N DNA content, adapted cells are similar to G1 cells in that they have upregulated cyclin E expression and hypophosphorylated Rb protein. The mechanism of the p53-dependent arrest in nocodazole-treated adapted cells requires the cyclin-dependent kinase inhibitor p21, as p21−/− fibroblasts fail to arrest in response to nocodazole treatment and become polyploid. Moreover, p21 is required to a similar extent to maintain cell cycle arrest after either nocodazole treatment or irradiation. Thus, the p53-dependent checkpoint following spindle disruption functionally overlaps with the p53-dependent checkpoint following DNA damage.
The proper execution of events in the eukaryotic cell cycle is regulated by a number of different checkpoints. For example, to ensure stable maintenance of the genome, cells arrest in G1 or G2 upon detection of DNA damage, providing time for DNA repair before the initiation of DNA synthesis or entry into mitosis (11). Other checkpoints function during mitosis to monitor successful assembly of the spindle and control the initiation of metaphase, thus protecting the cell from chromosome missegregation (37). Genes that are required for cellular arrest at different checkpoints have been identified, demonstrating that each checkpoint is a genetic pathway activated by specific signals. The inactivation of checkpoint genes leads to increased mutation rate, chromosome loss, or changes in ploidy, depending on the genes affected (34). Interestingly, some genes that are mutated in human cancers are involved in checkpoint functions, suggesting that without such controls in place, the resulting genetic damage predisposes cells to malignancy (38).
The p53 tumor suppressor gene is mutated in over half of all sporadic human cancers. p53 has an essential role in the G1 checkpoint in response to DNA-damaging agents such as radiation (20, 21, 23). Functional analysis of the p53 protein has shown that it is a transcription factor with sequence-specific DNA binding activity (12, 22, 42). After DNA damage, p53 activates the transcription of several downstream target genes, including p21, an inhibitor of cyclin-dependent kinases (CDKs) (10). The induction of p21 causes subsequent arrest in the G1 phase of the cell cycle by binding of cyclin-CDK complexes (14, 15, 41). Additional p53 target genes are likely to cooperate with p21 in implementing G1 arrest, as p21-deficient mouse cells are only partially defective in their DNA damage arrest response, while p53-deficient cells are completely defective (2, 7).
Recently, p53 has been proposed to have an additional, novel function as a checkpoint at mitosis. Wild-type fibroblasts arrest when mitotic spindle assembly is disrupted by the addition of drugs which bind microtubules. However, studies have shown that p53-deficient fibroblasts fail to arrest under such conditions but instead undergo a new round of DNA synthesis in the absence of cell division, becoming polyploid (5, 8, 33). This phenotype is very similar to that observed in yeast strains that have inactivated spindle assembly checkpoints. Saccharomyces cerevisiae strains with mutations in the MAD or BUB genes, which monitor spindle assembly, become polyploid when treated with spindle inhibitors (17, 27). Based on this similarity, one might conclude that, like the MAD and BUB gene products, p53 monitors spindle integrity and can thus be termed a mitotic checkpoint. Also, additional data have suggested that the spindle components themselves may be under p53 regulation, as p53 has been shown to localize to the centrosome (1) and p53−/− mouse embryonic fibroblasts (MEFs) contain abnormal numbers of centrosomes (13). However, later work has shown that in cells treated with spindle inhibitors, p53 is neither expressed during mitosis nor required for mitotic arrest (33). Instead, expression of p53 protein occurs only after cells exit mitotic arrest and progress to an interphase state while retaining 4N DNA content. These data argue that p53 does not function as a mitotic checkpoint but acts at some subsequent point in the cell cycle to induce arrest following a defect in M phase.
In this study, we have characterized in greater detail the p53-dependent checkpoint following disruption of the mitotic spindle with microtubule-destabilizing drugs. Upon examining the response of wild-type and p53-deficient mouse embryo fibroblasts to spindle inhibitors, we observed that cells of either genotype underwent a transient arrest at mitosis and subsequently progressed into a G1-like state without ever completing cell division. Wild-type MEFs remained arrested in this state, while p53−/− MEFs initiated another round of DNA synthesis. Using time-lapse videomicroscopy, we were able to define the length of mitotic arrest and establish precisely the timing of S phase reentry in p53−/− MEFs after exit from mitotic arrest. We also demonstrated a requirement for the p53 target gene product, p21, in executing cell cycle arrest after spindle disruption. Upon further characterization of arrested cells, we determined that cells expressed molecular markers associated with the G1 phase of the cell cycle, despite having 4N DNA content. These data demonstrate that the p53-dependent checkpoint in cells with disrupted mitotic spindles has strong similarity to the p53-dependent checkpoint in G1 following DNA damage. Our data suggest that rather than having a novel role as a spindle checkpoint, p53 functions in the G1 phase of the cell cycle and via the same downstream pathway in murine cells which have sustained either DNA damage or microtubule disruption.
MATERIALS AND METHODS
Cells and cell culture.
p53+/+ and p53−/− MEFs were derived from 13.5-day-old embryos and used between passages 3 and 8 (18). MEFs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum supplemented with penicillin and streptomycin and grown in an atmosphere containing 5% CO2. For flow cytometry and immunoblot experiments, MEFs were plated at a density of 5 × 105 to 1 × 106 cells per 100-mm-diameter dish 24 h in advance of experiments. Nocodazole (Sigma) was kept as a 1 mg/ml stock in dimethylsulfoxide. Nocodazole treatment of MEFs was performed at a concentration of 0.125 μg of nocodazole/ml of medium.
NIH 3T3 cells were obtained from the American Type Culture Collection and grown in DMEM containing 10% calf serum supplemented with penicillin and streptomycin. For flow cytometry and immunoblot experiments, NIH 3T3 cells were plated at a density of 5 × 105 cells per 100-mm-diameter dish. Twenty-four hours after plating, cells were serum synchronized by being washed twice with DMEM and then being placed in DMEM plus 0.5% calf serum for 48 h. Nocodazole treatment of NIH 3T3 cells was performed at a concentration of 0.5 μg of nocodazole/ml of medium.
Flow cytometry.
Approximately 106 cells per 100-mm-diameter dish were detached in 0.25% trypsin and washed in phosphate-buffered saline (PBS). Following centrifugation, cells were resuspended in 0.5 ml of cold PBS. Cells were fixed by adding 4.5 ml of cold 100% ethanol dropwise to the sample while vortexing gently and then placing at −20°C for at least 8 h. After fixation, cells were pelleted out of ethanol, washed once with PBS, and resuspended in 20 μg of propidium iodide (Sigma) plus 200 μg of RNase (Sigma)/ml of PBS. Cells were incubated at 37°C for 30 min and then allowed to stain for at least 8 h at 4°C. Samples were analyzed for DNA content on a Becton Dickinson FACScan.
Immunofluorescence.
A total of 105 MEFs were plated onto glass coverslips in a 34-mm-diameter tissue culture well. MEFs were allowed to adhere to coverslips for 24 h and were then changed into medium with or without nocodazole (0.125 μg/ml) and incubated for 24 h. Bromodeoxyuridine (BrdU) and fluorodeoxyuridine (FdU) (Sigma) were added to the medium from a 1,000× stock solution in H2O, for a final concentration of 3 μg of BrdU plus 0.3 μg of FdU/ml of medium; cells were incubated for an additional 4 h. Coverslips were then fixed in 4% paraformaldehyde-PBS for 20 min at room temperature, washed in PBS, permeabilized for 15 min in 0.25% Triton X-100–PBS, and washed again with PBS. For BrdU detection, slips were denatured in 1.5 N HCl for 10 min and washed several times with PBS. Slips were incubated with a murine monoclonal antibody to BrdU (1:50 dilution; Becton Dickinson) in 10% goat serum-PBS for 30 min at 37°C. After two washes in PBS, fluorescein-conjugated anti-mouse antibody (1:200 dilution; Jackson Immunoresearch Laboratories) was added in 10% goat serum-PBS and the slips were incubated for 30 min at 37°C. Slips were washed with PBS and stained for 5 min in 0.2 μg of 4′,6-diamidino-2-phenylindole (DAPI; Sigma)/ml of PBS. Coverslips were mounted on glass slides with Mowiol and analyzed via fluorescence microscopy.
Time-lapse videomicroscopy.
MEFs were grown on glass coverslips as for immunofluorescence. A single coverslip was placed in a dish containing 2 ml of HEPES-buffered DMEM plus 10% fetal bovine serum, with or without nocodazole (0.125 μg/ml). Each coverslip was observed for up to 24 h under a Nikon microscope. Recording was done by taking one picture frame every 8 s with a GYYR TLC2100 time-lapse videocassette recorder. During recording, MEFs were maintained at 37°C with a Delta temperature controller and kept in an environment of 5% CO2 by bubbling gas across the surface of the media. Pictures were printed out with a Sony color video printer. All equipment was obtained from Micro Video Instruments, Inc.
For time-lapse videomicroscopy followed by immunofluorescence, MEFs were grown on gridded coverslips (Bellco) placed in 34-mm-diameter wells, with 105 MEFs plated per well. A single square of the grid was followed by time-lapse videomicroscopy for 18 to 24 h. For BrdU incorporation assays, 2 μl of BrdU-FdU 1,000× stock solution was added directly to the 2 ml of media in the dish 4 h before the end of the experiment, for a final concentration of 3 μg of BrdU plus 0.3 μg of FdU/ml of medium. After recording was complete, the gridded slip was fixed in 4% paraformaldehyde–PBS and immunofluorescence was performed. The cells which had been recorded on video were identified during immunofluorescence analysis by locating the same square of the grid.
Immunoblotting.
Whole-cell extracts were made by scraping approximately 106 cells in 200 μl of boiling lysis buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 1% sodium dodecyl sulfate [SDS]). Lysates were heated at 100°C for 10 min, quick frozen in a dry ice-ethanol bath, and stored at −80°C. Protein concentration of lysates was determined with a bicinchoninic acid kit (Pierce). For cyclin E, cyclin B1, and p21 immunoblots, 100 μg of protein was loaded per lane on an SDS–10% polyacrylamide gel (29:1). Gels were electrophoresed at 150 V for 4 h and then transferred to Immobilon-P membrane (Millipore) in low-molecular-weight transfer buffer (25 mM Tris, 190 mM glycine, 20% methanol) for 12 h at 25 V at 4°C. Blots were blocked in 0.01% Tween 20-PBS (PBS-T) plus 5% dried milk for 1 h at room temperature and then probed with primary antibody diluted in block solution for 1 h at room temperature. Antibodies and dilutions were as follows: cyclin E (Santa Cruz M-20), 1:200 dilution; cyclin B1 (Pharmingen GNS-1), 1:200 dilution; and p21 (Santa Cruz C19-G), 1:100 dilution. Blots were then washed with PBS-T three times for 10 min. For p21 detection, blots were put through an additional incubation step in mouse anti-goat antibody (1:12,000; Jackson Immunoresearch Laboratories), diluted in block solution for 1 h at room temperature, and washed three times in PBS-T. All blots were then incubated in peroxidase-linked secondary antibody (1:5,000 dilution; Amersham) for 1 h at room temperature and washed three times in PBS-T. Blots were developed with chemiluminescent reagents and exposed to Kodak X-Omat 5 film for 1 to 20 min.
For pRB immunoblots, 350 μg of protein was loaded per lane on an SDS–6% PAGE gel (29:1). Gels were electrophoresed at 20 mA overnight and then transferred to Immobilon-P membrane in high-molecular-weight transfer buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, 20% methanol) for 14 h at 25 V at 4°C and for an additional hour at 40 V. Blots were blocked in 0.03% Tween 20 in 10 mM Tris (pH 7.5)–150 mM NaCl (TBS-T) plus 5% dried milk for 1 h at room temperature and then probed with primary antibody diluted 1 to 100 in block solution (Pharmingen G3-245) for 1 h at room temperature. After three 10-min washes in TBS-T, blots were incubated in rabbit anti-mouse antibody (1:14,000; Jackson Immunoresearch Laboratories) for 1 h at room temperature. Blots were washed three times in TBS-T, incubated in peroxidase-linked tertiary antibody (1:5,000 dilution; Amersham) for 1 h at room temperature, and washed three times in TBS-T. Blots were developed as described above.
RESULTS
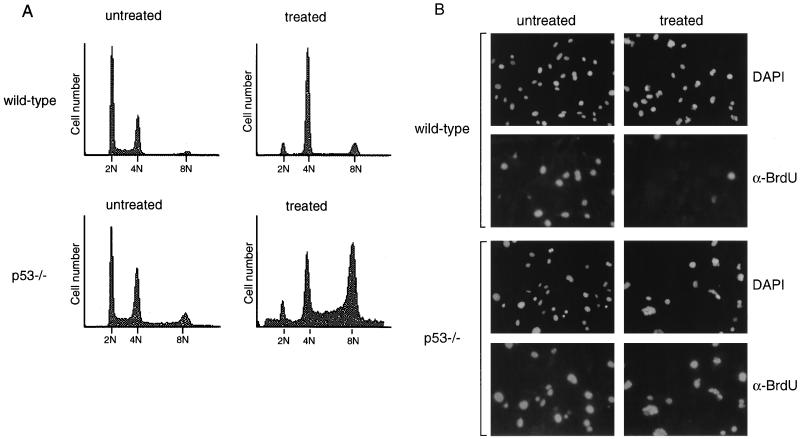
Because previous results regarding p53 function at mitosis were contradictory, we chose to characterize in greater detail the effect of p53 deficiency on cells treated with spindle inhibitors. To confirm a requirement for p53 following spindle disruption, wild-type and p53-deficient MEFs were grown in the presence of the microtubule-destabilizing drug nocodazole. After 24 h of treatment, cells were fixed and stained with propidium iodide for analysis by flow cytometry (Fig. 1A). Wild-type MEFs treated with nocodazole arrested with primarily a 4N DNA content. A small subpopulation of wild-type cells that had 8N DNA content was observed, perhaps reflecting the presence of cycling tetraploid cells in the normal population, which would be predicted to arrest with 8N DNA content upon spindle disruption. As described previously, p53−/− fibroblasts did not arrest with a 4N DNA content upon drug treatment but continued through an additional round of S phase to become 8N (Fig. 1A) (5, 8, 33).
FIG. 1.
p53 is required for cell cycle arrest following nocodazole treatment. (A) FACS profiles of wild-type and p53−/− MEFs untreated or treated with nocodazole for 24 h. DNA content is represented on the x axis; number of cells counted is represented on the y axis. Data shown are representative of multiple experiments performed on two different wild-type and p53−/− clones. (B) Immunofluorescent staining of wild-type and p53−/− MEFs. MEFs were treated with nocodazole for 24 h, pulsed with BrdU in the presence of nocodazole for an additional 4 h, and fixed. Immunofluorescence was performed to detect BrdU incorporation (α-BrdU) and nuclear staining (DAPI). Immunofluorescence was performed on two different wild-type and p53−/− clones.
In order to examine specifically the fraction of cells capable of S phase reentry following nocodazole treatment, cells were treated with nocodazole for 24 h, pulsed with BrdU for an additional 4 h while still in the presence of drug, and analyzed by immunofluorescence for anti-BrdU staining (Fig. 1B). Untreated wild-type and p53−/− MEFs had similar proportions of cells in S phase. After 28 h of nocodazole treatment, very few BrdU-positive cells were observed in the wild-type MEF population, consistent with their ability to induce cell cycle arrest after nocodazole treatment. However, many p53−/− MEFs incorporated BrdU while in the presence of nocodazole, indicating that they were able to reenter the cell cycle and initiate DNA synthesis, despite having failed to undergo proper mitosis. Both wild-type and p53−/− cells were observed to contain micronuclei following nocodazole treatment, resulting from chromosome decondensation and subsequent reformation of the nuclear membrane (24). These results confirm previous observations (5, 8, 33) describing a p53-dependent arrest following destabilization of the mitotic spindle.
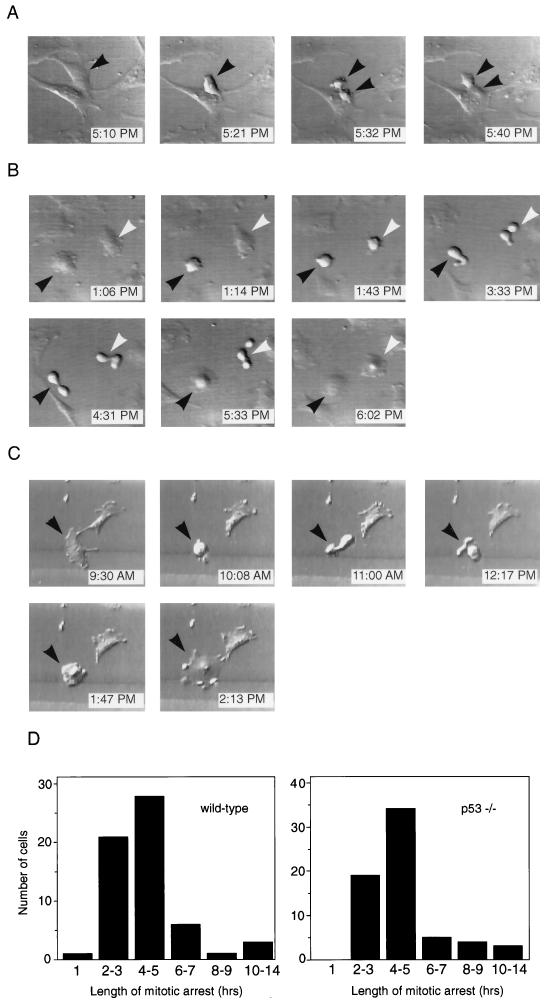
One interpretation of the results described in Fig. 1 is that p53 functions in a checkpoint at mitosis, based on the fact that p53−/− MEFs behave abnormally when the mitotic spindle is disrupted. To test this hypothesis directly, we used time-lapse videomicroscopy to observe p53+/+ and p53−/− MEFs during mitosis. If p53 were part of a mitotic checkpoint, then we would expect to see a difference in the behavior of nocodazole-treated p53+/+ and p53−/− MEFs at mitosis. Figure 2A shows a representative untreated p53−/− fibroblast during normal cell division. The cell rounded up from the coverslip (Fig. 2A, 5:21 PM), underwent cytokinesis (Fig. 2A, 5:32 PM), and completed division into two daughter cells (Fig. 2A, 5:40 PM). Thus, the length of mitosis, as determined visually by rounding of the parent cell and separation into two daughter cells, was approximately 20 min. Normal mitosis lasted 26 ± 8 (mean ± standard deviation) min in wild-type MEFs and 27 ± 7 min in p53−/− MEFs, based on observations made for at least 15 cells of each genotype (data not shown). For our next experiments, we used time-lapse techniques to determine the fate of fibroblasts that initiated mitosis while in the presence of the spindle inhibitor nocodazole. Figure 2B shows two nocodazole-treated p53+/+ fibroblasts that entered mitosis within the same 30-min window. Each cell rounded up from the coverslip as was observed for normal mitosis (Fig. 2B, 1:14 PM and 1:43 PM). However, the cells then changed shape rapidly in apparent attempts to complete cell division over a period of approximately 4 h (Fig. 2B, 3:33 PM and 4:31 PM). Strikingly, after this time, the cells flattened again without completing mitosis (Fig. 2B, 5:33 PM and 6:02 PM). This behavior of exiting from mitotic arrest into an apparent interphase state without actually completing mitosis, which we will refer to as adaptation, has been previously described (24, 35). We also examined the behavior of p53−/− fibroblasts that entered mitosis while in the presence of nocodazole. A representative p53−/− fibroblast that initiated mitosis while growing in nocodazole is shown in Fig. 2C. Like the p53+/+ cells, the p53−/− fibroblast rounded up (Fig. 2C, 10:08 AM), changed morphology over a 4-h period (Fig. 2C, 11:00 AM and 12:17 PM), and then flattened back out again onto the coverslip (Fig. 2C, 1:47 PM and 2:13 PM).
FIG. 2.
Time-lapse videomicroscopy of wild-type and p53−/− MEFs at mitosis in the presence or absence of nocodazole. (A) Control p53−/− MEF that underwent mitosis in normal media. Each photograph lists the time when the video recording occurred. The cell entered mitosis at 5:21 PM (arrowhead), cytokinesed at 5:32 PM (two arrowheads), and completed division by 5:40 PM. The cell shown is representative of over 30 cells (wild-type and p53−/−) observed undergoing normal mitosis. (B) Two wild-type MEFs that initiated mitosis in the presence of nocodazole. Each cell entered mitosis (1:14 PM and 1:43 PM), remained arrested at mitosis for several hours (3:33 PM and 4:31 PM), and then adapted (5:33 PM and 6:02 PM). Black and white arrowheads indicate the two different cells. (C) p53−/− MEF that entered mitosis in the presence of nocodazole (10:08 AM, arrowhead). It arrested at mitosis for several hours (11:00 AM and 12:17 PM) and eventually adapted (1:47 PM and 2:13 PM). (D) Quantitation of length of time that individual wild-type and p53−/− MEFs spent at mitotic arrest. Length of mitotic arrest was determined morphologically by time-lapse videomicroscopy, beginning when a cell first became rounded and refractile and ending when it flattened back onto the coverslip. Wild-type MEFs spent an average of 4.4 ± 2.4 h at mitotic arrest, while p53−/− MEFs spent an average of 4.6 ± 2.2 h at mitotic arrest. At least 60 cells were observed for each genotype.
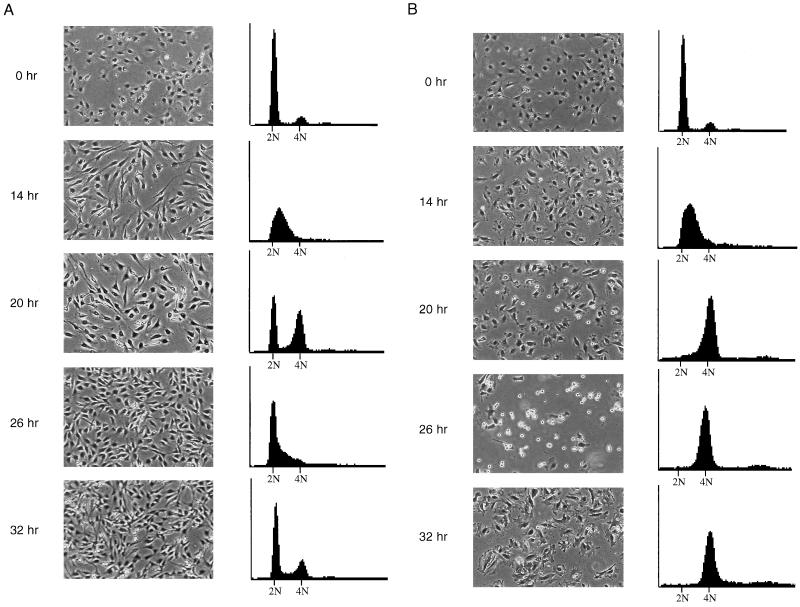
FIG. 5.
Mitotic arrest and adaptation occur in synchronized, nocodazole-treated NIH 3T3 cells. (A) Untreated NIH 3T3 cells. Cells were synchronized in low serum for 48 h and then released into high serum and monitored for 32 h. At each time point, cells were photographed (left panels) and then prepared for FACS analysis for DNA content (right panels). Time points are measured in hours postrelease from serum starvation. Cells were initially in G0 (0 h), entered S phase at 14 h, and completed cell division by 26 h. (B) Nocodazole-treated NIH 3T3 cells. Cells were synchronized in low serum for 48 h and then released into high serum plus nocodazole and monitored for 32 h. Progression of cells through cell cycle was monitored as above. Cells exited G0 and entered S phase by 14 h, remained arrested at mitosis through 26 h, and adapted by 32 h with a 4N DNA content.
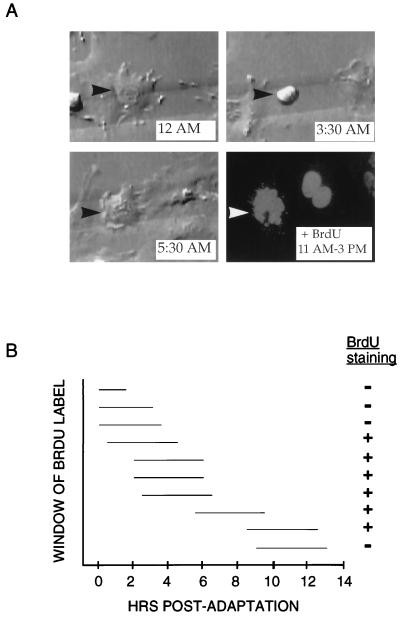
FIG. 3.
Timing of S phase entry in p53−/− MEFs treated with nocodazole. (A) Time-lapse videomicroscopy and subsequent immunofluorescence of representative p53−/− MEF that arrested at mitosis in the presence of nocodazole. The cell initiated mitotic arrest starting shortly after 12 AM and then adapted at 5:30 AM (top panels and lower left panel). At 11 AM, the BrdU label was added to the media; the cell was then recorded for an additional 4 h and fixed. Immunofluorescence was performed to detect BrdU incorporation (lower right panel). As indicated by arrowheads, the same cell was identified during video recording and immunofluorescence by its position on a gridded coverslip. (B) Measurement of time of S phase entry relative to time of adaptation from mitotic arrest. Ten p53−/− MEFs on gridded coverslips were treated with nocodazole, monitored by time-lapse videomicroscopy, and pulsed with BrdU for 4 h at various times following adaptation. For each cell, the horizontal line indicates the period during which BrdU was present (measured in hours) relative to the time elapsed since the cell underwent adaptation. The + or − indicates whether the cell stained positive for BrdU incorporation by immunofluorescence.
Figure 2D shows quantitation of the time spent at mitotic arrest for individual p53+/+ and p53−/− MEFs that were followed by time-lapse videomicroscopy. The length of arrest was determined for at least 60 cells of each genotype. Both p53+/+ and p53−/− MEFs spent a much longer period of time in a rounded mitotic state when they were grown in nocodazole-containing medium than when they were grown in normal medium. The average time spent arrested at mitosis under these conditions was 4.4 h for p53+/+ MEFs and 4.6 h for p53−/− MEFs compared to 26 to 27 min normally spent at mitosis in untreated cells. Cells of each genotype subsequently exited from mitosis and flattened onto the coverslip without completing cytokinesis. Interestingly, several cells of each genotype were observed to undergo prolonged mitotic arrest of 8 to 14 h prior to adaptation (Fig. 2D). These data demonstrate that p53 does not act as a mitotic checkpoint per se, because p53+/+ and p53−/− MEFs behave identically at mitosis in the presence of nocodazole.
Given that p53 does not affect mitotic arrest kinetics, we wished to identify the stage at which p53 is required for cell cycle arrest following nocodazole treatment. To address this question, individual p53−/− MEFs treated with nocodazole were followed through mitotic entry and arrest, mitotic exit/ adaptation, and S phase entry. Cells were plated onto gridded coverslips and followed by time-lapse videomicroscopy as described above. After 18 to 22 h of treatment, cells were pulsed with BrdU for an additional 4 h in the presence of nocodazole and then analyzed by immunofluorescence. Figure 3A shows a representative p53−/− MEF that attempted mitosis, adapted, and then incorporated BrdU. The cell entered mitosis at 12 AM, remained in mitotic arrest for several hours, and then exited mitosis at 5:30 AM. BrdU label was added at 11 AM, and the cell was fixed 4 h later. Immunofluorescent detection for BrdU uptake revealed that the cell had been in S phase during the time the BrdU label was present, which was 5.5 to 9.5 hr after adaptation had occurred. Ten individual p53−/− fibroblasts were tracked with this method to determine the number of hours which elapsed between morphological adaptation and S phase entry (Fig. 3B). Importantly, cells did not enter S phase immediately after exiting mitosis, as cells labeled with BrdU 0 to 3.5 h postadaptation failed to stain positively. The p53−/− MEFs began to incorporate BrdU in a time window approximately 4 to 6 h postadaptation. One p53−/− MEF incubated with BrdU for 9 to 13 h after it had undergone adaptation failed to incorporate label. The behavior of this fibroblast does not necessarily indicate the time limit for the reentry of p53−/− MEFs into S phase, however, because the fibroblast may either already have completed S phase at the time of the BrdU label or may not yet have initiated it. These data show that p53 is required to prevent entry into S phase beginning in a specific time interval after nocodazole-treated MEFs have attempted mitosis and subsequently adapted into an interphase state.
p53 has been shown to prevent cells in G1 from entering S phase following DNA damage (20, 21, 23). This arrest is mediated in large part by p21, a transcriptional target of the p53 protein which inhibits CDKs and prevents phosphorylation of the Rb protein, a requirement for S phase entry (2, 7). To test whether a similar mechanism might be functioning to prevent S phase entry in nocodazole-treated adapted cells, we performed immunoblot analysis for p21 protein on wild-type and p53−/− MEFs treated with nocodazole (Fig. 4A). In wild-type cells, levels of p21 protein increased after 16 h of nocodazole treatment and remained elevated over a 48-h period. No p21 protein was detectable in nocodazole-treated p53−/− MEFs. These observations suggested a possible role for p21 in maintaining arrest following nocodazole treatment. To establish whether p21 was required for the arrest following spindle disruption, wild-type, p53−/−, and p21−/− MEFs were treated with nocodazole for 24 h and analyzed by flow cytometry (Fig. 4B). As was observed in Figure 1, wild-type MEFs arrested with 4N DNA content, while p53−/− MEFs continued to increase in ploidy. p21−/− MEFs were observed to have a phenotype similar to that of p53−/− MEFs, with a significant fraction of cells undergoing an additional round of S phase to become 8N in DNA content.
FIG. 4.
Requirement for p21 in cell cycle arrest following nocodazole treatment. (A) Immunoblot analysis for p21 protein on extracts from wild-type and p53−/− MEFs treated with nocodazole. Unsynchronized cells were treated with nocodazole for times indicated and harvested for protein analysis. (B) FACS profiles of p21−/− MEFs untreated or treated with nocodazole for 24 h. DNA content is represented on the x axis; number of cells counted is represented on the y axis. Data shown are representative of three experiments performed on two different p21−/− clones. (C) Immunofluorescent staining of p21−/− MEFs. Cells were treated with nocodazole for 24 h, pulsed with BrdU in the presence of nocodazole for an additional 4 h, and fixed. Immunofluorescence was performed to detect BrdU incorporation (α-BrdU) and nuclear staining (DAPI). Immunofluorescence was performed on two different p21−/− clones. (D) Quantitation of number of cells in S phase in wild-type, p21−/−, and p53−/− MEFs following nocodazole treatment. Cells were treated with nocodazole, and immunofluorescence was performed as described for panel C. Data shown are the averages from three different experiments, with standard deviations as indicated. In each experiment, 100 cells of each genotype were examined and the number of BrdU-positive nuclei was counted to determine the percentage of S phase cells. Two different clones were tested for each genotype.
To demonstrate further that p21−/− cells were capable of reentry into S phase following nocodazole treatment, p21−/− MEFs were treated with nocodazole for 24 h, pulsed with BrdU while still in the presence of nocodazole for another 4 h, and analyzed by immunofluorescence (Fig. 4C). A high percentage of nocodazole-treated p21−/− MEFs were found to incorporate BrdU under these conditions, indicating inappropriate S phase entry. Similar results were observed when wild-type, p53−/−, and p21−/− MEFs were treated with the microtubule-destabilizing drug colcemid, demonstrating the generality of this response (data not shown). Quantitation of the percentage of wild-type, p53−/−, and p21−/− MEFs that stained positively for BrdU incorporation after 28 h of nocodazole treatment is presented in Fig. 4D. Approximately 54% of p21−/− MEFs incorporated BrdU during the labeling period compared to 65% of p53−/− MEFs and 14% of wild-type MEFs. Untreated MEFs of all three genotypes had similar percentages of cells in S phase (approximately 50 to 60%; data not shown). Thus, the p21−/− MEFs had an abnormal arrest phenotype which was similar to that of p53−/− MEFs, although not as severe. Given these results, we concluded that p21 is required to fully prevent cells treated with spindle drugs from reentering S phase.
Thus far, our data demonstrated that p53 functions to prevent S phase entry following mitotic arrest and adaptation. However, while inappropriate S phase entry had been observed by FACS analysis to occur in thousands of cells, mitotic arrest and adaptation had been observed in only approximately 100 cells in these experiments. To demonstrate that mitotic arrest and adaptation also occurred on the level of entire populations of cells, we turned to methods other than time-lapse videomicroscopy. NIH 3T3 cells were used in these experiments for ease of synchronization and to show that our previous observations did not reflect a phenotype specific to our MEF clones. The NIH 3T3 cell line has been shown to contain wild-type p53 (16, 29, 31, 36, 40). NIH 3T3 cells were synchronized for 48 h in low serum medium and then released into high serum medium with or without nocodazole. At 6-h time points, cells were photographed and then fixed for flow cytometric analysis of DNA content (Fig. 5). At the time of release into high serum, cells were synchronized with a 2N DNA content (Fig. 5A, untreated, and B, nocodazole-treated; 0 h). Cells entered S phase synchronously (Fig. 5A and B, 14 h) and completed DNA synthesis by 20 h after release into high-serum-containing medium (Fig. 5A and B, 20 h). The presence of nocodazole did not noticeably delay progression into or completion of S phase, as judged by DNA content. The first mitoses were visible at the same time point in photographs of untreated and nocodazole-treated cells as rounded refractile cells (Fig. 5A and B, 20 h). Within 6 h, all of the cells in the untreated population had completed mitosis, as the 4N population had been replaced by cells with 2N and S phase DNA content (Fig. 5A, 26 h). In contrast, all of the cells in the nocodazole-treated population had a 4N DNA content at 26 h, and most appeared to be rounded up and arrested at mitosis (Fig. 5B, 26 h). By the next time point, however, these cells had flattened out again but still had a 4N DNA content, indicating their failure to complete mitosis (Fig. 5B, 32 h). In the adapted cells, microtubules were depolymerized and chromatin was decondensed, as determined by indirect immunofluorescence for tubulin and nuclear staining with DAPI, respectively (data not shown). Thus, as observed with wild-type MEFs, NIH 3T3 cells did not undergo an additional round of DNA synthesis during nocodazole treatment. These results indicate that the great majority of murine fibroblasts in a synchronized population undergo mitotic arrest and adaptation when treated with nocodazole.
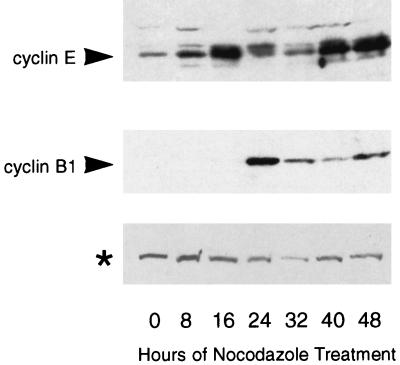
We observed some similarities between the p53-dependent checkpoint following nocodazole treatment and the p53-dependent G1 checkpoint following irradiation. Our data showed that a primary effector of the G1 checkpoint response, p21, was also required for arrest after spindle disruption. Also, adapted cells had the flattened appearance and decondensed chromatin characteristic of cells in G1. To determine whether adapted cells were similar to G1 cells at the molecular level, we measured expression of cyclin B1, a mitotic cyclin, and cyclin E, a G1 cyclin, in nocodazole-treated cells. Cyclin B1 is expressed at high levels in mitotic cells and declines when cells enter G1, while cyclin E expression is highest in cells in the late G1 phase of the cell cycle (9, 26). NIH 3T3 cells were synchronized in low serum, released into medium with high serum plus nocodazole, and collected at time points for immunoblotting. The blot was probed with antibodies to cyclin B1 and cyclin E (Fig. 6). Cyclin B1 levels peaked at 24 h and decreased afterward. The appearance of this peak corresponded with the presence of mitotic cells (data not shown). Cyclin E levels were elevated at 16 h as cells progressed from G1 into S phase and then decreased with the onset of mitosis. At 32 h, mitotic-arrested cells had just adapted into a flattened state (data not shown) and cyclin E levels remained low (Fig. 6). Cyclin E levels then increased again in the adapted cells at 40 h (Fig. 6). Thus, adapted cells express high levels of cyclin E, a G1 marker, despite having a DNA content characteristic of cells in the G2 or M phase of the cell cycle. The delayed expression of cyclin E relative to adaptation may reflect the fact that cyclin E is normally expressed late in G1 and that adapted cells take several hours to progress to this state.
FIG. 6.
Expression of cyclin E protein is upregulated in nocodazole-treated, adapted cells. Immunoblot analysis of cyclin B1 and cyclin E expression. NIH 3T3 cells were synchronized in low serum for 48 h and released into high serum plus nocodazole. Extracts were prepared at 8-h time points for the next 48 h and analyzed by immunoblotting for cyclin B1 and cyclin E expression. Cyclin E expression was highest in cells entering S phase (16 h) and in cells that had adapted (40 and 48 h), while cyclin B1 expression was highest in mitotic cells (24 h) and declined thereafter. The asterisk indicates a cross-reacting protein detected by cyclin E antibody that serves as an internal loading control.
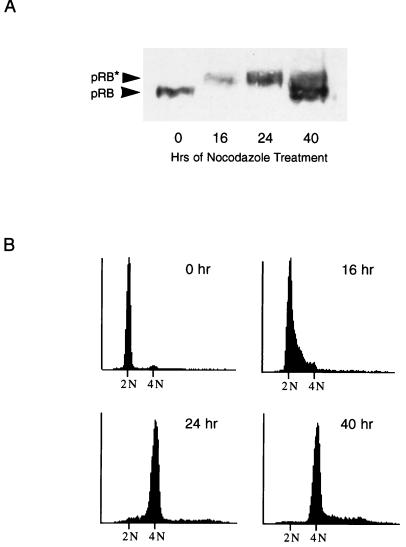
To characterize further the nocodazole-induced adapted state, we examined adapted cells for another marker of G1, hypophosphorylated Rb protein. In the G1 phase of the cell cycle, the Rb protein (pRB) is in a hypophosphorylated state (3, 4, 6, 32). As part of the G1-to-S-phase transition, pRB becomes hyperphosphorylated on multiple sites and remains so until anaphase of mitosis (30). We analyzed serum-synchronized, nocodazole-treated NIH 3T3 cells at various time points for their pRB phosphorylation status (Fig. 7A) and DNA content (Fig. 7B). At time 0, cells had been serum starved for 48 h and were 2N in DNA content. All pRB was hypophosphorylated, as would be expected for cells in G0. Sixteen hours after release into medium containing high serum plus nocodazole, pRB had become hyperphosphorylated, corresponding with the entry of cells into S phase. At 24 h postrelease, cells had completed DNA synthesis and were entering mitotic arrest, from which they later adapted (Fig. 7B and data not shown). By 40 h of treatment, cells had adapted and pRB was predominantly in the hypophosphorylated state. Therefore, like cyclin E expression, pRB phosphorylation status in adapted cells resembles the pattern seen in G1 cells. Because pRB is present in adapted cells in the active, or hypophosphorylated, state, it is possible that like p53 and p21, pRB functions to prevent S phase reentry during nocodazole treatment. Such a mechanism seems likely, given that p21 is required for arrest in nocodazole-treated cells and that p21 induces cell cycle arrest in part by inhibiting pRb phosphorylation.
FIG. 7.
RB protein is hypophosphorylated in nocodazole-treated, adapted cells. (A) Immunoblot analysis of pRB. NIH 3T3 cells were synchronized in low serum for 48 h and released into high serum plus nocodazole. Extracts were prepared at time points for the next 48 h and analyzed by immunoblotting for pRB. Hypophosphorylated pRB (pRB) and hyperphosphorylated pRB (pRB*) are indicated by arrows. (B) DNA content of NIH 3T3 cells. Cells were treated as described in panel A. At each time point, a duplicate plate was collected and prepared for FACS analysis of DNA content. Cells were initially synchronized in G0 (0 h), progressed to S phase (16 h), and arrested at mitosis (24 h).
DISCUSSION
Many functions have been attributed to the p53 protein, which is clearly a critical cell cycle regulator in mammalian cells (25). In this study, we have addressed the question of whether the ability of cells to initiate and maintain the mitotic spindle checkpoint is affected by their p53 status. We have attempted to clarify the role of p53 at mitosis by monitoring the response of individual p53+/+ and p53−/− MEFs to spindle inhibitors. In order for p53 to qualify as a true mitotic checkpoint, p53+/+ and p53−/− MEFs would have to behave differently at mitosis. However, we observed that both wild-type and p53-deficient MEFs arrested at mitosis following disruption of the spindle and that cells of either genotype arrested for the same length of time and behaved identically during the mitotic arrest. This behavior is opposite to that of mammalian cells in which the MAD2 or BUB1 mitotic checkpoint gene products, which monitor the integrity of the mitotic spindle, have been inhibited. Recent studies have identified a human homolog for the S. cerevisiae MAD2 checkpoint gene, hsMAD2, as well as a murine homolog of the S. cerevisiae BUB1 checkpoint gene. As would be predicted from experiments with S. cerevisiae mad strains, mammalian cells electroporated with an anti-hsMAD2 antibody did not undergo arrest at mitosis when the cells were treated with nocodazole (28). Similarly, when treated with nocodazole, mammalian cells which expressed a dominant negative version of the murine Bub1 gene arrested at mitosis much less frequently than their normal counterparts (39). Thus, when contrasted with genes which are known to act at mitotic checkpoints in mammalian cells, p53 acts in a different manner. We can conclude that, unlike the hsMAD2 and murine Bub1 genes, the presence or absence of p53 has no impact on the ability of cells to detect microtubule disruption and initiate metaphase arrest. Thus, we have shown definitively that p53 does not act to implement a checkpoint at mitosis in mouse embryonic fibroblasts.
We observed that p53 does have a checkpoint function in MEFs following nocodazole treatment, specifically, to prevent S phase reentry following adaptation from mitotic arrest. Consistent with this finding is the observation that levels of p53 protein and its downstream target gene product p21 both increase following treatment with nocodazole or other microtubule drugs (Fig. 4A) (33, 40). Importantly, we have found that the downstream arrest mechanism activated by p53 requires the CDK inhibitor p21, as p21−/− MEFs fail to arrest in response to nocodazole treatment and enter another round of S phase, in contrast to the results of an earlier study (7). Although our data were obtained with cells treated with nocodazole, we obtained similar results with the spindle inhibitor colcemid (data not shown), which was the microtubule-destabilizing drug used in the earlier study. We eliminated the possibility that the p21 protein itself was acting at a mitotic checkpoint, because like wild-type and p53−/− MEFs, p21−/− MEFs treated with nocodazole arrested at mitosis for 4 to 5 h and then adapted (data not shown). The simplest explanation for our observations is that in adapted cells, activation of p53 leads to increases in the level of p21 protein, which then initiates cell cycle arrest by inhibiting cyclin-CDK activity. Interestingly, the contribution of p21 to the arrest response following nocodazole treatment and adaptation is proportionately similar to the contribution of p21 to the G1 arrest response following DNA damage. In nocodazole-treated, adapted MEFs, p21 is responsible for about 80% of the p53-mediated arrest response, while in irradiated MEFs, p21 accounts for approximately 70 to 80% of the p53-dependent arrest in G1 (2, 7). These data demonstrate not only that p21 is required for the arrest following nocodazole treatment but also that the requirement for p21 is quantitatively similar to the requirement for p21 in the G1 arrest following DNA damage.
Taken together, our results suggest that p53 could be acting as a component of a G1 checkpoint in nocodazole-treated cells. First, p53 function is required to prevent reentry into S phase following checkpoint activation, as occurs in the G1 checkpoint following DNA damage. Second, p21 is required to execute this arrest and to an extent similar to its role in the G1 arrest checkpoint. Finally, cells which have exited mitotic arrest have the flattened appearance and decondensed chromatin of G1 cells and express the G1-specific markers cyclin E and hypophosphorylated pRB. Thus, p53 could be implementing a G1-like arrest in 4N adapted cells.
An interesting and as-yet-unanswered question is the nature of the upstream signal which triggers p53-dependent arrest following nocodazole treatment. One possibility is that as during the normal G1 checkpoint, p53 detects DNA damage. It is readily conceivable that a cell which attempted to undergo chromosome separation at metaphase when its spindle was destabilized by nocodazole could incur DNA damage, which would then be detected following exit from mitotic arrest. In this case, both the upstream and downstream signals for arrest following nocodazole treatment would be identical to those for G1 arrest following irradiation. Alternatively, p53 could be activated by other types of signals, such as the presence of two centrosomes in a G1-like cell. Another possible signal could be the presence of excess chromosomes in a G1-like cell, which would trigger a p53-dependent checkpoint to prevent endoreduplication. Our data do exclude one possibility, however, which is that p53 is activated in response to a loss of microtubule stability induced by nocodazole treatment. Our data on NIH 3T3 cells, which contain functional p53, show that like other cell types, they progress unhindered from G1 to mitosis while in the constant presence of nocodazole (Fig. 5) (19). Were p53 detecting a loss of microtubule stability in G1 or G1-like cells, these cells would never have progressed through S phase but instead would have arrested shortly after entry into G1.
While it has been speculated to act at multiple different cell cycle checkpoints, the p53 protein appears to have overlapping checkpoint functions following irradiation and spindle disruption. After irradiation, cells undergo p53-dependent arrest in G1 (20, 21, 23); after nocodazole-induced disruption of the mitotic spindle, cells arrest transiently at mitosis, adapt into a G1-like state, and then undergo p53-dependent arrest (33). In both instances, the p53-mediated arrest requires the CDK inhibitor p21 (2, 7). Thus, both the stage of the cell cycle at which p53 induces arrest and the downstream mechanism by which it implements arrest are similar after either irradiation or nocodazole treatment. In cells which lack functional p53 protein, failure to arrest following irradiation or nocodazole treatment would be predicted to lead to two distinctly different outcomes of chromosomal damage and polyploidy, respectively. Thus, the loss of p53 checkpoint function during tumorigenesis could lead to decreased genomic stability by multiple mechanisms.
ACKNOWLEDGMENTS
We thank J. Brugarolas for p21−/− MEFs and L. Attardi for critical reading of the manuscript.
J.S.L. was supported in part by a predoctoral fellowship from the Office of Naval Research. T.J. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Brown C R, Doxsey S J, White E, Welch W J. Both viral (adenovirus E1B) and cellular (hsp70, p53) components interact with centrosomes. J Cell Physiol. 1994;160:47–60. doi: 10.1002/jcp.1041600107. [DOI] [PubMed] [Google Scholar]
- 2.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 3.Buchkovich K, Duffy L A, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 4.Chen P L, Scully P, Shew J Y, Wang J Y, Lee W H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 5.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 6.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 7.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 8.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRB function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 9.Dunphy W G, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 10.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 12.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 13.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Turck C W, Morgan D O. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 17.Hoyt M A, Totis L, Roberts B T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 18.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 19.Jordan M A, Thrower D, Wilson L. Effects of vinblastine, podophyllotoxin, and nocodazole on mitotic spindles. J Cell Sci. 1992;102:401–416. doi: 10.1242/jcs.102.3.401. [DOI] [PubMed] [Google Scholar]
- 20.Kastan M, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 21.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 22.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 23.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung A L, Sherwood S W, Schimke R T. Cell-line specific differences in the control of cell cycle progression in the absence of mitosis. Proc Natl Acad Sci USA. 1990;87:9553–9557. doi: 10.1073/pnas.87.24.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 26.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Burbidge S A, Griffin S, Smith H M. Discordance between accumulated p53 protein level and its transcriptional activity in response to u.v. radiation. Oncogene. 1996;13:413–418. [PubMed] [Google Scholar]
- 30.Ludlow J W, Glendening C L, Livingston D M, DeCaprio J A. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michieli P, Chedid M, Lin D, Pierce J H, Mercer W E, Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 32.Mihara K, Cao X R, Yen A, Chandler S, Driscoll B, Murphree A L, T’Ang A, Fung Y K. Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science. 1989;246:1300–1303. doi: 10.1126/science.2588006. [DOI] [PubMed] [Google Scholar]
- 33.Minn A J, Boise L H, Thompson C B. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 34.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 35.Rieder C L, Palazzo R E. Colcemid and the mitotic cycle. J Cell Sci. 1992;102:387–392. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- 36.Rittling S R, Denhardt D T. p53 mutations in spontaneously immortalized 3T12 but not 3T3 mouse embryo cells. Oncogene. 1992;7:935–942. [PubMed] [Google Scholar]
- 37.Rudner A D, Murray A W. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 38.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 39.Taylor S S, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 40.Tishler R, Calderwood S, Coleman C, Price B. Increases in sequence specific DNA binding by p53 following treatment with chemotherapeutic and DNA damaging agents. Cancer Res. 1993;53:2212–2216. [PubMed] [Google Scholar]
- 41.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 42.Zambetti G P, Bargonneti J, Walker K, Prives C, Levine A J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]