Abstract
The c-jun proto-oncogene encodes a transcription factor which is activated by mitogens both transcriptionally and by phosphorylation by Jun N-terminal kinase (JNK). We have investigated the cellular signalling pathways involved in epidermal growth factor (EGF) induction of the c-jun promoter. We find that two sequence elements, which bind ATF1 and MEF2D transcription factors, are required in HeLa cells, although they are not sufficient for maximal induction. Activated forms of Ras, RacI, Cdc42Hs, and MEKK increased expression of the c-jun promoter, while dominant negative forms of Ras, RacI, and MEK kinase (MEKK) inhibited EGF induction. These and previously published results suggest that EGF activates the c-jun promoter by a Ras-to-Rac-to-MEKK pathway. This pathway is similar to that used for posttranslational activation of c-jun by JNK.
The proto-oncogene c-jun is activated in mitogen-treated cells by two mechanisms. The first is phosphorylation of the N-terminal region of the c-Jun protein by Jun N-terminal kinase (JNK) (23, 62). Activation of JNK is mediated by activation of a signalling pathway including the small GTPase Rac and protein kinases MEK kinase (MEKK) and JNKK (also known as MKK4 and SEK) (reviewed in references 30 and 62). The second mechanism for c-jun activation is induction of c-jun transcription (34, 52, 55). c-jun is a cellular immediate-early gene whose transcription is increased rapidly in response to external stimuli such as growth factors. This increase does not require new protein synthesis and thus should be due to a limited number of posttranslational events (34, 52).
c-jun is rapidly induced in cultured cells in response to certain stimuli such as epidermal growth factor (EGF), serum, 12-O-tetradecanoyl phorbol-13-acetate (TPA), nerve growth factor, and UV (4, 12, 34, 52, 55, 65). It is as yet unclear whether these agents use a common pathway. Induction of c-jun is important for cell cycle progression since antibodies to the c-jun product blocked progression of NIH 3T3 cells through the cell cycle (31). The c-jun gene encodes a component of the AP1 transcription factor, which binds DNA elements termed TPA-responsive elements or AP1 sites (reviewed in reference 3). Earlier studies showed that an AP1-like element in the c-jun promoter mediated positive autoregulation of the c-jun gene (2).
We have previously shown that a site situated at −59 of the c-jun promoter and, to a lesser extent, the AP1-like site at −72 are important for serum and EGF induction of the c-jun promoter (20). This latter site is bound by AP1 and ATF family members, both basic-leucine zipper DNA binding families, but it has been unclear which factors bind this site in cells (2, 22, 58). The −59 site in the c-jun promoter binds members of the MEF2 family of proteins, which include MEF2A, -B, -C, and -D (21). These proteins are part of the MADS box family of transcription factors, which include serum response factor and the yeast protein MCM1. The MEF2 proteins share extensive homology in their MADS box domains and in a short MEF2-specific domain following the MADS box (reviewed in reference 45). These regions include their DNA binding and dimerization domains. There is little or no similarity among the family members outside the MADS box-MEF2 domain. We found that the predominant type of MEF2 factor binding to the c-jun MEF2 site in HeLa cells was MEF2D, with minor binding by MEF2A (21).
The intracellular pathways which link cell surface receptors such as the EGF receptor to the c-jun promoter are unknown. We have investigated the role of a number of signalling components known to be activated by EGF. The best characterized of these is Ras (41, 62). Ras can activate a number of effectors, including the protein kinase Raf and phosphatidylinositol 3-kinase (PI3K) (39). Ras can also activate Rac and Rho, members of the Rho family of small GTPases (53). Rac can in turn activate a protein kinase cascade which leads to the activation of JNK. This cascade has not been completely elucidated but includes the protein kinase MEKK, which phosphorylates JNKK, which then phosphorylates and activates JNK (13, 36, 57). JNK can phosphorylate and activate several transcription factors, including the c-jun product, ATF2, and Elk-1 (16, 23, 61, 68). Cdc42Hs is another member of the Rho family that can also activate JNK, but it does not appear to be activated by Ras (11, 32, 44, 48). The Rho family GTPases can also activate the c-fos serum response element (SRE) independently of Elk-1 (or its related family members), though the mechanism is still unknown (24).
In this report we show that EGF uses the Ras-to-Rac-to-MEKK pathway to activate the c-jun promoter. We also show that the c-jun AP1-like site binds ATF1 and CREB proteins in HeLa cells and that both the ATF and MEF2 sites are critical for EGF and Rac/Cdc42 responsiveness of the c-jun promoter.
MATERIALS AND METHODS
Plasmids. (i) Luciferase reporter genes.
Plasmids pJC6GL3, pJSXGL3, pJTXGL3, and pJSTXGL3 contain positions −225 to +150 of the murine c-jun promoter. The HindIII-XhoI fragments of pJC6, pJSX, pJTX, and pJSTX (20) were subcloned into the pGL3-luciferase vector (Promega) upstream of the firefly luciferase gene. pJSXGL3, pJTXGL3, and pJSTXGL3 contain mutations at the MEF2 site, ATF (AP1-like) site, and both sites, respectively, as described previously (20). pJC7GL3 and pJC9GL3 were generated by subcloning the HindIII-XhoI fragment of pJC7 and pJC9 (20) upstream of the luciferase gene in pGL3. pJC7GL3 contains positions −133 to +150 of the c-jun promoter, while pJC9GL3 contains positions −80 to +150.
pOFLucGL3 contains the minimal promoter of the human c-fos gene (−53 to +42) upstream of the luciferase gene in pGL3. The c-jun promoter fragments of pJF6, pJF7, pJF9, and pJF10 were generated with PCR primers that flanked their respective ends, digested with HindIII and BglII, and subcloned upstream of the c-fos promoter in pOFLucGL3. pJF6 contains positions −225 to −31 of the c-jun promoter; pJF7 contains −133 to −31, pJF9 contains −77 to −31, and pJF10 contains −225 to −80. pFos-GL3 contains −356 to +109 of the murine c-fos promoter upstream of the luciferase gene in pGL3.
(ii) Expression vectors.
The following mammalian expression vectors for activated signalling molecules were used: RacI(V12) in pCGT (29), Cdc42Hs(V12) in pCMV5 (32), RhoA(V14) in pEXV (51), and Ras(V12) in pSVSPORT (provided by C. Chandra Kumar). pRafBXB was used as an activated form of c-Raf (7), and pMEKE in pcDNA3 was used as an activated form of MEK1 (11). Overexpression of an N-terminal truncated MEKK1 in pCMV5 was used to increase MEKK activity in cells (42). p110* in pCG (25) was used as an activated form of the catalytic subunit of PI3K.
For dominant negatives, we used Cdc42(N17) in pCMV5 (44), RhoA(N19) in pEXV (51), RacI(N17) in pEXV (53), MEKKΔ(K432M) in pSRα (42), and Ras(N17) in pSRα (44); for c-Raf, we used Raf324FH6 in pLNCX (gift of C. Chandra Kumar).
Transfections and luciferase assays.
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% newborn calf serum. Duplicates of a 60-mm-diameter plate were transfected by the calcium phosphate coprecipitation method (56). The c-jun promoter reporter plasmids (1 μg), the internal control pCMV-β-galactosidase (2 μg), and the indicated amounts of expression vectors were transfected, keeping the total DNA at 10 μg per plate with herring sperm DNA. At 16 to 20 h after transfection, the medium was changed; 8 to 12 h later, the cells were serum starved in Dulbecco’s modified Eagle’s medium with 0.2% serum for 24 to 30 h. Cells were either untreated or treated with EGF at 100 ng/ml for 3 h prior to lysis of cells for luciferase and β-galactosidase assays. Preparation of cell extracts were performed as specified by the manufacturer (Promega) and as described previously (28). Luciferase and β-galactosidase assays were described previously (28). All luciferase values were normalized to β-galactosidase activities. The p38 inhibitor SB203580 (Calbiochem) was added at 10 μM 30 min before EGF addition to cells transfected with pJC6GL3. TPA at 100 ng/ml was used to induce cells transfected with pFos-GL3 (1 μg) as described above for EGF induction of pJC6GL3.
Antibodies.
The anti-c-Jun/AP1(D) (sc-44) and anti-c-Fos(K25) (sc-253) sera were affinity-purified rabbit polyclonal antibodies from Santa Cruz Biotechnology. The anti-c-Jun serum AJ2 was an affinity-purified rabbit polyclonal antibody from Oncogene Research Products, Calbiochem (PC06L). For the anti-c-Jun serum AJ1, histidine-tagged rat c-jun cDNA in the expression vector pDS56 (a gift from Tom Curran) was expressed in Escherichia coli MC15 cells and isolated by nickel affinity chromatography under denaturing conditions as described previously (1). Purified protein was loaded on a sodium dodecyl sulfate (SDS)-polyacrylamide gel, and the gel slice was injected into rabbits to generate sera.
The anti-CREB antiserum was kindly provided by Michael E. Greenberg and was described previously (14). The anti-ATF3 sera were generated by injection of recombinant ATF3 to rabbits as described previously (69). For the anti-ATF1 serum, histidine-tagged human ATF1 (in pET3b vector) was expressed in E. coli BL21(DE3)LysS, isolated by nickel affinity chromatography under denaturing conditions, and loaded on an SDS-polyacrylamide gel. The gel slice was injected into rabbits to generate antisera. For the anti-ATF4 serum, a partial human ATF4 cDNA encoding amino acids 207 to 351 was cloned in the pET3b vector and expressed in E. coli BL21(DE3)LysS as described by Studier et al. (60). Because the recombinant protein had a high expression level but low solubility, the inclusion body which contained mostly the recombinant protein was resuspended in SDS-polyacrylamide gel electrophoresis loading buffer and loaded on an SDS-polyacrylamide gel without further purification. The gel slice was injected into rabbits to generate antiserum. The ATF-2 antibody (9222) was an affinity-purified rabbit polyclonal antibody from New England Biolabs.
Oligonucleotides.
The jATF probe was a 25-bp double-stranded oligonucleotide containing the sequence spanning the murine c-Jun ATF site, previously called the c-Jun AP-1 site (20) (5′-TCGAGCTCGGGGTGACATCATGGGA-3′ and 5′-GATCTCCCATGATGTCACCCCGAGC-3′). The AP1 probe was a 22-bp double-stranded oligonucleotide containing the AP-1 consensus recognition sequence (5′-TCGAGCGTGACTCAGCGCGCGA-3′ and 5′-GATCTCGCGCGCTGAGTCACGC-3′).
Gel mobility shift assays.
For in vitro expression, ATF1, ATF2, and ATF4 open reading frames were inserted in the pTM1 expression vector (47), while c-jun was inserted in pGEM. Coupled transcription-translation using the reticulocyte lysate system was performed as specified by the manufacturer (Promega).
Nuclear extracts from HeLa cells were prepared and gel mobility shift assays were performed as described previously (50). Double-stranded oligonucleotides used as probes were labeled with polynucleotide kinase and [γ-32P]ATP. The DNA binding reactions were performed at room temperature with 1 ng of the 32P-labeled probe for 30 min. In each assay, 2 μg of herring sperm DNA was included as nonspecific competitor. For antibody supershift experiments, the binding reaction mixtures were incubated with 0.2 to 2 μl of nonimmune or immune serum for 30 min at room temperature before addition of the probe. The DNA-protein complexes were separated by electrophoresis on 4% polyacrylamide–0.25× TBE (25 mM Tris base, 25 mM boric acid, 1 mM EDTA) gels.
RESULTS
Sequence elements required for EGF induction of the c-jun promoter.
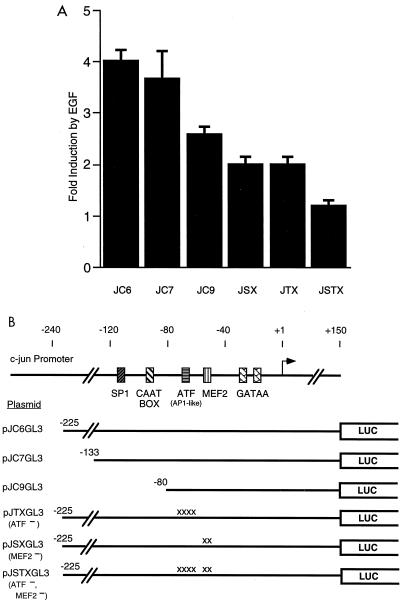
Using c-jun–chloramphenicol acetyltransferase (CAT) reporter genes and RNase protection assays, it was previously shown that EGF induction of the c-jun promoter requires a MEF2 site at −59 in the c-jun promoter (20). An AP1-like element at −72 was also found to be required for the general level of Jun-CAT expression but was not clearly involved in EGF induction. To analyze the c-jun promoter, we have transfected HeLa cells with c-jun promoter-luciferase reporter genes. EGF treatment of cells transfected with pJC6GL3, which contains positions −225 to +150 of the mouse c-jun promoter, resulted in fourfold induction of expression of the jun-luciferase gene (Fig. 1). The use of the pGL3-luciferase vector (Promega) was critical in our experiments since the vector was designed to remove a number of potential binding sites for site-specific transcription factors. We obtained anomalous results with c-jun promoter mutants using other forms of luciferase genes, presumably due to these cryptic sequence elements (data not shown).
FIG. 1.
(A) MEF2 and ATF sites are required for EGF induction of the c-jun promoter. HeLa cells were transiently transfected with the c-jun pGL3-luciferase reporter plasmids, as indicated, and pCMV-β-galactosidase as an internal control. After transfection, the cells were serum starved and treated with or without EGF (100 ng/ml) for 3 h before preparing cell lysates for luciferase and β-galactosidase assays. The fold induction of luciferase activity in EGF-treated cells relative to untreated cells is shown. Values shown are the averages of at least two separate experiments done in duplicate ± standard errors of the means. (B) c-jun promoter constructs. The positions of binding sites for the transcription factors SP1, CTF, ATF, and MEF2 are indicated. The ATF site was previously referred to as an AP1-like element. The regions of the c-jun promoter in each construct are indicated. Point mutations in the ATF or MEF2 sites are indicated (x). LUC, luciferase.
We tested the requirements of c-jun promoter elements for EGF induction by assaying different deletion mutants. A Jun-luciferase construct with 1.6 kb of the human c-jun promoter behaved similarly to pJC6GL3 (data not shown). Deletion to −133 of the mouse c-jun promoter in pJC7GL3 also had no effect on induction. Deletion to −80 (pJC9GL3) decreased induction to about 2.5-fold (Fig. 1). pJC9GL3 contains both the MEF2 and AP1-like elements, suggesting that sequence elements upstream of −80 may also function in EGF-induced expression.
To determine their individual contributions to EGF inducibility, we made mutations in the MEF2 and AP1-like sites of pJC6GL3 (plasmids pJSXGL3 and pJTXGL3, respectively). Mutations in either site reduced EGF induction by about 50% (Fig. 1). A construct containing a double mutation at these sites, pJSTXGL3, almost completely abolished EGF induction. These results suggest that both the MEF2 and AP1-like sites of the c-jun promoter are required for EGF induction although partial induction can occur without either site.
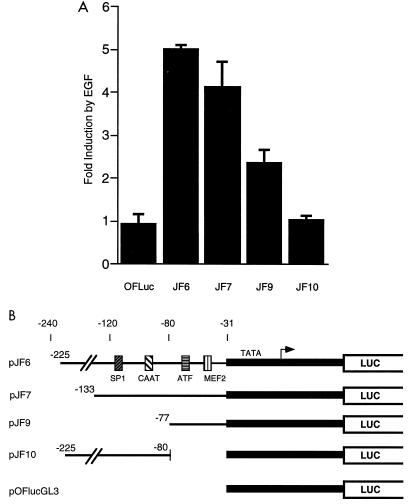
To determine which sequence elements in the c-jun promoter were sufficient for EGF induction, we assayed various segments upstream of a minimal c-fos promoter. This minimal promoter in pOFlucGL3 contains a TATA box but not other known elements and was not inducible by EGF (Fig. 2). We found that the region from −225 to −31 of c-jun was sufficient to cause the minimal promoter to be induced fivefold by EGF (pJF6 in Fig. 2). A smaller region of the c-jun promoter, −133 to −31, was similarly inducible (pJF7), but removal of 5′ sequences from −133 to −78 reduced induction to about 2.5-fold (pJF9 [Fig. 2]). This last construct contains both the MEF2 and AP1-like sites but was poorly inducible, suggesting that elements between −133 and −77 are required for full induction. This region contains both CAAT and SP1 elements. We tested whether this region was sufficient for EGF induction by placing the region from −225 to −80 on the minimal c-fos promoter. Expression from this construct, pJF10, was not inducible by EGF (Fig. 2). Together, the results in Fig. 1 and 2 suggest that the MEF2 and AP1-like elements function together to mediate EGF induction and that they require additional upstream element(s) for full induction.
FIG. 2.
(A) Upstream elements cooperate with ATF and MEF2 sites for EGF induction of the c-jun promoter. HeLa cells were transiently transfected with the indicated reporter plasmids (3 μg) and assayed for EGF induction of luciferase activity as described for Fig. 1. (B) Heterologous c-jun promoter constructs. The indicated fragments of the c-jun promoter were cloned upstream of a minimal c-fos promoter (shaded in grey) and a luciferase (LUC) reporter gene.
ATF1 is the predominant factor in HeLa nuclear extracts that binds to the c-jun AP1-like element.
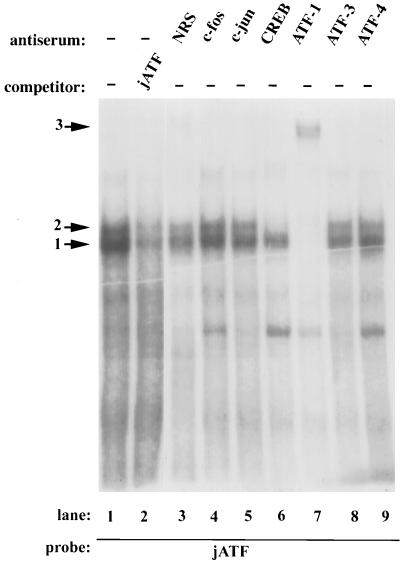
We used gel mobility shift assays with HeLa nuclear extracts to determine which factors bind to the c-jun AP1-like element. We observed two specific bands in gel mobility shift assays using a labeled oligonucleotide spanning the c-Jun AP1-like site (complexes 1 and 2 [Fig. 3, lane 1]). These bands were competed by excess unlabeled oligonucleotide probe (lane 2). We also tested extracts from HeLa cells treated with EGF for 5 to 60 min for binding activity to the c-jun AP1-like site. There was no change in the migration or amount of the complexes (data not shown). These results suggest that regulation is not exerted by changing the amount or type of these complexes.
FIG. 3.
ATF1 and CREB from HeLa nuclear extracts bind to the c-Jun ATF site. Gel mobility assays were performed with HeLa cell nuclear extracts and a 32P-labeled double stranded oligonucleotide spanning the c-jun ATF site as a probe. A 50-fold molar excess of unlabeled oligonucleotide (jATF) was included as a specific competitor as indicated. Other nonspecific oligonucleotides had no effect (data not shown). The addition of antisera to the indicated proteins is shown. NRS, normal rabbit serum; −, no serum added. The arrows indicate the DNA-protein complexes obtained.
While the site at −72 was previously termed an AP1-like site (2), its sequence, TGACATCA, is more similar to a consensus ATF site, TGACGTCA, than to a consensus AP1 site, TGACTCA. For this reason, we used antibodies against ATF family members as well as against Fos and Jun, components of AP1 complexes, to inhibit or supershift the bands in gel mobility shift assays. Nonimmune serum or antisera for Fos, Jun, ATF3, or ATF4 had no effect on the complexes (Fig. 3). Antiserum to CREB, a member of the ATF family, abolished the upper band while having little effect on the bottom complex (lane 6). Antiserum to ATF1 inhibited both complexes and caused a weak supershifted complex (complex 3 [lane 7]). The ATF1 and CREB antisera had no effect on a MEF2 gel mobility shift complex (data not shown). Since ATF1 and CREB can heterodimerize (26), these results are consistent with the bottom predominant band being an ATF1 homodimer while the upper band is an ATF1-CREB heterodimer. We cannot rule out, however, the possibility that ATF1 or CREB heterodimerize with other partners. We will henceforth refer to the c-jun AP1-like site as the jun ATF site.
As controls for the specificity and effectiveness of the antisera, we have found that the anti-ATF3 serum can specifically supershift in vitro translated ATF3 (69) and that the anti-ATF4 serum supershifted in vitro-translated ATF4 but not ATF1 (data not shown). The anti-Fos serum partially inhibited the gel shift of HeLa nuclear extract to a consensus AP1 site (data not shown). Two additional batches of anti-Jun serum also had no effect on the gel shift of HeLa nuclear extract to the jun ATF site. These two sera (AJ1 and AJ2) were able to completely supershift in vitro-translated c-Jun but not ATF1 (data not shown).
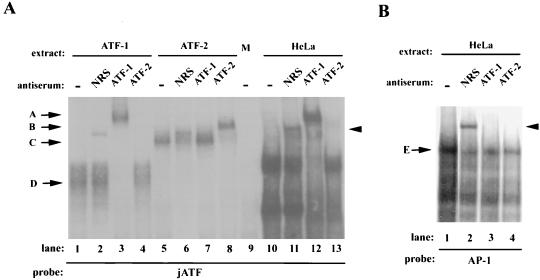
ATF1 and CREB cannot heterodimerize with either c-Jun or ATF2 in vitro (4, 17, 18). We further tested whether ATF2 is in the complex, however, since it can be activated by Rac and JNK and can bind to ATF sites (16). Antiserum to ATF2 had no effect on the complex from HeLa nuclear extracts, while anti-ATF1 completely supershifted the complexes (Fig. 4A, lanes 12 and 13). The anti-ATF2 serum was able to supershift in vitro-translated ATF2 (compare lanes 5 and 8) but had no effect on in vitro-translated ATF1 (lane 4). Conversely, the anti-ATF1 serum was able to supershift in vitro-translated ATF1 but not ATF2 (lanes 3 and 7), further demonstrating its specificity. We used a consensus AP1 site to test whether this site is similar to the jun ATF site. We detected a complex (band E [Fig. 4B, lane 1]) which could be specifically competed by the AP1 site (data not shown). This band migrated slightly below the bands binding to the jun ATF site (data not shown). The AP1 site complex was not affected by the anti-ATF1 serum (lane 3), demonstrating that the Jun ATF and AP1 sites bind distinct factors. We observed a complex in all lanes containing the control normal rabbit serum due to a nonspecific factor in this serum preparation.
FIG. 4.
Antisera to ATF1, but not ATF2, specifically affect the c-Jun ATF complexes. (A) Gel mobility shift assays were performed with the c-jun ATF site probe and in vitro-translated ATF1 (lanes 1 to 4), in vitro-translated ATF2 (lanes 5 to 8), mock in vitro translation extract (lane 9), or HeLa cell nuclear extracts (lanes 10 to 13). Either no serum (−), nonimmune serum (NRS), or anti-ATF1 or anti-ATF2 serum was added as indicated. (B) HeLa nuclear extracts were assayed with an AP1 consensus site probe and the indicated sera. Arrows D and C mark complexes obtained with in vitro-translated ATF1 and ATF2. Antibody-supershifted complexes are marked with arrows A and B. Complex E shows the complex binding to the consensus AP1 site. The arrowheads to the right indicate a nonspecific complex obtained with nonimmune serum.
The small G proteins Rac1 and Cdc42 can activate the c-jun promoter.
We were interested in identifying intracellular signalling pathways involved in EGF signalling from its cell surface receptor to the c-jun promoter. We first used a number of activated forms of known signalling molecules. EGF is known to activate Ras (41, 62), and we found that activated Ras [Ras(V12)] increased expression from the c-jun reporter gene pJC6GL3 (Fig. 5). One direct target of Ras is Raf, which activates a protein kinase cascade leading to activation of the Erk mitogen-activated protein kinases (MAPKs) (46, 66, 67). An activated form of Raf (RafBXB), however, did not increase expression from pJC6GL3 (Fig. 5). In addition, an activated form of MEK, a protein kinase activated by Raf, did not increase expression from pJC6GL3 (data not shown). Another direct effector of Ras is PI3K. An activated form of the p110 subunit of PI3K (25), however, did not significantly activate the jun reporter pJC6GL3 (data not shown). As controls, we found that the Raf, MEK1, and p110 constructs increased expression of a c-fos promoter reporter gene (data not shown).
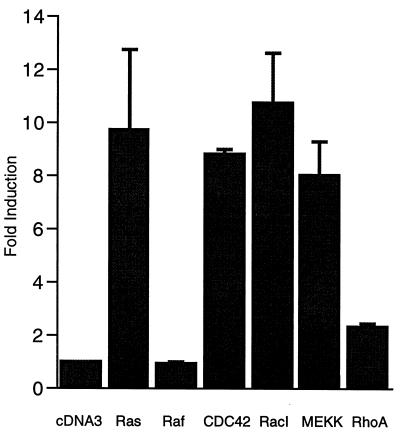
FIG. 5.
Activation of the c-jun promoter by activated forms of the small G proteins Ras, Rac, and Cdc42Hs. HeLa cells were transiently transfected with the c-jun–luciferase reporter pJC6GL3, pCMV-β-galactosidase as an internal control, and the following expression vectors (3 μg of each except for 2 μg for MEKK): pcDNA3 (empty vector control), Ras(V12), RafBXB, Cdc42Hs(V12), RacI(V12), RhoA(V14), or MEKK1. Cells were serum starved overnight prior to lysis for luciferase and β-galactosidase assays. The fold induction of luciferase activity is shown relative to activity in cells transfected with an empty expression vector (pcDNA3). The luciferase activities were normalized to the β-galactosidase activities except for assays with RasV12. With RasV12, the activity from pCMV-β-galactosidase was consistently induced fourfold, and this change was compensated for in normalizing the effect of Ras on pJC6GL3. The values shown are the averages of at least two separate experiments done in quadruplicate ± standard errors of the means.
The small GTPases RacI and RhoA were reported to be activated by Ras (53). Cdc42Hs is highly related to RacI, and both can activate the protein kinase JNK (11, 44). Both activated RacI and Cdc42Hs activated the c-jun promoter, while activated RhoA only weakly increased expression (Fig. 5). The RhoA vector was active since it was able to increase expression of a c-fos SRE reporter gene (data not shown).
Since Cdc42Hs and RacI are known to activate a pathway leading to activation of JNK, we tested an upstream component of the JNK pathway, MEKK. Overexpression of this protein kinase is sufficient to activate JNK (42) and also resulted in activation of the c-jun promoter (Fig. 5).
None of the activators except Ras significantly affected the internal control plasmid pCMV-β-galactosidase, suggesting that their effect on the c-jun promoter is specific. Ras(V12) increased pCMV-β-galactosidase up to fourfold, but we compensated for this increase in the data presented in Fig. 5.
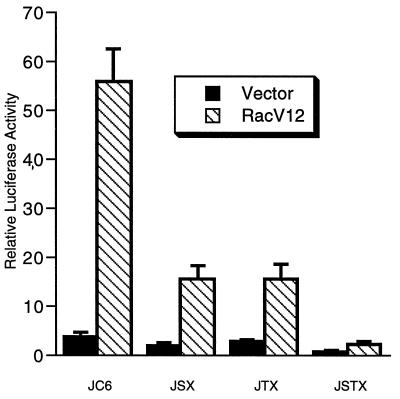
We tested whether activation of the c-jun promoter by RacI required the MEF2 and ATF sites. We found that RacI did not activate pJF10, which lacks the MEF2 and ATF sites (shown in Fig. 2), while pJF6 was strongly induced (data not shown). In addition, as seen in Fig. 6, mutation of either of the MEF2 or ATF sites in pJC6GL3 strongly reduced activation of the promoter by Rac. Mutation of both sites caused a further reduction of activity. Similar results were obtained with Cdc42Hs (data not shown). These results are similar to those obtained with EGF (Fig. 1) and suggest that RacI and Cdc42Hs act through both the MEF2 and ATF sites.
FIG. 6.
Requirement of the MEF2 and ATF sites for Rac activation of the c-jun promoter. HeLa cells were transiently transfected with the indicated reporter genes, pCMV-β-galactosidase as an internal control, and either 2.5 μg of empty expression vector (pcDNA3) or racI(V12). After transfection, cells were serum starved in 0.2% newborn calf serum overnight and then lysed for luciferase and β-galactosidase assays. Relative luciferase activities normalized to the β-galactosidase activities are shown. The values shown are the averages of two separate experiments done in duplicate ± standard errors of the means.
Dominant negative Ras, RacI, and MEKK can block EGF induction of the c-jun promoter.
We used dominant negative forms of the signalling molecules to test whether Ras, RacI, and MEKK are required for EGF induction of the c-jun promoter. We transfected expression vectors containing the dominant negative constructs together with the c-jun reporter plasmid pJC6GL3. Expression of dominant negative mutants of MEKK and Ras strongly reduced EGF induction from about fivefold to twofold (Fig. 7A). In contrast, dominant negative forms of Cdc42Hs (Fig. 7A) and of RhoA and Raf (data not shown) had no effect on EGF-induced Jun-luciferase expression.
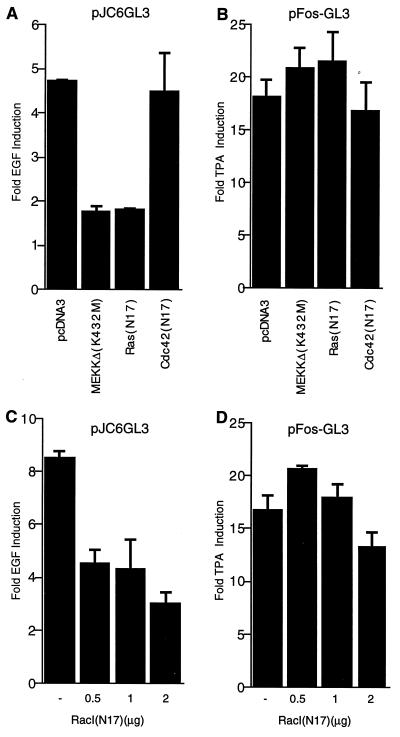
FIG. 7.
Dominant negative Ras, RacI, and MEKK block EGF induction of the c-jun promoter. (A) HeLa cells were transiently transfected with pJC6GL3, pCMV-β-galactosidase, and 1 μg of the empty expression vector pcDNA3 or the indicated dominant negative vectors. After transfection, cells were serum starved for 30 h and then treated with or without EGF (100 ng/ml) for 3 h prior to lysis for luciferase and β-galactosidase assays. The fold induction in EGF-treated relative to untreated cells is shown. The results are the means of duplicates ± standard errors of the means. (B) HeLa cells were transiently transfected with the c-fos reporter plasmid pFos-GL3 (1 μg), pCMV-β-galactosidase (1.5 μg), and the indicated dominant negative vectors as for panel A except that the cells were treated with or without TPA (100 ng/ml) for 3 h. The fold induction by TPA relative to untreated cells is shown. (C) Increasing amounts of dominant negative RacI were transfected as for panel A with the c-jun reporter gene except that EGF was added at 200 ng/ml. (D) Increasing amounts of RacI(N17) were transfected with the c-fos reporter gene and assayed for the effect on TPA induction as for panel B.
To demonstrate the specificity of the dominant negative mutants, we tested their ability to inhibit TPA induction of a c-fos–luciferase reporter gene. TPA induction of the c-fos promoter acts primarily through an Erk MAPK pathway which is activated by protein kinase C downstream of Ras (reviewed in reference 64). As expected, TPA induction of the c-fos reporter was not significantly affected by the dominant negative mutants (Fig. 7B).
To test the effect of dominant negative RacI [RacI(N17)] on EGF induction of the c-jun promoter, we titrated the amount of transfected construct since we observed some nonspecific inhibition on TPA induction of the c-fos promoter when transfecting high amounts of RacI(N17) (data not shown). In the experiment shown in Fig. 7C, increasing amounts of dominant negative RacI inhibited EGF induction by greater than 50%. No effect was observed on TPA induction of the c-fos reporter with the lower amounts of RacI(N17) used, while 2 μg of RacI(N17) caused a slight reduction (Fig. 7D). These results with dominant negative inhibitors suggest that Ras, RacI, and MEKK are required for EGF signalling to the c-jun promoter.
Recently the MAPK p38 and its activator MEK6 were found to activate the c-jun promoter (19). In addition, expression of MEKK at high levels can activate p38 (59, 72). We found, however, that the p38 inhibitor SB205380 had no effect on EGF induction of the c-jun promoter, suggesting that p38 is not required (data not shown). However, two homologs of p38, p38γ and SAPK4, are insensitive to this inhibitor (33) such that they or other related kinases could be involved in activation of the c-jun promoter.
DISCUSSION
We have shown that the Ras-RacI-MEKK pathway is necessary and sufficient for EGF induction of the c-jun promoter. This pathway appears to operate predominantly through two sequence elements, ATF and MEF2 sites, which are bound by ATF1 and MEF2D in HeLa cells.
Sequence elements for EGF induction of the c-jun promoter.
The MEF2 site at −59 and an ATF site at −72 in the c-jun promoter were found to be required for EGF induction. Mutation of either site reduced induction but did not abolish it, suggesting that each element can at least partially function without the other. The mutation of both sites abolished induction, yet these sites do not completely account for induction since they were not sufficient for maximal induction.
An additional region of the promoter, from −133 to −77, was needed for full induction. This region includes potential SP1, CAAT box, and ets sites, although we have not determined which, if any, of these sites are required for maximal induction. This region of the promoter was not sufficient for even partial EGF induction on a heterologous promoter but rather increased induction by the MEF2-ATF segment of the c-jun promoter. Factors binding between −133 and −77 upstream of the MEF2 and ATF sites may interact with the MEF2 and ATF factors to increase transcriptional activation. It is also possible, however, that the upstream factors are regulated by EGF but are not sufficient to activate the promoter without ATF and/or MEF2 factors.
We previously found that the MEF2 and ATF sites were critical for EGF induction (20) but have found slightly different results here. The first difference from our previous work (20) is in the role of the ATF site. Using RNase protection assays to measure expression, we observed no effect of mutation of the ATF site in the context of a −225 c-jun promoter–CAT reporter gene, although reduced expression was found following mutation of the ATF site in a −80 c-jun promoter construct. A second difference is in the ability of the MEF2 and ATF sites to give maximal induction without additional sequence elements. Here we have found that the MEF2-ATF sites give only modest induction which was increased when upstream sequence was present (Fig. 1 and 2). We believe that the differences are due to cryptic transcription factor binding sites in the original reporter genes used. In this study, we used the pGL3-luciferase vector which has been specifically designed to remove potential ATF, AP1, AP2, and SP1 sites in the luciferase gene. Consistent with this notion, comparison of EGF induction of the c-jun promoter in the previous luciferase vector with that in pGL3 showed that mutations in the MEF2 and ATF sites have much more pronounced effects in the context of the pGL3 vector (data not shown).
The c-fos promoter also contains an ATF site which is involved in nerve growth factor induction of the promoter (5). While the c-fos ATF site (termed a cyclic AMP response element at −60) is not inducible alone, it can accentuate induction together with other c-fos elements (such as the SRE) (5). This situation is similar to that with c-jun, where the ATF site alone gives weak induction but is required for maximal induction of the promoter.
Factors binding at the ATF and MEF2 sites.
The ATF site at −72 of the c-jun promoter was originally termed an AP1 site based on its binding to recombinant c-jun (2). As noted by Smith et al., however, this site more closely resembles a consensus ATF site (58). Using specific antisera, we have shown that the predominant factor binding this site in HeLa cell nuclear extracts is ATF1, with lesser binding by CREB. ATF1 and CREB are closely related proteins that can form heterodimers (26) such that the complex affected by the anti-CREB serum is likely to be a heterodimer of ATF1 and CREB. Our results are consistent with those of Hurst et al., who showed that in HeLa cell extracts, ATF1 homodimers along with ATF1-CREB heterodimers bound to ATF sites (26). Smith et al. detected four complexes binding to the c-jun ATF site in CCL64 cell extracts (58). Similar to our results, one complex was affected by anti-CREB serum, but two other complexes were affected by antiserum to another ATF family member, ATFa (58). This difference could be due to the different cell types used. Inconsistent with our results, Herr et al. found in HeLa extracts that Fos and Jun along with some ATF2 bound to the jun ATF site (which they termed jun1TRE) (22). The different results may be due to differences in the batches of HeLa cells, the exact experimental conditions, or the specificity of the antisera used.
While our results suggest that ATF1 homodimers and ATF1-CREB heterodimers bind to the c-jun ATF site in HeLa cells, we cannot rule out the possibility that other factors heterodimerize with either of these factors. Since c-Jun and ATF2 can be phosphorylated and activated by JNK, which in turn can be activated by RacI (16, 23, 61), we tested whether either of these proteins might be in the complex. Antisera to both had no effect on the complex. In addition, ATF1 and CREB did not heterodimerize with either c-Jun or ATF2 when examined in vitro (4, 17, 18), suggesting that c-Jun and ATF2 are most likely not in the complex.
Overexpression of c-jun was previously found to activate the c-jun promoter through the c-jun AP1-like site, suggesting a direct positive autoregulatory loop (2). The finding that ATF family members preferentially bind to this site, rather than c-Jun itself, suggests that the autoregulation is not direct but involves activation of ATF1 or CREB by an indirect mechanism. It is possible, however, that when c-jun is overexpressed it will directly occupy the jun ATF site even though this does not appear to occur under physiological conditions.
Four MEF2 family members, MEF2A to MEF2D, can bind to MEF2 sites. We previously found, using specific sera, that MEF2D is the predominant factor in HeLa cell extracts that binds to the c-jun MEF2 site, with MEF2A accounting for about 10% of the complex (21). The proportion of these and other family members is likely to vary in different cell types. Ornatsky and McDermott found that MEF2A is the predominant factor in C2C12 myotubes (49). Using our sera, they also found that MEF2A was as abundant as MEF2D in HeLa cells. This result likely reflects different batches of HeLa cells and demonstrates the variability of expression of these family members.
Signalling pathways.
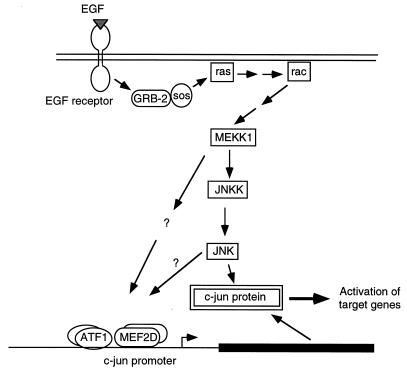
Using activated and dominant negative forms of signalling molecules, we have found that EGF induction of the c-jun promoter acts through Ras, Rac, and MEKK. The proposed pathway is shown in Fig. 8. EGF induction of Ras is mediated by EGF receptor binding to GRB2 and Sos, a guanine nucleotide exchange factor for Ras (9, 35). Ras can then activate Rac, though the mechanism is still unknown (53). MEKK functions downstream of Rac (44), although there is likely at least one intermediary between Rac and MEKK. A number of protein kinases have been found to bind Rac, but it is unclear which if any of these is involved in activation of MEKK (37, 38, 40). MEKK activates the protein kinase JNKK, which in turn activates JNK (13, 42, 57, 73). EGF can efficiently activate this pathway, as shown by EGF induction of JNKKinase activity (8, 43). We have demonstrated that the Ras-Rac-MEKK pathway is involved in EGF signalling to the c-jun promoter but have not determined which proteins downstream of MEKK are involved. Activated and dominant negative forms of JNK are either not available or not very effective, and thus we were not able to clearly determine its involvement. Therefore, activation of the c-jun promoter could involve JNK or a separate pathway downstream of MEKK, as indicated by the question marks in Fig. 8. Investigation of how the transcription factors on the c-jun promoter are regulated will help clarify this point.
FIG. 8.
Model of signalling pathways for EGF activation of c-jun. Components for signalling pathways to the c-jun promoter and for phosphorylation of c-Jun protein are shown and are described in the text. The pathway for induction of the c-jun promoter downstream of MEKK is unknown and may involve JNK and/or a separate pathway as indicated by the question marks. Two arrows between components indicate that the activation of the protein is not direct.
Cdc42Hs was also able to activate the c-jun promoter, but it is unlikely to be involved in the EGF-stimulated pathway. First, dominant negative Cdc42Hs did not inhibit EGF induction. Second, studies with dominant negative Cdc42Hs mutants suggest that Cdc42 does not operate between Ras and Rac (32, 48). The closely related GTPase Rho also does not appear to be involved in signalling to the c-jun promoter, since activated Rho strongly activated the fos promoter but not the jun promoter. Likewise, two other molecules downstream of Ras, Raf and the p110 subunit of PI3K, do not appear to be involved since activated forms of these molecules did not activate the c-jun promoter.
It has been reported that activators of the MAPK p38 can activate the c-jun promoter (19). Rac can also activate p38, although MEKK activates only p38 when it is expressed at high levels (36, 59, 72, 74). An inhibitor of p38α and -β had no effect on EGF induction, suggesting that these kinases are involved, though we cannot rule out that other homologs or related kinases mediate activation. There could also be redundancy of p38 and JNKK such that inhibition of p38 had no effect.
It is unclear as yet how the factors on the c-jun promoter are regulated. MEF2C was recently shown to be regulated by p38 phosphorylation (19). However, MEF2C is not present in HeLa cells (49), the region of MEF2C that is phosphorylated is not conserved in MEF2D, and as mentioned above, a p38 inhibitor did not inhibit EGF induction. Our preliminary experiments did not reveal EGF-induced changes in MEF2D phosphorylation (21), but a more careful analysis is required. In vivo footprinting studies have shown that the c-jun MEF2 site is occupied before and after serum treatment (54). We have also found that MEF2D DNA binding activity does not change in EGF-treated HeLa cell nuclear extracts, suggesting that regulation is likely to be on MEF2D’s transcriptional activation function rather than its DNA binding activity (21). This may be due to posttranslational modifications or complexing with regulatory proteins.
We did not find a change in binding to the jun ATF site in extracts from EGF-treated cells. One other potential mechanism for regulation of ATF1 and CREB is phosphorylation. Phosphorylation of CREB and ATF1 was induced by fibroblast growth factor, nerve growth factor, or EGF in various cell lines (5, 27, 63). This phosphorylation was found to be either p38 dependent (27, 63) or MEK1 (an activator of ERKs) dependent (71). These requirements are different from what we have found here for EGF induction of the c-jun promoter in HeLa cells. ATF1 and CREB can be phosphorylated and activated by a number of protein kinases, including cyclic AMP-dependent protein kinase (protein kinase A), MAPKAP kinase-2, and RSK2 (15, 63, 71). Further study will be needed to determine whether phosphorylation of ATF1 or CREB is required for induction of the c-jun promoter and, if so, which protein kinases are involved.
ACKNOWLEDGMENTS
We thank Michael Greenberg, C. Chandra Kumar, and J. Silvio Gutkind for their kind gifts of antisera and plasmids.
This work was supported by grant BE-261 from the American Cancer Society.
REFERENCES
- 1.Abate C, Luk D, Curran T. A ubiquitous nuclear protein stimulates the DNA-binding activity of fos and jun indirectly. Cell Growth Differ. 1990;1:455–462. [PubMed] [Google Scholar]
- 2.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Karin M. The role of Jun, Fos, and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 4.Benbrook D M, Jones N C. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 5.Bonni A, Ginty D D, Dudek H, Greenberg M E. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D A, O’Hara M, Angel P, Chojkier M, Karin M. Prolonged activation of jun and collagenase genes by tumor necrosis factor-α. Nature. 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 7.Bruder J T, Heidecker G, Rapp U R. Serum-, TPA-, and ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 8.Cano E, Hazzalin C A, Mahadevan L C. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol Cell Biol. 1994;14:7352–7362. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 10.Chen B P C, Wolfgang C D, Hai T. Analysis of ATF3, a transcription factor induced by physiological stress and modulated by gadd153/Chop10. Mol Cell Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 12.de Groot R P, Pals C, Kruijer W. Transcriptional control of c-jun by retinoic acid. Nucleic Acids Res. 1991;19:1585–1591. doi: 10.1093/nar/19.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 14.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Campbell D, Derijard B, Davis R. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 17.Hai T, Liu F F, Coukos W, Green M. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 18.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 20.Han T, Lamph W W, Prywes R. Mapping of epidermal growth factor-, serum-, and phorbol ester-responsive sequence elements in the c-jun promoter. Mol Cell Biol. 1992;12:4472–4477. doi: 10.1128/mcb.12.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han T, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herr I, Van Dam H, Angel P. Binding of promoter-associated AP-1 is not altered during induction and subsequent repression of the c-jun promoter by TPA and UV irrandiation. Carcinogen. 1994;15:1105–1113. doi: 10.1093/carcin/15.6.1105. [DOI] [PubMed] [Google Scholar]
- 23.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 24.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 25.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol 3-kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 26.Hurst H C, Totty N F, Jones N C. Identification and functional characterisation of the cellular activating transcription factor 43 (ATF-43) protein. Nucleic Acids Res. 1991;19:4601–4609. doi: 10.1093/nar/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansen F, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Rac regulation of actin polymerization and proliferation by a pathway distinct from JUN kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 30.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. Philos Trans R Soc Lond Ser B. 1996;351:127–134. doi: 10.1098/rstb.1996.0008. [DOI] [PubMed] [Google Scholar]
- 31.Kovary K, Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, McDonnell P C, Young P R. Novel homologues of CSBP/p38 MAP kinase: activation, substrate specificity and sensitivity to inhibition by pyridinyl imidazoles. Biochem Biophys Res Commun. 1997;235:533–538. doi: 10.1006/bbrc.1997.6849. [DOI] [PubMed] [Google Scholar]
- 34.Lamph W W, Wamsley P, Sassone-Corsi P, Verma I M. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 35.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 36.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 37.Manser E, Leun T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 38.Manser E, Leung T, Salihuddin H, Zhao Z S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 39.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 40.Martin G A, Bollag G, McCormick F, Abo A. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medema R H, Bos J L. The role of p21ras in receptor tyrosine kinase signaling. Crit Rev Oncogen. 1993;4:615–661. [PubMed] [Google Scholar]
- 42.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 43.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 45.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 47.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 48.Nobes C D, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 49.Ornatsky O I, McDermott J C. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and non-muscle cells. J Biol Chem. 1996;271:24927–24933. doi: 10.1074/jbc.271.40.24927. [DOI] [PubMed] [Google Scholar]
- 50.Prywes R, Roeder R G. Inducible binding of a factor to the c-fos enhancer. Cell. 1986;47:777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- 51.Qiu R, Chen J, McCormick F, Symons M. A role for rho in ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quantin B, Breathnach R. Epidermal growth stimulates transcription of the c-jun proto-oncogene in rat fibroblast. Nature. 1988;334:538–539. doi: 10.1038/334538a0. [DOI] [PubMed] [Google Scholar]
- 53.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 54.Rozek D, Pfeifer G P. In vivo protein-DNA interactions at the c-jun promoter in quiescent and serum-stimulated fibroblasts. J Cell Biochem. 1995;57:479–487. doi: 10.1002/jcb.240570313. [DOI] [PubMed] [Google Scholar]
- 55.Ryder K, Nathans D. Induction of protooncogene c-jun by serum growth factors. Proc Natl Acad Sci USA. 1988;85:8464–8467. doi: 10.1073/pnas.85.22.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 58.Smith S E, Papavassiliou A G, Bohmann D. Different TRE-related elements are distinguished by sets of DNA binding proteins with overlapping sequence specificity. Nucleic Acids Res. 1993;21:1581–1585. doi: 10.1093/nar/21.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stein B, Brady H, Yang M X, Young D B, Barbosa M S. Cloning and characterization of MEK6, a novel member of the mitogen-activated protein kinase kinase cascade. J Biol Chem. 1996;271:11427–11433. doi: 10.1074/jbc.271.19.11427. [DOI] [PubMed] [Google Scholar]
- 60.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 61.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 62.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 63.Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb M J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 64.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 65.Unlap T, Franklin C C, Wagner F, Kraft A S. Upstream regions of the c-jun promoter regulate phorbol ester-induced transcription in U937 leukemic cells. Nucleic Acids Res. 1992;20:897–902. doi: 10.1093/nar/20.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 68.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 69.Wolfgang C D, Chen B P C, Martindale J L, Holbrook N J, Hai T. Gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu B, Fodor E J B, Edwards R H, Rutter W J. Nerve growth factor induces the proto-oncogene in PC12 cells. J Biol Chem. 1989;264:9000–9003. [PubMed] [Google Scholar]
- 71.Xing J, Ginty D D, Greenberg M E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 72.Xu S, Robbins D J, Christerson L B, English J M, Vanderbilt C A, Cobb M H. Cloning of rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 74.Zanke B W, Rubie E A, Winnett E, Chan J, Randall S, Parsons M, Boudreau K, McInnis M, Yan M, Templeton D J, Woodgett J R. Mammalian mitogen-activated protein kinase pathways are regulated through formation of specific kinase-activator complexes. J Biol Chem. 1996;271:29876–29881. doi: 10.1074/jbc.271.47.29876. [DOI] [PubMed] [Google Scholar]