Abstract
Multiple or pleiotropic drug resistance in the yeast Saccharomyces cerevisiae requires the expression of several ATP binding cassette transporter-encoding genes under the control of the zinc finger-containing transcription factor Pdr1p. The ATP binding cassette transporter-encoding genes regulated by Pdr1p include PDR5 and YOR1, which are required for normal cycloheximide and oligomycin tolerances, respectively. We have isolated a new member of the PDR gene family that encodes a member of the Hsp70 family of proteins found in this organism. This gene has been designated PDR13 and is required for normal growth. Overexpression of Pdr13p leads to an increase in both the expression of PDR5 and YOR1 and a corresponding enhancement in drug resistance. Pdr13p requires the presence of both the PDR1 structural gene and the Pdr1p binding sites in target promoters to mediate its effect on drug resistance and gene expression. A dominant, gain-of-function mutant allele of PDR13 was isolated and shown to have the same phenotypic effects as when the gene is present on a 2μm plasmid. Genetic and Western blotting experiments indicated that Pdr13p exerts its effect on Pdr1p at a posttranslational step. These data support the view that Pdr13p influences pleiotropic drug resistance by enhancing the function of the transcriptional regulatory protein Pdr1p.
Multiple drug resistance refers to a limited number of genetic alterations giving rise to a complex spectrum of tolerance to cytotoxic compounds having different intracellular targets (32). In human tumor cells, multiple drug resistance is often associated with overproduction of ATP binding cassette (ABC) transporter-encoding genes, such as MDR1 (22) or MRP (11). The overproduction of these gene products leads to enhanced efflux of toxic drugs across the plasma membrane and permits tolerance of otherwise lethal dosages.
In the yeast Saccharomyces cerevisiae, a similar multidrug tolerance phenotype can be observed and is referred to as pleiotropic drug resistance. The first PDR gene identified and cloned was designated PDR1 (2). DNA sequence analysis of this locus indicated that Pdr1p was a zinc finger-containing protein that showed strong sequence similarity to other fungal transcriptional regulatory proteins (2). Strains lacking PDR1 were hypersensitive to a broad range of drugs, including cycloheximide and oligomycin. Sequence analysis of S. cerevisiae chromosome II indicated that a homolog of PDR1, PDR3 (16), was present at this location and encoded a protein showing 36% amino acid identity with Pdr1p (15). Genetic and biochemical experiments demonstrated that Pdr1p and Pdr3p act to influence pleiotropic drug resistance (15, 33). However, mutants lacking PDR3 were not observed to have a pronounced defect in drug resistance, unlike Δpdr1 strains, suggesting that Pdr1p was the major contributor of drug tolerance (15, 33).
PDR1 was originally identified on the basis of semidominant mutant alleles that produced high-level resistance to cycloheximide and oligomycin, among other compounds (reviewed in reference 3). Epistasis and Northern blot experiments demonstrated that PDR1 conferred cycloheximide resistance through the transcriptional activation of the PDR5 gene (43). PDR5 was shown to encode an ABC transporter protein (5, 6, 28) that is located in the plasma membrane (5, 17) and that can act as a drug efflux pump (36, 38). Deletion mapping and DNase I footprinting analysis indicated that both Pdr1p and Pdr3p bound to several sites upstream of the PDR5 transcription start site and activated the expression of this gene (33, 34). These binding sites were named Pdr1p/Pdr3p response elements (PDREs).
While semidominant PDR1 mutants required PDR5 to mediate cycloheximide resistance, the loss of PDR5 did not affect PDR1-mediated oligomycin resistance. We screened a high-copy-number plasmid library for sequences that would elevate oligomycin resistance and recovered several different genes (35). One of these loci was found to encode an ABC transporter protein resembling the MRP gene product and was designated YOR1 (35). The loss of YOR1 leads to a large decrease in Pdr1p-mediated oligomycin resistance but has no effect on cycloheximide resistance. A second gene identified in this screen elevated both oligomycin and cycloheximide tolerance. We refer to this gene as PDR13.
In this study, we demonstrate that PDR13 encodes an Hsp70 homolog that acts to elevate the function of Pdr1p, leading to increased expression of PDRE-containing genes and drug resistance. Strains lacking PDR13 are compromised for growth and induce the expression of stress-responsive genes. A gain-of-function mutant form of Pdr13p is able to complement the growth defect of a Δpdr13 strain and elevates both PDRE-containing gene expression and drug resistance. Taken together, these data implicate Pdr13p as an upstream modulator of the expression of PDR genes through control of the transcription factor Pdr1p.
MATERIALS AND METHODS
Yeast strains and media.
The genotypes of the yeast strains used in this study are listed in Table 1. Yeast transformations were performed with the lithium acetate procedure of Ito et al. (29) or a high-efficiency technique (21). Standard YPD medium and minimal medium were used for the growth of cells and drug resistance assays (49). Drug resistance assays were performed by spot tests (53). β-Galactosidase activity was measured as previously described (24).
TABLE 1.
S. cerevisiae strains used
| Strain designation | Description | Source or reference |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− | Scott Emr |
| PB2 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr3-Δ1::hisG | 33 |
| PB3 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr1-Δ2::hisG | 33 |
| PB4 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr1-Δ2::hisG pdr3-Δ1::hisG | 33 |
| DKY2.1 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr5-Δ1::hisG | 33 |
| DKY7 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−yor1-1::hisG | 35 |
| YRT9 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr13-Δ1::hisG | This study |
| TCH4 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr13-Δ1::hisG pdr3-Δ1::hisG PDR5-HIS3-PDR5-lacZ | This study |
| TCH1 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel−pdr13-Δ1::hisG pdr3-Δ1::hisG | This study |
Plasmids.
An integrating PDR5-lacZ fusion gene was constructed by transferring an EcoRI/SalI fragment from pKV2 (33) into pRS303 (50) to form pTH120. Strain TCH4 was generated by transforming TCH1 with StuI-cut pTH120, which directs recombination to PDR5. A SalI/NotI fragment carrying the wild-type PDR13 gene was cloned into the 2μm-containing vector pRS424 (10) to form pTH87. The same PDR13 fragment was also inserted into pRS316 (50) to generate pTH86 and into pRS314 (50) to produce pTH143. The original YEp24-based recombinant carrying PDR13 was designated pDOC10-2. The GAL1-PDR1 fusion gene contained a PDR1 fragment (extending from the ATG to a SalI site located 700 bp downstream of the translation stop codon) cloned downstream of the GAL1 promoter carried in the low-copy-number plasmid pSEYC68-GAL (44). The resulting construct was named pTH7. The YOR1-lacZ fusion plasmid was previously described (26), and a mutated variant of this clone that lacked the PDREs was constructed.
Construction of a PDR13 mutant library and selection of hyperactive PDR13 alleles.
pTH86 DNA (20 μg) was mutagenized with 100 μl of 45% formic acid for 1 min. The DNA was recovered by ethanol precipitation, and the PDR13 gene was amplified by PCR from this mutagenized template by use of the flanking T3 and T7 universal primers. The PCR product was cleaved with SalI/NotI and cloned into similarly digested pRS314. The library was amplified in bacteria, and plasmid DNA was prepared.
Strain TCH4 was transformed by a high-efficiency method (21) with the mutant PDR13 library and plated on SD plates (49) containing 0.25 μg of cycloheximide per ml. Survivors were tested for the presence of elevated levels of PDR5-lacZ expression, and plasmids were recovered from appropriate transformants. TCH4 cells were retransformed with each plasmid to confirm that the elevated cycloheximide tolerance and PDR5 expression phenotypes were linked to the plasmid. The sequence of the entire PDR13 gene carried in each plasmid was determined by the University of Iowa DNA Core Facility by use of a set of custom oligonucleotide primers.
Immunological methods.
Rabbit polyclonal antisera were generated by standard immunization techniques (27) against Pdr1p and Pdr13p expressed in bacteria. Both S. cerevisiae proteins were produced as fusion proteins with glutathione S-transferase (GST) carried in plasmid pGEX-KG (23). The GST-Pdr13p fusion protein was constructed by cloning into SacI/NotI-cleaved pGEX-KG full-length Pdr13p downstream of the GST cassette carried in pGEX-KG as a SacI/NotI fragment. The GST-Pdr1p fusion protein was generated by cloning an EcoRI fragment encoding Pdr1p residues 768 to 1063 into the EcoRI site of pGEX4. Both fusion proteins were purified through the use of glutathione-agarose columns as described previously (19).
Protein extraction and analysis.
Cells were grown in minimal medium to an A600 of 0.5 to 0.7, harvested at 4°C by centrifugation, and resuspended in sorbitol breaking buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris [pH 7.4]), with protease inhibitors. Cells were broken by agitation in the presence of glass beads for 25 min at 4°C and centrifuged (Eppendorf 5415C) at 12,000 rpm for 5 min. Protein concentrations were determined by the method of Lowry et al. (41). Equal amounts of protein were resuspended in Laemmli sodium dodecyl sulfate (SDS) loading buffer (37) and analyzed by Western blotting with the anti-Pdr1p or anti-Pdr13p antisera. Antigen-antibody complexes were visualized with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin secondary antibody and enhanced chemiluminescence reagents (Pierce Supersignal).
Gene disruption.
A PDR13 disruption allele was produced by deletion of a BglII fragment from the wild-type PDR13 gene carried as a SalI/NotI fragment in pRT5. The deleted BglII fragment was replaced with the hisG-URA3-hisG fragment from pNKY51 (1). The resulting plasmid replaced PDR13 DNA from 41 bp upstream of the putative ATG to residue 203 with hisG-URA3-hisG. This plasmid was designated pRT6 and was cleaved with SalI/NotI prior to transformation into wild-type cells (SEY6210). URA3 transformants were selected, and correct integration was confirmed by Southern blotting (47). The URA3 gene was removed from a selected disruptant by treatment with 5-fluoro-orotic acid (7), and the resulting pdr13-Δ1::hisG strain was designated YRT9.
RESULTS
Identification of PDR13 as a pleiotropic drug resistance gene.
We previously reported a high-copy-number plasmid library screen in which colonies that exhibited elevated oligomycin resistance were identified by replica plating (35). Two different loci that were both capable of consistently elevating oligomycin tolerance were recovered in this screen. Characterization of the YOR1 structural gene, encoding an ABC transporter protein, has already been reported (35). The second locus found was designated PDR13.
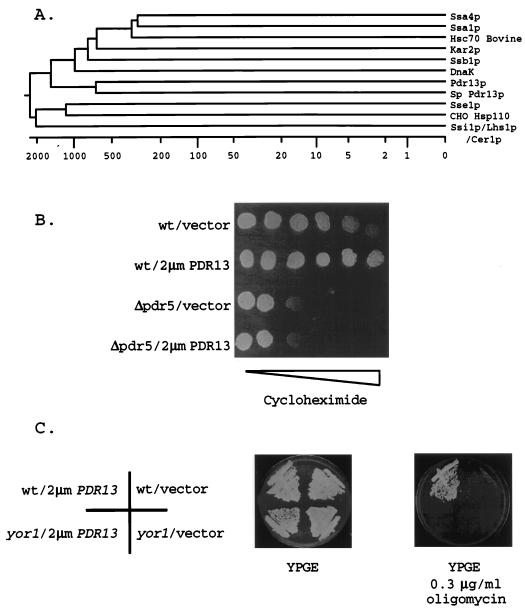
High-copy-number plasmids carrying PDR13 elevated both oligomycin and cycloheximide resistance. This effect was not seen with the 2μm YOR1 clone, which increased oligomycin tolerance only (35). DNA sequence analysis of the S. cerevisiae insert in the PDR13 clone indicated that this fragment came from chromosome VIII. Subcloning analysis established that the PDR13 gene corresponded to the YHR064c locus that was found to encode an Hsp70 homolog during the sequencing of chromosome VIII (31). Alignment of Pdr13p with other Hsp70 proteins from S. cerevisiae, Schizosaccharomyces pombe, and bovine sources indicated that Pdr13p showed the highest degree of sequence similarity (40%) to an S. pombe Hsp70 protein and lower levels of homology to other Hsp70 proteins (e.g., 27% similarity to Ssa4p). This sequence similarity is relatively low compared to that seen between some other Hsp70 proteins, which can be as high as 81% (Ssa1p and Ssa4p). However, the sequence similarity of the large Hsp70 proteins (Ssi1p/Lhs1p/Cer1p, Sse1p, and Sse2p) to the other family members is at least as low as that exhibited by Pdr13p (45). Based on these alignment data, Pdr13p is a unique member of the Hsp70 family in S. cerevisiae.
Having determined that this Hsp70 homolog was able to increase both oligomycin and cycloheximide resistance, we examined the influence of PDR13 on strains lacking either PDR5 or YOR1. A large body of data implicates PDR5 as encoding an ABC transporter protein that is required for cycloheximide tolerance (5, 6, 28, 39), while we have shown that YOR1 is a key determinant of oligomycin resistance (35). To evaluate if PDR13 acted through these loci, a 2μm plasmid containing PDR13 was introduced into isogenic wild-type and Δpdr5 and yor1 mutant strains. Cycloheximide resistance and oligomycin resistance were then assayed in each genetic background.
The presence of the Δpdr5 allele eliminated the ability of high-copy-number PDR13 to stimulate cycloheximide tolerance, even at very low concentrations of cycloheximide (Fig. 1). High-copy-number PDR13 was also unable to increase oligomycin resistance to normal levels in a strain lacking YOR1. These findings strongly suggested that Pdr13p required the presence of PDR genes to normally elevate drug resistance and suggested two possible models for the action of this Hsp70 protein. First, perhaps Pdr13p influences the activity of the transcription factors that regulate the expression of PDR genes. Second, since Hsp70 proteins are known to influence protein folding and translocation across membranes (12), perhaps Pdr13p influences the insertion of Pdr5p and Yor1p into membranes. Below we provide evidence that Pdr13p acts to stimulate expression of the PDR5 and YOR1 loci.
FIG. 1.
Drug resistance conferred by the Hsp70 homolog Pdr13p requires the presence of PDR genes. (A) A phylogenetic tree showing the relationship of the Pdr13p sequence to sequences of other Hsp70 homologs was generated by use of the Megalign routine of Lasergene software (DNAstar). Sequences were aligned by use of the Jotun-Hein algorithm. The Hsp70 proteins are from S. cerevisiae, with the following four exceptions: Hsc70 (bovine), DnaK (E. coli), Sp Pdr13p (S. pombe Pdr13p homolog), and CHO Hsp110 (Cricetulus griseus). The numbers on the bottom of the panel indicate the evolutionary distances between the sequences. (B) The functional dependence of Pdr13p on the presence of PDR5 was tested by placing 1,000 cells of the indicated genotype on a YPD (49) plate containing a gradient of cycloheximide (indicated by the bar of increasing width). Isogenic wild-type (wt) or Δpdr5 cells were transformed with a high-copy-number vector plasmid (pRS426) or the same plasmid carrying the PDR13 structural gene. (C) The indicated transformants were streaked on YPGE medium (49) or YPGE medium plus oligomycin at 0.3 μg/ml and incubated at 30°C. Wild-type (wt) cells are SEY6210, while the yor1 (DKY7) mutant is an isogenic derivative. Plasmids were as in panel B.
High-copy-number PDR13 elevates the expression of Pdr1p target genes.
To determine if high-copy-number PDR13 influenced the level of expression of PDR loci, we used gene fusions between the PDR5 and YOR1 structural genes and lacZ. Each of these plasmid-borne fusion genes has been demonstrated to respond to the same transcriptional control signals as its chromosomal counterpart (33, 35). A TRP5-lacZ fusion gene was used as a control for a locus unrelated to the PDR network. Each fusion gene was introduced into a wild-type cell along with either a high-copy-number plasmid containing PDR13 or the empty vector alone. β-Galactosidase activities were then determined for transformants carrying these plasmids.
Both the PDR5-lacZ and the YOR1-lacZ fusion genes produced higher levels of β-galactosidase enzyme activity in the presence of high-copy-number PDR13 (Table 2). PDR5-dependent enzyme activity increased from 36 to 170 U/unit of optical density at 600 nm (OD600) upon introduction of the 2μm plasmid carrying PDR13. Similarly, YOR1-dependent enzyme activity increased from 3.5 to 14 U/OD600 in the presence of high-copy-number PDR13. TRP5-lacZ expression was not affected by changes in PDR13 gene dosage.
TABLE 2.
Expression of Pdr1p-regulated genes is elevated in the presence of high-copy-number PDR13a
| Fusion gene | β-Galactosidase activity (U/OD600) in the presence of:
|
|
|---|---|---|
| Vector | 2μm PDR13 | |
| PDR5-lacZ | 36 ± 13 | 170 ± 34 |
| mPDR5-lacZ | 0.5 ± 0.3 | 0.6 ± 0.3 |
| YOR1-lacZ | 3.5 ± 0.5 | 14 ± 2 |
| mYOR1-lacZ | 2 ± 0.1 | 3 ± 0.2 |
| SNQ2-lacZ | 13 ± 3 | 40 ± 8 |
| TRP5-lacZ | 37 ± 8 | 35 ± 7.5 |
A wild-type strain (SEY6210) was transformed with the indicated lacZ fusion genes carried on low-copy-number plasmids along with either the 2μm plasmid pRS424 (vector) (10) or pRS424 carrying PDR13. Plasmids containing altered versions of the PDR5 and YOR1 promoters lacking the normal PDREs are designated mPDR5-lacZ and mYOR1-lacZ, respectively. These mutant plasmids have been described elsewhere (26, 34). Appropriate transformants were grown in minimal medium with necessary supplements, and β-galactosidase activities were assayed as described previously (24). The values reported represent the averages of at least three independent determinations ± standard errors.
These data indicated that the most likely mechanism of action of Pdr13p was to stimulate the transcription of the PDR5 and YOR1 structural genes, which would result in increased cycloheximide and oligomycin resistances, respectively. Previous studies of the function of the PDR5 and YOR1 promoters indicated that the action of the closely related zinc finger-containing transcription factors Pdr1p and Pdr3p was required for normal expression of each of these loci (15, 33, 35). Pdr1p and Pdr3p were shown to bind to DNA elements designated PDREs (34). To determine if the PDREs were required for the observed effect of Pdr13p, we used mutant derivatives of the PDR5-lacZ and YOR1-lacZ fusion genes lacking normal copies of the PDREs. These fusion genes were then evaluated with regard to β-galactosidase levels in response to changes in PDR13 gene dose as described above for the wild-type lacZ fusion genes (Table 2).
Loss of the PDREs from the PDR5 promoter led to a precipitous decrease in β-galactosidase activity produced by the resulting fusion gene. This low level of expression was unaffected by the presence of high-copy-number PDR13. Loss of the single PDRE from the YOR1 promoter eliminated the ability of the resulting gene fusion to respond to an increase in PDR13 gene dose. These data suggest that Pdr13p acts through the PDREs in both promoters and further that Pdr1p and/or Pdr3p is a target for the action of Pdr13p. To address this possibility, we examined the activity of Pdr13p in strains lacking PDR1 and/or PDR3.
PDR13 affects the function of Pdr1p but not Pdr3p.
To test the requirement of Pdr1p and/or Pdr3p for normal Pdr13p activity, we used a series of isogenic strains lacking PDR1, PDR3, or both genes (33). Each strain was transformed with a 2μm vector plasmid or the same plasmid carrying PDR13. Transformants were then tested for their cycloheximide resistance phenotypes by spot test assays (Fig. 2).
FIG. 2.
Elevation of cycloheximide resistance by high-copy-number PDR13 requires the presence of PDR1. A strain lacking both the PDR1 and the PDR3 structural genes (PB4) was transformed with the indicated plasmids, resulting in low- or high-copy-number forms of these genes. Along with the plasmids expressing PDR1 and/or PDR3, a 2μm clone of PDR13 or a high-copy-number vector was introduced into the cells. Transformants were grown in media to select for the presence of each plasmid and then assayed for growth on rich medium (YPD) or this same medium containing the concentrations of cycloheximide shown.
High-copy-number PDR13 was only able to increase cycloheximide resistance if the PDR1 gene was present. Removal of the PDR3 gene did not affect Pdr13p-stimulated cycloheximide tolerance. The cycloheximide resistance of mutant strains lacking either PDR1 or PDR1 and PDR3 was not enhanced upon introduction of a 2μm plasmid carrying PDR13. We interpret these data as indicating that the action of Pdr13p is specific to Pdr1p and that Pdr3p is dispensable.
To further examine the specificity of Pdr13p for Pdr1p, high-copy-number plasmids carrying either the PDR1 or the PDR3 structural gene were introduced into a Δpdr1 pdr3 strain, and the gene dosage of PDR13 was varied as described above. Transformants were again assayed for their ability to respond to cycloheximide challenge by spot test assays (Fig. 2).
A Δpdr1 pdr3 strain containing a 2μm plasmid carrying the PDR3 gene exhibited more growth in the presence of cycloheximide than the same strain transformed with a vector plasmid alone. We believe that this increase in cycloheximide resistance is due to the overproduction of Pdr3p when its structural gene is present in high copy numbers, as described before (33). However, this elevated cycloheximide resistance was unchanged when the PDR13 gene dose was increased. The presence of a 2μm clone of the PDR1 gene was highly responsive to increases in PDR13 copy number and conferred the highest level of cycloheximide-resistant growth. This finding suggests that even elevated Pdr3p levels are not responsive to Pdr13p and provides further evidence for the specificity of Pdr13p for Pdr1p.
To confirm that the observed increases in cycloheximide resistance represented enhanced expression of PDR5, we introduced a PDR5-lacZ fusion gene into the set of isogenic strains and varied the gene dosages of PDR1 and/or PDR3. A high-copy-number plasmid carrying PDR13 or the empty vector alone was also introduced, and PDR5-dependent β-galactosidase levels were determined.
Expression of the PDR5-lacZ fusion gene responded to the presence of high-copy-number PDR13 only if the PDR1 gene was intact (Table 3). Both the wild-type and the Δpdr3 strains exhibited an approximately threefold increase in PDR5-lacZ expression when the high-copy-number PDR13-containing plasmid was present. The Δpdr1 strain expressed PDR5-dependent β-galactosidase activity at 22.5 U/OD600; this level was not increased upon introduction of the high-copy-number PDR13-containing plasmid. The Δpdr1 pdr3 strain produced very low levels of enzyme activity that were not increased by the high-copy-number PDR13-containing plasmid.
TABLE 3.
Elevation of PDR5 expression by high-copy-number PDR13 requires the presence of PDR1a
| Fusion gene | β-Galactosidase activity (U/OD600) in the following strains in the presence of the indicated plasmid:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type
|
Δpdr1
|
Δpdr3
|
Δpdr1 pdr3
|
|||||
| Vector | 2μm PDR13 | Vector | 2μm PDR13 | Vector | 2μm PDR13 | Vector | 2μm PDR13 | |
| PDR5-lacZ | 39 ± 12 | 147 ± 33 | 22.5 ± 5 | 22.5 ± 6 | 44 ± 11 | 186 ± 45 | 2 ± 0.7 | 2 ± 0.5 |
| TRP5-lacZ | 29 ± 2 | 30 ± 4 | 31 ± 6 | 32.5 ± 6 | 35.5 ± 2 | 36 ± 10 | 41 ± 3 | 47 ± 8 |
A set of isogenic strains containing the indicated alleles of PDR1 and PDR3 were transformed with PDR5-lacZ and TRP5-lacZ fusion genes on low-copy-number plasmids. In addition, either the 2μm plasmid pRS424 (vector) or pRS424 carrying the wild-type PDR13 locus was introduced into each transformant. Transformants were grown and assayed for β-galactosidase activities as described in Table 2, footnote a.
These data are consistent with the hypothesis that Pdr13p acts as an upstream regulator of Pdr1p. However, these experiments evaluated the effect of PDR13 when this gene was present on a high-copy-number plasmid. To examine the role of the chromosomal PDR13 gene, we prepared a strain containing a disrupted allele of this gene.
Growth defects of cells lacking PDR13.
A gene disruption allele of PDR13 was prepared by replacing genomic DNA from 41 bp upstream of the potential PDR13 ATG to amino acid 203 of the coding sequence with hisG. The resulting allele, pdr13-Δ1::hisG, has a deletion of 203 amino acids from the conserved Hsp70 ATPase domain of Pdr13p. The PDR13 locus was disrupted in both haploid and diploid cells. As we found that PDR13 is not an essential gene (see below), we focused our analysis on the isogenic wild-type and pdr13-Δ1::hisG strains.
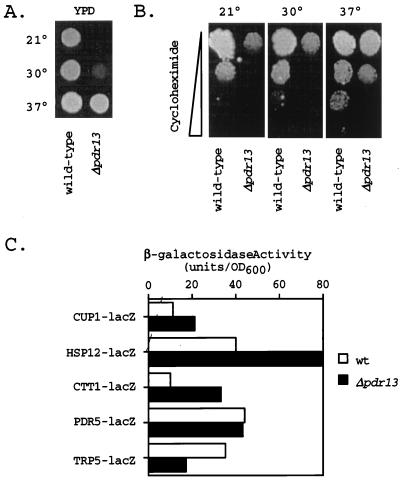
Introduction of the pdr13-Δ1::hisG allele produced a cell that was viable but unable to grow at a normal rate (Fig. 3). The Δpdr13 strain exhibited a cold-sensitive growth phenotype. The doubling times of wild-type cells in YPD medium were 94, 85, and 81 min when measured at 21, 30, and 37°C, respectively. The isogenic Δpdr13 strain showed doubling times of 320, 189, and 122 min at the same three temperatures in the same medium.
FIG. 3.
Cells lacking PDR13 are defective in growth. (A) One thousand cells each of isogenic wild-type and Δpdr13 strains were placed on YPD medium and incubated at the indicated temperatures (Celsius) for 2 days. (B) Isogenic wild-type and Δpdr13 cells were tested as in panel A for growth on a YPD plate containing a gradient of cycloheximide increasing from 0 to 0.25 μg/ml (indicated by the width of the bar on the left). Plates were incubated at 21, 30, or 37°C for 4 days. (C) Cells of the wild type (wt) and the isogenic Δpdr13 derivative were transformed with the indicated lacZ fusion genes carried on low-copy-number plasmids. Transformants were grown in minimal media at 30°C, and β-galactosidase activities were determined. Values are the averages of at least two determinations.
The temperature-dependent ability of the Δpdr13 strain to tolerate cycloheximide was also evaluated by spot test assays. Δpdr13 cells were defective for growth in the presence of cycloheximide, and the reduction in growth in the presence of this drug closely paralleled the general growth defect of the Δpdr13 strain. We were not able to demonstrate a convincing drug resistance-specific defect in Δpdr13 cells and attribute this inability to the general poor growth of these cells.
Expression of several different lacZ fusion genes was also assayed in the Δpdr13 background. A PDR5-lacZ, TRP5-lacZ, CUP1-lacZ, HSP12-lacZ, or CTT1-lacZ fusion plasmid was introduced into wild-type and Δpdr13 cells, and β-galactosidase activities were determined. The CTT1 and HSP12 genes are stress-responsive loci that are regulated by the stress response element-binding proteins Msn2p and Msn4p (42, 46, 48, 51). CUP1 is regulated by the heat shock transcription factor in response to heat shock and oxidative stress (40). These reporter genes were assayed to evaluate the possibility that the loss of the PDR13 gene would elicit a generalized stress response mediated through either the stress response element- or the heat shock transcription factor-dependent systems. Consistent with this interpretation, CTT1-lacZ expression increased by 300% in comparison with that in isogenic Δpdr13 and wild-type cells. HSP12-lacZ expression and CUP1-lacZ expression both increased by 200% in the absence of the PDR13 locus. PDR5-lacZ expression was not significantly affected but TRP5-lacZ expression was decreased to 50% normal in Δpdr13 cells.
One interpretation consistent with the data given above is that Δpdr13 cells are constitutively stressed due to the absence of Pdr13p. This idea is reflected in the diminished rate of growth of these cells as well as the enhanced expression of CTT1, HSP12, and CUP1. The generalized stress caused by the loss of PDR13 made the analysis of specific defects in PDR gene regulation impractical in this genetic background. Previous experiments showed that the loss of multiple PDR genes, including PDR1, does not detectably influence cell growth rate (see reference 4 for a review). This result argues that Pdr13p has other target proteins that contribute to normal cell growth. We explored this possibility by isolating mutant forms of Pdr13p that support normal growth of cells but are hyperresistant to drugs.
Isolation of a hyper-drug-resistant form of Pdr13p.
To dissect the action of Pdr13p on the pleiotropic drug resistance and growth phenotypes, we selected mutant forms of this protein that were able to confer elevated drug resistance but still provided normal growth. This was accomplished by constructing a mutant library of low-copy-number plasmids carrying the PDR13 gene. This mutant library was then transformed into Δpdr13 pdr3 cells containing an integrated PDR5-lacZ fusion gene. We selected transformants that grew normally, were hyperresistant to cycloheximide, and exhibited elevated levels of β-galactosidase activity. Plasmids were recovered from these transformants and retested to confirm that the gene responsible for the mutant phenotypes was linked to the plasmid. The DNA sequence alteration of the PDR13 gene was then determined for several plasmids that exhibited linkage to the mutant phenotypes.
The sequences of eight isolates indicated a single amino acid change: serine at position 295 was replaced with phenylalanine (S295F). The relative drug resistances of a Δpdr13 pdr3 strain transformed with a high-copy-number PDR13-containing plasmid or with low-copy-number plasmids expressing the wild-type or S295F forms of Pdr13p were evaluated by spot test assays (Fig. 4).
FIG. 4.
Enhanced drug resistance of cells expressing S295F Pdr13p. A strain lacking the PDR13 gene (TCH1) was transformed with low-copy-number plasmids expressing the wild type (PDR13) or the S295F (S295F PDR13) form of PDR13 or a high-copy-number plasmid carrying PDR13 (2μm PDR13). Appropriate transformants were tested for their relative resistance to the presence of the indicated concentrations of cycloheximide or oligomycin by spot test assays.
The defective growth of the Δpdr13 pdr3 strain was restored to normal by transformation with all of these versions of the PDR13 gene (data not shown). However, differences in drug resistance were clearly apparent. The presence of either high-copy-number PDR13 or low-copy-number S295F PDR13 produced a large increase in tolerance to cycloheximide and oligomycin compared to that in low-copy-number wild-type PDR13 transformants. The single amino acid change in S295F Pdr13p was able to elevate cycloheximide and oligomycin resistance to a level at least equal to that conferred by a high-copy-number clone of the wild-type locus. Other experiments demonstrated that the S295F allele was dominant over the wild-type PDR13 locus (data not shown).
To confirm that S295F Pdr13p also acted to elevate the expression of Pdr1p target genes, the Δpdr13 pdr3 strain was transformed with low-copy-number plasmids expressing either the wild-type or the S295F form of Pdr13p as well as a high-copy-number plasmid carrying wild-type PDR13. In addition to these plasmids expressing Pdr13p, a TRP5-lacZ, CTT1-lacZ, or PDR5-lacZ fusion gene was introduced into each background. The expression of each fusion gene was assayed in these three genetic backgrounds (Table 4).
TABLE 4.
The S295F allele of PDR13 elevates Pdr1p target gene expressiona
| PDR13 allele | β-Galactosidase (U/OD600) activity in the presence of:
|
||
|---|---|---|---|
| PDR5-lacZ | TRP5-lacZ | CTT1-lacZ | |
| pdr13-Δ1 | 70 ± 17 | 16 ± 2 | 23 ± 3 |
| PDR13 | 58 ± 16.5 | 49 ± 5.5 | 9 ± 2 |
| 2μm PDR13 | 299 ± 84 | 50 ± 2 | 5 ± 1 |
| S295F PDR13 | 182 ± 38 | 51 ± 7 | 7 ± 1 |
A Δpdr13 strain was transformed with a low-copy-number vector plasmid (pRS314), pRS314 carrying the wild-type or S295F form of the PDR13 gene, or a high-copy-number plasmid containing the wild-type PDR13 locus (2μm PDR13). Along with these plasmids giving rise to the different PDR13 alleles, low-copy-number plasmids carrying PDR5-lacZ, TRP5-lacZ, or CTT1-lacZ fusion genes were introduced into the cells. Transformants of interest were grown in minimal medium, and β-galactosidase activities were determined as described previously (24).
The presence of the S295F allele of PDR13 elevated the expression of PDR5-lacZ to levels similar to those produced by introduction of the 2μm plasmid carrying PDR13. Transformants containing any of the three versions of the PDR13 gene produced equivalent levels of TRP5-lacZ expression, indicating that Pdr13p did not globally enhance gene expression. Finally, the presence of the S295F allele of PDR13 returned CTT1-lacZ expression to wild-type levels. These data indicated that the gain-of-function S295F mutant form of Pdr13p reproduced what was previously seen with the high-copy-number wild-type gene. The S295F form of Pdr13p appeared to be a more effective activator of drug resistance than the high-copy-number wild-type protein, although both produced similar levels of PDR5-dependent β-galactosidase activity. The reason for this effect is unknown but may be related to the different replication properties of the two plasmid vectors used in this experiment.
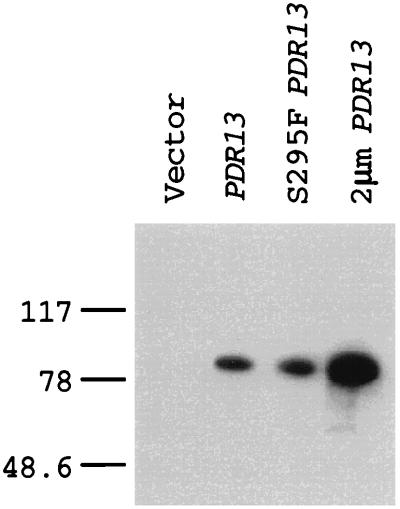
To further explore the nature of Pdr13p control of Pdr1p function, we produced a polyclonal antiserum directed against Pdr13p. This antiserum was used in Western blot analysis of protein extracts prepared from Δpdr13 cells and transformants carrying the low-copy-number wild-type or S295F form of PDR13 or a 2μm plasmid expressing wild-type Pdr13p (Fig. 5). The anti-Pdr13p antiserum detected a single polypeptide with a molecular mass of 81 kDa in cells transformed with plasmids expressing the three different PDR13 alleles. The 81-kDa protein was absent from protein extracts prepared from the Δpdr13 cells. Importantly, both the wild-type and the S295F forms of Pdr13p were expressed at equivalent levels, while the high-copy-number plasmid carrying PDR13 produced approximately threefold more protein than either of the low-copy-number plasmids. This result confirms that the elevated Pdr1p function seen in the presence of the S295F Pdr13p derivative is due to a change in the activity of Pdr13p rather than to a change in its steady-state level.
FIG. 5.
Equivalent expression of wild-type and S295F forms of Pdr13p. Whole-cell protein extracts were prepared from a Δpdr13 mutant strain carrying a vector plasmid (pRS314), a low-copy-number plasmid containing either wild-type PDR13 or the S295F allele of PDR13, and a high-copy-number plasmid (2μm PDR13) bearing the wild-type PDR13 gene. Protein (25 μg) was resolved by SDS-PAGE and analyzed by Western blotting with anti-Pdr13p antiserum. Numbers on the left are in kilodaltons.
Pdr13p acts posttranslationally to regulate Pdr1p.
A central issue that emerges from these studies concerns the details of the activation of Pdr1p by Pdr13p. Pdr13p may change the level of expression of Pdr1p or, alternatively, may regulate the activity of Pdr1p. To examine which of these explanations was likely to be correct, we carried out two different experiments. First, the PDR1 coding sequence was placed under the control of the GAL1 promoter, and the resulting GAL1-PDR1 chimera was examined for its response to different PDR13 gene dosages. Second, an antiserum was produced against Pdr1p and used to directly examine the steady-state levels of Pdr1p in strains containing different PDR13 gene doses.
The GAL1-PDR1 fusion gene was constructed by placing a fragment of PDR1 (extending from its ATG to the end of the coding sequence) downstream of the GAL1 promoter carried on a low-copy-number vector. This plasmid was introduced into a wild-type strain along with a high-copy-number plasmid expressing Pdr13p or the empty vector. Transformants were then tested for cycloheximide tolerance by spot test assays (Fig. 6).
FIG. 6.
Pdr13p does not act on the PDR1 promoter region. A wild-type strain was transformed with a low-copy-number plasmid containing a GAL1-PDR1 fusion gene. Along with this fusion gene, a high-copy-number plasmid containing the PDR13 gene (2μm PDR13) or the plasmid vector alone (vector) was introduced. Transformants containing each pair of plasmids were tested for their ability to grow on YPGE-galactose medium (YPGE-Gal) (49) lacking or containing cycloxheximide.
Pdr1p, produced from the GAL1 promoter, still responded to the presence of elevated PDR13 gene dose. Transformants carrying the GAL1-PDR1 fusion gene were more resistant to cycloheximide in the presence of multiple copies of PDR13 than in the presence of a single copy of this gene. This observation strongly argues that the effect of Pdr13p on Pdr1p function is not a consequence of control of PDR1 promoter function.
To directly examine the steady-state levels of Pdr1p in cells with various PDR13 gene doses, a polyclonal antiserum directed against Pdr1p was produced. Protein extracts were prepared from Δpdr1 pdr3 cells transformed with high-copy-number plasmids carrying PDR1 and PDR13 or carrying PDR1 alone or with the empty vector. The steady-state level of Pdr1p in each transformant was then evaluated by Western blot analysis of equal amounts of protein extracts resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 7). The anti-Pdr1p antiserum recognized a single polypeptide species of approximately 121 kDa in transformants carrying the PDR1 gene. The levels of Pdr1p were not altered by changes in PDR13 gene doses. This observation provides strong support for the hypothesis that Pdr13p control of Pdr1p occurs through posttranslational modulation of the ability of the zinc finger-containing transcription factor Pdr1p to activate gene expression.
FIG. 7.
Pdr13p regulates Pdr1p function at a posttranslational step. Whole-cell protein extracts (100 μg) were electrophoresed by SDS-PAGE. Protein extracts were made from Δpdr1 pdr3 cells containing (+) or lacking (−) the indicated high-copy-number plasmids. After electrophoresis, the polypeptides were transferred to nitrocellulose and analyzed by blotting with anti-Pdr1p antiserum.
DISCUSSION
While the role of Hsp70 proteins in polypeptide metabolism has long been appreciated, recent work has indicated that Hsp70 proteins can act as regulatory factors that modulate the activity of transcription factors (20). One of the earliest examples of Hsp70 proteins influencing the activity of a transcription factor came from analysis of expression of the SSA4 gene in S. cerevisiae (8). S. cerevisiae strains lacking the Hsp70-encoding loci SSA1 and SSA2 overproduce the SSA4 transcript and protein through activation of the heat shock transcription factor (25). Detailed analysis of the maturation of the progesterone receptor has shown that Hsp70 proteins must act on this factor so that it can achieve its active conformation (9). Escherichia coli Hsp70 DnaK acts to negatively regulate its own transcription (reviewed in reference 13).
The data presented here argue that Pdr13p is a positive regulator of Pdr1p. Increases in the level of Pdr13p lead to an enhancement in the activity of Pdr1p. While the action of Pdr13p is clearly not restricted to its effect on Pdr1p (see below), Pdr13p does not affect the function of Pdr3p. Pdr3p shows 36% sequence identity to Pdr1p, binds to the same DNA element as Pdr1p, and shows extensive functional overlap with Pdr1p (15, 33). Δpdr1 cells containing a 2μm clone of PDR3 exhibit higher cycloheximide resistance than wild-type cells (Fig. 2). However, the function of even this presumably elevated level of Pdr3p cannot be regulated by Pdr13p, further supporting the notion that this Hsp70 protein controls Pdr1p but not Pdr3p activity.
This observation is important in light of what is known about the phenotype of cells lacking either the PDR1 or the PDR3 gene. Loss of PDR1 from the cell has a pronounced phenotypic effect on cycloheximide resistance, with a Δpdr1 cell exhibiting hypersensitivity to a broad range of compounds (2, 15, 33). In opposition to the effect of PDR1 removal, a Δpdr3 cell is not markedly different from a wild-type cell in terms of cycloheximide tolerance (15, 33). Surprisingly, expression studies with PDR5, the principal Pdr1p-Pdr3p target gene for cycloheximide resistance, indicate that both mRNA and PDR5-lacZ expression levels are similar in Δpdr1 and Δpdr3 strains (33). A key difference in these two experiments is that one is done in the presence of cycloheximide (phenotype testing), while one is performed in its absence (expression measurements). A hypothesis to explain these data is that drug exposure acts through Pdr13p to stimulate Pdr1p activity, and this activation of Pdr1p is required for normal drug resistance. The fact that Pdr1p but not Pdr3p can respond to Pdr13p would explain the observed phenotypes of strains individually lacking either of these transcription factors. This hypothesis is currently being evaluated.
While Pdr13p does not affect the function of Pdr3p, there are clearly targets of Pdr13p action other than Pdr1p. The reduction in growth rate exhibited by Δpdr13 cells indicates that Pdr1p cannot be the only downstream target of Pdr13p. Strains lacking Pdr1p, Pdr1p and Pdr3p, and downstream Pdr1p-Pdr3p target genes have not been found to exhibit growth phenotypes in the absence of drugs (reviewed in reference 4). In contrast, a Δpdr13 strain displays a growth phenotype that is partially suppressed at 37°C (Fig. 3). This finding suggests the possibility that some other Hsp70 protein expressed at 37°C is able to provide some degree of Pdr13p function. There is precedence for Hsp70 gene disruptions leading to cold-sensitive phenotypes, as mutant strains lacking both SSB1 and SSB2 exhibit a cold-sensitive phenotype that can be suppressed by elevating the growth temperature (14).
An intriguing possibility suggested by the appearance of a growth defect in Δpdr13 cells is that Pdr13p may act as a link between essential cellular functions and the PDR pathway. Perhaps Pdr13p coordinates the activity of the PDR genes with fundamental cell metabolism and activates Pdr1p function in response to an as-yet-unknown signal. It is important to bear in mind that drugs such as cycloheximide and oligomycin are unlikely to represent the natural substrates of the PDR genes, making the true function of these genes a still-unresolved question.
The identification of the S295F form of Pdr13p as a hyperactive regulator of Pdr1p function is interesting for three different reasons. First, this particular mutation provides a new class of Hsp70 mutants with altered function. Mutant Hsp70 derivatives with increased function have not been reported before. Isolation of this class of mutant Hsp70 proteins likely has been slowed by the complex, pleiotropic nature of the roles of these proteins. Our ability to mutationally separate the essential function(s) of Pdr13p from its role in the control of Pdr1p activity will allow genetics to be used to analyze the molecular details underlying this regulatory interaction. Second, the S295F mutation is located in the most highly conserved segment of Hsp70 proteins, the ATPase domain (18). This idea is consistent with the work of James et al. (30), who showed that at least part of the specificity of action of S. cerevisiae Hsp70 proteins was determined by this N-terminal ATPase domain. dnaK mutants with a partial loss of function also mapped to this domain (52). Finally, the change from serine to phenylalanine suggests the possibility that phosphorylation is involved. Alternatively, the large change in amino acid identity between serine and phenylalanine may lead to the observed phenotype. Discrimination between these models requires direct biochemical experiments aimed at addressing the regulation and activity of Pdr13p.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM49825 to W.S.M. and DK25295 to the University of Iowa Diabetes and Endocrinology Center. W.S.M. is an established investigator of the American Heart Association.
We thank Elizabeth Craig and David Toft for helpful discussions.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 3.Balzi E, Goffeau A. Multiple or pleiotropic drug resistance in yeast. Biochim Biophys Acta. 1991;1073:241–252. doi: 10.1016/0304-4165(91)90128-4. [DOI] [PubMed] [Google Scholar]
- 4.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 5.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. PDR5: a novel yeast multidrug resistance transporter controlled by the transcription regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 6.Bissinger P H, Kuchler K. Molecular cloning and expression of the S. cerevisiae STS1 gene product. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 7.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine 5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 8.Boorstein W R, Craig E A. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J Biol Chem. 1990;265:18912–18921. [PubMed] [Google Scholar]
- 9.Chen S, Prapapanich V, Rimerman R A, Honore B, Smith D F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins Hsp90 and Hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deely R G. Overexpression of a transporter gene in multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 12.Craig E A, Gambill B D, Nelson R J. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 14.Craig E A, Jacobsen K. Mutations in cognate genes of Saccharomyces cerevisiae HSP70 result in reduced growth rates at low temperatures. Mol Cell Biol. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 16.Delaveau T, Jacq C, Perea J. Sequence of a 12.7 kb segment of yeast chromosome II identifies a PDR-like gene and several new open reading frames. Yeast. 1992;8:761–768. doi: 10.1002/yea.320080909. [DOI] [PubMed] [Google Scholar]
- 17.Egner R, Mahe Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaherty K M, DeLuca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 19.Frangioni J V, Neel B G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 20.Frydman J, Hohfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:75–106. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- 21.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 23.Guan K-L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 24.Guarente L. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 25.Halladay J T, Craig E A. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol Cell Biol. 1995;15:4890–4897. doi: 10.1128/mcb.15.9.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallstrom, T. C., and W. S. Moye-Rowley. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem., in press. [DOI] [PubMed]
- 27.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 28.Hirata D, Yano K, Miyahara K, Miyakawa T. Saccharomyces cerevisiae YDR1, which encodes a member of the ATP-binding cassette (ABC) superfamily, is required for multidrug resistance. Curr Genet. 1994;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- 29.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James P, Pfund C, Craig E A. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 31.Johnston M, Andrews S, Brinkman R, Cooper J, Ding H, Dover J, Du Z, Favello A, Fulton L, Gattung S, Geisel C, Kirsten J, Kucaba T, Hillier L, Jier M, Johnston L, Langston Y, Latreille P, Louis E J, Macri C, Mardis E, Menezes S, Mouser L, Nhan M, Rifkin L, Riles L, St. Peter H, Trevaskis E, Vaughan K, Vignati D, Wilcox L, Wohldman P, Waterston R, Wilson R, Vaudin M. Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII. Science. 1994;265:2077–2082. doi: 10.1126/science.8091229. [DOI] [PubMed] [Google Scholar]
- 32.Kane S E, Pastan I, Gottesman M M. Genetic basis of multidrug resistance of tumor cells. J Bioenerg Biomembr. 1990;22:593–618. doi: 10.1007/BF00762963. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann D J, Burnett P E, Golin J, Mahe Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katzmann D J, Hallstrom T C, Mahe Y, Moye-Rowley W S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 35.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowski A, Soumillion J-P, Konings W N, Goffeau A. Anticancer drugs, ionophoric peptides and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Leonard P J, Rathod P K, Golin J. Loss-of-function mutation in the yeast multiple drug resistance gene PDR5 causes a reduction in chloramphenicol efflux. Antimicrob Agents Chemother. 1994;38:2492–2494. doi: 10.1128/aac.38.10.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leppert G, McDevitt R, Falco S C, Van Dyk T K, Ficke M B, Golin J. Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics. 1990;125:13–20. doi: 10.1093/genetics/125.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X-D, Thiele D J. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- 41.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 42.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 43.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 44.Moye-Rowley W S, Harshman K D, Parker C S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- 45.Rassow J, von Ahsen O, Bomer U, Pfanner N. Molecular chaperones: towards a characterization of the heat-shock protein 70 family. Trends Cell Biol. 1997;7:129–133. doi: 10.1016/S0962-8924(96)10056-8. [DOI] [PubMed] [Google Scholar]
- 46.Ruis H, Hamilton B. Regulation of yeast catalase genes. In: Scandalios J G, editor. Molecular biology of free radical scavenging systems. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 153–172. [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 50.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wild J, Kamath-Loeb A, Ziegelhoffer E, Lonetto M, Kawasaki Y, Gross C A. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci USA. 1992;89:7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu A, Wemmie J A, Edgington N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]