Abstract
 In DNA-encoded library
synthesis, amine-substituted building blocks
are prevalent. We explored isocyanide multicomponent reactions to
diversify DNA-tagged amines and reported the Ugi-azide reaction with
high yields and a good substrate scope. In addition, the Ugi-aza-Wittig
reaction and the Ugi-4-center-3-component reaction, which used bifunctional
carboxylic acids to provide lactams, were explored. Five-, six-, and
seven-membered lactams were synthesized from solid support-coupled
DNA-tagged amines and bifunctional building blocks, providing access
to structurally diverse scaffolds.
In DNA-encoded library
synthesis, amine-substituted building blocks
are prevalent. We explored isocyanide multicomponent reactions to
diversify DNA-tagged amines and reported the Ugi-azide reaction with
high yields and a good substrate scope. In addition, the Ugi-aza-Wittig
reaction and the Ugi-4-center-3-component reaction, which used bifunctional
carboxylic acids to provide lactams, were explored. Five-, six-, and
seven-membered lactams were synthesized from solid support-coupled
DNA-tagged amines and bifunctional building blocks, providing access
to structurally diverse scaffolds.
Introduction
DNA-encoded chemical libraries (DECLs) are collections of combinatorial compounds where each structure is coded with a DNA sequence.1 This technology seamlessly integrates efficient compound handling with selection-based screening methodologies. In recent years, the combination of innovative DEL synthetic and screening methods has emerged as a routine platform for the identification of small molecules in drug discovery in both academic and industrial settings. The power of DEL technology rests in the efficient handling of large compound numbers and in the inexpensive and rapid affinity selection against a wide variety of protein targets for bioactive compound identification.2 However, the scope of split-pool and DNA-compatible reactions is limited, especially with respect to cyclization reactions, calling for reaction development that makes novel scaffold architectures accessible. A DEL synthesis route often starts from N-protected amino acids that are coupled to an amino-modified DNA strand by using amide coupling reactions. Usually, these building blocks are then deprotected and N-substituted by carbonyl chemistry such as amide-bond formation or by nucleophilic aromatic substitution (Figure 1A).3
Figure 1.
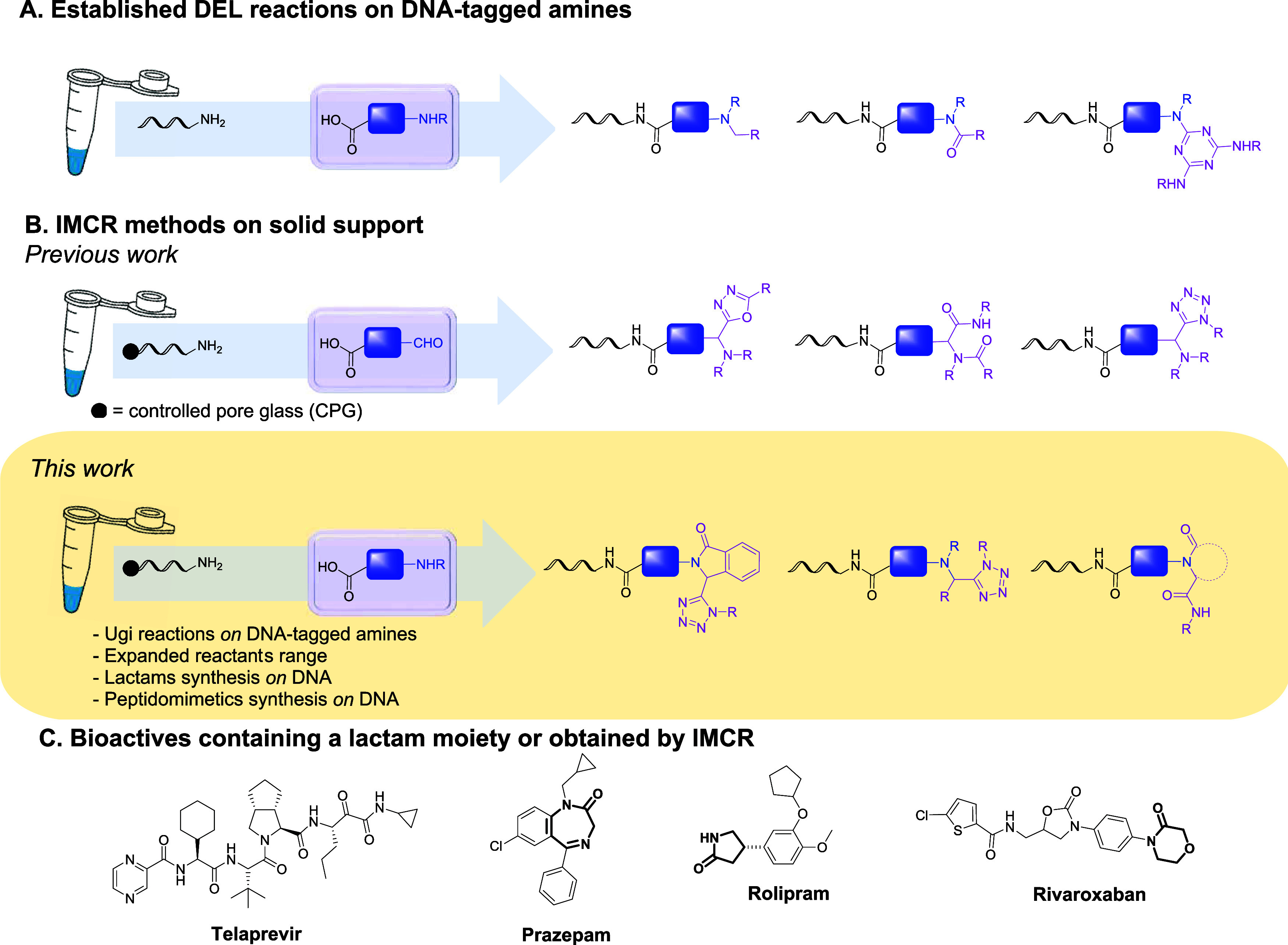
(A) Classical DEL reactions on DNA-tagged amines. (B) Expanded DNA-tagged amine functionalization through the implementation of IMCRs on solid-phase supported DNA. (C) Examples of biologically active molecules containing a lactam moiety obtained through IMCR.
Alternatively, DNA-tagged amines could be substituted by isocyanide multicomponent reactions (IMCRs). The combination of DEL and IMCR offers a rapid approach to scaffold diversity in a drug development platform. Structurally diverse target compounds are accessible through a single reaction step and simple starting materials (Figure 1B).4 In a recent publication, Dömling et al. have explored the potential of IMCRs to provide straightforward access to common bioactive scaffolds such as isoquinolines or isoindolines and their application in “scaffold hopping”.5 Furthermore, in the synthesis of telaprevir, an IMCR is efficiently used to increase overall atom economy by reducing reaction steps.6 Additionally, they have been used in the synthesis of lactams, a privileged class of heterocycles in medicinal chemistry and drug design.7 These scaffolds are part of the core structure of a large number of natural and non-natural compounds covering a broad spectrum of antidepressant, antitumor, and anti-inflammatory biological activities (Figure 1C).8
Our group demonstrated the compatibility of four different isocyanide MCR chemistries with DNA-encoded combinatorial synthesis, leading to a broad scope of pharmaceutically relevant scaffolds, such as tetrazoles and oxadiazoles, starting from DNA-tagged aldehydes on solid support (CPG controlled pore glass) (Figure 1B).9 We opted for initiating DEL synthesis with CPG-coupled DNA strands because the DNA is base-protected, conferring greater chemical stability against deamination reactions, a free choice of solvents for reaction optimization, and simple washing procedures to remove excess reagents. Recently, the group of Ding published the synthesis of DNA-tagged lactams from DNA-coupled isocyanides and aldehydes via the Ugi-4-center-3-component reaction.10 However, the potential of these chemistries for producing DNA-encoded libraries was limited by the restricted choice of starting materials for coupling to the DNA (e.g., bifunctional carboxylic acid-aldehyde building blocks). To overcome this limitation, we explored the possibility of attaching the amine moiety to the DNA instead, which would expand the range of reactants to abundantly available amino acids and primary amines (see the Supporting Information).
Herein, we report our efforts to expand the scope of reactions on DNA-tagged amines; they included the exploration of the Ugi-azide reaction (UA-4CR), the Ugi-aza-Wittig reaction (U-4CR/aza-Wittig), and the Ugi-4-center-3-component reaction (U-4C-3CR). Both the UA-4CR and the U-4C-3CR reaction were designed to provide medicinally interesting five-, six-, and seven-membered lactams via IMCRs from DNA-tagged amines, thus expanding the scope of scaffold architectures for DEL design.
Results and Discussion
We started our investigations of UA-4CR on DNA-tagged secondary amine 1a. Initial exploration with the previously reported reaction conditions, using methanol as the solvent, an excess of aldehyde, TMSN3, and isocyanide (1000 equiv each) at a reaction temperature of 60 °C,9a gave trace amounts of the tetrazole product (Table 1; entry 2). In order to optimize the reaction conditions, we first tested different solvents (Table 1; entries 3–4). The polar protic solvent, methanol, was exchanged by polar aprotic and nonpolar solvents, such as dimethylformamide, 1,2-dichloroethane, and chloroform, or their mixtures. The use of chloroform improved the product conversions to 34% (see Table 1; entry 4 and Supporting Information). In order to boost the condensation step, different equivalents of benzaldehyde were tested (Table 1, entries 1, 5, 6). Increasing the benzaldehyde concentration did not improve conversion rates. During the course of our optimization experiments, it was found that the moderately polar solvent tetrahydrofuran gave better product conversions compared to chloroform (Table 1, entry 1). Testing the reaction temperature, time, and reagent concentration in tetrahydrofuran increased the product conversion to 55% (Table 1, entry 9).
Table 1. Optimization of UA-4CR on CPG-Coupled Conjugates 1a and 1b.
| entry | deviation from above | conversion (%) |
|---|---|---|
| 1a | none | 46 |
| 2 | MeOH at 60 °C | 4 |
| 3 | DMF | 31 |
| 4 | CHCl3 | 34 |
| 5 | 100 equiv of 2a in CHCl3 | 17 |
| 6 | 10,000 equiv of 2a in CHCl3 | 2 |
| 7a | 25 °C | 28 |
| 8a | 40 °C | 51 |
| 9a | 45 °C, 5 h in the 2nd reaction step | 55 |
CPG-bound conjugates 1a (20 nmol) and 2a (1000 equiv), 50 μL of solvent, rt, 3 h, then 3a (1000 equiv) and TMSN3 (1000 equiv), 50 °C, 16 h. AMA cleavage (30% aqueous ammonia/40% aqueous methylamine 1:1 vol/vol) at rt for 0.5 h, 10merTC = 5′-TTC CTC TCC T-3′-CPG, reaction performed using CPG-bound conjugates 1b and 3b.
Having optimized reaction conditions in hand, we set out to investigate the reaction scope with a diverse set of aldehydes (2) that were reacted with the DNA-N-methyl glycine conjugate 1b (Scheme 1). Aliphatic aldehydes, such as acetal, propional, and isobutyral aldehydes, gave the target tetrazoles (4c–e) with good yields, while the sterically hindered pivalaldehyde did not yield the desired product (4f). Heteroaromatic 2-picolinaldehyde and 1-methylimidazole-2-aldehyde were evaluated next, affording products 4i and 4j with high conversions of 66 and 59%, respectively. Ortho- (4h, 4k) and para-substituted (4l, 4m) benzaldehydes were generally well tolerated, with a preference for the latter, which gave higher yields (55 and 51%).
Scheme 1. Scope of UA-4CR on CPG-Coupled DNA Amine Conjugate (1) with Aldehyde 2 and Isocyanide 3b.

Next, we explored the amine scope of the reaction by testing five additional DNA-secondary amine conjugates (1a, c–f), which were all reacted with picolinaldehyde (2h, see the Supporting Information). The desired products 4o–s were obtained with moderate conversions ranging from 32 to 45%.
Notably, conjugates 1a, c–f were prepared by nucleophilic substitution of the respective primary amines on the DNA-coupled chloroacetate conjugate (see the Supporting Information), following a procedure previously reported in the literature.11 We then turned our interest to the reactivity of DNA-primary amine conjugates (1g–i; see the Supporting Information), which were prepared by coupling the respective α-amino acids to the amino-modified DNA strand. Using the optimized UA-4CR conditions with picolinaldehyde (2h), product formation of DNA conjugates 4t–v with moderate conversions of 14, 34, and 31%, was observed, thus significantly expanding the reactant scope for the UA-4CR. Overall, the UA-4CR was successfully performed on 19 examples with DNA-tagged primary and secondary amines. The focus was shifted to the UA-4C reaction that can be used to obtain medicinally interesting lactams. We explored the possibility of obtaining these scaffolds on DNA tags using a diverse set of DNA-coupled α-amino acids (1g–i, k, l) and bifunctional aldehydes (2n–p) as reaction partners (see the Supporting Information). When DNA-tagged glycine 1j was reacted with methyl 2-acetylbenzoate (2n) under previously optimized UA-4CR conditions, we observed spontaneous condensation to the desired isoindolinone (4w) with a moderate conversion of 38%. Unfortunately, no product was obtained when using methyl 3-formyl-1H-indole-2-carboxylate or methyl 4-oxobutanoate (2o, 2p; see the Supporting Information). Exchanging 1j with a diverse set of DNA-coupled five α-amino acids (1g–i, k, and l) did not lead to the formation of the expected lactamization products under UA-4CR conditions, except for the methionine derivative 4x.
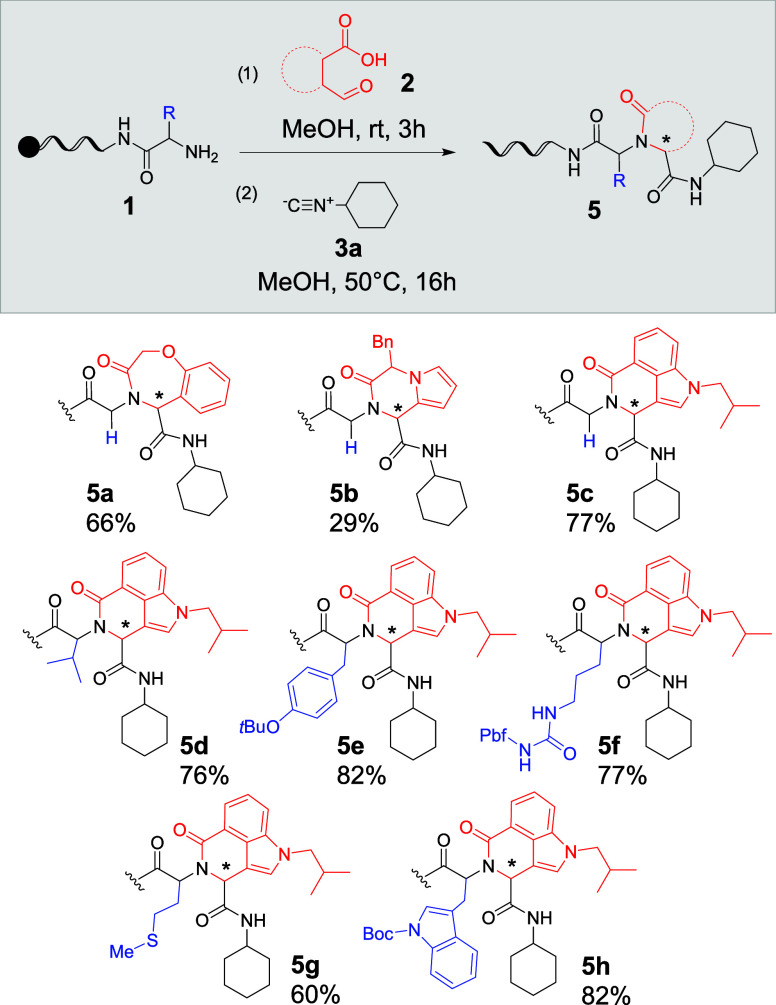
Previously, the U-4CR/aza-Wittig reaction was reported for DNA-tagged aldehydes. Based on the established reaction conditions using dichloroethane as the solvent, the possibility of the formation of oxadiazoles was tested in a small temperature-dependent study (50–80 °C; see the Supporting Information). Unfortunately, no product formation could be detected. Instead, a side product is observed by mass spectrometry that could correspond to a reaction product among the DNA-tagged amine, the aldehyde, and carboxylic acid (see the Supporting Information). It was considered that an excess of aldehyde could inhibit the reactivity of the specific phospharane-substituted isonitrile (Pinc)12 used in the U-4CR/aza-Wittig reaction. Having successfully established the UA-4CR-condensation sequence, we then tested the lactam formation using U-4C-3CR. Conjugate 1j was reacted with eight different aromatic carboxylic acid-aldehyde building blocks (2q–x; see the Supporting Information) in methanol (Scheme 2).
Scheme 2. Expanding the UA-4CR and −3CR Scope via the Reaction of DNA-Bound α-Amino Acids (1g–i, k, l) with Carboxylic Acid-Aldehyde Building Blocks (2q–s).
Newly formed stereocenter, Pbf: 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl.
Three of the tested bifunctional reactants (2q–s; see the Supporting Information) afforded the expected products in moderate to very high conversion (5a–c), while for the remainder, no desired product mass peak was observed by MALDI-MS analysis (see the Supporting Information). As a requirement to obtain the final lactam, an intermediate of the U-4C-3CR reaction has to undergo a rearrangement, which could be inhibited for particular bifunctional carboxylic acid-aldehyde building blocks. However, to our surprise, when 1g–i, k, l were reacted with the bifunctional carboxylic acid-aldehyde building block 2s and the isocyanide 3b in methanol (U-4C-3CR reaction conditions), the respective products (5d–h) were isolated in good conversions ranging from 60 to 82%.
Conclusions
In summary, we have reported the extension of IMCRs for the diversification of solid-supported DNA-tagged primary and secondary amines for encoded library synthesis. UA-4CR was successfully performed on a variety of secondary amines that can be obtained on DNA-tagged substrates by a simple nucleophilic substitution. All products gave retention time shifts in the HPLC traces that allow for isolation of the products from unreacted starting materials, thus improving DEL purity. The extension of the IMCR to a diverse range of DNA-tagged amines introduces a new avenue for synthesizing a DNA-encoded library on the basis of the CPG-based strategy. This approach leverages a wide array of commercially available building blocks, including amino acids, thereby expanding the scope of library synthesis. Given their relevance in drug design, we focused our efforts on the preparation of lactam scaffolds via U-4C-3CR and demonstrated the successful transformation of DNA-coupled primary amines, derived from amino acids, to five-, six-, and seven-membered lactams using bifunctional reagents such as carboxylic acid-aldehyde conjugates.
Methods
Ugi-Azide Four-Component Reaction on CPG-Bound DNA
Prior to use, all solid materials were dried in vacuo for 30 min. The aldehyde (1500 equiv, 30 μmol) and the CPG-bound oligonucleotide conjugate (20 nmol) were dissolved/suspended in 50 μL of dry THF, and the reaction mixture was shaken at RT for 3 h to effect imine formation. Afterward, the isocyanide (1000 equiv, 20 μmol) previously dissolved in 20 μL of dry THF and TMSN3 (1000 equiv, 20 μmol) were added, and the mixture was shaken for an additional 5 h at 40 °C. The CPG-coupled DNA conjugate was filtered over a filter column, washed three times with each 200 μL of DMF, MeOH, ACN, and CH2Cl2, and dried in vacuo. Success of the reaction was controlled by cleaving a small portion of the CPG-coupled oligonucleotide conjugate (0.7–0.9 mg, ∼20 nmol) with 500 μL of AMA solution (AMA, aqueous ammonia (30%)/aqueous methylamine (40%), 1:1, vol/vol) for 30 min at ambient temperature. To this solution, 20 μL of 1 M Tris-buffer (pH = 7.5) was added, and the mixture was dried in a SpeedVac and dissolved in 200 μL of distilled water. The crude was analyzed by RP-HPLC and MALDI-MS. The product was purified by preparative RP-HPLC.
Ugi Four-Center Three-Component Reaction on CPG-Bound DNA
Prior to use, all solid materials were dried in vacuo for 30 min. The bifunctional aldehyde-acid (1000 equiv, 20 μmol) was either directly added to the CPG-bound oligonucleotide conjugate (20 nmol) and it was filled up to a final volume of 50 μL with MeOH or the bifunctional aldehyde-acid (1000 equiv, 20 μmol) was dissolved in 50 μL of MeOH and then added to the CPG-bound oligonucleotide conjugate. The reaction mixture was shaken at ambient temperature for 3 h to effect imine formation. Afterward, the isocyanide (1000 equiv, 20 μmol) was added and the reaction mixture was shaken for 16 h at 50 °C. The CPG-coupled DNA conjugate was filtered over a filter column, washed three times with each 200 μL of DMF, MeOH, ACN, and CH2Cl2, and dried in vacuo. Success of the reaction was controlled by cleaving a small portion of the CPG-coupled oligonucleotide conjugate (0.7–0.9 mg, ∼20 nmol) with 500 μL of AMA solution (AMA, aqueous ammonia (30%)/aqueous methylamine (40%), 1:1, vol/vol) for 30 min at ambient temperature. To this solution, 20 μL of 1 M Tris-buffer (pH = 7.5) was added, and the mixture was dried in a SpeedVac and dissolved in 200 μL of distilled water. The crude was analyzed by RP-HPLC and MALDI-MS. The product was purified by preparative RP-HPLC.
Ugi-Aza-Wittig Reaction on CPG-Bound DNA
Prior to use, all solid materials were dried in vacuo for 30 min. The aldehyde (1000 equiv, 20 μmol) and the CPG-bound oligonucleotide conjugate (20 nmol) were dissolved/suspended in 30 μL of 1,2-dichloroethane. The reaction mixture was shaken at ambient temperature for 3 h to effect imine formation. Then, the acid (1000 equiv, 20 μmol) was dissolved in 80 μL of 1,2-dichloroethane and transferred to (isocyanoimino)triphenylphosphorane (1000 equiv, 20 μmol), and this mixture was added to the CPG-bound conjugate. The reaction mixture was shaken for 16 h at the indicated temperature. The CPG-coupled DNA conjugate was filtered over a filter column, washed three times with each 200 μL of DMF, MeOH, ACN, and CH2Cl2, and dried in vacuo. Success of the reaction was controlled by cleaving a small portion of the CPG-coupled oligonucleotide conjugate (0.7–0.9 mg, ∼ 20 nmol) with 500 μL of AMA solution (AMA, aqueous ammonia (30%)/aqueous methylamine (40%), 1:1, vol/vol) for 30 min at ambient temperature. To this solution, 20 μL of 1 M Tris-buffer (pH = 7.5) was added, and the mixture was dried in a SpeedVac and dissolved in 200 μL of distilled water. The crude was analyzed by RP-HPLC and MALDI-MS.
Acknowledgments
A.B. gratefully acknowledges financial support by the DFG (BR 5049/7-1) and by the Wellcome Trust-funded project “Academic DNA-Encoded Library Resource”.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07136.
Synthesis protocols and analytical data of the reaction products (PDF)
Author Contributions
∥ S.W. and E.D. contributed equally to this work. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): AB declares a competing financial interest as co-founder of Serengen GmbH, a company that provides DEL services.
Supplementary Material
References
- a Gironda-Martínez A.; Donckele E. J.; Samain F.; Neri D. DNA-Encoded Chemical Libraries: A Comprehensive Review with Successful Stories and Future Challenges. ACS Pharmacol. Transl. Sci. 2021, 4, 1265–1279. 10.1021/acsptsci.1c00118. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Satz A. L.; Brunschweiger A.; Flanagan M. E.; Gloger A.; Hansen N. J. V.; Kuai L.; Kunig V. B. K.; Lu X.; Madsen D.; Marcaurelle L. A.; Mulrooney C.; O’Donovan G.; Sakata S.; Scheuermann J. DNA-encoded chemical libraries. Nat. Rev. Methods Primers 2022, 2, 3 10.1038/s43586-021-00084-5. [DOI] [Google Scholar]
- a Clark M. A.; Acharya R. A.; Arico-Muendel C. C.; Belyanskaya S. L.; Benjamin D. R.; Carlson N. R.; Centrella P. A.; Chiu C. H.; Creaser S. P.; Cuozzo J. W.; Davie C. P.; Ding Y.; Franklin G. J.; Franzen K. D.; Gefter M. L.; Hale S. P.; Hansen N. J. V.; Isreal D. I.; Jiang J.; Kavarana M. J.; Kelley M. S.; Kollmann C. S.; Li F.; Lind K.; Mataruse S.; Medeiros P. F.; Messer J. A.; Myers P.; O’Keefe H.; Oliff M. C.; Pise C. E.; Satz A. L.; Skinner S. R.; Svendsen J. L.; Tang L.; van Vloeten K.; Wagner R. W.; Yao G.; Zhao B.; Morgan B. A. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009, 5, 647–654. 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]; b Satz A. L.; Cai J.; Chen Y.; Goodnow R.; Gruber F.; Kowalczyk A.; Petersen A.; Naderi-Oboodi G.; Orzechowski L.; Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjugate Chem. 2015, 26, 1623–1632. 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]; c Wrenn S. J.; Weisinger R. M.; Halpin D. R.; Harbury P. B. J. Am. Chem. Soc. 2007, 129, 13137–13143. 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d MacConnell A. B.; McEnaney P. J.; Cavett V. J.; Paegel B. M. DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb. Sci. 2015, 17, 518–534. 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fitzgerald P. R.; Paegel B. M. DNA-Encoded Chemistry: Drug Discovery from a Few Good Reactions. Chem. Rev. 2021, 121, 7155–7177. 10.1021/acs.chemrev.0c00789. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Sun Z. M.; Yang S. G.; Xue L. J.; Zhang J.; Yang K.; Hu Y. J. N-Alkyl Linkers for DNA-Encoded Chemical Libraries. Chem.- Asian J. 2022, 17, e202200016 10.1002/asia.202200016. [DOI] [PubMed] [Google Scholar]; c Sun Z.; Zhang J.; Zhang H.; Cao H.; Xiao L.; Yang K.; Hu Y. J. DNA Compatible Oxidization and Amidation of Terminal Alkynes. Bioconjugate Chem. 2022, 33, 1585–1594. 10.1021/acs.bioconjchem.2c00340. [DOI] [PubMed] [Google Scholar]
- a Ugi I.; Steinbrueckner C. Über ein neues Kondensations-Prinzip. Angew. Chem. 1960, 72, 267–268. 10.1002/ange.19600720709. [DOI] [Google Scholar]; b Dömling A. Innovations and Inventions: Why Was the Ugi Reaction Discovered Only 37 Years after the Passerini Reaction?. J. Org. Chem. 2023, 88, 5242–5247. 10.1021/acs.joc.2c00792. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Buskes M. J.; Coffin A.; Troast D. M.; Stein R.; Blanco M. J. Accelerating Drug Discovery: Synthesis of Complex Chemotypes via Multicomponent Reactions. ACS Med. Chem. Lett. 2023, 14, 376–385. 10.1021/acsmedchemlett.3c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Zarganes-Tzitzikas T.; Neochoritis C. G.; Dömling A. Atorvastatin (Lipitor) by MCR. ACS Med. Chem. Lett. 2019, 10, 389–392. 10.1021/acsmedchemlett.8b00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Wang Q.; Zheng Q.; Kurpiewska K.; Kalinowska-Tluscik J.; Dömling A. Access to Isoquinolin-2(1H)-yl-acetamides and Isoindolin-2-yl-acetamides from a Common MCR Precursor. J. Org. Chem. 2022, 87, 14463–14475. 10.1021/acs.joc.2c01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znabet A.; Polak M. M.; Janssen E.; de Kanter F. J. J.; Turner N. J.; Orru R. V. A.; Ruijter E. A highly efficient synthesis of telaprevir by strategic use of biocatalysis and multicomponent reactions. Chem. Commun. 2010, 46, 7918–7920. 10.1039/c0cc02823a. [DOI] [PubMed] [Google Scholar]
- Pharande S. G. Synthesis of Lactams via Isocyanide-Based Multicomponent Reactions. Synthesis 2021, 53, 418–446. 10.1055/s-0040-1706297. [DOI] [Google Scholar]
- Saldívar-González F. I.; Lenci E.; Trabocchi A.; Medina-Franco J. L. Exploring the Chemical Space and the Bioactivity Profile of Lactams: a Chemoinformatic Study. RSC Adv. 2019, 9, 27105. 10.1039/C9RA04841C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kunig V. B. K.; Ehrt C.; Dömling A.; Brunschweiger A. Isocyanide Multicomponent Reactions on Solid-Phase-Coupled DNA Oligonucleotides for Encoded Library Synthesis. Org. Lett. 2019, 21, 7238–7243. 10.1021/acs.orglett.9b02448. [DOI] [PubMed] [Google Scholar]; b Kunig V. B. K.; Potowski M.; Akbarzadeh M.; Klika Skopic M.; Dos Santos Smith D.; Arendt L.; Dormuth I.; Adihou H.; Andlovic B.; Karatas H.; Shaabani S.; Zarganes-Tzitzikas T.; Neochoritis C. G.; Zhang R.; Groves M.; Gueret S. M.; Ottmann C.; Rahnenfuhrer J.; Fried R.; Domling A.; Brunschweiger A. TEAD-YAP Interaction Inhibitors and MDM2 Binders from DNA-Encoded Indole-Focused Ugi Peptidomimetics. Angew. Chem., Int. Ed. 2020, 59, 20338–20342. 10.1002/anie.202006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantell M. A.; Marcaurelle L.; Ding Y. One Reaction Served Three Ways: The On-DNA Ugi 4C-3C Reaction for the Formation of Lactams. Org. Lett. 2023, 25, 1241–1245. 10.1021/acs.orglett.2c04043. [DOI] [PubMed] [Google Scholar]
- Potowski M.; Esken R.; Brunschweiger A. Translation of the copper/bipyridine-promoted Petasis reaction to solid phase-coupled DNA for encoded library synthesis. Bioorg. Med. Chem. 2020, 28, 115441 10.1016/j.bmc.2020.115441. [DOI] [PubMed] [Google Scholar]
- a Weinberger B.; Fehlhammer W. P. N-Isocyanoiminotriphenylphosphorane: Synthesis, Coordination Chemistry, and Reactions at the Metal. Angew. Chem., Int. Ed. 1980, 19, 480–481. 10.1002/anie.198004801. [DOI] [Google Scholar]; b Frost J. R.; Scully C. C. G.; Yudin A. K. Oxadiazole grafts in peptide macrocycles. Nat. Chem. 2016, 8, 1105–1111. 10.1038/nchem.2636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.