Abstract
Multimorbidity represents an increasingly important public health challenge with far-reaching implications for health management and policy. Mental health and metabolic diseases have a well-established epidemiological association. In this study, we investigate the genetic intersection between type 2 diabetes and schizophrenia. We use Mendelian randomization to examine potential causal relationships between the two conditions and related endophenotypes. We report no compelling evidence that type 2 diabetes genetic liability potentially causally influences schizophrenia risk and vice versa. Our findings show that increased body mass index (BMI) has a protective effect against schizophrenia, in contrast to the well-known risk-increasing effect of BMI on type 2 diabetes risk. We identify evidence of colocalization of association signals for these two conditions at 11 genomic loci, six of which have opposing directions of effect for type 2 diabetes and schizophrenia. To elucidate these colocalizing signals, we integrate multi-omics data from bulk and single-cell gene expression studies, along with functional information. We identify putative effector genes and find that they are enriched for homeostasis and lipid-related pathways. We also highlight drug repurposing opportunities including N-methyl-D-aspartate (NMDA) receptor antagonists. Our findings provide insights into shared biological mechanisms for type 2 diabetes and schizophrenia, highlighting common factors that influence the risk of the two conditions in opposite directions and shedding light on the complex nature of this comorbidity.
Subject terms: Schizophrenia, Target identification
Introduction
Multimorbidity, the coexistence of two or more chronic health conditions within an individual, has been of growing public health concern in recent years1. The simultaneous presence of multiple medical conditions poses substantial challenges for healthcare systems worldwide, affecting disease management, patient outcomes, and healthcare costs. Multimorbidity is not simply the sum of individual diseases but rather represents a complex interplay of interacting factors, including genetic predisposition, environmental influences, and shared pathophysiological pathways. Understanding the underlying mechanisms of multimorbidity is essential for guiding effective treatment strategies for patient-centered care. Yet, most health-related and drug development research is focused on treating or preventing individual diseases2. Treating each condition separately is inefficient and increases the patient’s treatment burden, possibly leading to adverse effects.
Individuals with mental health disorders are at higher risk of having multimorbid physical health conditions than those without psychiatric disorders, which contribute to lower life quality and premature death3,4. Here, we study the comorbidity between type 2 diabetes and schizophrenia, two conditions that commonly co-occur and have been described to be genetically correlated5. Dissecting the shared genetic etiology of these diseases can help identify risk variants and effector genes that could be used as biomarkers or druggable targets for their treatment and prevention.
Type 2 diabetes is characterized by persistent elevated glucose levels and insulin resistance, with typical onset of symptoms during middle adulthood. In 2021, over 536 million people were affected by type 2 diabetes worldwide6. The heritability of type 2 diabetes has been estimated as ~50%7. Schizophrenia is a major psychiatric disorder typically characterized by problems with perception, cognitive function and behavior and presents with hallucinations, delusion, disorganized thinking and speech8. Unlike type 2 diabetes, schizophrenia affects young people with onset during late adolescence or early adulthood. The prevalence of schizophrenia is ~1%9, and of schizophrenia and related psychotic disorders is ~3%. Genetic epidemiological studies have shown that schizophrenia has an estimated general heritability of ~80%8.
Observational studies have yielded indications of an epidemiologically positive association between type 2 diabetes and schizophrenia10. An evaluation of multiple observational studies derived a pooled relative risk of type 2 diabetes in schizophrenia patients versus healthy controls of 1.82 (95% confidence interval [CI] = 1.56–2.13)11. Sociodemographic and lifestyle factors are also considered key in the observed association. In addition, treatment with antipsychotic medication is associated with greater weight gain risk12 and increased risk for diabetes among individuals with schizophrenia13. However, hyperinsulinemia and impaired glucose tolerance have also been observed in first-episode schizophrenia patients who are antipsychotic naïve compared to healthy controls matched for age, sex and body mass index (BMI)14,15. Genetic studies point to a partial shared genetic aetiology between type 2 diabetes and schizophrenia16–18. For instance, a polygenic risk score for schizophrenia onset has been found to be positively correlated with insulin resistance among first-episode, drug naïve patients with schizophrenia19. Causal inference analysis using Mendelian randomization has not provided evidence of a causal relationship between type 2 diabetes and schizophrenia20. Elevated fasting insulin levels have been shown to have a causal effect on schizophrenia20, albeit with inconsistent results21. This suggests that insulin might exert an impact on this condition via a direct modulation of brain function instead of impaired glucose metabolism.
Beyond the role of increased adiposity, which is, in part, influenced by some antipsychotic drugs, possible additional biological pathways underlying the comorbidity between type 2 diabetes and schizophrenia may include immune dysfunction, particularly autoimmunity and chronic low-grade systemic inflammation21,22. Emerging evidence suggests that this interplay may be partly mediated by insulin resistance. Insulin has an anti-inflammatory effect, with inflammatory markers experiencing elevation in contexts of insulin resistance and diabetes23,24. Increased levels of inflammatory mediators IL-1β, IL-6, TNF-α and pro-inflammatory adiponectin have been observed in drug naïve, first episode schizophrenia patients with normal weight compared to overweight individuals without schizophrenia25. The role of inflammation in schizophrenia has been validated by Mendelian randomization analyses that show evidence of potential causal connections between inflammatory biomarkers and schizophrenia26.
Genome-wide correlation analyses show evidence of a weak negative genetic correlation between type 2 diabetes and schizophrenia18,27. Due to the difference in age-of-onset between the two conditions, and the diabetogenic side-effects of antipsychotic drugs, it is difficult to ascertain from observational studies whether type 2 diabetes and schizophrenia share underlying biological mechanisms. In this study, we aim to disentangle the shared genetic aetiology of type 2 diabetes and schizophrenia by resolving colocalizing signals to provide greater insight into the well-known comorbidity.
Methods
Datasets
GWAS summary statistics
For type 2 diabetes, we used the latest published multi-ancestry GWAS summary statistics not adjusted for BMI from the Diabetes Meta-Analysis of Trans-Ethnic (DIAMANTE) consortium that encompass 180,834 cases and 1,159,055 controls28. The distribution of effective sample sizes across ancestries included 51.1% European ancestry, 28.4% East Asian ancestry, 8.3% South Asian ancestry, 6.6% African ancestry including admixed African American populations and 5.6% Hispanic ancestry. In our analyses utilizing the multi-ancestral findings, we integrated the p-values emanating from the meta-analysis conducted via MR-MEGA29 and the effect size estimation and standard error from a fixed-effects model. We set the threshold for genome-wide significance at p-value < . For schizophrenia, we used the latest published multi-ancestry GWAS summary statistics comprising data from 74,776 cases and 101,023 controls30. The distribution across ancestries indicated European ancestry at 74.3%, East Asian ancestry at 17.5%, African American ancestry at 5.7%, and Latino ancestry at 2.5%. Genome-wide significance was set at p-value < .
In some analyses, we also used GWAS summary data from endophenotypes of type 2 diabetes, including glycaemic and adiposity-related traits. We used the following glycaemic traits data from MAGIC including up to 281,416 individuals without diabetes: fasting glucose, fasting insulin, HbAc1 and 2h-glucose-post-challenge31. Only summary statistics adjusted for BMI were made publicly available. Multi-ancestry summary statistics from MAGIC did not include effect size estimates, so we restricted the analyses to European ancestry summary statistics only. Adiposity-related traits were defined as BMI, body fat percentage, whole body fat mass and waist-to-hip ratio ratio (WHR) unadjusted for BMI. For BMI (N = 806,834) and WHR (N = 697,734), we used the recent meta-analysis combining data from the GIANT consortium and the UK Biobank32. The inverse rank normalized GWAS summary statistics for whole body fat mass (N = 330,762) and body fat percentage (N = 331,117) from the UK Biobank were taken from the Neale Lab website (http://www.nealelab.is/uk-biobank/).
Molecular quantitative trait loci summary statistics
We used various datasets of molecular quantitative trait loci (QTLs) from relevant tissues for each condition, namely brain (adult and fetal) for schizophrenia and adult brain, pancreatic islets, liver and subcutaneous/visceral adipose tissue for type 2 diabetes. We used bulk and single-cell type data from different molecular levels, including expression (eQTL), protein (pQTL), chromatin accessibility (caQTL), methylation (mQTL) and splicing (sQTL). For the eQTLs from the brain cortex, summary statistics were available for different ancestries separately. GTEx v8 expression and splicing QTL datasets consist of multi-ancestry samples, the majority (85.3%) of which are of European ancestry. All other data sets are from individuals of European ancestry only. A detailed list of the employed QTL datasets including sample size can be found in Table 1.
Table 1.
Overview of molecular quantitative trait loci (QTL) summary statistics employed in this work.
| Tissue/cell type | QTL type | Sample size | Reference |
|---|---|---|---|
| Cortex Europeans | Expression | 2683 | MetaBrain95 |
| Cortex East Asians | Expression | 208 | MetaBrain95 |
| Cortex Africans | Expression | 319 | MetaBrain95 |
| Basal ganglia | Expression | 208 | MetaBrain95 |
| Hippocampus | Expression | 168 | MetaBrain95 |
| Cerebellum | Expression | 492 | MetaBrain95 |
| Brain (prefrontal cortex)* | Chromatin accessibility | 292 | PsychENCODE51 |
| Brain (prefrontal cortex) | Protein | 330 | PsychENCODE51 |
| Brain (dorsolateral prefrontal cortex) | Methylation | 411 | Ng et al.96 |
| Brain amygdala | Splicing | 129 | GTEx v897 |
| Brain cerebellum | Splicing | 209 | GTEx v897 |
| Brain cortex | Splicing | 205 | GTEx v897 |
| Brain frontal cortex | Splicing | 175 | GTEx v897 |
| Brain hippocampus | Splicing | 165 | GTEx v897 |
| Brain hypothalamus | Splicing | 170 | GTEx v897 |
| Brain amygdala | Expression | 129 | GTEx v897 |
| Brain hypothalamus | Expression | 170 | GTEx v897 |
| Brain cells (single cell data) | Expression | 192 | Bryois et al.98 |
| Fetal brain (prefrontal cortex, striatum and cerebellum)* | Methylation | 166 | Hannon et al.99 |
| Fetal brain (no specific region) | Expression | 120 | O’Brien et al.100 |
| Pancreatic islets | Expression | 420 | InsPIRE101 |
| Pancreatic islets* | Splicing | 399 | Atla et al.102 |
| Liver | Expression | 208 | GTEx v897 |
| Liver* | Protein | 287 | He et al.103. |
| Liver | Splicing | 208 | GTEx v897 |
| Subcutaneous adipose tissue | Expression | 581 | GTEx v897 |
| Subcutaneous adipose tissue | Expression | 434 | METSIM54 |
| Subcutaneous adipose tissue | Splicing | 426 | METSIM104 |
| Subcutaneous adipose tissue | Splicing | 581 | GTEx v897 |
| Visceral adipose tissue | Expression | 469 | GTEx v897 |
| Visceral adipose tissue | Splicing | 469 | GTEx v897 |
For the brain caQTL, fetal brain mQTL, pancreatic islets sQTL and liver pQTL, only genome-wide significant or nominally significant results were available. We performed a lift-down from GRCh38 to GRCh37 using the R package CrossMap (version 0.5.4) for all eQTL datasets from MetaBrain and the fetal brain eQTL dataset33. We extracted regional expression and splicing QTL data from GTEx v8 by querying the eQTL catalogue’s RESTful application programming interface (API) v2 in R34. Subsequently, leveraging the liftOver function from the R package rtracklayer (version 1.22.0)35, we conducted a genomic mapping from GRCh38 to GRCh37. For GTEX v8, splicing QTLs were defined as leafcutter splice junction QTLs. Leafcutter quantifies RNA splicing variation using short-read RNA-seq data.
Quantification and statistical analysis
Genetic overlap between type 2 diabetes and schizophrenia
We quantified genetic correlation between type 2 diabetes and schizophrenia by conducting a linkage disequilibrium (LD) score regression analysis using the LDSC software (v1.0.1) with –rg flag36. We used the multi-ancestry GWAS summary statistics from the meta-analyses for type 2 diabetes and schizophrenia to enhance our discovery power. Since the European ancestry proportion is the largest one, we used the pre-computed LD scores from the 1000 Genomes European ancestry haplotypes37. A sensitivity analysis was conducted using exclusively European subset summary statistics. The complete results for the primary and the sensitivity analyses can be found in Table S1.
Causal inference analysis between schizophrenia and type 2 diabetes
To assess whether schizophrenia and type 2 diabetes have a causal relationship, we performed bi-directional two-sample Mendelian randomization analyses using the multi-ancestry GWAS summary statistics to enhance predictive power38 (Table S2A). We used the TwoSampleMR R package (version 0.5.7), which is curated by MR-Base39. We selected independent genome-wide significant (p-value < ) variants as instrumental variables (IVs). Independence was defined as LD-based clumped variants with a strict LD threshold of R2 = 0.001 over a 10 Mb window on either side of the index variant. Since the largest proportion of the samples in both type 2 diabetes and schizophrenia GWAS summary statistics are from European ancestry, we used the European reference LD panel from 1000 Genomes. As a sensitivity analysis for the estimated direction of effect, we applied Steiger-filtering to ensure that the IVs were more strongly associated with the exposure than the outcome (Table S2B). Additionally, we performed sensitivity analyses on the European ancestry subset only to compare the direction of effect (Table S2C).
To assess whether the IVs were strongly associated with the exposure, we calculated the F-statistic for each IV and selected only those that were larger than ten for the Mendelian randomization analyses to minimise bias due to weak IVs. F-statistic was estimated from summary level data as , where beta is the effect size and se is the standard error38. After the removal of weak IVs, we calculated the overall F-statistic as . We applied the inverse variance weighted (IVW) method, which performs a random-effects meta-analysis of the Wald ratios for each SNP. As a first sensitivity analysis, we applied the weighted median (WM) and the MR-Egger regression methods to ensure consistency of the effect size direction. The intercept of MR-Egger regression was used to assess horizontal pleiotropy. Finally, we tested for heterogeneity using the Q-statistic, which was calculated using the mr_heterogeneity function from the TwoSampleMR R package. To account for multiple testing, we corrected the p-values separately for each Mendelian randomization method employed by using the Bonferroni method. We extended the causal inference analyses to other psychiatric traits (Table 1) and endophenotypes related type 2 diabetes including adiposity-related traits.
Causal inference using childhood and adulthood body mass index as exposures
We performed causal inference analysis using univariate and multivariate two-sample Mendelian randomization with childhood and adulthood BMI as exposures. For childhood BMI, we used the IVs derived from a genetically predicted early life body size GWAS calculated based on recall data, from the UK Biobank40. Firstly, we estimated the total effect of each life-stage BMI value by conducting univariate Mendelian randomization analyses using each genetically predicted BMI value as a separate exposure and type 2 diabetes and schizophrenia as outcomes (Table S3). For this analysis, we applied two-sample Mendelian randomization with the IVW, WM and MR-Egger regression methods using the TwoSampleMR R package, as described above39.
Subsequently, we performed a multivariate Mendelian randomization analysis to estimate the direct effects of each life-stage BMI value using both childhood and adulthood BMI simultaneously as exposures (Table S4). We used the LD independent IVs from the univariate analyses. We applied the IVW method from the MVMR R package (version 0.4)41. To assess the effect of heterogeneity due to pleiotropy on the causal estimates, the MVMR R package calculates an adapted heterogeneity Q-statistic and estimates the causal effect of each exposure on the outcome accounting for heterogeneity, pleiotropy, and weak instrument bias. For both univariate and multivariate analyses we conducted sensitivity analyses for the direction of effect using only the European subset of the type 2 diabetes and schizophrenia GWAS (Table S3B and Table S4B).
Genetic colocalization analysis
For both type 2 diabetes and schizophrenia, we defined genomic regions spanning 2 Mb windows centered on independent genome-wide significant lead SNPs from the individual GWAS summary statistics. Within each identified region, we performed statistical colocalization analysis between type 2 diabetes and schizophrenia using the estimated regression coefficients (effect sizes) and standard errors (Table S5). We used the coloc.abf function from the coloc R package (version 3.2.1) for the analyses42. This function calculates the posterior probability for a set of five association hypotheses under the assumption of a single causal variant per trait in the region. Our investigative emphasis lay primarily upon the broader genetic landscape encompassing these regions, rather than a focused endeavour to identify a precise causal variant. Thus, the single-variant assumption of coloc.abf was not an issue here. The hypotheses are summarized below:
H0: no trait has a genetic association in the region
H1: trait 1 has a genetic association in the region
H2: trait 2 has a genetic association in the region
H3: both traits have a genetic association in the region, but with different causal variants
H4: both traits share a genetic association (single causal variant) in the region
We used the default prior probabilities of the coloc R package for our analyses. We considered evidence for colocalization if the posterior probability of H4 (PP4) > 0.8. In addition to the posterior probability, we calculated a 95% credible set for the causal variant by taking the cumulative sum of the variants’ posterior probabilities to be causal, conditional on H4 being true. LD between the estimated lead causal variant and the other variants in the region was calculated using PLINK (version 2.0 alpha)43 based on the UK Biobank data32 and was used for visualizing the results in regional association plots. In regions where we find evidence of colocalization (PP4 > 0.8), we ran sensitivity analyses using GWAS summary statistics for type 2 diabetes and schizophrenia derived from samples of European ancestry only.
Knockout mouse phenotypes
We performed a schematic search for each gene located within the vicinity (2 Mb window) of the genomic loci that colocalized between type 2 diabetes and schizophrenia to screen for knockout mice showing phenotypes related to each of the conditions. The databases used in this scope were the International Mouse Phenotyping Consortium (IMPC) (https://www.mousephenotype.org/), Mouse Genome Informatics (MGI) (http://www.informatics.jax.org/) and Rat Genome Database (RGD) (https://rgd.mcw.edu/) databases. For IMPC and RGD we extracted the knockout mice phenotypes for each analysed gene using the programmatic data access via their API. For MGI, we used the MGI batch query.
For type 2 diabetes, we looked for insulin and diabetes-related phenotypes that included the following terms: insulin, glucose, diabetes, hyperglycaemia, pancreas, pancreatic, obesity, BMI, body weight, body mass, body fat, beta cell, and glucosuria. For schizophrenia, we included neuropsychiatric-related phenotypes including brain and craniofacial morphology44–46, behaviour and psychiatric traits. As a sensitivity analysis we have extended the list of phenotypes related to schizophrenia to motoric phenotypes. A full list of the included phenotypes can be found in Table S9A and Table S9B.
Rare and syndromic human diseases
For every gene located within the vicinity of genomic regions that show evidence of genetic colocalization between type 2 diabetes and schizophrenia, we extracted data from the Online Mendelian Inheritance in Man (OMIM) (https://omim.org/) via its API. We looked up whether any rare and syndromic diseases linked to those genes showed any phenotype related to type 2 diabetes or schizophrenia. We defined association with schizophrenia if the disease showed any neurological phenotype. The full list of rare and syndromic human diseases associated with type 2 diabetes and schizophrenia can be found in Table S9A and Table S9B respectively.
Differential gene expression
For the genes in the regions of colocalization between type 2 diabetes and schizophrenia, we conducted a lookup on publicly available summary statistics of differential expression RNA-seq datasets related to the studied conditions. For type 2 diabetes, we used RNA-seq data from surgical pancreatic tissue samples from 57 metabolically phenotyped pancreatectomized patients, from which 39 were previously diagnosed with type 2 diabetes and 18 were non-diabetes patients47. The differential expression analysis was based on a linear model with age, sex, and BMI as covariates. We defined genes as differentially expressed if they changed more than 1.5 fold in either direction and had an adjusted p-value < 0.05, as in the original publication. For schizophrenia, we retrieved differential gene expression data based on RNA-seq and genotype data of post-mortem brains (frontal and temporal cortex) from PsychENCODE that includes 559 schizophrenia patients and 936 controls48. Differential expression was assessed using a linear mixed effects model accounting for known biological, technical and surrogate variables as fixed effects as well as subject-level technical replicates as random effects. Genes were defined as differentially expressed at a false discovery rate (FDR) < 0.05.
Multi-trait colocalization analysis with QTL data
Within the genomic loci exhibiting evidence of colocalization between type 2 diabetes and schizophrenia, we performed multi-trait colocalization analyses between type 2 diabetes GWAS summary statistics, schizophrenia GWAS summary statistics and each molecular QTL summary statistics from disease-relevant tissues or cell types summarized in Table 2. The analyses were performed within a 2 Mb window around the lead variant of the 95% credible set from the colocalization analysis between type 2 diabetes and schizophrenia.
Table 2.
Direction of effect of the six variants included in the 95% credible set from the colocalization between type 2 diabetes and schizophrenia.
| Effect allele | T2D | SCZ | Cortex | Cerebellum | Islets | |
|---|---|---|---|---|---|---|
| rs72951506 | T | - | + | + | + | + |
| rs72951548 | T | - | + | + | + | + |
| rs80196932 | C | - | + | + | + | + |
| rs9401019 | G | - | + | + | + | + |
| rs1501474 | T | - | + | + | + | + |
| rs10782188 | G | - | + | + | + | + |
The table depicts the direction of effect of six variants for type 2 diabetes (T2D), schizophrenia (SCZ), gene expression in the cortex, cerebellum and pancreatic islets.
To perform the multi-trait colocalization analyses, we used the R package HyPrColoc (version 1.0)49. Similarly to the coloc.abf function from the coloc R package, HyPrColoc estimates posterior probabilities for the above-mentioned colocalization hypotheses and identifies a putative shared causal variant. To assess whether all traits colocalize together without grouping them, we switched off the Bayesian divisive clustering algorithm (bb.alg = FALSE). As instructed by the developers, we assumed no sample overlap between all the input summary statistics. Evidence of colocalization was defined for a PP4 > 0.8. LD between the lead causal variant and the other variants in the genomic locus was calculated using PLINK (version 2.0 alpha)43 based on the UK Biobank data32.
Since we know a priori that schizophrenia and type 2 diabetes signals colocalize in the analysed genomic loci, we adapted the prior probabilities of the HyPrColoc algorithm accordingly. For the first parameter, prior.1, which denotes the probability of a variant being associated with a single trait, we kept the default value of since most regions that colocalize between type 2 diabetes and schizophrenia are genome-wide significant (p-value < ) for only one of the conditions. We slightly increased the conditional colocalization prior parameter, prior.c, from the default of 0.02–0.05 due to the evidence of colocalization between type 2 diabetes and schizophrenia in the region. This parameter represents the prior probability that a variant is associated with an additional trait given that it is associated with one trait.
As for the colocalization between type 2 diabetes and schizophrenia, we calculated a 95% credible set for the causal variant for each colocalized genomic locus by taking the cumulative sum of the variants’ posterior probabilities to be causal conditional on H4 being true. We considered relevant evidence of colocalization between distinct QTL datasets and both type 2 diabetes and schizophrenia if the 95% credible set of the HyPrColoc colocalization overlapped at least by one variant with the 95% credible set of the type 2 diabetes-schizophrenia colocalization analysis.
Scoring of potential effector genes
Firstly, we scored all genes within the vicinity (2 Mb window centered on the lead causal variant) of genome loci that colocalize (PP4 > 0.8) between type 2 diabetes and schizophrenia using four biological lines of evidence based on multi-omics and functional biological data:
Colocalization with at least one molecular QTL from disease-relevant tissues/cell types
Differential gene expression from pancreatic islets or brain
Knockout mice with phenotypes related to type 2 diabetes or neuropsychiatric traits
Rare and syndromic human diseases with phenotypes related to the type 2 diabetes or schizophrenia
For colocalization with methylation QTLs, we used the getMappedEntrezIDs function from the missMethyl R package (version 1.32.1) using annotation data from the Illumina’s HumanMethylation450 platform to map CpG sites to genes50. We excluded the following genes from the analysis due to inconsistent gene identifiers: SNORA40, snoU13 and Y_RNA.
We constructed one score for each condition. Each score was extended by information on established effector genes for the corresponding condition extracted from the literature. For type 2 diabetes, we retrieved a list of 135 high-confidence effector genes from the curated T2D Effector Prediction Summary from the Type 2 Diabetes Knowledge Portal (https://t2d.hugeamp.org/method.html?trait=t2d&dataset=egls) that scored at least 3. For schizophrenia, we used a list of 321 prioritized effector genes from PsychENCODE that were supported by more than two evidence sources51. Since our analysis overlaps with criteria used to define a gene as effector genes for the individual conditions, we followed an orthogonal approach to incorporate this information: if a gene is an prioritized effector gene for a condition, but scored zero in our analysis, we updated the respective condition score to one.
As an additional line of evidence, we looked up whether any variant in the 95% credible set from the colocalization analysis between type 2 diabetes and schizophrenia was a missense variant for any of the analyzed genes on Ensembl GRCh37 release 11052. The total score was defined as the sum of the type 2 diabetes, schizophrenia and missense score. If the missense variant score was the only line of evidence for a gene, the total score was kept at zero since this information is not directly related to the studied comorbidity.
We scored the genes based on the outlined six biological lines of evidence that were integrated in an orthogonal manner (Table S6). Genes that scored at least one point in the type 2 diabetes score and one point in the schizophrenia score and had a total score of at least 3 in the total score were defined as genes showing evidence of involvement in both conditions simultaneously. A subset of those that scored at least 4 were defined as putative effector genes.
Causal inference analysis using multi-omics data
To query whether the putative genes have a causal effect on type 2 diabetes or schizophrenia, we performed two-sample Mendelian randomization analyses between the expression QTL summary statistics and each disease GWAS summary statistics (Table S7A). We conducted Mendelian randomization analyses with different molecular QTL of tissues or cell types that show evidence of colocalization with both conditions simultaneously. We used the same software and sensitivity analyses described above for the causal inference analysis between type 2 diabetes and schizophrenia. If only one IV was available after clumping, harmonizing the data and filtering weak IVs through the F-statistics, we employed the Wald ratio method. We were only able to perform MR-Egger regression in instances where there were more than three independent IVs. If the IVs from the QTL datasets were absent in the disease GWAS, we ran the Mendelian randomization analysis with a LD proxy variant calculated on the European ancestry 100 genome reference panel instead. To search for LD proxies, we used the R package LDlinkR that queries the web-based LDlink tool and based the calculations in the European ancestry 100 genome reference panel53. The adjusted significance threshold for the eQTL data from adipose tissue from METSIM was set to p-value < , as defined in the original publication54. We performed sensitivity analyses within the European ancestry subset only to compare the direction of effect (Table S7B).
Pathway analysis
On the derived genes showing evidence of involvement in both type 2 diabetes and schizophrenia, we performed gene set enrichment analysis using the human resources and the enrichment software from the ConsensusPathDB (http://cpdb.molgen.mpg.de/)55 (Table S8). We did not perform the analysis on the putative effector genes due to the small number of genes on this set. For the enrichment analysis, we used the Gene Ontology human networks including following subcategories up to level 5: molecular function, biological processes, and cellular component. Finally, we required a minimum overlap of 2 genes for enrichment and set the significance threshold at FDR < 0.05.
Druggable genome
We queried the druggability status of the genes showing evidence of involvement in type 2 diabetes and schizophrenia using two databases. Firstly, we used the Druggable Genome database that consists of 4479 genes classified into three tiers based on their progress in the drug development pipeline56. Tier 1 consists of 1427 genes that are targets or clinical-phase drug candidates of already approved small molecules and biotherapeutic drugs. Tier 2 includes 682 genes that encode targets with known bioactive drug-like small-molecule binding partners and genes with ≥ 50% identity (over ≥ 75% of the sequence) with approved drug targets. Tier 3 comprise 2370 genes that encode secreted or extracellular proteins, proteins with more distant similarity to approved drug targets, and members of key druggable gene families that were not included in Tier 1 or 2. Tier 3 was further subdivided to prioritize genes in proximity (+−50 kbp) to a GWAS risk variant based on data from the GWAS catalog and had an extracellular location (Tier 3 A). Tier 3B consists of the remaining genes.
For genes showing evidence of involvement in type 2 diabetes and schizophrenia included in Tier 1 from the Druggable Genome, we further examined the approved or clinical trial drugs using the DrugBank database (https://www.drugbank.com, accessed on the 9th of August 2023) and the Open Targets platform57.
Results
Insights into shared genetics between type 2 diabetes and schizophrenia
We assessed the genetic correlation between type 2 diabetes (, ) and schizophrenia (, ) on a genome-wide scale using multi-ancestry data from the largest published GWAS summary statistics for the individual conditions. Using the same data, we assessed the potential causal relationship via Mendelian randomization analyses between type 2 diabetes and schizophrenia on a genome-wide scale. Genetic instruments for non-continuous exposures capture liability to the exposure rather than variation58. Hence, in Mendelian randomization, if the exposure is a disease, i.e. a non-continuous trait, the results should be interpreted in terms of liability to the disease. We find nominal evidence for a negative genetic correlation (rg = −0.04, standard error = 0.02, p-value = 0.023) (Table S1) and no evidence of a causal relationship between type 2 diabetes and schizophrenia (Table S2A). Insulin resistance has been shown to play a role in schizophrenia pathogenesis15,19 and weight gain, a risk factor for type 2 diabetes, is a consequence of some antipsychotic medications12. Hence, to assess the effect of different biological mechanisms involved in type 2 diabetes on schizophrenia, we expanded the Mendelian randomization analysis to endophenotypes of type 2 diabetes including adiposity-related traits. Interestingly, we find no evidence of causality between insulin-related traits and schizophrenia. We find evidence that schizophrenia liability is protective against increased BMI (schizophrenia → BMI: OR = 0.98, 95% CI = (0.96,0.99), p.adj = 0.03). The protective direction of effect remained consistent in the Steiger-filtered analysis (Table S2B) and in the analysis using only the European subset of the schizophrenia GWAS summary statistics (Table S2C). Although we find evidence that increased BMI is protective against schizophrenia (BMI → schizophrenia: odds ratio (OR) = 0.81, 95% CI = (0.74,0.89), adjusted p-value (p.adj)=), the direction of effect did not remain consistent after Steiger-filtering (Table S2B). The results of the Steiger-filtered analysis tackle the issue of reverse causation and suggest that the Mendelian randomization analysis has robust evidence of potential causality only for the direction schizophrenia liability to low BMI.
To further dissect the effect between BMI and type 2 diabetes as well as schizophrenia, we performed causal inference analyses using Mendelian randomization in a univariate and multivariate setting with childhood and adulthood BMI as exposures (Fig. 1). For type 2 diabetes, we find that both childhood and adulthood BMI are potentially causal for the disease when studied individually (childhood BMI: OR = 2.59, 95% CI = (2.23,3.01), p.adj = ; adulthood BMI: OR = 3.58, 95% CI = (3.09,4.16), p.adj=) (Table S3). We replicated previous results59 and show that the effect of childhood BMI is completely attenuated in the multivariate setting (childhood BMI: OR = 1.19, 95% CI = (0.95,1.5), p.adj = 0.18; adulthood BMI: OR = 3.27, 95% CI = (2.68,3.99), p.adj =) (Table S4). For schizophrenia, we find that only adulthood BMI has a significant protective effect against the condition in both the univariate (childhood BMI: OR = 0.88, 95% CI = (0.72,1.06), p.adj = 0.17; adulthood BMI: OR = 0.75, 95% CI = (0.64,0.87), p.adj = ) (Table S3) and multivariate setting (childhood BMI: OR = 1.04, 95% CI = (0.8,1.34), p.adj = 0.79; adulthood BMI: OR = 0.77, 95% CI = (0.61,0.96), p.adj = 0.046) (Table S4). All directions of effect remained consistent in the Stiger-filtered analysis.
Fig. 1. Mendelian randomization results of childhood versus adulthood BMI analysis.
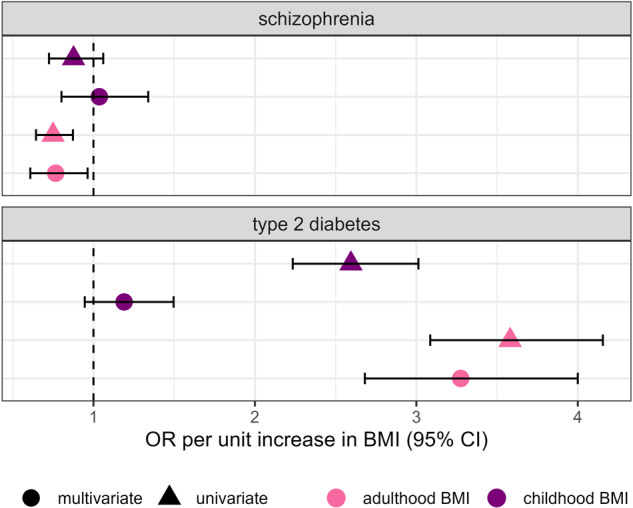
Forest plot depicting the direct and indirect effects for genetically predicted childhood and adulthood BMI on type 2 diabetes and schizophrenia. The effect is shown in odds ratio (OR) per unit increase in the BMI category, namely childhood and adulthood BMI. The results of the univariate analysis are represented by triangles and the multivariate results are represented by circles.
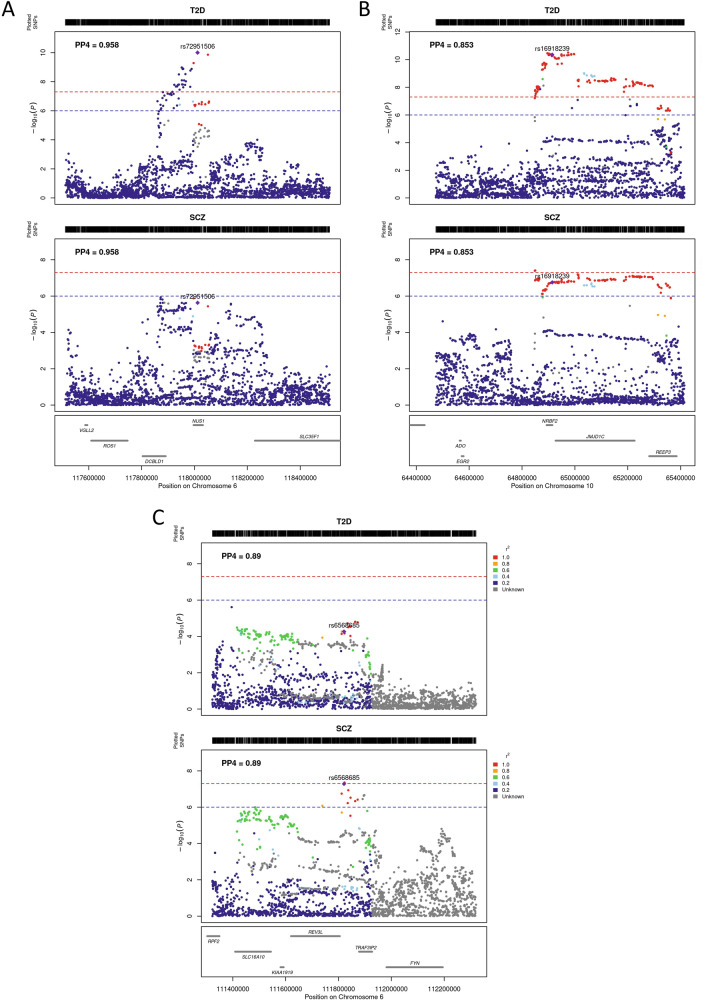
To identify shared genetic signals, we performed Bayesian colocalization analysis on genome-wide significantly associated type 2 diabetes and schizophrenia risk loci (p-value < ) using the GWAS summary statistics for each condition. We find evidence of colocalization (posterior probability of a shared causal variant (PP4) > 0.8) in 11 genomic loci (Table S5). Six of these genomic loci have opposing directions of effect for type 2 diabetes and schizophrenia. For two loci, the 95% credible set for the causal variant of the colocalization analysis consists of only one genetic variant, narrowing down the common genetic risk to a single variant.
To resolve colocalizing signals, we incorporated multi-omics and functional biology information to score the 444 genes located within the 11 genomic loci that colocalized between type 2 diabetes and schizophrenia (Fig. 2). We performed multi-trait colocalization analyses with bulk and single-cell molecular QTL from disease-relevant tissues and cell-types, namely several brain regions for schizophrenia, and pancreatic islets, liver, and subcutaneous adipose tissue for type 2 diabetes, respectively (Fig. 2). Our analyses included data from expression, protein, splicing and chromatin accessibility QTLs. In addition, we assessed whether these genes were differentially expressed in pancreatic islets or brain. We further conducted an extensive phenotypic search on knockout mice and rare and syndromic human diseases databases. Finally, we annotated missense variants in the 95% credible sets from the colocalization. 37 genes showed at least one line of evidence linking them to type 2 diabetes and at least one linking them to schizophrenia and were defined as potential effector genes for the comorbidity. Of these, we defined 15 genes showing evidence of involvement in both type 2 diabetes and schizophrenia with a total score of at least three, and three putative effector genes that displayed at least four different lines of evidence, namely EGR2, LAMA4 and NUS1 (Table S6). None of the three putative effector genes has been previously defined as effector gene for either type 2 diabetes or schizophrenia.
Fig. 2. Study design for scoring genes around colocalized regions.
Created with BioRender.com.
Insights gained from putative effector genes
NUS1
NUS1 (nuclear undecaprenyl pyrophosphate synthase 1 homolog) is the highest-scoring effector gene. This genomic locus colocalizes between type 2 diabetes and schizophrenia with a PP4 of 0.96 and includes six variants in the 95% credible set for the shared causal variant from the colocalization (Fig. 3A). All six variants have opposing risk-increasing alleles for type 2 diabetes and schizophrenia. The lead causal variant, rs72951506, is located in the intron of NUS1 and reaches nominal significance for schizophrenia (p-value = ) and genome-wide significance for type 2 diabetes (p-value = ). Additionally, type 2 diabetes and schizophrenia colocalize with genetic variants associated with the expression of NUS1 in the cerebellum (PP4 = 0.94), cortex (PP4 = 0.89) and pancreatic islets (PP4 = 0.96). The variants in the 95% credible set from the colocalization are associated with increased expression of NUS1 in these tissues, increased risk of schizophrenia and decreased risk of type 2 diabetes (Table 2).
Fig. 3. Regional plots of colocalized regions that yielded a putative effector gene.
A NUS1 region, B EGR2 region, C LAMA4 region.
NUS1 encodes a type I single transmembrane domain receptor, which is a subunit of cis-prenyltransferase, and serves as a specific receptor for the neural and cardiovascular regulator Nogo-B60. It is involved the regulation of intracellular LDL-derived cholesterol trafficking61. Variants in the NUS1 gene are associated with type 2 diabetes, epilepsy and intellectual disability. NUS1 has not been previously defined as a putative effector gene for type 2 diabetes nor schizophrenia and is not a target of any approved drug.
NUS1 shows significantly higher expression in brains from schizophrenia patients compared to controls48. Nus1 knockout mice show increased circulating triglyceride levels, abnormal brain vasculature morphology and abnormal lipid homeostasis. Increased levels of circulating triglycerides have been shown to be associated with increased risk of type 2 diabetes in humans62. Two rare and syndromic human diseases are associated with mutations in NUS1. The first, congenital disorder of glycosylation type 1aa, is a severe neurodevelopmental disorder with symptoms including seizures, developmental delay and hypotonia60 caused by a homozygous mutation in the NUS1 gene. The mutation results in an Arg290His substitution at a residue in the highly conserved C-terminal domain. The second syndromic disease associated with NUS1 is intellectual development disorder-55 with seizures. It is caused by de-novo heterozygous mutations in the NUS1 gene63. The mutations leading to both disorders result in autosomal-dominant loss-of-function variants in NUS164,65, which contradicts the findings from the differential gene expression analysis in brains of schizophrenia patients compared to controls. Fibroblasts with silenced NUS1 show increased accumulation of free cholesterol60.
We performed Mendelian randomization between the expression of putative effector genes and type 2 diabetes or schizophrenia (Table S7A). Our analysis results suggest an opposing direction of causal effect of NUS1 expression on type 2 diabetes and schizophrenia risk. Increased expression of NUS1 in the brain and in pancreatic islets has a potentially causal effect on schizophrenia (cortex: OR = 1.22, 95% CI = (1.09,1.37), p.adj = 0.002; cerebellum: OR = 1.05, 95% CI = (1.02,1.09), p.adj = 0.002; islets: OR = 1.21, 95% CI = (1.12,1.3), p.adj ). Differential expression analysis in brain supports this evidence, as NUS1 is over-expressed in schizophrenia patients. Increased expression of NUS1 in brain and pancreatic islets is potentially protective against type 2 diabetes (cortex: OR = 0.92, 95% CI = (0.85,0.99), p.adj = 0.06; cerebellum: OR = 0.94, 95% CI = (0.94,0.97), p.adj < ; islets: OR = 0.86, 95% CI = (0.82,0.9), p.adj = ).
EGR2
A genomic locus on chromosome 10 that encompasses a putative effector gene colocalizes between type 2 diabetes and schizophrenia with a PP4 of 0.84 (Fig. 3B). This region shows PP4 = 0.082 and PP3 = 0.82 in the European ancestry-only sensitivity analysis. The 95% credible set from the colocalization analysis contains 56 variants. Synthesis of the orthogonal lines of evidence from functional genomics data point to EGR2 (early growth response-2) as the putative effector gene for this region. For all shared causal candidate variants, the risk-increasing alleles for type 2 diabetes and schizophrenia are opposite.
Egr2 knockout mice show decreased body weight, weight loss, decreased nerve conduction velocity, decreased neuron number, delayed eye opening, and abnormal neuron physiology and differentiation. In humans, these phenotypes are associated with a lower risk of type 2 diabetes and a higher risk of schizophrenia. EGR2 shows significantly lower expression in the brain of schizophrenia patients compared to healthy controls48.
Mutations in the EGR2 gene are associated with three rare and syndromic human diseases: Charcot-Marie-tooth disease type 1D, a sensorineural peripheral polyneuropathy that affects both motor and sensory nerve function; Dejerine-Sottas syndrome, a demyelinating peripheral neuropathy with onset in infancy that results in delayed motor development; and Congenital hypomyelinating neuropathy, which is characterized clinically by onset of hypotonia at birth, areflexia, distal muscle weakness, and very slow nerve conduction velocities. In all these diseases, the mutation of EGR2 leads to a decrease or a complete loss of gene function.
EGR2 encodes a transcription factor that is a prime regulator of Schwann cell myelination66. It is involved in the development of the jaw opener musculature by playing a role in its innervation through trigeminal motor neurons67. EGR2 has been reported to also play a role in hindbrain segmentation and development66 and in adipogenesis, possibly by regulating the expression of CEBPB68. Variants in EGR2 gene are associated with serum triglycerides and cholesterol levels, temperament and adventureness.
Mendelian randomization analysis shows evidence of potential causality between increased expression of EGR2 in the cortex and schizophrenia (OR = 1.21, 95% CI = (1.09,1.35), p.adj = 0.002) (Table S7A). There was no evidence of a causal effect of EGR2 levels in disease-relevant tissues on type 2 diabetes.
LAMA4
We identify LAMA4 (laminin, alpha 4) as a putative effector gene underpinning a colocalizing genomic locus on chromosome 6 (PP4 = 0.89). The 95% credible set for the causal variant from the colocalization analysis consists of 36 variants (Fig. 3C). All variants show opposing risk-increasing alleles for type 2 diabetes and schizophrenia. Variants in the 95% credible set for the causal variant from the colocalization are associated with trunk fat mass as well as depressive and maniac episodes in bipolar disorder69,70.
Laminins are a family of extracellular matrix glycoproteins that constitute basement membranes71. They have been implicated in multiple biological processes including attachment, migration and organization of cells into tissues during embryonic development. LAMA4 encodes the laminin subunit alpha-4 protein, a structural component that contributes to cell adhesion and tissue organization in various biological processes72. Heterozygous mutations in LAMA4 are causal to the syndromic disease dilated cardiomyopathy-1JJ. In addition, variants in this gene are associated with various phenotypes including BMI, insulin, bipolar disorder and myocardial infarction. LAMA4 is the target of ocriplasmin, a drug indicated for the treatment of symptomatic vitreomacular adhesion73.
Lama4 knockout mice show decreased body size and weight and abnormal neuromuscular synapse morphology and lethargy. In humans, decreased body weight and lethargy are associated with decreased risk of type 2 diabetes and increased risk of schizophrenia, respectively. LAMA4 shows differential expression in pancreatic islets from healthy versus diabetes patients and in brains from schizophrenia patients versus controls47,48. It is over-expressed in diabetes patients and down-regulated in schizophrenia patients. The observed direction of effect of LAMA4 on each investigated condition in knockout mice and in differential gene expression is concordant.
Insights into disease biology and treatment targets
We conducted pathway enrichment analyses on the set of genes showing evidence of involvement in both type 2 diabetes and schizophrenia (Table S8) and find “homeostatic process” and “intracellular lipid transport” to be the most significantly enriched biological processes. These genes exhibited enrichment in further pathways related to lipids, for instance “lipid transport”, “lipid localization” and “lipid homeostasis”, as well as pathways related to the regulation of metabolic processes (Fig. 4). These findings point toward a potential involvement of lipid metabolism in the comorbidity of type 2 diabetes and schizophrenia.
Fig. 4.
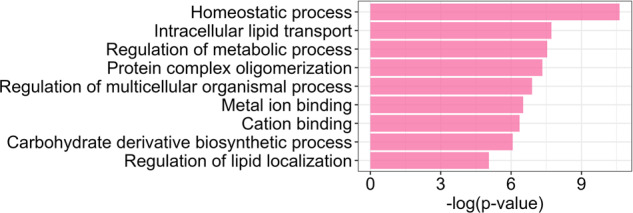
Results of enrichment analysis on the set of genes showing evidence of involvement in type 2 diabetes and schizophrenia.
The 15 genes showing evidence of involvement in both type 2 diabetes and schizophrenia represent relevant candidates for further functional and clinical investigation. Thus, we queried the druggability status of these genes and find that five are included in the druggable genome56. Of these, four genes (ATP1A1, GRIN2B, LAMA4 and TK1) are tier 1 druggable targets, i.e., their products are targets of existing drugs with market authorization or in clinical development, and one (ACACA) is a tier 2 druggable target, i.e., its product has known bioactive drug-like binding partners.
ACACA (acetyl-CoA carboxylase alpha) codes for the enzyme acetyl-CoA carboxylase alpha (ACC-alpha), which plays a role in fatty acid synthesis in humans, a process that impacts energy balance and lipid metabolism. In cases of impaired ACC-alpha function, biotin supplementation can help support proper enzyme activity as a cofactor, leading to better overall metabolic health74. An ACC inhibitor was tested in a phase II clinical trial for type 2 diabetes and there is evidence that the ACC-alpha pathway in pancreatic alpha cells could be a therapeutic strategy in type 2 diabetes by limiting glucagon secretion75.
ATP1A1 (ATPase Na + /K+ transporting subunit alpha 1) encodes a subunit of the sodium-potassium pump, which plays a vital role in maintaining cell membrane potential and various physiological processes, including nerve impulse transmission76. Cardiac glycosides drugs, such as digoxin, inhibit the sodium-potassium pump and have been used to treat heart-related conditions by enhancing the heart muscle contractions77. Digoxin was also tested in a phase I clinical trial for epilepsy, depressive disorder and type 2 diabetes. In type 2 diabetes patients, it increased glucose intolerance78. Digoxin augmented the effect of antiepileptic drugs in mice, increased the chances of depression after a myocardial infarction and was linked to increased risk of psychosis79–81.
GRIN2B (glutamate ionotropic receptor N-methyl-D-aspartate type subunit 2B) encodes a subunit of the N-methyl-D-aspartate (NMDA) receptor ion channel in the brain that plays a role in synaptic plasticity, learning, memory, and various neurological functions82. There are several approved drugs that modulate NMDA receptor activity, including felbamate and haloperidol. Felbamate, an anticonvulsant used to treat severe epilepsy, has an antagonistic effect on NMDA receptors, reducing their activity83. Haloperidol is an antipsychotic used in the treatment of schizophrenia that has been shown to inhibit the NMDA receptor84. Two further clinically approved NMDA antagonists, ketamine and acamprosate, have been tested for schizophrenia treatment with inconclusive results85,86. Although GRIN2B is not rated as a putative effector gene for schizophrenia, it has been shown that variants in this gene are associated with this condition in a Siberian and a Han Chinese population87,88.
LAMA4 is classified as a Tier 1 target in the druggable genome as the protein product of this gene is a target of the proteolytic enzyme ocriplasmin. Ocriplasmin is approved to treat symptomatic vitreomacular adhesion and was in clinical trial for stroke and diabetic macular edema treatment73 (www.clinicaltrials.gov). LAMA1, LAMA3 and LAMA5, other laminins, are targets of the approved drug lanoteplase, used in the treatment of myocardial infarction but with unknown mechanisms of action89. In an analysis aimed at identifying potential drug candidates based on perturbed transcriptomic pathways in Parkinson’s disease, lanoteplase emerged as the candidate with strongest enrichment90.
Discussion
In this work, we dissect the genetic etiology shared between type 2 diabetes and schizophrenia. In line with smaller-scale previous studies, we find evidence of a weak negative genetic correlation and no evidence of a causal relationship between the two comorbid conditions18,27. Considering the sample sizes of the disease GWAS employed in this study, our study provides robust evidence of a non-causal relationship between type 2 diabetes and schizophrenia. These results point to common pathways underlying type 2 diabetes and schizophrenia that act in opposite direction on each condition. Most schizophrenia patients have had long-term exposure to antipsychotics. We show that Mendelian randomization results using schizophrenia GWAS data not adjusted for antipsychotic use finds no evidence of a causal effect of schizophrenia liability on type 2 diabetes. More research taking data on medication use into account is needed to further dissect the effect of antipsychotics on type 2 diabetes.
Type 2 diabetes and schizophrenia are epidemiologically positively correlated. However, it has been previously shown, and we have validated this observation here, that the conditions have a negative genetic correlation. There are multiple potential reasons behind this. Firstly, there is a large gap between the age of onset of type 2 diabetes and schizophrenia. Secondly, comorbidities are often underdiagnosed among schizophrenia patients91. In addition, analyses aimed at dissecting the association between schizophrenia and BMI yield inconsistent results partly due to the weight gain effect of some antipsychotics. Our analyses rely on integrating data from population-based studies for type 2 diabetes and smaller clinical studies for schizophrenia. For type 2 diabetes, GWAS results are biased towards individuals with very prevalent symptoms with a formal disease diagnosis. This liability threshold bias can lead to different and even opposing patterns between observational and GWAS-based studies. It is worth noting that our findings support the estimated negative genetic correlation between type 2 diabetes and schizophrenia, and that the derived putative effector genes as well as high BMI have opposite direction of effect on the studied conditions.
We provide evidence of a potential protective effect between schizophrenia liability and BMI. This association is in the opposite direction to the well-established causal effect between BMI and type 2 diabetes, pointing to potentially different adiposity-related mechanisms underpinning each condition. We replicated previous results showing that the causal effect of genetically predicted childhood BMI on type 2 diabetes is almost entirely attenuated when adjusting for adulthood BMI59. When looking at the causal effect of life-stage BMI to schizophrenia, we show evidence that adulthood BMI is protective against schizophrenia, but not childhood BMI. In this analysis, we find no evidence of reverse causation. Schizophrenia is associated with low late-pregnancy maternal BMI, low birth weight and being thin during childhood92. Thus, we hypothesize that the effect of body size on schizophrenia might be mediated earlier in development and is not fully captured by our analysis using genetically predicted data for early life BMI at 10 years old. Unhealthy lifestyle and medication use might be underlying causes of the shown relationship between adulthood BMI and schizophrenia.
By leveraging recent large-scale GWAS for type 2 diabetes and schizophrenia, we find evidence of colocalization at 11 genomic loci. We score genes in the vicinity of the colocalized regions by incorporating multi-omics and functional genomics information and derive a list of 15 genes showing evidence of involvement in both type 2 diabetes and schizophrenia, which are enriched for biological pathways associated with lipid regulation. We highlight the putative effector genes, which show the highest score of involvement in both type 2 diabetes and schizophrenia in our data-based statistical genetic approach. For these genes, we have derived the effect on each condition mainly based on causal inference analysis using expression QTL data from relevant tissues. The putative effector genes involved simultaneously in type 2 diabetes and schizophrenia derived in this work have not been established as effector genes for the individual conditions based on previous publications. There are various factors that can lead to a gene not having been defined as a high confidence effector gene for a disease previously, for example due to a lack of comprehensive post-GWAS analysis or insufficient evidence in single-disorder discovery efforts. We show that the three putative effector genes have opposite direction of effect on type 2 diabetes and schizophrenia risk. These results are in support of a causal effect of BMI on both conditions in opposite directions.
We have run the genetic colocalization with the assumption of a single causal variant per region because we are primarily interested in finding regions where type 2 diabetes and schizophrenia share signals. To relax the single variant assumption an LD reference panel is mostly needed, which is not straight-forward in the case of multi-ancestry data with different ancestry compositions. This has an influence on downstream analyses that rely on the variants included in the 95% credible set of the colocalization analysis, which is a limitation of the present work. We have defined regions to run genetic colocalization analysis based on at least one of the studied conditions having a genome-wide risk signal (p-value < 5e-8). Hence, we also ran the analysis on regions where only one of the conditions shows evidence of association. This approach was selected to address potential power limitations stemming from varying sample sizes in the individual GWAS. Subsequently, we embellish the colocalization findings by delving deeply into biological lines of evidence that indicate the involvement of putative effector genes within these colocalized regions.
The biological function of the putative effector genes NUS1 and LAMA4 as well as the result of the enrichment analysis highlight adipogenesis and cholesterol trafficking as potential biological mechanisms influencing the comorbidity between type 2 diabetes and schizophrenia. These mechanisms might reflect the presence of an underlying metabolic vulnerability in a subset of individuals with schizophrenia beyond the side effects of antipsychotics.
The genomic locus that yields LAMA4 as a putative effector gene does not reach nominal significance (p-value < 1) in the multi-ancestry GWAS for type 2 diabetes (Fig. 3C). The lead causal variant rs6568685 is an intron variant of TRAF3IP2-AS1. This variant reaches a p-value of 1.7 in the European-ancestry type 2 diabetes GWAS and a p-value of 0.26 in the East Asian ancestry GWAS. Hence, this may be a European-specific association or a low frequency variant in East Asians. Alternatively, the association in this locus may reach genome-wide significance in diverse non-European ancestry datasets once sample sizes are large enough.
Human genetics evidence has been shown to support the majority of FDA-approved drugs in 202193. We investigated the potential for drug development or repurposing for genes prioritized by our analyses, for the comorbidity between type 2 diabetes and schizophrenia. We highlight two interesting candidates for drug repurposing opportunities that could be investigated for schizophrenia treatment: lanoteplase, which targets laminins and is currently used in the treatment of myocardial infarction; and felbamate, an NMDA receptor inhibitor used to treat severe epilepsy. On the one hand, digoxin, a drug targeting ATP1A1, has been shown to increase glucose tolerance in type 2 diabetes patients78, but on the other hand increase risk of psychosis81,94. To avoid complications, drugs targeting genes with opposing direction of effect for different diseases should be administered with caution and alongside with monitoring of comorbidity risk and progression.
As a result of great efforts from the GWAS community, both the type 2 diabetes and schizophrenia GWAS summary statistics are meta-analyses that expand beyond European-centric data, by including data from diverse global populations. However, the vast majority of primary tissue molecular QTL data is derived from European-ancestry individuals. Hence, our analyses with statistical genetics methods that rely on LD estimations, such as Mendelian randomization, were constrained to using LD from European-ancestry populations, which still constitutes the majority (>70%) of samples contributing to GWAS. It will be important for future studies to generate molecular QTL data from a wide array of tissues across multiple diverse populations.
Mental health disorders are often accompanied by metabolic disorders that ultimately lead to premature death. Here we have studied the shared genetic underpinning between type 2 diabetes and schizophrenia and provide insights into the potential common biological mechanisms and shared effector genes. We have shown that the derived set of shared putative effector genes have opposing direction of effect on the individual conditions. Our findings emphasize that treatments targeting those genes should be tempered with caution regarding exasperation of the comorbidity.
Supplementary information
Acknowledgements
Data on glycaemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. G.M.K. and G.D.S. work in a unit funded by the Medical Research Council (MRC) and the University of Bristol (MC_UU_00032/01, MC_UU_00032/06). G.M.K. acknowledges funding support from the Wellcome Trust (grant no: 201486/Z/16/Z and 201486/B/16/Z), the UK Medical Research Council (grant no: MC_UU_00032/06; MR/W014416/1; and MR/S037675/1), and the UK National Institute of Health Research Bristol Biomedical Research Centre (grant no: NIHR 203315). LMH acknowledges funding from the National Institutes of Health: NIMH (R01MH124839, R01MH118278) and NIEHS (R01ES033630).
Author contributions
E.Z. and A.L.A. conceptualized the project. G.M.K., A.P.M., G.D.S. and L.M.H. gave valuable input to the methodological approach. A.L.A. conducted the data analyses. All authors assisted in interpreting the results. A.L.A. drafted the initial manuscript. All authors contributed to revising the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Code availability
All original code used to generate the results presented in this paper is available at GitHub and deposited on Zenodo (10.5281/zenodo.10016486).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41537-024-00445-5.
References
- 1.Skou ST, et al. Multimorbidity. Nat. Rev. Dis. Primers. 2022;8:48. doi: 10.1038/s41572-022-00376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langenberg C, Hingorani AD, Whitty CJM. Biological and functional multimorbidity—from mechanisms to management. Nat. Med. 2023;29:1649–1657. doi: 10.1038/s41591-023-02420-6. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues M, Wiener JC, Stranges S, Ryan BL, Anderson KK. The risk of physical multimorbidity in people with psychotic disorders: a systematic review and meta-analysis. J. Psychos. Res. 2021;140:110315. doi: 10.1016/j.jpsychores.2020.110315. [DOI] [PubMed] [Google Scholar]
- 4.Deste, G. & Lombardi, C. M. Editorial: Cardiometabolic disease and psychiatric disorders. Front. Psychiatry.10.3389/fpsyt.2023.1174055 (2023). [DOI] [PMC free article] [PubMed]
- 5.Fanelli G, et al. Insulinopathies of the brain genetic overlap between somatic insulin-related and neuropsychiatric disorders. Transl. Psychiatry. 2022;12:59. doi: 10.1038/s41398-022-01817-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun, H. et al. IDF Diabetes atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 10.1016/j.diabres.2021.109119 (2022). [DOI] [PMC free article] [PubMed]
- 7.Avery AR, Duncan GE. Heritability of Type 2 diabetes in the washington state twin registry. Twin Res. Hum. Genet. 2019;22:95–98. doi: 10.1017/thg.2019.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuki Y, et al. Mechanisms underlying the comorbidity of schizophrenia and type 2 diabetes mellitus. IJNP. 2021;24:367–367. doi: 10.1093/ijnp/pyaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward M, Druss B. The epidemiology of diabetes in psychotic disorders. Lancet Psychiat. 2015;2:431–451. doi: 10.1016/S2215-0366(15)00007-3. [DOI] [PubMed] [Google Scholar]
- 11.Stubbs B, Vancampfort D, De Hert M, Mitchell AJ. The prevalence and predictors of type two diabetes mellitus in people with schizophrenia: a systematic review and comparative meta-analysis. Acta. Psychiatr. Scand. 2015;132:144–157. doi: 10.1111/acps.12439. [DOI] [PubMed] [Google Scholar]
- 12.Mamakou V, Thanopoulou A, Gonidakis F, Tentolouris N, Kontaxakis VP. Schizophrenia and type 2 diabetes mellitus. Psychiatrike = Psychiatriki. 2018;29:64–73. doi: 10.22365/jpsych.2018.291.64. [DOI] [PubMed] [Google Scholar]
- 13.Smith M, et al. First- v. second-generation antipsychotics and risk for diabetes in schizophrenia: systematic review and meta-analysis. Br. J .Psychiatry. 2008;192:406–411. doi: 10.1192/bjp.bp.107.037184. [DOI] [PubMed] [Google Scholar]
- 14.Guest PC, et al. Increased levels of circulating insulin-related peptides in first-onset, antipsychotic naïve schizophrenia patients. Mol. Psychiatry. 2010;15:118–119. doi: 10.1038/mp.2009.81. [DOI] [PubMed] [Google Scholar]
- 15.Pillinger T, et al. Impaired glucose homeostasis in first-episode schizophrenia: a systematic review and meta-analysis. JAMA. Psychiatry. 2017;74:261–269. doi: 10.1001/jamapsychiatry.2016.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackinger S, et al. Evidence for genetic contribution to the increased risk of type 2 diabetes in schizophrenia. Transl. Psychiatry. 2018;8:1–10. doi: 10.1038/s41398-018-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry BI, et al. Common mechanisms for type 2 diabetes and psychosis: findings from a prospective birth cohort. Schizophr. Res. 2020;223:227–235. doi: 10.1016/j.schres.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry, B. I. et al. Evidence for shared genetic aetiology between schizophrenia, cardiometabolic and Inflammation-related traits: genetic correlation and colocalization analyses. Schizophr. Bulletin Open10.1093/SCHIZBULLOPEN/SGAC001 (2022). [DOI] [PMC free article] [PubMed]
- 19.Tomasik J, et al. Association of insulin resistance With schizophrenia polygenic risk score and response to antipsychotic treatment. JAMA. Psychiatry. 2019;76:864–867. doi: 10.1001/jamapsychiatry.2019.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, et al. Glucose and insulin-related traits, type 2 diabetes and risk of schizophrenia: a Mendelian randomization study. EBioMedicine. 2018;34:182–188. doi: 10.1016/j.ebiom.2018.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry BI, et al. The potential shared role of inflammation in insulin resistance and schizophrenia: a bidirectional two-sample mendelian randomization study. PLoS Med. 2021;18:e1003455. doi: 10.1371/journal.pmed.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khandaker GM, Dantzer R, Jones PB. Immunopsychiatry: important facts. Psychol. Med. 2017;47:2229–2237. doi: 10.1017/S0033291717000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 2004;25:4–7. doi: 10.1016/j.it.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Reinehr T. Inflammatory markers in children and adolescents with type 2 diabetes mellitus. Clin. Chim. Acta. 2019;496:100–107. doi: 10.1016/j.cca.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Song X, et al. Elevated levels of adiponectin and other cytokines in drug naïve, first episode schizophrenia patients with normal weight. Schizophr. Res. 2013;150:269–273. doi: 10.1016/j.schres.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 26.Hartwig FP, Borges MC, Horta BL, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample mendelian randomization study. JAMA. Psychiatry. 2017;74:1226–1233. doi: 10.1001/jamapsychiatry.2017.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai L, et al. Evidence that the pituitary gland connects type 2 diabetes mellitus and schizophrenia based on large-scale trans-ethnic genetic analyses. J. Transl. Med. 2022;20:1–12. doi: 10.1186/s12967-022-03704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan A, et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 2022;54:560–572. doi: 10.1038/s41588-022-01058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mägi R, et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trubetskoy V, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021;53:840–860. doi: 10.1038/s41588-021-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bycroft C, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinf. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerimov N, et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 2021;53:1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinf. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015 47:3. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson E, et al. Mendelian randomization. Nat. Rev. Methods Primers. 2022;2:1–21. doi: 10.1038/s43586-021-00092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani, G. et al. The MR-base platform supports systematic causal inference across the human phenome. eLife.10.7554/ELIFE.34408 (2018). [DOI] [PMC free article] [PubMed]
- 40.Richardson TG, Sanderson E, Elsworth B, Tilling K, Davey Smith G. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: Mendelian randomisation study. BMJ. 2020;369:m1203. doi: 10.1136/bmj.m1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanderson E, Spiller W, Bowden J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable mendelian randomization. Stat. Med. 2021;40:5434–5452. doi: 10.1002/sim.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383–e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. AJHG. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huckins LM, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 2019;51:659–674. doi: 10.1038/s41588-019-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deutsch CK, et al. Quantitative measures of craniofacial dysmorphology in a family study of schizophrenia and bipolar illness. Schizophr. Bull. 2015;41:1309–1316. doi: 10.1093/schbul/sbv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Mol. Psychiatry. 2004;10:27–39. doi: 10.1038/sj.mp.4001563. [DOI] [PubMed] [Google Scholar]
- 47.Wigger L, et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat. Metab. 2021;3:1017–1031. doi: 10.1038/s42255-021-00420-9. [DOI] [PubMed] [Google Scholar]
- 48.Gandal MJ, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foley CN, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 2021;12:764–764. doi: 10.1038/s41467-020-20885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinf. 2016;32:286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 51.Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science10.1126/SCIENCE.AAT8464/SUPPL_FILE/AAT8464-WANG-SM.PDF (2018). [DOI] [PMC free article] [PubMed]
- 52.Cunningham F, et al. Ensembl 2022. Nucleic Acids Res. 2021;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers, T. A., Chanock, S. J. & Machiela, M. J. LDlinkR: An R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front. Genet.10.3389/fgene.2020.00157 (2020). [DOI] [PMC free article] [PubMed]
- 54.Raulerson CK, et al. Adipose tissue gene expression associations reveal hundreds of candidate genes for cardiometabolic traits. Am. J. Hum. Genet. 2019;105:773–787. doi: 10.1016/j.ajhg.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamburov A, et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med.10.1126/SCITRANSLMED.AAG1166/SUPPL_FILE/AAG1166_TABLE_S1.ZIP (2017). [DOI] [PMC free article] [PubMed]
- 57.Ochoa D, et al. The next-generation open targets platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 2022;51:D1353–D1359. doi: 10.1093/nar/gkac1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howe LJ, Tudball M, Davey Smith G, Davies NM. Interpreting Mendelian-randomization estimates of the effects of categorical exposures such as disease status and educational attainment. Int. J. Epidemiol. 2021;51:948–957. doi: 10.1093/ije/dyab208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson TG, et al. Childhood body size directly increases type 1 diabetes risk based on a lifecourse Mendelian randomization approach. Nat. Commun. 2022;13:2337. doi: 10.1038/s41467-022-29932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park EJ, et al. Mutation of Nogo-B receptor, a subunit of <em>cis</em>-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 2014;20:448–457. doi: 10.1016/j.cmet.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrison KD, et al. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 2009;10:208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian S, Chait A. Hypertriglyceridemia secondary to obesity and diabetes. Biochim. Biophys. Acta. (BBA) Mol. Cell Biol. Lipids. 2012;1821:819–825. doi: 10.1016/j.bbalip.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Yu SH, et al. Lysosomal cholesterol accumulation contributes to the movement phenotypes associated with NUS1 haploinsufficiency. Genet. Med. 2021;23:1305–1314. doi: 10.1038/s41436-021-01137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Den K, et al. Recurrent NUS1 canonical splice donor site mutation in two unrelated individuals with epilepsy, myoclonus, ataxia and scoliosis—a case report. BMC Neurol. 2019;19:253. doi: 10.1186/s12883-019-1489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamdan FF, et al. High rate of recurrent De Novo mutations in developmental and epileptic encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szigeti K, et al. Functional, histopathologic and natural history study of neuropathy associated with EGR2 mutations. Neurogenetics. 2007;8:257–262. doi: 10.1007/s10048-007-0094-0. [DOI] [PubMed] [Google Scholar]
- 67.Turman JE, Jr, Chopiuk NB, Shuler CF. The Krox-20 null mutation differentially affects the development of masticatory muscles. Dev. Neurosci. 2001;23:113–121. doi: 10.1159/000048703. [DOI] [PubMed] [Google Scholar]
- 68.Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005;1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 69.Fabbri C, Serretti A. Genetics of long-term treatment outcome in bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016;65:17–24. doi: 10.1016/j.pnpbp.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 70.Neale, B. UKBB GWAShttps://www.nealelab.is/uk-biobank (2018).
- 71.Aumailley M. The laminin family. Cell Adh. Migr. 2013;7:48–55. doi: 10.4161/cam.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shan N, et al. Laminin α4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis and is lowered in preeclamptic placentas. Placenta. 2015;36:809–820. doi: 10.1016/j.placenta.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Stalmans P, et al. Enzymatic vitreolysis with ocriplasmin for vitreomacular traction and macular holes. N. Engl. J. Med. 2012;367:606–615. doi: 10.1056/NEJMoa1110823. [DOI] [PubMed] [Google Scholar]
- 74.Tong L. Structure and function of biotin-dependent carboxylases. Cell Mol. Life Sci. 2013;70:863–891. doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veprik A, et al. Acetyl-CoA-carboxylase 1 (ACC1) plays a critical role in glucagon secretion. Commun. Biol. 2022;5:238. doi: 10.1038/s42003-022-03170-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horisberger, J. D. & Geering, K. Brain Na,K-ATPase. in Encyclopedia of Neuroscience 3rd edn (ed. Squire, L. R.) (Academic Press, Oxford, 2009).
- 77.Bluschke V, Bonn R, Greeff K. Increase in the (Na+ + K+)-ATPase activity in heart muscle after chronic treatment with digitoxin or potassium deficient diet. Eur. J. Pharmacol. 1976;37:189–191. doi: 10.1016/0014-2999(76)90021-2. [DOI] [PubMed] [Google Scholar]
- 78.Spigset O, Mjörndal T. Increased glucose intolerance related to digoxin treatment in patients with type 2 diabetes mellitus. J. Intern. Med. 1999;246:419–421. doi: 10.1046/j.1365-2796.1999.00587.x. [DOI] [PubMed] [Google Scholar]
- 79.Schleifer SJ, et al. Digitalis and β-blocking agents: effects on depression following myocardial infarction. Am. Heart J. 1991;121:1397–1402. doi: 10.1016/0002-8703(91)90144-7. [DOI] [PubMed] [Google Scholar]
- 80.Tsyvunin V, Shtrygol S, Shtrygol D. Digoxin enhances the effect of antiepileptic drugs with different mechanism of action in the pentylenetetrazole-induced seizures in mice. Epilepsy Res. 2020;167:106465. doi: 10.1016/j.eplepsyres.2020.106465. [DOI] [PubMed] [Google Scholar]
- 81.Eisendrath, T. S. J, Sweeney, M. A. Toxic neuropsychiatric effects of digoxin at therapeutic serum concentrations. Am. J. Psychiatry.144, 506-7 (1987). [DOI] [PubMed]
- 82.Endele S, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 83.Kleckner NW, Glazewski JC, Chen CC, Moscrip TD. Subtype-selective antagonism of N-methyl-D-aspartate receptors by felbamate: insights into the mechanism of action. J. Pharmacol. Exp. Ther. 1999;289:886–894. [PubMed] [Google Scholar]
- 84.Zhuravliova E, Barbakadze T, Natsvlishvili N, Mikeladze DG. Haloperidol induces neurotoxicity by the NMDA receptor downstream signaling pathway, alternative from glutamate excitotoxicity. Neurochem. Int. 2007;50:976–982. doi: 10.1016/j.neuint.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 85.Beck K, et al. Association of ketamine with psychiatric symptoms and implications for Its therapeutic use and for understanding schizophrenia: a systematic review and meta-analysis. JAMA. Network Open. 2020;3:e204693–e204693. doi: 10.1001/jamanetworkopen.2020.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ralevski E, et al. Treatment with acamprosate in patients with schizophrenia spectrum disorders and comorbid alcohol dependence. J. Dual Diagn. 2011;7:64–73. doi: 10.1080/15504263.2011.569440. [DOI] [PubMed] [Google Scholar]
- 87.Poltavskaya, E. G. et al. Study of early onset schizophrenia: associations of GRIN2A and GRIN2B polymorphisms. Life10.3390/life11100997 (2021). [DOI] [PMC free article] [PubMed]
- 88.Yang Y, et al. Association study of N-Methyl-D-Aspartate receptor subunit 2B (GRIN2B) polymorphisms and schizophrenia symptoms in the han chinese population. PLoS ONE. 2015;10:e0125925. doi: 10.1371/journal.pone.0125925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhana N, Spencer CM. Lanoteplase. BioDrugs. 2000;13:217–224. doi: 10.2165/00063030-200013030-00006. [DOI] [PubMed] [Google Scholar]
- 90.van den Hurk M, et al. Druggable transcriptomic pathways revealed in Parkinson’s patient-derived midbrain neurons. npj Parkinson’s Dis. 2022;8:134. doi: 10.1038/s41531-022-00400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fleischhacker WW, et al. Comorbid somatic illnesses in patients with severe mental disorders: clinical, policy and research challenges. J. Clin. Psychiatry. 2008;69:514–519. doi: 10.4088/JCP.v69n0401. [DOI] [PubMed] [Google Scholar]
- 92.Wahlbeck K, Forsén T, Osmond C, Barker DJP, Eriksson JG. Association of schizophrenia with low maternal body mass index, small size at birth and thinness during childhood. AMA Arch. Gen. Psychiatry. 2001;58:48–52. doi: 10.1001/archpsyc.58.1.48. [DOI] [PubMed] [Google Scholar]
- 93.Ochoa, D. et al. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat. Rev. Drug Discov.10.1038/D41573-022-00120-3 (2022) [DOI] [PubMed]
- 94.Ashton CH. Psychiatric effects of drugs for other disorders. Medicine. 2008;36:501–504. doi: 10.1016/j.mpmed.2008.06.002. [DOI] [Google Scholar]
- 95.de Klein N, et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat. Genet. 2023;55:377–388. doi: 10.1038/s41588-023-01300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ng B, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Consortium TG, et al. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bryois J, et al. Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nat. Neurosci. 2022;25:1104–1112. doi: 10.1038/s41593-022-01128-z. [DOI] [PubMed] [Google Scholar]
- 99.Hannon E, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Brien HE, et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Viñuela, A. Genetic variant effects on gene expression in human pancreatic islets and their implications for T2D. Nat. Commun. 10.1038/s41467-020-18581-8 (2020). [DOI] [PMC free article] [PubMed]
- 102.Atla G, et al. Genetic regulation of RNA splicing in human pancreatic islets. Genome Biol. 2022;23:196. doi: 10.1186/s13059-022-02757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He B, Shi J, Wang X, Jiang H, Zhu H-J. Genome-wide pQTL analysis of protein expression regulatory networks in the human liver. BMC Biol. 2020;18:97. doi: 10.1186/s12915-020-00830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brotman SM, et al. Subcutaneous adipose tissue splice quantitative trait loci reveal differences in isoform usage associated with cardiometabolic traits. Am. J. Hum. Genet. 2022;109:66–80. doi: 10.1016/j.ajhg.2021.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original code used to generate the results presented in this paper is available at GitHub and deposited on Zenodo (10.5281/zenodo.10016486).