Abstract
PML is a nuclear protein with growth-suppressive properties originally identified in the context of the PML-retinoic acid receptor α (RARα) fusion protein of acute promyelocytic leukemia. PML localizes within distinct nuclear structures, called nuclear bodies, which are disrupted by the expression of PML-RARα. We report that PML colocalizes with the nonphosphorylated fraction of the retinoblastoma protein (pRB) within nuclear bodies and that pRB is delocalized by PML-RARα expression. Both PML and PML-RARα form complexes with the nonphosphorylated form of pRB in vivo, and they interact with the pocket region of pRB. The regions of PML and PML-RARα involved in pRB binding differ; in fact, the B boxes and the C-terminal region of PML, the latter of which is not present in PML-RARα, are essential for the formation of stable complexes with pRB. Functionally, PML abolishes activation of glucocorticoid receptor-regulated transcription by pRB, whereas PML-RARα further increases it. Our results suggest that PML may be part of transcription-regulatory complexes and that the oncogenic potential of the PML-RARα protein may derive from the alteration of PML-regulated transcription.
Acute promyelocytic leukemia (APL) is characterized by a clonal expansion of myeloid precursors blocked at the promyelocytic stage. A chromosome translocation involving the PML gene on chromosome 15 and the retinoic acid receptor α (RARα) gene on chromosome 17 is found in over 95% of APL cases (1, 7, 16). The resulting PML-RARα fusion gene encodes a PML-RARα fusion protein that is implicated in the pathogenesis of the disease (17, 41, 42, 58). In fact, PML-RARα transgenic mice develop abnormal myelopoiesis with phenotypic features of APL (8, 31), and in vitro, PML-RARα expression has biological activities which are consistent with the promyelocytic leukemia phenotype, e.g., block of terminal differentiation and increased survival (29).
PML is a ubiquitously expressed, matrix-associated nuclear phosphoprotein whose overexpression induces growth suppression (11, 23, 55). However, its physiological function and biochemical activities remain unknown. PML is a member of a growing family of proteins characterized by the presence of a RING domain, two additional Cys/His-rich regions (B1 and B2 boxes), and an α-helical coiled-coil domain (5, 6, 50, 61). Within the latter, four clusters of heptads of hydrophobic amino acids define a dimerization interface through which PML forms homodimers and, in APL cells, heterodimers with PML-RARα (42, 59).
PML is localized within discrete nuclear structures referred to as nuclear bodies (NBs), ND10, Kr bodies, or promyelocytic leukemia oncogenic domain (19, 45, 71). Other components of the PML NBs are Sp100 (68), NDP55 (2), Int-6 (15), and PIC-1 (4). The integrity of the PML NBs is lost in APL cells: PML-RARα localizes to novel nuclear structures (so-called microspeckles) and causes the delocalization of PML and other components of the NBs (19, 45, 71). The disorganization of the NB structure is thought to be relevant to the pathogenesis of the APLs since retinoic acid treatment, which reverts the differentiation block of the APL blasts in vitro and causes disease remission in vivo, induces degradation of the PML-RARα protein and the consequent assembly of the PML NBs (for a review, see reference 28).
Altered localization of PML and structural changes of the NBs have also been shown to occur during DNA virus infection (10, 18, 21, 43). Some viral proteins, such as herpes simplex virus type 1 Vmw110 and adenovirus E4-ORF3, have been described to be directly involved in the redistribution of NB components (10, 18, 21), whereas Epstein-Barr virus (EBV) EBNA-5 protein has been described to colocalize with PML within morphologically intact NBs (66). Interestingly, PML expression as well as the size and number of the PML NBs increase after treatment of cells with the antiviral agent interferon (12, 46). In summary, a number pieces of indirect evidence suggest that PML and/or the PML NBs are involved in growth control and are the targets of DNA viral infection. Identification of proteins that interact with PML within NBs might help in defining the role of PML and PML NBs in normal and leukemic cells. We investigated the physical and functional interactions of PML with the retinoblastoma gene product (pRB).
pRB regulates cell proliferation by controlling a set of transcription factors (the E2F family of proteins) that activate genes involved in the G1/S transition (70). In the early G1 phase of the cell cycle, pRB is unphosphorylated and stably complexed with E2F; as cells pass the G1/S boundary, pRB becomes phosphorylated, resulting in the functional release of E2F (70). pRB has also been described to regulate the activity of promoters that depend on other transcription factors, such as SP1 or glucocorticoid receptor (44, 65, 69). However, the physiological consequences of these activities of pRB remain unknown.
The analysis of pRB subcellular localization suggests that pRB is distributed in at least two distinct subnuclear compartments, one diffuse and one corresponding to circumscribed granules (54, 67) which morphologically resemble NBs. Interestingly, the EBNA-5 protein has been demonstrated to colocalize with pRB within distinct nuclear foci in EBV-infected lymphoblastoid cells (38).
The similarity in subnuclear localization, together with the shared property of inducing growth suppression, prompted us to investigate the physical and functional interactions between PML and pRB. We report here that PML and pRB colocalize within the PML NBs and that PML forms complexes with the unphosphorylated form of pRB. Functionally, PML and pRB do not appear to be mutually necessary to exert their respective growth suppressor activities, while PML displays an inhibitory effect on pRB-regulated transcriptional activation of glucocorticoid receptor (GR)-responsive promoters. PML-RARα expression causes the delocalization of pRB from the PML NB and shows stimulatory activity on the GR-responsive promoters.
MATERIALS AND METHODS
Plasmids and expression vectors.
pCMV-pRB, pCMV-RBΔ21, and pCMV-RBΔ22 expression vectors have been previously described (24, 60). The pCMV-PML3 expression vector was generated by cloning the previously described PML3 cDNA (22) in the BamHI site of the pCMV-Neo-BamHI vector (3). The pSG5-PML/RARα and pCDNA3-PML/RARα expression vectors were obtained by cloning the PML-RARα coding sequence (58) in the EcoRI site of the pSG5 and pCDNA3 expression vectors, respectively. For co-in vitro translation studies, PML3 (22), hemagglutinin epitope (HA)-tagged PML-RARα (P/R-HA [30]), and pRB were subcloned in the pGEM3 plasmid vector (Promega). The ΔH and ΔC mutants were derived from the PML3 clone M58 (22), using the strategy described elsewhere (30). The ΔRING mutant was obtained by deleting the 5′ PstI fragment of PML and generating an HA fusion cDNA, using BamHI linkers. The resulting fragment was cloned in the pCDNA1 vector and encodes an HA-PML fusion protein containing the PML3 sequences starting from amino acid 103. The ΔB1B2 in pSG5 (a gift of A. Dejean) was generated by PCR deletion of the sequence encoding PML amino acids 129 to 227. The ΔB1B2-PML3 construct used in this study was obtained by ligating the 5′ EcoRI-KpnI fragment of the ΔB1B2 construct in pSG5 to the 3′ KpnI-EcoRI fragment of PML3 in the pCDNA3 expression vector. All PML-RARα deletion mutants have been described elsewhere (30). For generation of overexpressing inducible cell lines, PML3, PML2, PML-P/R, PML-RARα, ΔH-P/R, and ΔC-P/R cDNAs were cloned in the Zn-inducible mouse metallothionein promoter expression vector (29). The pCDNA3-Sp100 expression vector was obtained by subcloning the entire Sp100 coding sequence from the pSG5 expression vector (a gift of H. de Thè) into the EcoRI site of pCDNA3.
Cell culture and transfection.
U937 human myeloid leukemia cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. For the production of overexpressing clones, cells were transfected with the PML3, PML2, PML-P/R, PML-RARα, ΔH-P/R, and ΔC-P/R inducible expression vectors by electroporation as described previously (29). After electroporation, the cells were selected in G418 and subcloned under limiting dilution conditions. Expression of the exogenous protein was evaluated by Western blotting after 6 to 12 h of induction with 100 mM ZnSO4, using the PG-M3 (26) or anti-RARα-F (gift from P. Chambon, Strasbourg, France) antibody, and revealed by the enhanced chemiluminescence (ECL) method (ECL kit; Amersham).
C33A human cervix carcinoma cells (ATCC HTB 31) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and transfected by the calcium phosphate precipitation procedure (63).
The EBV-immortalized IB4 cord blood cell line was maintained in Iscove’s medium containing 5% fetal calf serum.
Coimmunoprecipitation and in vitro binding experiments.
Two cellular systems were used for coimmunoprecipitation experiments.
(i) U937 cell clones overexpressing PML3, PML2, PML-P/R, PML-RARα, ΔC-PML/RARα, and ΔH-PML/RARα were used to assess the association of the corresponding overexpressed proteins with the endogenous pRB, p107, and p130. A total of 5 × 108 cells were diluted to a concentration of 5 × 105 cells/ml, and protein expression was induced with 100 μM ZnSO4 for 6 to 12 h. Cells were collected in E1A buffer (HEPES, 50 mM; NaCl, 250 mM, EDTA, 5 mM; dithiothreitol, 1 mM; Nonidet P40, 0.1%; phenylmethylsulfonyl fluoride [Sigma], 1 mM; leupeptin [Sigma], 1 mg/ml; aprotinin [Sigma], 1 mg/ml). The cell suspension was briefly sonicated, and the lysates were clarified by centrifugation. Lysates were precleared by incubation for 1 h with protein A-Sepharose (Pharmacia). Immunoprecipitation was obtained by adding to the precleared lysate protein A-Sepharose (Pharmacia) and the relevant serum. Sepharose beads were washed four times in 1× NET buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 0.25% gelatin, 0.02% sodium azide) and resuspended in sodium dodecyl sulfate (SDS) sample buffer. Immunoprecipitates were Western blotted with the indicated antisera by the ECL method.
(ii) C33A cells transiently transfected with 10 μg of expression vectors for PML3, ΔC, ΔH, ΔRING, ΔB1B2, pRB, RBΔ21, RBΔ22, and pSG5-PML/RARα were used for the remaining experiments. At 48 h after transfection, cells were washed in phosphate-buffered saline, collected in E1A buffer, and immunoprecipitated as described above.
For in vitro binding experiments, bacterially expressed glutathione S-transferase (GST) fusion proteins were prepared according to standard procedures (63). For each binding reaction, 5 μg of GST fusion protein bound to glutathione-Sepharose beads (Pharmacia) was incubated for 1 h in ice with 500 μg of relevant lysates prepared in E1A buffer or with in vitro-translated proteins. After 10 washes in 1× NET buffer, Sepharose beads were resuspended in SDS sample buffer and analyzed by Western blotting.
Antibodies.
PML antisera used in this study were monoclonal antibody PG-M3 (26), polyclonal antibody 2912A, directed against the PML N terminus, and polyclonal antibody 2417, raised against a peptide corresponding to amino acids 592 to 607 of the PML3 sequence (22). Immunoprecipitation of pRB was performed with monoclonal antibody XZ77 (35); Western blotting analysis was performed with G3-245 (Pharmingen). The anti-pRB monoclonal antibody aRB1C1 used in immunofluorescence studies has been previously described (67). It was raised against a TrpE-pRB fusion protein contain pRB sequences between amino acids 300 and 928. p107 was analyzed with the monoclonal antibody SD9 (20), and p130 was analyzed with polyclonal antibody C-20 (Santa Cruz). PML-RARα was immunoprecipitated either with monoclonal antibody PG-M3 or with anti-RARα polyclonal antibody C-20 (Santa Cruz); Western blotting analysis was performed with the anti-RARα-F antibody (gift from P. Chambon).
Immunofluorescence staining.
IB4 and U937 cells were used for PML-pRB staining; for PML-RARα-pRB immunofluorescence experiments, we used U937 clone PR9 after 12 h of 100 μM ZnSO4 treatment. The cells were cytocentrifuged and fixed in methanol at room temperature for 5 min followed by acetone at −20°C for 2 min. PML and PML-RARα stainings were performed with an anti-PML polyclonal antibody (2912A); pRB staining was performed with monoclonal antibody aRB1C1 (67). After extensive washes in phosphate-buffered saline, the cells were stained with fluorescein isothiocyanate (FITC)- or rhodamine-conjugated anti-mouse or anti-rabbit immunoglobulin antibodies (Southern Biotechnology Associates). Preparations were examined on an Olympus BX-60 fluorescence microscope equipped with a chilled digital color camera (C5810 3CCD; Hamamatsu Photonics). Images were captured with a 24-bit board (Image grabber 24; Neotech) on a 8100/80 Power Macintosh personal computer (Apple). For colocalization experiments, the same microscopic fields were examined with distinct cubes for fluorescein (excitation filter, 470 to 490 nm; diachronic mirror, 505 nm; barrier filter, 515 to 550 nm) and rhodamine (excitation filter, 510 to 550 nm; diachronic mirror 570 nm; barrier filter, 590 nm). The images were directly superimposed by the C5810 3CCD control unit.
Co-in vitro translation.
In vitro translation experiments were performed with the Promega TNT coupled reticulocyte lysate system as specified by the manufacturer. Of the 25 μl of the final reaction mixture, 1 μl was conserved in SDS loading buffer for a control, and two aliquots of 12 μl each were diluted to 100 μl with E1A buffer. Translation products were immunoprecipitated with the corresponding antisera and loaded on an SDS-acrylamide gel.
Colony formation assays.
C33A cells were transfected with 10 μg of pCMV-pRB or equimolar quantities of the other expression vectors, normalized to a total content of 20 μg of DNA per transfection with pGEM3 DNA. At 48 h after transfection, each plate was trypsinized and replated at dilutions 1:50, 1:100, and 1:500. After an additional 24 h, G418 was added to the medium at 750 μg/ml (active concentration). After 14 to 16 days of selection, G418-resistant colonies were colored with crystal violet and counted.
Transactivation experiments.
HeLa cells were plated on the day prior to transfection (2.5 × 105 cells per 60-mm-diameter dish) and were transfected by calcium phosphate precipitation (63) with the following expression vectors: 5 μg of reporter gene plasmid MTV-LTR-CAT (9, 53, 56), 1 μg of plasmid RSV-hGR (gifts of H. Samuels), and 500 ng of pCMV-β-gal (Clontech, Palo Alto, Calif.) with or without pSG5-pRB (8 μg), pSG5-PML/RARα (2 μg), or pMT2-PML3 (2 μg). Plasmid pGEM3 was used as carrier to bring the total amount of DNA to 17 μg. The medium was changed after 16 h, and transfected cells were collected 24 h later. One micromolar dexamethasone was added at the time of transfection and maintained after the medium was changed, for a total induction of 40 h before extracts were assayed for chloramphenicol acetyltransferase (CAT) activity. Results were normalized by measuring β-galactosidase activity. CAT activity was measured according to published procedures (27, 64).
RESULTS
PML and pRB colocalize within NBs.
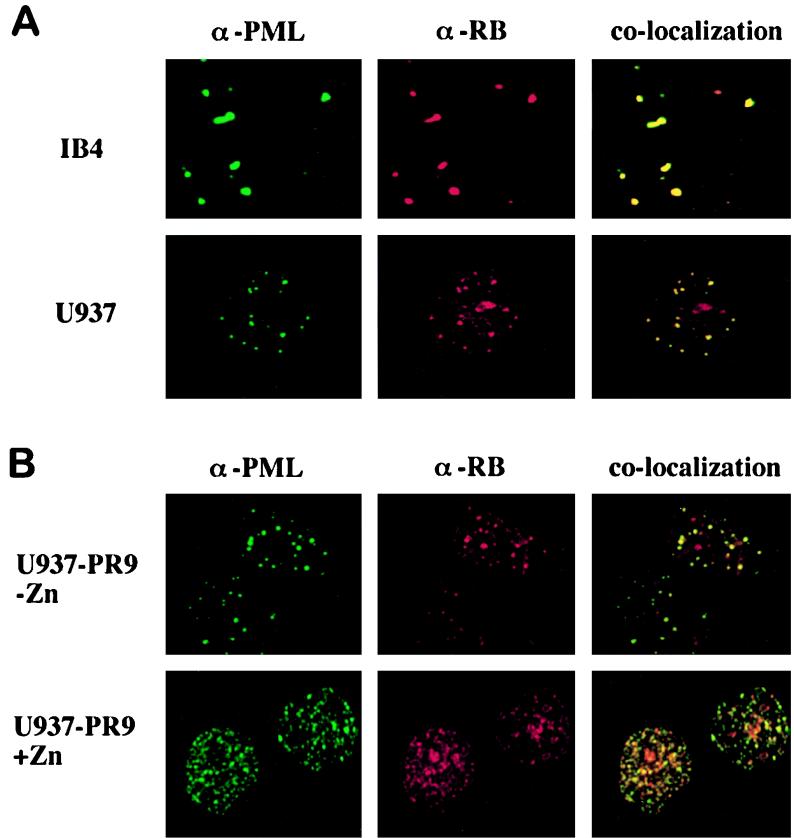
EBV EBNA-5 protein has been described to colocalize with PML within NBs (66). EBNA-5 protein has also been demonstrated to colocalize with pRB within distinct nuclear substructures, morphologically resembling PML NBs, in EBV-infected lymphoblastoid cells (38). We therefore investigated the possibility that pRB colocalizes with PML within the NBs by performing immunofluorescence experiments with the IB4 EBV-immortalized cord blood and U937 monocytic cell lines. Cells were fixed with methanol-acetone and stained with a polyclonal anti-PML antibody (2912A) revealed with a FITC-conjugated anti-rabbit secondary antibody and with anti-pRB monoclonal antibody aRB1C1, which has been reported to specifically stain the fraction of pRB which localizes within distinct nuclear foci (67), revealed with a rhodamine-conjugated anti-mouse secondary antibody. Figure 1A shows the results of immunofluorescence experiments performed with the IB4 and U937 cells. In both cell lines, anti-PML and anti-pRB stainings revealed specific labeling of nuclear bodies (Fig. 1A). Superimposition of PML and pRB staining showed that approximately 80% of PML NBs colocalized with the pRB-containing NBs. Similar experiments were performed in U937 cells with other proteins of the pRB family (p107 and p103), referred to as pocket proteins, which share with pRB a structural domain (pocket) through which they bind viral proteins and a series of cellular factors involved in the control of proliferation (36, 37, 39): p130 revealed a speckled nuclear pattern, while p107 appeared to be predominantly diffuse within the nucleus. No colocalization could be demonstrated with anti-PML and either anti-p130 or anti-p107 antibodies (data not shown).
FIG. 1.
pRB colocalizes with PML within nuclear bodies and is delocalized by PML-RARα expression. (A) Immunofluorescence experiments using an anti-pRB monoclonal antibody (α-RB; aRB1C1) revealed with a rhodamine-conjugated anti-mouse antibody and an anti-PML polyclonal antibody (α-PML; 2912A) revealed with an FITC-conjugated anti-rabbit antibody were performed with IB4 and U937 cells. Superimposition of PML and pRB staining is shown in the righthand panels, indicating colocalization of PML and pRB within NBs in both cell lines. (B) Similar experiments were performed with the U937 PR9 clone before and after induction of PML-RARα expression with ZnSO4. Superimposition of PML and pRB staining is shown in the righthand panels, revealing that pRB is delocalized into the PML-RARα microspeckles by the expression of the fusion protein. The anti-RB antibody aRB1C1 does not cross-react with PML-RARα, as revealed by Western blotting experiments and immunoprecipitations of in vitro-translated PML-RARα polypeptides (our unpublished results).
PML associates with the pRB complex.
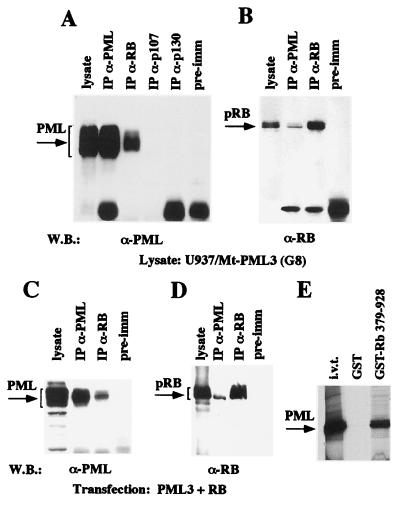
We next investigated if the observed colocalization of PML and pRB corresponded to the existence of nuclear complexes containing both PML and pRB. We also studied the possibility that PML could interact with p107 and p103. To investigate the possible associations, we performed coimmunoprecipitation experiments (Fig. 2A) using a U937 clone overexpressing the PML3 protein (clone G8), obtained by stably transfecting U937 cells with an expression vector containing the PML3 cDNA under the control of the zinc-inducible mouse metallothionein promoter. High levels of PML3 expression were obtained after treatment of the cells for 12 h with 100 μM ZnSO4. Lysates from zinc-induced G8 cells were precleared with protein A-Sepharose and immunoprecipitated with anti-PML (PG-M3), anti-pRB (XZ77), anti-p107 (SD9), and anti-p130 (C-20) antibodies or rabbit preimmune serum as described in Materials and Methods. The resulting immunoprecipitates were analyzed by Western blotting for the presence of PML3 with an anti-PML3 antibody (Fig. 2A). The presence of pRB, p107, and p130 proteins in the corresponding immunoprecipitates was controlled by decorating the same membrane with the antibodies (data not shown). PML was present only after immunoprecipitation with the anti-PML or anti-pRB antibody, suggesting that PML binds to pRB complexes. The PML-pRB complex was also revealed by anti-pRB Western blotting of an anti-PML immunoprecipitate (Fig. 2B). Similar results were obtained in coimmunoprecipitation experiments with lysates from C33A cells transiently cotransfected with expression vectors pCMV-PML3 and pCMV-pRB (Fig. 2C and D). Note that in both cellular systems, it appears that PML coprecipitates with the fastest-migrating form of pRB, which has been previously identified as the nonphosphorylated form of the protein (48).
FIG. 2.
PML associates with the pRB complex. (A) PML coimmunoprecipitates with pRB but not with other pocket proteins. Cell lysates from the PML3-overexpressing U937 G8 clone were immunoprecipitated (IP) with anti-PML3 (α-PML) polyclonal (2417), anti-pRB (α-RB) monoclonal (XZ77), anti-p107 (α-p107) monoclonal (SD9), and anti-p130 (α-p130) polyclonal (C-20) antibodies and a preimmune (pre-imm) rabbit serum. The arrow on the left indicates PML3, revealed by Western blotting (W.B.) with anti-PML3 polyclonal antibody 2417. (B) PML coprecipitates with the nonphosphorylated pRB. Anti-PML immunoprecipitates from U937 (G8) cell lysates were analyzed by Western blotting with anti-pRB monoclonal antibody G3-245. The arrow on the left indicates pRB. (C and D) PML and pRB coimmunoprecipitate in transiently transfected cells. C33A cells were transiently transfected with the pCMV-PML3 and pCMV-pRB expression vectors, and lysates were immunoprecipitated with the anti-PML monoclonal (PG-M3) or the anti-pRB polyclonal (XZ77) antibody. Membranes were decorated with anti-PML3 antibody 2417 (C) or anti-pRB antibody G3-245 (D). Brackets on the left correspond to PML3 (C) and pRB (D). (E) In vitro binding analysis of bacterially expressed GST or GST-RB 379-928 (Fig. 3A) with in vitro-translated (i.v.t.) PML3 polypeptides.
We next tried to reconstitute the PML-pRB complex in vitro by performing GST pulldown experiments using in vitro-translated PML3 and GST-pRB recombinant protein GST-RB 379-928 (Fig. 3A). These experiments demonstrated that GST-pRB efficiently binds in vitro-translated PML3 (Fig. 2E).
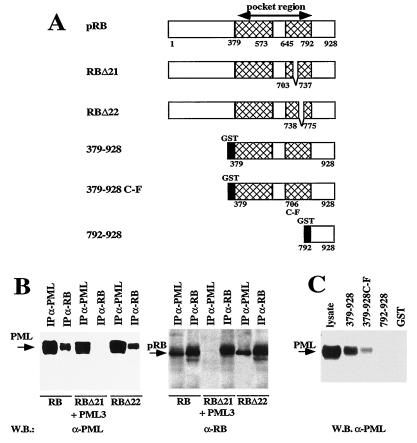
FIG. 3.
The PML binding domain of pRB is within the pocket region. (A) Schematic representation of the pRB mutants used for the analysis. RBΔ21 and RBΔ22 represent deletion mutants lacking pRB exons 21 and 22, respectively, cloned in eukaryotic expression vector pCMV-Neo-BamHI for in vivo association studies. 379-928, 379-928 C-F, and 792-928 are different portions of the pRB C-terminal region expressed in bacteria as GST fusion proteins used for in vitro binding experiments (40, 52). (B) In vivo analysis of the association between PML and pRB deletion mutants. Coimmunoprecipitation experiments were performed with the antisera indicated above the blots (abbreviations are as defined in the legend to Fig. 2), using lysates from C33A cells transiently transfected with pCMV-PML3 and either the pCMV-RB, pCMV-RBΔ21, or pCMV-RBΔ22 expression vector. Left, analysis with the anti-PML3 antibody 2417; right, analysis with anti-pRB antibody G3-245. (C) In vitro binding analysis of GST-RB fusion proteins with a lysate from U937 (G8) cells overexpressing PML3. The GST-pRB proteins are shown in panel A; the procedure is described in Materials and Methods. The membrane was decorated with the anti-PML3 antibody 2417.
Taken together, these results suggest that PML and nonphosphorylated pRB form stable complexes in vivo and that these complexes can be reconstituted in vitro.
The pocket region of pRB is necessary for PML association.
Many cellular and viral proteins have been shown to bind to the nonphosphorylated pRB, within a region defined as the T/E1A binding or pocket region (36, 37, 39) (Fig. 3A). To investigate if the pocket region of pRB is relevant for association of PML to the pRB complex we performed coimmunoprecipitation experiments using C33A cells transiently cotransfected with pCMV-PML3 and with two pRB mutants (RBΔ21 and RBΔ22), bearing deletions of the sequences corresponding to exons 21 and 22, respectively, which encode portions of the pRB pocket region (Fig. 3A and B). PML binds RBΔ22 as efficiently as wild-type pRB but loses completely the capacity to bind RBΔ21. These results were supported by in vitro binding experiments using GST-pRB fusion proteins containing various pRB domains (Fig. 3A) incubated with a lysate from the G8 clone overexpressing PML3 (Fig. 3C). GST-pRB 379-928, which bears the entire wild-type pocket region of pRB plus the C-terminal region, exhibited efficient binding to PML (Fig. 3C). GST-RB 379-928 C-F, which bears a point mutation within the pocket region that abolishes pRB binding to E1A, revealed a diminished binding to PML. GST-RB 792-928, which bears the C terminus only, did not bind PML at all. These results suggest that PML binds to the pRB pocket region with structural determinants within the region from amino acids 703 to 737.
Multiple regions of the PML protein are relevant for interaction with the pRB complex.
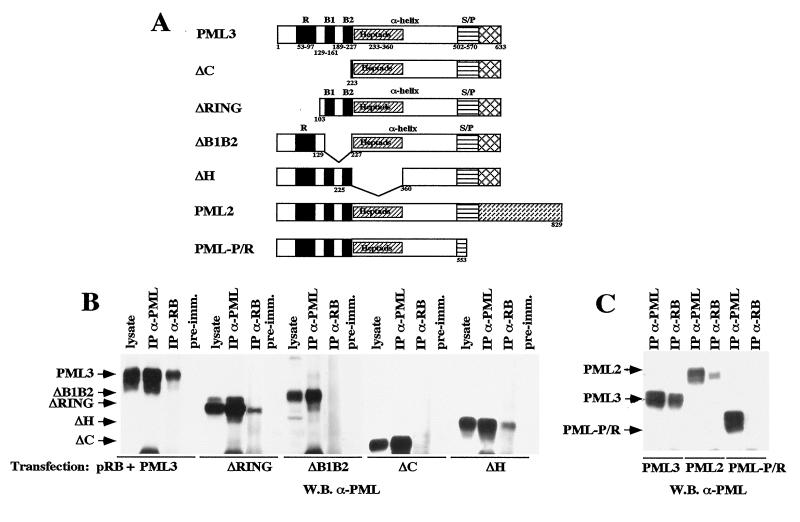
We next investigated the regions of PML involved in complex formation with pRB. PML contains an amino-terminal tripartite motif (RING region, B1-B2 boxes, and coiled-coil region) and variable C termini that define four PML isoforms (22). The tripartite motif is retained within the PML-RARα fusion protein (57).
To evaluate the role of the PML N-terminal region in the formation of complexes with pRB, we studied the potential of various PML mutants lacking the RING and B1-B2 (ΔC) regions or the coiled-coil (ΔH) region (Fig. 4A) to coimmunoprecipitate with pRB. These deletions do not grossly alter the function of PML, as shown by the fact that these mutants retain the capacity to inhibit cell growth (23). We performed coimmunoprecipitation experiments using lysates from C33A cells transiently transfected with pRB and either the ΔH or ΔC mutant. Deletion of the PML coiled-coil region reduced pRB binding, whereas deletion of the PML RING and B1-B2 regions completely abolished the association (Fig. 4B). To further map the PML amino-terminal region involved in pRB complex formation, we generated PML mutants lacking either the RING region (ΔRING) or the B1+B2 boxes (ΔB1B2) (Fig. 4A). Both mutants were cloned into a eukaryotic expression vector (pCDNA) and cotransfected with pCMV-RB into C33A cells. Anti-PML 2417 and anti-pRB XZ77 immunoprecipitates from the corresponding cellular lysates were decorated with the anti-PML3 antibody 2417. The ΔRING mutant revealed less capacity than the wild-type PML3 to complex pRB, while the ΔB1B2 mutant showed no binding (Fig. 4B). These results suggest that the B1 and B2 boxes are indispensable for the formation of the PML-RB complex and that integrity of the tripartite motif (RING, B1-B2, and coiled coil) is required for optimal stability of the complex.
FIG. 4.
Determination of the PML regions involved in the association with pRB. (A) Schematic representation of the PML mutants (ΔH, ΔC, ΔRING, ΔB1B2, and PML-PR) and the two PML isoforms (PML2 and PML3) used in this study. The major PML domains are shown above or within the maps; the relevant amino acid boundaries are shown below. (B) The PML N terminus is involved in pRB binding. Coimmunoprecipitation experiments were performed with lysates from C33A cells transfected with pRB and either the PML3, ΔH, ΔC, ΔRING, or ΔB1B2 expression vector. The membrane was decorated with an anti-PML3 antibody directed against the C terminus (see the legend to Fig. 2 for abbreviations). (C) Relevance of the PML C terminus in pRB binding. Coimmunoprecipitation experiments were performed as described in Materials and Methods, using lysates from U937 clones expressing the PML3 and PML2 isoforms, which have different C termini, and the PML-P/R mutant, which bears a C-terminal deletion. The membrane was decorated with anti-PML antibody PG-M3, directed against the PML N terminus. Specific bands are indicated by arrows on the left.
To evaluate the role of the PML C-terminal region, we then analyzed the capacity to form complexes with pRB of a PML mutant lacking the C terminus (PML-PR) and two PML isoforms with different C termini (PML2 and PML3). Coimmunoprecipitation experiments were performed with lysates from U937 clones 6, G8, and 13, overexpressing the PML-PR mutant and the PML3 and PML2 isoforms, respectively. The PML-PR mutant revealed no binding to pRB; the PML2 isoform exhibited a weaker interaction with pRB compared to PML3 (Fig. 4C). These results suggest that the PML C-terminal region also contributes to the stability of the PML-pRB complex and that different PML isoforms have different capacities to complex with pRB, probably due to their different C termini.
The PML-RARα fusion protein retains the ability of PML to form complexes with pRB in vivo.
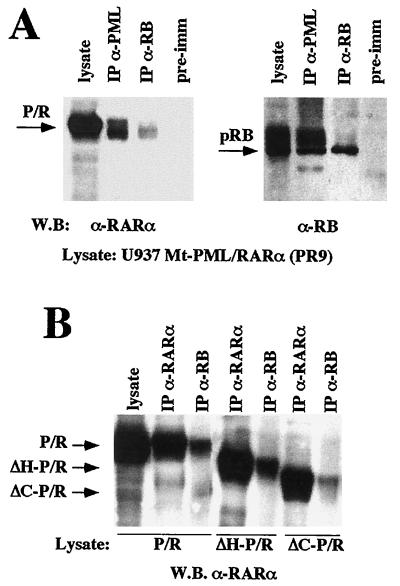
We then investigated whether the PML-RARα fusion protein maintains the capacity to form complexes with pRB by performing coimmunoprecipitation experiments using cell lysates from U937 clone PR9, which contains PML-RARα under the control of the zinc-inducible metallothionein promoter (29). These studies demonstrated that PML-RARα, like PML, also associates with the nonphosphorylated form of pRB (Fig. 5A). The PML-RARα-pRB association was unexpected since the mapping of the PML regions involved in pRB binding showed that the PML C terminus, which is lost in the fusion protein, is indispensable for complex formation. We therefore mapped the determinants of the PML-RARα-pRB interaction. Coimmunoprecipitation experiments using lysates of C33A cells cotransfected with pSG5-PML/RARα and mutant-encoding plasmids pCMV-RBΔ21 and pCMV-RBΔ22, and in vitro binding experiments using PML-RARα with the GST-pRB fusion proteins described above, demonstrated that PML-RARα binds to the pocket region of pRB (data not shown). To assess the relevance of the PML functional domains in the context of the PML-RARα fusion protein, we performed coimmunoprecipitation experiments with lysates of U937 clones overexpressing PML-RARα mutants bearing deletions of the portions described above for the PML mutants (ΔC-P/R and ΔH-P/R [30]) (Fig. 5B). The ΔH-P/R mutant, which lacks the PML coiled-coil region, was still capable of binding pRB. The ΔC-P/R mutant, which lacks the PML tripartite motif, binds pRB less efficiently.
FIG. 5.
PML-RARα associates with pRB. (A) Coimmunoprecipitation of pRB and PML-RARα was performed as described in Materials and Methods, using the lysate from a U937 clone expressing PML-RARα (PR9) (see the legend to Fig. 2 for abbreviations). Left, Western blot analysis of anti-PML (PG-M3) and anti-pRB (XZ77) immunoprecipitates with the anti-RARα anti-F antibody; right, Western blot analysis with the anti-pRB antibody G3-245. (B) PML-RARα mutants bearing deletions analogous to those described for PML in Fig. 3 within the PML moiety of PML-RARα (ΔC-PR and ΔH-PR [30]) were assayed for the capacity to bind pRB. Coimmunoprecipitation experiments were performed with lysates from U937 cell clones expressing the indicated PML-RARα mutants.
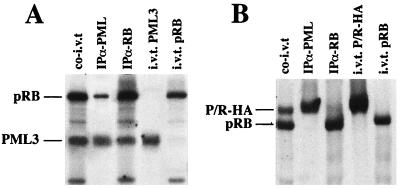
To further characterize these interactions, we performed in vitro binding studies. pRB protein was co-in vitro translated with PML3 (Fig. 6A) or P/R-HA (Fig. 6B). Translation products were then immunoprecipitated with anti-pRB (XZ77) and anti-PML (PG-M3) antibodies. PML3, but not PML-RARα, appeared to coprecipitate with pRB, indicating that the PML-pRB complex can be reconstituted in a reticulocyte lysate whereas the PML-RARα-pRB complex cannot. The PML2 isoform and the PML-PR mutant, like PML-RARα, failed to coimmunoprecipitate with pRB after co-in vitro translation (data not shown), confirming in vitro that the PML-pRB interaction depends on the C terminus of PML.
FIG. 6.
Reconstitution of the PML-pRB, but not of the PML-RARα-pRB, complex in rabbit reticulocyte lysates. pRB was in vitro translated together with PML3 (A) or P/R-HA (B), using rabbit reticulocyte lysates. The resulting products were immunoprecipitated with either anti-PML or anti-pRB antibodies, as indicated above the lanes (see the legend to Fig. 2 for abbreviations). A fraction of the co-in vitro translation product was loaded as a control (co-i.v.t.). In vitro translation products of the single proteins are loaded at the right of each panel, to exclude comigration of nonspecific bands.
Taken together, these data indicate that the PML-RARα-pRB complex involves some of the regions which are involved in the formation of the PML-pRB complex (the PML RING and B1-B2 regions and the pRB pocket) and leave open the question of how the PML-RARα-pRB complex is assembled, since the PML C terminus, which is indispensable for the formation of the PML-pRB complex, is not retained within the PML-RARα fusion protein.
PML-RARα disperses pRB from the NBs.
In APL cells, NBs are disrupted, and not only PML-RARα but also PML, Sp100, and other components of the NBs appear to be dispersed within the nucleus into a microspeckled pattern (19, 45, 71). To investigate whether PML-RARα expression is capable of dispersing pRB from the NBs, we performed immunofluorescence experiments using the U937-PR9 clone, before and after zinc induction of fusion protein expression (Fig. 1B). In uninduced cells, no PML-RARα protein is expressed and both PML and pRB staining reveals localization within NBs. After treatment of cells with 100 μM ZnSO4 for 12 h, PML-RARα protein is expressed at high levels and PML staining reveals a finely microspeckled pattern, while NBs disappear (Fig. 1B). aRB1C1 antibody staining of pRB revealed the same fine microspeckles, and superimposition of anti-PML and anti-pRB staining demonstrated apparent colocalization, suggesting that PML-RARα expression delocalizes pRB to the APL-specific microspeckles (Fig. 1B).
PML suppresses growth by a pRB-independent mechanism.
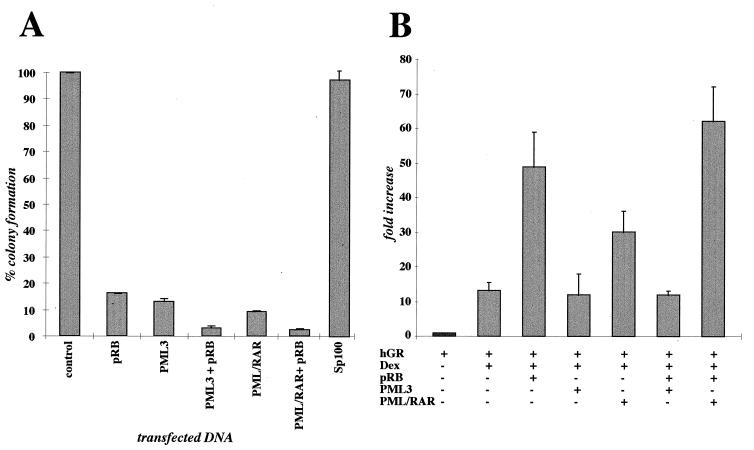
PML-RARα has a cell-type-specific effect on cell growth: it induces growth arrest of all nonhematopoietic cell lines and of the majority of the hematopoietic cell lines tested, while it promotes survival in a small subset of hematopoietic cell lines (23, 25). PML, instead, induces growth arrest in all the cell lines tested (23, 25). We investigated whether growth suppression induced by PML or PML-RARα was pRB dependent by performing colony formation assays in nonhematopoietic cells lacking functional pRB (C33A and SAOS-2). Another component of the NBs, Sp100 (68), was also tested. Briefly, cells defective for pRB were transfected with either pCMV-PML, pCDNA3-PML/RARα, pCMV-pRB, a combination of pCMV-pRB with either pCMV-PML or pCDNA3-PML/RARα, pCDNA3-Sp100, or the pCMV-Neo-BamHI vector (control). Cells were selected with G418 for 12 to 15 days, after which resistant colonies were scored. Figure 7A shows the mean values of four separate experiments performed with C33A cells (each experiment was performed in triplicate) where percentages of colonies formed in with plates transfected PML, pRB, PML plus pRB, PML-RARα, PML/RARα plus pRB, and Sp100 are compared to values for a vector-transfected control. PML, pRB, and PML-RARα expression alone greatly reduced colony formation (averages of 88, 82, and 90%, respectively). The combination of pRB and either PML or PML-RARα overexpression virtually abolished colony formation (<95%). Analogous results were obtained in three separate experiments performed with pRB−/− SAOS-2 cells (data not shown). No reduction in colony formation was detected in Sp100-transfected cells compared to empty vector-transfected cells. These results suggest that PML and PML-RARα inhibit growth by a pRB-independent mechanism.
FIG. 7.
Functional relevance of the PML-pRB association. (A) PML inhibits colony formation with a pRB-independent mechanism. C33A cells, which lack functional pRB, were transfected with equimolar quantities of the expression vectors indicated below the graph; G418-resistant colonies were scored after 12 to 15 days of selection. Results are shown as percentage of colonies with respect to C33A cells transfected with an empty vector (control). The results represent the average of four separate experiments, each performed in triplicate. (B) Effect of PML3 and PML-RARα on pRB regulation of GR-mediated transcription in HeLa cells. The data are expressed as fold activation of plasmid MTV-LTR-CAT with respect to the basal control value. Results are the average of four separate experiments, each performed in triplicate. hGR, human GR; Dex, dexamethasone.
PML, but not PML-RARα, abolishes pRB activation of GR-regulated transcription.
pRB has been described to play a role in the regulation of transcription from several promoters. We investigated the effect of PML expression on pRB regulation of three transcription factors: (i) E2F1, which is strongly repressed by pRB (34); (ii) SP1, which is enhanced by pRB (44, 69); and (iii) GR, which is also activated by pRB (65). PML expression had no detectable effect on pRB regulation of either E2F1- or SP1-dependent transcription (data not shown) but appeared to have a consistent inhibitory effect on pRB transactivation of GR-dependent transcription (Fig. 7B). HeLa cells were transiently transfected with reporter construct MTV-LTR-CAT and various combinations of human GR, pRB, and PML3 as indicated in Fig. 7B. A β-galactosidase expression vector was included in each transfection for normalization of transfection efficiency. Cells were treated with 10−6 M dexamethasone where indicated. At 40 h after transfection, cells were lysed, β-galactosidase activity was evaluated, and lysates equivalent to 3 U of β-galactosidase were assayed for CAT activity. The average values from four separate experiments (each performed in triplicate) are shown in Fig. 7B. PML expression in the absence of pRB appeared to have no significant effect on GR-regulated expression. However, pRB transactivation of GR-driven transcription was strongly inhibited by the coexpression of PML. Unlike PML expression, PML-RARα expression appears to moderately activate GR-regulated transcription. Coexpression of pRB and PML-RARα results in promoter activation which is greater than that determined for either protein alone. PML-RARα therefore appears to have a different effect with respect to wild-type PML in affecting pRB regulation of GR-driven promoters.
DISCUSSION
In this report, we have demonstrated that PML forms stable complexes with the nonphosphorylated form of pRB and that PML and pRB colocalize within the PML NBs. Whereas it was possible to study the localization of endogenous PML and pRB proteins by immunofluorescence experiments, biochemical analysis of this association was performed with overexpressed PML protein and endogenous pRB. This is due to the unavailability of antibodies which identify endogenous PML protein by Western blotting. This technical limitation prevented us from calculating precisely the stoichiometry of the PML-pRB complex in vivo. Based on the results obtained from coimmunoprecipitation of overexpressed PML and endogenous pRB, the stoichiometry of the complex appears rather low, approximately 0.5 to 1% for both PML and pRB.
Both PML and pRB proteins are present in different cellular compartments and in different functional forms. PML is localized within NBs, predominantly associated to the nuclear matrix, but it is also found in the soluble fraction of the nucleus and in the cytoplasm (11, 45, 55). pRB exists in a nonphosphorylated form, which is also found both in the nuclear matrix and in the nuclear soluble fraction, and in a phosphorylated form, mainly found in the soluble fraction of the nucleus, and it is localized in two subnuclear compartments, one diffuse and one corresponding to circumscribed granules (51, 54). However, we do not know the precise relative distributions of the matrix-associated or soluble fraction of both PML and nonphosphorylated pRB within the different nuclear compartments. We found that PML colocalizes with pRB within the NB and that it forms soluble complexes with hypophosphorylated pRB. These findings imply that fractions of the soluble PML and hypophosphorylated pRB form stable complexes within the NBs. However, we cannot exclude that similar complexes are also formed on the nuclear matrix, either within the NBs or in other nuclear compartments.
The pRB domain involved in binding PML corresponds to the pocket region. This observation suggests that the PML-pRB complex could be a target for the adenovirus E1A protein, which also binds pRB within the pocket region (36, 39). The formation of stable complexes between the RB protein and the E1A protein (14) determines inactivation of the growth-suppressive function of pRB and uncontrolled cell cycle progression (70). Interestingly, altered localization of PML and structural changes of the NBs have also been shown to occur during adenovirus infection (10, 18). Furthermore, PML expression as well as the size and number of the PML NBs increase after treatment of cells with the antiviral agent interferon (12, 46). It is therefore possible that the PML-pRB complex represents a functionally relevant target of E1A, and its eventual dissociation by the E1A protein may be important for the capacity of the virus to induce cell proliferation.
We next investigated the function of the PML-pRB complex. Since both PML and pRB are growth suppressors (49, 55, 70), the PML-pRB complex could have a role in determining growth arrest. We have found that PML exerts growth-suppressive activity in cells lacking functional pRB, suggesting that the interaction between the two proteins is not necessary for this effect, whereas coexpression of the two proteins further increases their individual growth-suppressive effects. We cannot, however, exclude that other proteins, such as p130 and p107, even though they do not physically associate with PML, may substitute for pRB in these cellular systems.
pRB has been described to regulate transcription from a set of promoters in an E2F-independent manner (44, 65, 69). One of the described systems of pRB transactivation is GR-mediated transcription (65). pRB has, in fact, been described to potentiate GR-mediated transcriptional activation with a mechanism which involves the presence of and the interaction with another transcription factor, hBrm (65). It has been recently reported that the transcriptional activity of GR, in the absence of pRB overexpression, is slightly (about twofold) increased by PML in Cos-7 cells (33). We did not observe any significant effect of PML on the transcriptional activity of GR in HeLa cells. However, our study demonstrates that PML has a clear inhibitory effect on pRB potentiation of GR-regulated transcription, therefore indicating one of the possible functions of the PML-pRB complex. Activated GR, pRB, and PML have all been described to be involved in the control of cellular differentiation, although with different effects. Activated GR is necessary for sustained self-renewal and arrest of differentiation in chicken hematopoietic precursors (72). This effect was proved to depend on the capacity of activated GR to act as a transcription factor. Nonphosphorylated pRB has been shown to be relevant in promoting differentiation of various cell types, such as adipocytes, myocytes, erythroid precursor cells, and neuronal cells, by synergizing with specific transcription factors (13, 32, 47, 62). PML-RARα, instead, blocks differentiation in hematopoietic precursor cell lines (29), and this function depends on both the PML and RARα portions of the protein (30). One could speculate that GR, pRB, and PML participate in determining the choice between differentiation and self-renewal and that the PML-pRB complex is part of this regulation. PML overexpression could result in inhibition of the pRB-GR synergism, therefore favoring differentiation.
We have demonstrated that the APL-specific fusion protein PML-RARα deregulates PML-pRB function. PML-RARα, in fact, retains the capacity to interact with pRB, and the expression of PML-RARα disperses pRB from the NBs. Functionally, however, PML-RARα does not inhibit pRB potentiation of GR-mediated transactivation but, on the contrary, appears to further enhance it. Even though the mechanism through which these interactions can ultimately result in opposite effects on GR-regulated promoters remains to be elucidated, one can speculate that the functional difference between the PML-RARα-pRB and PML-pRB complexes could contribute to the leukemogenic potential of PML-RARα by stimulating GR activity and ultimately favoring self-renewal and blocking differentiation.
The molecular mechanism underlying the formation of the PML-RARα-pRB complex in vivo remains unclear. We have demonstrated that the interaction between PML and pRB requires two regions of the PML protein, the B1 and B2 boxes and the C terminus, and can be reconstructed in a reticulocyte system, whereas the interaction of PML-RARα with pRB does not. Notably, the PML C terminus, which is crucial in the formation of the PML-pRB complex, is absent in the PML-RARα fusion protein. The RARα component of the fusion protein might functionally substitute, in vivo, for the PML C terminus. However, coimmunoprecipitation experiments demonstrated that RARα does not form stable complexes with pRB (data not shown). Other cellular proteins may, therefore, be involved in the formation of the PML-RARα-pRB complex in vivo and may be responsible for the alteration of transcriptional regulation of target promoters by PML-RARα.
In conclusion, our data demonstrate that PML and pRB form a complex in vivo within a specific cellular compartment, the NBs, and that such a complex could influence the function of pRB, for example, by regulating the activity of pRB on GR-mediated transcription, and they suggest that interactions between these molecules could regulate important processes such as differentiation and proliferation. PML-RARα could antagonize the effect of PML on pRB, thereby contributing to deregulation of differentiation and proliferation in APL cells.
ACKNOWLEDGMENTS
We thank Herb Samuels for the MTV-LTR-CAT construct and for the RShGRα expression vector, Anne Dejean for the ΔB1B2 plasmid, and Hughes de Thè for the Sp100 plasmid. We are thankful to Daniela Riganelli for help in the analysis of the immunofluorescence experiments.
This work was supported by grants from AIRC, CNR-ACRO, and EC (Biomed and Biotech programs).
REFERENCES
- 1.Alcalay M, Zangrilli D, Pandolfi P P, Longo L, Mencarelli A, Giacomucci A, Rocchi M, Biondi A, Rambaldi A, Lo Coco F, Diverio D, Donti E, Grignani F, Pelicci P G. Translocation breakpoint of acute promyelocytic leukemia lies within the retinoic acid receptor alpha locus. Proc Natl Acad Sci USA. 1991;88:1977–1981. doi: 10.1073/pnas.88.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S J, Markowitz S, Fearon E R, Willson K V, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 4.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 5.Borden K L, Boddy M N, Lally J, O’Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain of the promyelocytic leukemia proto-oncoprotein PML. Proc Natl Acad Sci USA. 1995;93:1602–1606. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borden K L B, Lally J, Martin S, O’Reilly N J, Solomon E, Freemont P S. In vivo and in vitro characterization of the B1 and B2 zinc binding domains from the acute promyelocytic leukemia proto-oncoprotein PML. Proc Natl Acad Sci USA. 1996;93:1602–1606. doi: 10.1073/pnas.93.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow J, Goddard A, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 8.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. A PML/RARa transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruggemeier U, Rogge L, Winnacker E-L, Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 1990;9:2233–2239. doi: 10.1002/j.1460-2075.1990.tb07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonesca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang K S, Fan Y H, Andreef M, Liu J, Mu Z M. The PML gene encodes for a phosphoprotein associated with the nuclear matrix. Blood. 1995;85:3646–3653. [PubMed] [Google Scholar]
- 12.Chelby-Alix M K, Pelicano L, Quignon F, Koken M H M, Venturini L, Stadler M, Pavlovic J, Degos L, de Thè H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- 13.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 16.de Thè H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukemia fuses the retinoic acid receptor a gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 17.de Thè H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARa fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 18.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 19.Dyck J A, Maul G G, Miller W H, Don Chen J, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K. Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1. J Virol. 1993;67:7641–7647. doi: 10.1128/jvi.67.12.7641-7647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagioli M, Alcalay M, Pandolfi P P, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci P G. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- 23.Fagioli, M., M. Alcalay, L. Tomassoni, P. F. Ferrucci, A. Mencarelli, D. Riganelli, F. Grignani, T. Pozzan, I. Nicoletti, F. Grignani, and P. G. Pelicci. Cooperation between the RING+B1+B2 and coiled-coil domains of PML is necessary for its effects on cell survival. Oncogene, in press. [DOI] [PubMed]
- 24.Fattaey A R, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrucci P F, Grignani F, Fagioli M, Grignani F, Nicoletti I, Pelicci P G. Cell death induction by the acute promyelocytic leukemia specific PML/RARa fusion protein. Proc Natl Acad Sci USA. 1997;94:10901–10906. doi: 10.1073/pnas.94.20.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani F, Casini T, Ferrucci P F, Martelli M F, Pelicci P G, Falini B. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood. 1995;85:1871–1880. [PubMed] [Google Scholar]
- 27.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grignani F, Fagioli M, Alcalay M, Longo L, Pandolfi P P, Donti E, Biondi A, Lo Coco F, Grignani F, Pelicci P G. Acute promyelocytic leukemia: from genetics to treatment. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- 29.Grignani F, Ferrucci P F, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, Pelicci P G. The acute promyelocytic leukaemia specific PML/RARa fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 30.Grignani F, Testa U, Riganelli D, Ferrucci P F, Samoggia P, Pinto A, Aldinucci D, Gelmetti V, Fagioli M, Alcalay M, Seeler J, Grignani F, Nicoletti I, Peschle C, Pelicci P G. Effects on differentiation by the promyelocytic leukemia PML/RARa protein depend on the fusion of the PML protein-dimerization and RARa DNA binding domains. EMBO J. 1996;15:4949–4958. [PMC free article] [PubMed] [Google Scholar]
- 31.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Altered myeloid development and acute leukemia in transgenic mice expressing PML/RARa under control of Cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 32.Gu W, Scneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 33.Guiochon-Mantel A, Savouret J F, Quignon F, Delabre K, Milgrom E, De Thè H. Effect of PML and PML/RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol Endocrinol. 1995;9:1791–1803. doi: 10.1210/mend.9.12.8614415. [DOI] [PubMed] [Google Scholar]
- 34.Helin K, Harlow E, Fattaey A R. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Q, Bautista C, Edwards G, Defeo-Jones D, Jones R, Harlow E. Antibodies specific for the human retinoblastoma protein identify a family of related polypeptides. Mol Cell Biol. 1991;11:5792–5799. doi: 10.1128/mcb.11.11.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Q J, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H-J S, Wang N-P, Tseng B Y, Lee W-H, Lee E Y-H P. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W-Q, Szekely L, Wendel-Hansen V, Ringertz N, Klein G, Rosen A. Co-localization of the retinoblastoma protein and the Epstein-Barr virus-encoded nuclear antigen EBNA-5. Exp Cell Res. 1991;197:314–318. doi: 10.1016/0014-4827(91)90438-z. [DOI] [PubMed] [Google Scholar]
- 39.Kaelin W G J, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaelin W G J, Pallas D C, De Caprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 41.Kakizuka A, Miller W H J, Umesono K, Warrel R P J, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARa with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 42.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor a fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly C, Van Driel R, Wilkinson G W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 44.Kim S J, Onwuta U S, Lee Y I, Botchan L R, M R, Robbins P D. The retinoblastoma gene product regulates Sp-1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koken M H M, Puvio-Dutilleul F, Guillemin M C, Viron A, Cruz-Linares G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de Thè H. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;14:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The acute promyelocytic leukemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 47.Lee E Y, Yuan S S, Cox L A, Bradley A, Lee W H, Helin K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 48.Lee W-H, Shew J-Y, Hong F D, Sery T W, Donoso L A, Young L J, Bookstein R, Lee E Y-H P. The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature. 1987;329:642–645. doi: 10.1038/329642a0. [DOI] [PubMed] [Google Scholar]
- 49.Liu J H, Mu Z M, Chang K S. PML suppresses oncogenic transformation of NIH/3T3 cells activated by neu. J Exp Med. 1995;1815:1965–1973. doi: 10.1084/jem.181.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lovering R, Hanson I M, Borden K L B, Martin S, O’Reilly N J, Evan G I, Rahman D, Pappin D J C, Trowsdale J, Freemont P. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA. 1993;90:2112–2116. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mancini M A, Shan B, Nickerson J A, Penman S, Lee W H. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miesfeld R, Rusconi S, Godowski P J, Maler B A, Okret S, Wilkstrom A C, Gustaffson J A, Yamamoto K R. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell. 1986;46:389–399. doi: 10.1016/0092-8674(86)90659-8. [DOI] [PubMed] [Google Scholar]
- 54.Mittnacht S, Weinberg R A. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 55.Mu Z-M, Chin K-V, Liu J-H, Lozano G, Chang K-S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandolfi P P, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F, Rochette-Egly C, Gaube M-P, Chambon P, Pelicci P G. Genomic variability and alternative splicing generate multiple PML/RARa transcripts that encode aberrant PML proteins and PML/RARa isoforms in acute promyelocytic leukemia. EMBO J. 1992;11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, Lo Coco F, Grignani F, Pelicci P G. Structure and origin of the acute promyelocytic leukemia myl/RARa cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 59.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heterodimeric interaction with RXR. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin X-Q, Chittenden T, Livingston D, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 61.Reddy B A, Etkin L D, Freemont P S. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 62.Richon V M, Rifkind R A, Marks P A. Expression and phosphorylation of the retinoblastoma protein during induced differentiation of murine erythroleukemia cells. Cell Growth Differ. 1992;3:413–420. [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 64.Seed B, Sheed J Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 65.Singh P, Coe J, Hong W. A role for the retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 66.Szekely L, Pokrovskaja K, Jiang W G, de Thè H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szekely L, Uzvolgyi E, Jiang W-Q, Durko M, Wiman K G, Klein G, Sumegi J. Subcellular localization of the retinoblastoma protein. Cell Growth Differ. 1991;2:287–295. [PubMed] [Google Scholar]
- 68.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of a cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 69.Uvadia A J, Rogers K T, Higgins P D R, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the pRB protein and Sp-1 mediated transcription is stimulated by pRB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 71.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARa in acute promyelocytic leukemia cells. Cell. 1994;76:345–358. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 72.Wessely O, Deiner E M, Beug H, von Lindern M. The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. EMBO J. 1997;16:267–280. doi: 10.1093/emboj/16.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]