Abstract
We have identified longevity-associated genes in a long-lived Caenorhabditis elegans daf-2 (insulin/IGF receptor) mutant using serial analysis of gene expression (SAGE), a method that efficiently quantifies large numbers of mRNA transcripts by sequencing short tags. Reduction of daf-2 signaling in these mutant worms leads to a doubling in mean lifespan. We prepared C. elegans SAGE libraries from 1, 6, and 10-d-old adult daf-2 and from 1 and 6-d-old control adults. Differences in gene expression between daf-2 libraries representing different ages and between daf-2 versus control libraries identified not only single genes, but whole gene families that were differentially regulated. These gene families are part of major metabolic pathways including lipid, protein, and energy metabolism, stress response, and cell structure. Similar expression patterns of closely related family members emphasize the importance of these genes in aging-related processes. Global analysis of metabolism-associated genes showed hypometabolic features in mid-life daf-2 mutants that diminish with advanced age. Comparison of our results to recent microarray studies highlights sets of overlapping genes that are highly conserved throughout evolution and thus represent strong candidate genes that control aging and longevity.
Aging is a complex process, and it is commonly believed that a variety of genes and metabolic pathways contribute to the deterioration of cells, tissues, and organisms during aging. The complexity of aging is mirrored by an extensive literature describing the diverse mechanisms involved. Molecular mechanisms commonly associated with aging include free radical damage of macromolecules, shortening of telomeres (replicative senescence), accumulation of mutations due to impaired DNA repair mechanisms, low levels of stress resistance, and high levels of apoptosis. System theories of aging include the hormonal regulation of specific or general metabolic functions and immune system functions (for review, see Weinert and Timiras 2003).
Caenorhabditis elegans is widely used as a model system to study aging and longevity-related processes. A number of genes have been demonstrated to play major roles in determining the lifespan of this nematode. The insulin/insulin-like growth factor 1 receptor (I/IGF-1R) homolog DAF-2 is one of the principal components affecting lifespan in C. elegans. The insulin/IGF-1 system also regulates reproduction and lipid metabolism, as well as entry into a state of developmental diapause, called the dauer larva (Kimura et al. 1997; Gems et al. 1998). Dauer larvae are metabolically arrested, long-lived juvenile forms that function in long-term survival and dispersal (Klass and Hirsh 1976). Entry into the dauer state is environmentally induced by food limitation, high population density, and high temperatures (Golden and Riddle 1984). Additionally, mutations in genes that reduce insulin or TGF-β signaling also facilitate entry into the dauer state. DAF-2 signals through a conserved PI3-kinase/Akt pathway, ultimately acting to inhibit the activity of DAF-16, a FOXO family transcription factor (Lin et al. 1997; Ogg et al. 1997). Many mutations within genes in the insulin/IGF-1 pathway have been shown to significantly extend both mean and maximum adult lifespan (Dorman et al. 1995; Ailion et al. 1999; Paradis et al. 1999). This lifespan extension, the “Age” phenotype, is dependent on wild-type function of DAF-16. daf-2 mutants appear youthful and healthy and live more than twice as long as normal worms (Kenyon et al. 1993). Recent studies analyzed the transcriptional targets of DAF-16 in C. elegans and suggested that the insulin/IGF-1 pathway exerts its effect on lifespan by altering the expression of a variety of genes involved in cellular stress response, metabolism, and energy generation (McElwee et al. 2003; Murphy et al. 2003).
We used serial analysis of gene expression (SAGE) (Velculescu et al. 1995) to comprehensively analyze the transcriptomes of C. elegans daf-2(m41) mutant and control worms, which also carried a temperature-sensitive fer-15 (fertility) mutation to prevent the production of offspring that would confound expression profiling of adult worms.
SAGE involves the isolation of unique sequence tags from individual mRNA transcripts. We compared expression profiles of SAGE tags in aging daf-2 adults and between daf-2 mutants and controls. Genes corresponding to these tags were identified and grouped according to their biological function. Here we describe potential new lifespan-regulating genes and gene families that are differentially expressed between daf-2 mutants and controls.
Results
We examined three SAGE libraries of adult daf-2(m41) mutants and two control libraries for lifespan-extending genes. We reasoned that genes important for longevity would include those that were either progressively up-regulated (perhaps induced over time in order to protect against age-associated degradative processes) or down-regulated (repressed in order to minimize deleterious processes) from early to late adulthood. We also reasoned that longevity genes would be differentially expressed between long-lived daf-2 mutants and controls with normal lifespans. To assess physiological and metabolic longevity-related mechanisms, we concentrated on genes with known or inferred functions available at WormBase, an annotated database for C. elegans genes, proteins, and genomic sequences (http://www.wormbase.org/; Harris et al. 2004). Selected genes were grouped according to biological function and their associated biological processes. Strikingly, gene groups (e.g., heat shock protein genes) or gene families (e.g., hsp-16 genes) showed uniform and consistent expression patterns. We describe three different analyses comparing gene expression in daf-2 and control worms, and report fundamental metabolic changes that may contribute to the increased lifespan of daf-2 mutants.
daf-2(m41) mutations trigger dauer larvae formation constitutively at 25°C, as well as thermo-tolerance and lifespan extension. This allele was chosen because it is temperature-sensitive and less pleiotropic than other alleles such as daf-2(e1370) (Gems et al. 1998). Hence, changes in gene expression not associated with longevity should be minimized. Because daf-2 delays the onset of aging (Gems and Riddle 2000), we interpreted 10-d-old daf-2 adults to be the biological equivalent of 6-d-old controls. Throughout this manuscript, 6-d-old daf-2 and control nematodes were considered being of same “chronological age,” whereas 10-d-old daf-2 mutants and 6-d-old controls are referred to as being of comparable “biological age.” This assumption is based on the lifespan evaluation assessed within this study (Fig. 1) and the mortality rate data of Gems and Riddle (2000).
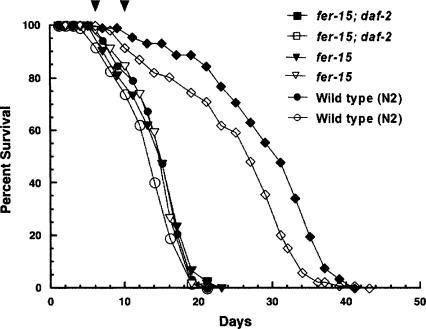
Figure 1.
Survival curves for double mutant daf-2(m41); fer-15(b26ts) (DR1993) and infertile control fer-15(b26ts) (DR1830) adults at 25.5°C. The double mutant lived significantly longer than control strains (P < 0.0001, log rank test). The two arrowheads at the top of the graph correspond to days 6 and 10 of the study, the times at which 100% and 95 ± 3% of the control and daf-2 populations were alive, respectively. Wild type (N2), circles; fer-15, inverted triangles; fer-15; daf-2, diamonds. Filled and open symbols represent data from two independent trials. The mean lifespan (±SE) of wild type was 14.3 ± 0.5 d (n = 222); fer-15, 14.9 ± 0.1d(n = 232); and fer-15; daf-2, 27.3 ± 1.7d(n = 235). Maximum lifespans for the strains were 20 d, 22 d, and 41 ± 1 d, respectively.
Lifespan evaluation of wild-type N2, fer-15, and fer-15; daf-2 adults
The life cycle of C. elegans includes four larval stages, L1–L4, which are separated by molts. After exiting the L4 stage, adult lifespan is ∼2 wks. The daf-2(m41) strain used in our experiments had a mean lifespan (±SE) of 27.3 ± 1.7 d (n = 235) and maximum lifespan of 41 ± 1 d compared to fer-15 control worms that had a mean lifespan of 14.9 ± 0.1 d (n = 232) and maximum lifespan of 22 ± 1 d (all at 25.5°C) that was not significantly different from the wild-type N2's mean lifespan of 14.3 ± 0.5 d (n = 222) and maximum lifespan of 20 ± 1 d (Fig. 1). Days 10 (daf-2) and 6 (control) mark the latest time points at which 100% of the respective populations were still alive. Henceforth, daf-2; fer-15 double mutant worms will be abbreviated as “daf-2,” and fer-15 controls will be referred to as “controls.”
Gene expression analysis of aging daf-2 mutants
SAGE libraries were prepared from daf-2 worms at days 1, 6, and 10 of adulthood. The day 6 library represents gene expression in mid-adult life, whereas day 10 marks the latest time before the occurrence of dead animals in the population.
We analyzed daf-2 libraries from days 1, 6, and 10 to identify tags that are either progressively up- or down-regulated (Fig. 2A) as adult worms age. We hypothesize that some of these genes may be important for increased life expectancy. daf-2 libraries were analyzed for SAGE tags according to the following criteria: tag frequencies must be either progressively increased or decreased from day 1 (d1) to day 10 (d10); either d1 > d6 > d10, or d1 < d6 < d10. We also required that the sum of tag counts for a specific transcript in all three libraries be ≥14 and that the fold difference in expression between days 1 and 10 be greater than 2.5 for at least one member of each gene family. The selection for tags that had an abundance of at least 14 minimized spurious effects due to low tag numbers. This relatively stringent tag abundance cutoff value was reasonable because we generated “deep” SAGE tag sequences from each SAGE library, ∼70,000 tags per library. Although these relatively stringent selection criteria may exclude some rare transcripts, it ensures high data quality.
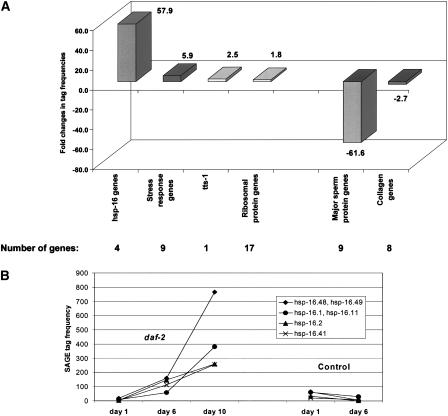
Figure 2.
Expression analysis of daf-2 SAGE libraries. Gene expression was analyzed in daf-2 worms at days 1, 6, and 10 of adulthood. Tags that met our selection criteria were grouped according to biological function. Mean fold differences of tag abundance were calculated between day 1 and day 10 and are shown in (A). The number of genes within a given group or gene family is noted. (B) The expression profile of four hsp-16-like genes in daf-2 and control worms. Expression levels of these genes drastically increase (∼60-fold) over daf-2 lifespan but are only marginally expressed in controls. Genes that are plotted together are closely related and their SAGE tags are indistinguishable; these tag numbers represent the sum of SAGE tags for both genes.
Tags of interest were mapped to a virtual transcriptome of known C. elegans genes (Pleasance et al. 2003; McKay et al. 2004; see also Methods). Selected genes were, if possible, clustered into gene families that were then grouped according to their biological functions. Table 1 gives an overview of the identified genes and gene families, their biological function, tag counts, and fold changes of gene expression. Figure 2A illustrates a strong increase of gene expression in daf-2 adults, from day 1 to day 10, for genes in the hsp-16-like (heat shock protein) and ribosomal protein (rps, rpl, rpa) gene families, and for genes belonging to general stress response pathways (hsp, mtl-1, gst-1, sip-1) as well as tts-1, a gene referred to as transcribed telomere-like sequence (Jones et al. 2001). In contrast, tag numbers for sperm-associated proteins (msp, ssq) and collagen (col) genes decreased with increasing age of daf-2 worms.
Table 1.
Progressive increase of gene expression in daf-2 mutants
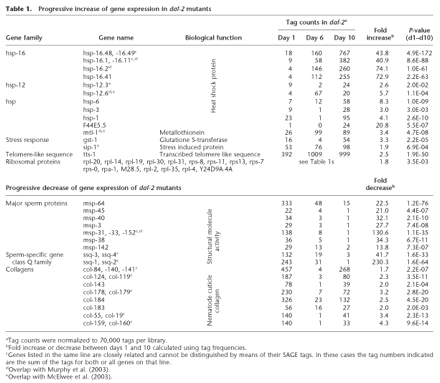
The large increase in hsp-16 expression may not be surprising, as DAF-2 is known to inhibit the forkhead transcription factor DAF-16, a potent activator of hsp-16 transcription (Hsu et al. 2003). We detected an ∼60-fold mean increase of four hsp-16-like gene transcripts; hsp-16.48 (hsp-16.49), hsp-16.1 (hsp-16.11), hsp-16.2 and hsp-16.41 (Fig. 2B). The hsp-16.48 and hsp-16.49 as well as the hsp-16.1 and hsp-16.11 gene pairs share high homologies at their 3′ ends, and their corresponding 14-base pair SAGE tags cannot be unambiguously mapped to the corresponding genes. Throughout this manuscript, closely related genes that ambiguously match the same SAGE tag (e.g., hsp-16.48 and hsp-16.49 or col-84, col-140 and col-141; see also Table 1) are counted together, as they cannot be distinguished from each other by SAGE.
Figure 2B shows a comparison of these genes' expression levels in daf-2 and control worms. While daf-2 mutants increase their stress response potential drastically via activation of hsp-16 genes, control worms have a low overall expression of these genes. Furthermore, daf-2 worms also up-regulate other heat shock proteins including hsp-12.3 and hsp-12.6, hsp-1, hsp-3, and hsp-6, as well as stress response proteins such as mtl-1, gst-1, and sip-1 (Table 1).
The expression of genes associated with sperm function, especially major sperm proteins (msp-like) and sperm-specific Q-proteins (ssq-like) was decreased in daf-2 adults. Sperm-specific proteins are expected to decrease with age as reproduction decreases and stops. Interestingly, the abundance of these msp and ssq tags is initially much higher in daf-2 than in control worms. The expression in daf-2 mutants is reduced from day 1 to day 6 to a level approximately seen on day 1 in control worms (Supplemental Table 1s). In both groups the expression of these sperm protein-associated tags stops altogether at advanced age. It can be hypothesized therefore that daf-2 regulates genes involved in reproduction such as these sperm-specific genes.
Another group of genes that is strongly affected by the daf-2 mutation is the large family of collagen (col) genes. Eight distinguishable col gene groups were shown to be progressively down-regulated throughout adulthood in daf-2 worms (Table 1). These genes code for proteins that share a common nematode cuticle collagen N-terminal domain. A comprehensive gene list including expression profiles in control worms and tag sequences can be found in Supplemental Table 1s.
Comparison of 6-d-old daf-2 adults to 6-d-old controls
To identify gene expression differences and metabolic changes that may lead to the increased life expectancy of daf-2 adults, we analyzed daf-2 and control worms at the same chronological age at day 6. Live control worms, free of contaminating dead animals, could be safely harvested at the latest on day 6. daf-2 mutants, in contrast, live another 4 d before dead worms first appear in the population. SAGE libraries were screened for tags that had an abundance of at least 10 in one of the libraries and were differentially expressed by ≥2.5-fold between daf-2 and controls, with a P-value < 0.05. Table 2 lists gene families affected in daf-2 mutants, their biological function, tag counts, and the fold changes in transcript levels. Supplemental Table 2s shows a comprehensive list of all genes for which expression was significantly altered. Figure 3 summarizes the comparison between daf-2 and control day 6 SAGE libraries. We detected a small number of gene families whose expression was higher in daf-2 than in controls. These include the hsp-16-related genes (hsp-16.48, hsp-16.1, hsp-16.2, hsp-16.41), hsp-12.6, tts-1, neuropeptide-like protein (nlp) genes, transthyretin-like genes, and ribosomal protein genes. nlp genes (nlp-27, -28, -29, -31) are expressed exclusively in neurons or “endocrine/secretory” cells and may encode functional neuropeptides. Some nlp genes are expressed in the amphid ASI neurons that play specific roles in chemosensation and dauer formation (Bargmann and Horvitz 1991; Ren et al. 1996). Furthermore, six genes belonging to the transthyretin-like gene family were expressed at higher levels in daf-2 than in control worms (Fig. 3). Transthyretin-like proteins are deduced to have carrier activity by binding and transporting steroid hormones; therefore, the extent or distribution of hormonal signaling may be enhanced in long-lived worms. Hormonal signaling through the DAF-12 nuclear receptor has been implicated in dauer formation and longevity (Antebi et al. 2000; Hsu et al. 2003).
Table 2.
Comparison of gene expression between daf-2 and control worms on day 6
| Gene family | Gene name | Biological function | Mean fold increasea | P-value | |
| Stress response | |||||
| hsp-16 genes | hsp-16.48, -16.49b | 31.2 | 5.7E-40 | ||
| hsp-16.1, -16.11b,c | 1.8 | 6.0E-03 | |||
| hsp-16.2c | Heat shock protein | 142.3 | 4.2E-29 | ||
| hsp-16.41 | 12.1 | 1.9E-21 | |||
| hsp-12 gene | hsp-12.6c,d | 65.3 | 1.3E-15 | ||
| tts transcript | tts-1 | Transcribed telomere-like sequence | 21.4 | 8.9E-232 | |
| Protein metabolism | |||||
| Ribosomal proteins | rpl-43, rpl-20, rpl-29, rpl-30, rpl-23, rpl-31, rpl-35, rpl-8, rpl-27, rpl-22, rps-11, rps-13, rps-7, rps-17, rps-0, rpa-1, Y105E8A. 16, M28.5, Y48G8AL.8a, Y41D4B.5 | 2.8 | 1.9E-03 | ||
| Cellular signaling | |||||
| Transthyretin-like proteins | ZK697.8, F22A3.2, ZC64.2, C40H1.5c, Y5F2A.2, Y51A2D.10 | Signaling and transport | 7.1 | 1.2E-03 | |
| nlp genes | nlp-27, nlp-28, nlp-29, nlp-31 | Neuropeptide-like protein | 8.2 | 9.2E-05 | |
| Gene family | Gene name | Biological function | Mean fold decreasea | P-value | |
| Lipid metabolism | |||||
| Vitellogenins | vit-4c | Lipid transport | 39.3 | 1.3E-12 | |
| vit-1, vit-2b,c,d | 385.1 | 8.5E-115 | |||
| vit-3, vit-5b,c,d | 100.9 | 4.6E-32 | |||
| vit-6d | 122.4 | 2.2E-37 | |||
| Stress response | |||||
| Cold shock | cey-1, cey-2, cey-3 | Cold-shock DNA-binding domain | 15.1 | 4.3E-06 | |
| Structure | |||||
| Tubulin genes | tbg-1, tbb-1, tbb-2, tba-2 | Microtubule-based movement | 30.1 | 4.5E-08 | |
| Actin genes | act-1, act-2, act-3, act-4, act-5d | Cytoskeleton organization | 26.6 | 5.0E-06 | |
| DNA/RNA metabolism | |||||
| Pumilio genes | puf-3d, puf-5 | RNA binding protein | 19.6 | 4.4E-06 | |
| puf-10, puf-6, puf-7b | 19.6 | 1.7E-06 | |||
| mcm genes | mcm-2, mcm-3, mcm-4, mcm-5, mcm-6 | Initiation of DNA replication | 19.4 | 1.1E-03 | |
| Protein metabolism | |||||
| asp genes | asp-1, asp-2, asp-6b,d, asp-3c, asp-4, asp-5 | Proteolysis and peptidolysis | 8.4 | 4.1E-02 | |
| spp genes | spp-1, ssp-10, spp-14 | 3.2 | 6.6E-02 | ||
| rpn genes | rpn-2, rpn-8, rpn-9, rpn-11 | Proteasome regulatory particle | 8.1 | 2.3E-03 | |
| eft genes | eft-2, eft-3, eft-4 | Translation elongation | 28.2 | 3.2E-14 | |
| F17C11.9a, Y41E3.10 | 5.9 | 5.7E-03 | |||
| cct genes | cct-1, cct-4, cct-6 | Cytosolic chaperone activity | 6.3 | 7.2E-04 | |
Fold increase or decrease calculated using tag frequencies.
Genes listed in the same line are closely related and cannot be distinguished by means of their SAGE tags. In these cases the tag numbers indicated are the sum of the tags for both or all genes on that line.
Overlap with Murphy et al. (2003).
Overlap with McElwee et al. (2003).
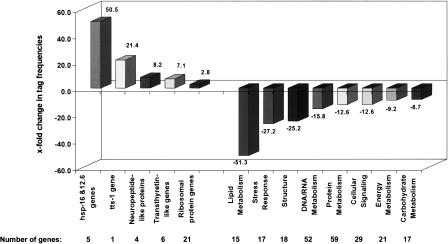
Figure 3.
Expression analysis of 6-d-old daf-2 and control worms. Genes matching our selection criteria were classified according to biological function. Mean fold differences for these genes are shown. The numbers of genes constituting these groups are indicated. Genes associated with major metabolic pathways are expressed at lower levels in daf-2 mutants.
Our findings shed light on how metabolic rate may influence longevity (Vanfleteren and De Vreese 1996; Van Voorhies 2003: for review, see Van Voorhies 2002 and Braeckman et al. 2002a). A large number of genes were expressed at lower levels in 6-d-old daf-2 mutants than in control worms. Grouping these genes according to their biological functions indicates repression of major metabolic pathways in daf-2 mutants (Fig. 3). Transcripts associated with lipid metabolism showed the most extreme reduction (mean decrease 51-fold). A total of 15 genes (see Supplemental Table 2s) including the complete vitellogenin (vit) gene family (vit-1 to vit-6), the fat-2 desaturase, and other lipid metabolism-associated transcripts were significantly decreased in daf-2 at day 6. Vitellogenin genes, which encode yolk proteins that provide essential nutrients to the developing embryo, have similarities to the human apolipoprotein B-100 precursor protein. Since alterations in lipid metabolism are thought to be associated with aging and aging-related diseases in humans (Keidar et al. 1990; Corder et al. 1993; Cohen et al. 2002), decreased activity of lipid processing pathways may contribute to the observed longevity phenotype in daf-2.
Nucleic acid metabolism (transcriptional regulation, initiation of DNA replication, chromosome organization) was also affected by the daf-2 mutation as evidenced by decreased gene expression of histones (his-24, -41, -62 and -72), histone deacetylases (hda-1, pqn-28) mcm genes (mcm-2 to 6), helicases (hel-1, cgh-1), and RNA binding proteins (puf-3, -5 and -6). Tags for 52 nucleic acid metabolism-associated genes showed an average 16-fold decrease in daf-2 adults (see Supplemental Table 2s).
Tags derived from 59 genes regulating protein turnover were expressed at lower levels in daf-2, with a mean difference of ∼13-fold. These include 22 genes regulating proteolysis and peptidolysis (e.g., asp and spp genes), translation initiation (e.g., T27F7.3b and E04D5.1) and elongation (e.g., eft genes, F17C11.9a, and Y41E3.10) as well as proteins with chaperone activity (cct-1, -4, and -6).
Transcripts related to energy metabolism (electron transport and ATP synthesis), cellular signaling (cell-cycle regulation, G-protein coupled signaling, kinases, and phosphatases), carbohydrate metabolism (glycolysis and gluconeogenesis), and structural proteins, specifically actins (act-1 to act-5) and tubulins (tbb-1, tbb -2, tba-2, and tbg-1) were also expressed at lower levels in daf-2.
We conclude that daf-2 worms at the same chronological age as controls appear strongly hypometabolic. Major metabolic systems appear to be repressed, and this metabolic repression may contribute to the observed increase in daf-2 life expectancy. Within this study we use the terms “metabolism” and “hypometabolic” to describe the sum of all biosynthetic and catabolic activities in an organism. Accordingly, we define that overall metabolic activity (turnover of proteins and lipids and DNA/RNA metabolism) rather than respiration rate is decreased in daf-2 mutants compared to control animals. Because we compared worm populations that genetically differ only in daf-2 expression, we conclude that these metabolic differences result from reduced daf-2 signaling.
Surprisingly, a number of cellular stress response genes were also expressed at lower levels in daf-2 mutants. Members of the cold-shock DNA binding family (cey-1, -2, and -4), certain heat shock proteins (hsp-1 and -3, sip-1, dnj-29, T05E11.3), and genes that scavenge free radicals (Y54G11A.13, F09E5.15, sod-1, gst-7) show lower expression levels in daf-2. Potentially, the observed hypometabolism in daf-2 worms reduces cellular stress levels, analogous to calorically restricted animals, by producing lower levels of reactive oxygen species (Sohal and Weindruch 1996). Perhaps such a decrease in internal (cellular) stress resulting from reduced metabolic activity may render unnecessary particular aspects of stress response.
Comparison of 10-d-old daf-2 adults with 6-d-old controls
We compared SAGE libraries prepared from daf-2 (day 10) versus control (day 6) worms. These libraries represent worms of the same “biological age” at times at which 100% of the respective populations remained alive (Fig. 1). The same tag selection criteria as described for the day 6 comparison were used.
Figure 4 gives an overview of the differential gene expression for this comparison. The apparent metabolic repression seen on day 6 in daf-2 worms was reversed; only transcripts involved in DNA/RNA metabolism (mcm, puf, and cey genes) and cellular signaling (cell-cycle regulation, signaling, and transport) remained lower in daf-2 worms (Table 3). Other daf-2-specific hypometabolic distinctions, such as protein turnover, energy management, and lipid metabolism were back to control levels at day 10.
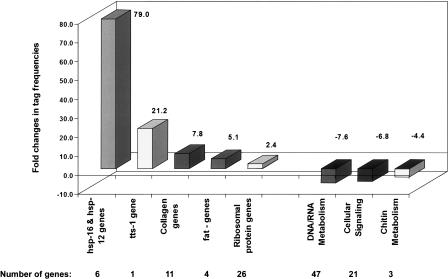
Figure 4.
Expression analysis of 10-d-old daf-2 and 6-d-old control worms. Genes matching our selection criteria were classified based on biological function. Mean fold differences for all genes within a group are shown. The numbers of genes constituting these groups are indicated. Differences in metabolic activities seen in daf-2 on day 6 are noticeably reversed.
Table 3.
Comparison of gene expression between 10-day old daf-2 and 6-day old control worms
Similar to the day 6 comparison (Fig. 3) we found that hsp-16 and hsp-12, like heat shock proteins, tts-1, and ribosomal proteins remained higher in daf-2 at day 10. Expression of small heat shock proteins was increased by a mean of 79-fold. The fat gene family (fat-1 to -3 and fat-6), encoding fatty acid desaturases that catalyze rate-limiting steps in the synthesis of mono-unsaturated fatty acids, showed a fivefold increase. Eleven collagen genes that constitute the nematode cuticle were increased in daf-2 by a mean of ∼eightfold. Another group of structural genes involved in chitin metabolism (cej-1, B0280.5, H02I12.1) was reduced in daf-2. Table 3 lists major gene families that are differentially expressed in the mutant versus control worms at biologically equivalent ages. A comprehensive list of all genes that show significant expression differences in this comparison is summarized in Supplemental Table 3s.
Statistical analysis of gene ontology classes
To test whether the specific gene classes summarized in Tables 1, 2, 3 are significantly overrepresented compared to the whole transcriptome, we used the GoMiner software (Zeeberg et al. 2003). GoMiner organizes lists of genes of interest for biological interpretation in the context of Gene Ontology (GO) and provides quantitative and statistical output files. We compared a nonredundant list of genes from Tables 1, 2, 3 (406 genes) to a list of all known C. elegans genes (WS134; 22,404 genes). GoMiner compared the two gene lists and returned GO categories within which genes were significantly overrepresented. The three GO categories showing the lowest P-values were exopeptidase activity (P = 0.0009), metabolism (P = 0.0036), and peptidase activity (P = 0.0053). Additional GO categories with significant gene enrichments were proteolysis and peptidolysis (P = 0.0153), protein catabolism (P = 0.0156), carbohydrate metabolism (P = 0.0158), response to oxidative stress (P = 0.021), oxygen and reactive oxygen species metabolism (P = 0.0234), and response to stress (P = 0.0348). This statistical analysis supports our findings that certain functional groups (gene families) are overrepresented compared to the whole transcriptome. Specifically, genes related to the GO term “metabolism” were considered to be enriched.
Global expression analysis of metabolism-associated genes
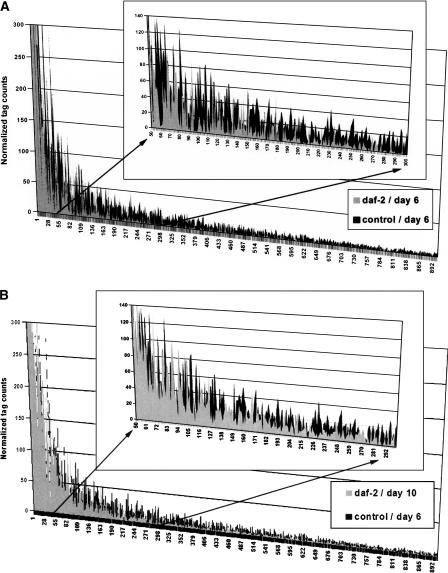
To assess the global expression profile of metabolism-associated genes in our SAGE libraries, we identified all C. elegans genes associated with the Gene Ontology term “metabolism” and analyzed the expression profile of tags that matched these metabolism-associated genes. Figure 5A illustrates the higher abundance of metabolism-associated transcripts in control compared to daf-2 animals at day 6 (same “chronological age”); the inset shows a section between genes #50 and #300 (high significance) at a higher magnification. Moreover, 4.4% of transcripts within this set (total of 1860 genes) are significantly increased and 15.4% are decreased in daf-2 compared to controls. This represents a 3.5-fold decrease in metabolism-related transcripts, which are expressed significantly less abundantly in 6-d-old daf-2 mutants compared to 6-d-old controls. Figure 5B, in contrast, shows the comparison of the same gene set in daf-2 (day 10) versus control (day 6) SAGE libraries (same “biological age”). In daf-2, 5.6% of these transcripts are significantly increased and 9.1% are decreased, representing a 1.6-fold reduction of metabolism-related transcripts, which are less abundantly expressed in mutant worms. This general trend confirms our earlier findings that at day 6, daf-2 and controls differ significantly in their metabolic activity (Fig. 3) but that these differences are much less pronounced by day 10.
Figure 5.
Global expression analysis of metabolism-associated genes in daf-2 and control worms. Genes associated with the GO term “metabolism” were analyzed. We compared expression profiles of the 900 most abundant genes in 6-d-old daf-2 vs. 6-d-old control worms (A) and 10-d-old daf-2 vs. 6-d-old control worms (B). The insets in both graphs show the section between tag #50 and #300 at a greater resolution. (A) At day 6, metabolism-associated tags in the control library are more abundant than in the daf-2 library (3.5-fold). (B) Comparison of daf-2 mutants (day 10) and controls (day 6) revealed a less distinct differential expression profile (1.6-fold lower expression of metabolism-associated tags in daf-2).
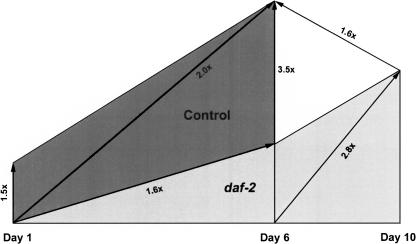
Analysis of 6-d-old versus 10-d-old daf-2 worms (“aging mutant”) confirms that the expression profile shown in Figure 4 is largely due to partial restoration of metabolic rate with advanced age in daf-2 rather than a decreasing rate in controls (Supplemental Fig. 5Cs). Comparison of these metabolism-associated genes in young, day-1 adults demonstrated that daf-2 mutants already display lower transcript levels in early adulthood (Supplemental Fig. 5Ds). These analyses show that the overall expression level of metabolism-associated genes is higher in control worms throughout the lifespan compared to daf-2 mutants. The fold differences of gene expression levels of metabolism-associated genes in daf-2 and control worms are illustrated in Figure 6. Adult controls start with a higher metabolic rate (1.5-fold at day 1); this difference increases progressively until day 6 (3.5-fold).
Figure 6.
Global gene expression differences of metabolism-associated genes in daf-2 and control worms. The graph schematically illustrates metabolic activity in mutant and control worms. Fold differences of all differentially expressed metabolism-associated genes between SAGE libraries are indicated. Arrowheads point to the SAGE library with the higher metabolic activity in a given comparison. Generally, metabolism-associated transcripts were more abundant in controls than in daf-2, and metabolic activity also increased with age.
Gene expression profiling of daf-2 mutants: Comparison of SAGE and cDNA microarray data
Two groups recently reported gene expression analyses of daf-2 mutants using cDNA microarrays (McElwee et al. 2003; Murphy et al. 2003). These analyses focused mainly on the identification of DAF-16 transcriptional targets that could explain the increased lifespan of daf-2 mutants. Similar to our study, Murphy et al. (2003) looked at gene expression changes over time by using RNA interference (RNAi) to phenocopy daf-2 and daf-2; daf-16 mutant alleles, and analyzed differential gene expression between these groups from days 1 to 8 of adulthood. We found a small but very interesting overlap between our results and the gene list presented by Murphy and colleagues. Only a small number of genes (26) appear in both lists, and 10 of these genes fall into three gene families: hsp-16 genes (three), fat genes (four), and vit genes (three). As this set of overlapping genes was validated by two different experimental approaches, it represents particularly appealing gerontogenes for further aging/longevity-related studies.
McElwee et al. (2003) compared gene expression profiles of daf-2 with daf-2; daf-16 double mutants at day 1 of adulthood. Genes differentially expressed between the single and double mutants primarily represent DAF-16 downstream targets that do not necessarily contribute to the daf-2 longevity phenotype. We identified considerable overlap of genes and gene families between our results and the data provided by McElwee and colleagues. Genes represented in both studies included stress response genes (hsp-12.6, hsp-12.3, sip-1, mtl-1), lipid metabolism genes (vit-1, vit-2, vit-5, vit-6, fat-3), structural genes (col-183, act-5, nid-1), and protein metabolism genes (e.g., asp-2, asp-6). Besides these exact gene matches we identified several gene families, but not necessarily the same specific members within these families, that were represented in both analyses; including col genes (15), gst genes (10), his genes (eight), srh genes (six), and nlp genes (three).
McElwee et al. (2003) also analyzed their microarray results using a global gene expression map that contains 44 gene clusters. Many of these clusters are enriched for genes that have a common biological function, or that are expressed in specific tissues. They reported overrepresentation of genes involved in metabolism. Amino acid, lipid and fatty acid metabolic genes, and genes associated with energy generation were strongly down-regulated in daf-2 versus daf-2; daf-16 worms. The combination of our results and the McElwee data highlights DAF-16 targets that have a high likelihood of governing longevity. Overlaps between our results and the Murphy and McElwee microarray studies are noted in Tables 1, 2, 3 and the Supplemental Tables.
To address whether SAGE detects more transcripts than microarray analysis, we constructed a C. elegans Long SAGE (21-base pair-tag) Meta library from 10 individual libraries that together encompasses 9 × 105 tags. Even after elimination of tags observed only once, for which there is some concern that they do not represent true transcripts, tag-to-gene mapping detected 20,900 transcripts, with 11,900 tags mapping adjacent to the major polyA site of the gene. This demonstrates the variety of multiple splice variants that can be detected with SAGE but would be missed using a conventional gene expression chip. When this Meta library was mapped to the genome rather than the transcriptome, we detected 34,200 transcripts, implying that 13,300 un-annotated transcripts can be predicted. This vast number of putative unannotated transcripts, presumably novel splice variants, will be subject to further analysis.
Wormbase (version WS134) contains 22,404 known transcripts, of which only 18,455 transcripts are spotted on the cDNA microarrays used by Murphy et al. (2003) and McElwee et al. (2003). A considerable number of tags from our SAGE libraries mapped to annotated transcripts that are not represented on the microarray, including Y105E8A.16 (rps-20), Y48G8AL.8a (rpl-17), and T09B4.5a. SAGE is outstanding for detecting putative new splice variants, whereas cDNA microarrays are efficient for the assessment of known transcriptional variants, and at present are often limited to one transcript per gene.
Discussion
Dozens of genes are already known to modulate adult lifespan in various organisms (for review, see Tatar et al. 2003). Remarkably, many of these are involved in hormonal signaling, and both the genes and their endocrine systems are conserved among eukaryotes. Insulin-like peptides, insulin-like growth factor (IGF), lipophilic signaling molecules, and sterols are all candidate effectors of aging in organisms as diverse as C. elegans, Drosophila melanogaster, and Mus musculus. Suppression of these hormonal signaling pathways can increase lifespan and delay age-dependent functional decline. This regulation is likely to be adaptive because, at least among invertebrates, these hormones regulate the organism's capacity to survive during states of reduced metabolism coupled with high stress resistance and arrested development. Mutations that increase lifespan by means of hormone-related mechanisms are thought to initiate elements of this survival program independent of the environmental stimuli. Because mechanisms underlying enhanced survival often oppose the aging process, they can illuminate cellular and molecular causes of senescence.
In C. elegans, mutations in the dauer formation (Daf) pathway were found to greatly increase adult longevity (Kenyon et al. 1993). Dauer diapause is a nonfeeding, stress-resistant larval state well suited for endurance and dispersal under adverse conditions. Animals with weak alleles of dauer-constitutive mutants (e.g., age-1 and daf-2) can bypass the dauer state and become long-lived adults in a manner dependent on the gene daf-16 (Kenyon et al. 1993; Larsen et al. 1995). The products of these genes and others indicate that insulin/IGF-like signal transduction is a central regulator of dauer formation and of adult aging (Finch and Ruvkun 2001). Temperature-sensitive daf-2 mutants can be divided into two overlapping classes that differ in their mutant phenotypes. Class 1 mutants such as m41 display dauer-constitutive, Age (increased longevity), and increased intrinsic thermotolerance (Itt) phenotypes, whereas class 2 mutants are more pleiotropic, exhibiting additional developmental and behavioral defects (Gems et al. 1998). It was recently demonstrated that daf-2(e1370) class 2 mutants that were also subjected to gonad ablation and daf-2 RNAi treatment had a very long life-span but were lethargic and dauer-like. When daf-2(e1368) class 1 mutants were treated in the same way, however, they had longer mean lifespans and were active for most of their lives with no apparent loss in health or vitality (Arantes-Oliveira et al. 2003).
We constructed our SAGE libraries with mRNA from daf-2(m41) mutants, a class 1 daf-2 allele that is similar to but slightly more severe than e1368. This mutant shows daf-constitutive, thermotolerance, and Age phenotypes. Our results are in agreement with the hypothesis of a tradeoff between longevity and a high metabolic, active life. By comparing daf-2 with controls of equal chronological age (day 6) we show that daf-2 mutants are hypometabolic; i.e., reduced in biosynthetic and catabolic activities. A clear reduction in transcripts for general metabolic activities including DNA/RNA, lipid, protein, and energy metabolism as well as cellular signaling is demonstrated in this study.
Gems and Riddle (2000) analyzed mortality rates of daf-2(m577) mutants and concluded that the m577 allele delays the onset of aging. m577 as well as the m41 allele used in the present study are class 1 alleles, resulting in Daf-c and Age traits, but not the numerous pleiotropic traits seen in class 2 mutants. Whether the mortality rates of these two alleles are truly identical, which also depends strongly on the experimental (culturing) conditions, is not known for certain. However, the lifespan evaluation of control and daf-2(m41) worms presented here is consistent with that of Gems and Riddle (2000). Mean and maximum lifespan were doubled and the onset of death was delayed in mutants. Because increasing mortality occurred 4 d apart, we considered 6-d-old controls and 10-d-old mutants to be of equivalent biological age.
The relationship between aging and metabolic rate is controversial. Several general theories of aging, including the rate of living and oxidative stress theories, correlate high metabolic rate (and high levels of reactive oxygen species) with decreased lifespan (Rubner 1908; Harmann 1956; Sohal and Weindruch 1996). On the other hand, Speakman et al. (2004) recently reported a positive association between metabolic intensity and lifespan due to higher mitochondrial uncoupling in a cohort of outbred mice.
Van Voorhies and Ward (1999) described a reduced metabolic rate for long-lived C. elegans mutants. They state that metabolic rates in age-1, daf-2, and clk-1; daf-2 mutants are reduced compared to wild type, and that a daf-16 mutation in a daf-2; daf-16 double mutant restored normal metabolic rates. They concluded that the increased longevity of some long-lived C. elegans mutants is caused by a reduced metabolism. Conversely, when changes in the metabolic activity in aging nematodes were assessed by a number of methods (oxygen consumption, CO2 production, heat output and ATP/ADP levels), the metabolic rate of long-lived clk and daf mutants were found to be essentially equivalent to that of wild-type C. elegans (Vanfleteren and De Vreese 1996; Braeckman et al. 1999; Braeckman et al. 2002b). Our data are consistent with the findings of Van Voorhies and Ward (1999), who showed that metabolic rates are significantly reduced in young daf-2 adults but leveled off during aging. We show that daf-2 mutants indeed express lower levels of metabolism-associated transcripts in midlife (Figs. 3, 5A).
Interestingly, we were able to classify an extensive number of these transcripts into particular gene families. Differential expression of whole gene families rather than single genes emphasizes their relevance to longevity-related processes. Categorizing these families into function-related groups revealed a metabolically repressed daf-2 phenotype in midlife. Genes regulating lipid, DNA/RNA, protein and energy metabolism, as well as cellular signaling and cell structure-related genes were expressed at much lower levels in daf-2 mutants, but these differences mostly faded at advanced age (Figs. 4, 5B). We speculate, with regard to the “rate of living” theory, that the metabolic repression in early and midlife adults (i.e., days 1 and 6) contributes substantially to the observed longevity of daf-2.
Three groups of genes were progressively increased over daf-2 adulthood or more highly expressed in daf-2 compared to controls; small heat shock proteins of the hsp-16 and hsp-12 family, the transcribed telomere-like sequence tts-1, and a number of ribosomal proteins (rps, rpl, rpa genes; Fig. 2A). Avoidance of stress-induced damage through expression of hsp-16 and hsp-12 like genes might be a major contributor to the observed longevity (Hsu et al. 2003). The cellular function of tts-1 is still under investigation; its high expression levels in dauer larvae (Jones et al. 2001), long-lived daf-2 adults, and in starved L1 larvae (data not shown) support a role for this transcribed telomere-like sequence in stress response-related activities. daf-2 mutants were previously described to display a higher stress response than wild-type animals (Henderson and Johnson 2001; Johnson et al. 2001). In fruit flies and mice, IGF-1 receptor knockouts and caloric restriction regimes also increase the cellular stress response (Sohal and Weindruch 1996; Tatar et al. 2001; Holzenberger et al. 2003; Koubova and Guarente 2003).
The increased expression of stress-response factors in aging daf-2 worms (hsp, mtl-1, gst-1, sip-1) is consistent with the age-specific increase of catalase and Cu/Zn superoxide dismutase activity levels in long-lived age-1 mutants reported by Vanfleteren (1993). Furthermore, Lund et al. (2002) analyzed gene expression profiles of sterile nematodes and reported that many heat shock genes (including hsp-16.1 and hsp-16.2) shared a common expression profile with a rise of expression until day 14 and a decrease of expression thereafter. These findings are consistent with current theories of aging that argue that protection against cellular stress is associated with successful aging and longevity.
The high levels of ribosomal protein transcripts suggest increased levels of protein biosynthesis in daf-2. Six-d-old daf-2 worms, however, showed decreases in some aspects of protein metabolism, particularly proteolytic enzymes and translational regulators, whereas ribosomal proteins are increased. Physiologically, high expression levels and turnover rates of small heat shock proteins could require increased levels of ribosomal proteins.
Downstream transcriptional targets of DAF-16 were analyzed in recent DNA microarray studies (McElwee et al. 2003; Murphy et al. 2003). Murphy et al. (2003) analyzed pooled data from 60 microarrays and suggested that the insulin/IGF pathway exerts its effect on lifespan by up-regulating a variety of genes, including stress response, antimicrobial and metabolic genes, and by down-regulating specific life-shortening genes. Similarly, McElwee et al. (2003) suggested that DAF-16 action regulates a wide range of physiological responses by altering the expression of genes involved in metabolism, energy generation, and cellular stress responses. Both studies detected hsp-16-like genes, marking the importance of these small heat shock proteins for longevity. Indeed, Walker and Lithgow (2003) showed that introduction of extra copies of hsp-16 genes conferred stress resistance and longevity in both wild type and long-lived mutants. Furthermore, Hsu et al. (2003) described potential DAF-16 binding sites upstream of these small heat shock proteins. The continuous increase of hsp-16-like genes during daf-2 aging shown in Figure 2B is consistent with the earlier data. Remarkably, the mean expression difference of hsp-16 and hsp-12 genes between daf-2 and control worms is ∼80-fold (day 10 vs. day 6; Fig. 3) and 50-fold (day 6; Fig.4), respectively.
Murphy et al. (2003) calculated Pearson correlation coefficients for expression profiles between different time points of aging daf-2 mutants and controls. Their analysis of pooled microarray data showed a high correlation of gene expression between 6-d-old controls and 6- or 8-d-old daf-2 mutants. The use of RNAi to phenocopy a knock out allele by Murphy et al. (2003), versus the daf-2(m41) mutant strain used in the present study and our specific analysis of metabolism-associated genes may account for some differences in global gene expression patterns.
Our approach differs markedly from the DNA microarray studies mentioned above. A major advantage of SAGE compared to microarray analysis is that it requires no a priori knowledge of the genes of interest. SAGE facilitates access to essentially the whole C. elegans transcriptome and hence supports a more consistent and comprehensive analysis of gene family members. Therefore, SAGE is better suited for detection of co-expressed functional classes of genes than is microarray analysis.
Our results are potentially useful for understanding human aging. Many processes involved in nematode aging may have fundamental and analogous roles in humans. The human orthologs of various gene family members identified in this study have been associated with human age-related diseases. For instance, the human ortholog of some vit genes (vit-3, vit-4, and vit-5) is apolipoprotein B (APOB). APOB gene polymorphisms are associated with coronary artery diseases and myocardial infarction (Chiodini et al. 2003). Furthermore, the human ortholog of the hsp-16 and hsp-12 -like genes is crystallin alpha B (CRYAB). The brains of Alzheimer's disease patients, with an aging-associated condition, express increased CRYAB transcript levels (Link et al. 2003).
The combination of SAGE and microarray data (McElwee et al. 2003; Murphy et al. 2003) highlights six genes that are reproducibly involved in nematode longevity: hsp-12.6, vit-2, vit-5, fat-3, nid-1, and mtl-1. Evidence of hypometabolism in daf-2 mutants and the identification of aging-associated genes and gene families in C. elegans will contribute substantially to the understanding of aging and longevity in nematodes as well as provide valuable clues to the basis of human aging.
Methods
Lifespan determinations
Lifespan studies were performed as described (Larsen et al. 1995), with animals raised at 15°C and placed at 25.5°C as L4 larvae. The first day of adulthood is designated as day 1 of the survival curve. Animals were scored every other day and transferred as necessary to avoid competition for food. Animals not moving or responding to prodding with the tip of a 0.2-mm-diameter wire were scored as dead. Statistical analyses were performed using SPSS Version 11.5.
Culture methods and strains
Nematodes were grown on NG agar medium seeded with Escherichia coli strain OP50 (Brenner 1974). We used two different strains of C. elegans: fer-15 (b26ts), a sterile mutant of the C. elegans N2 (Brenner 1974) wild-type strain, and the long-lived fer-15 (b26ts); daf-2(m41) double mutant. Sterile strains were used instead of N2 wild-type worms to avoid internal hatching of eggs, which eventually kills the mother. Additionally, early embryonic development takes place in the mother; therefore, embryonically expressed transcripts would obscure the expression profiles of adult worms of both long-lived mutants and controls. Both strains were raised at 15° C (permissive temperature) and shifted to 25.5°C (restrictive temperature) as L4 larvae or young adults. This temperature shift avoids dauer larvae formation of the otherwise dauer constitutive daf-2(m41) mutants, and constitutes an increased adult lifespan. fer-15 and fer-15; daf-2 strains were constructed as described (Larsen et al. 1995). Throughout this manuscript the fer-15 (b26ts) strain is referred to as control and the fer-15; daf-2 double mutant as daf-2.
Time points for SAGE library construction
fer-15 worms were harvested on days 1 and 6, whereas daf-2 worms were collected on days 1, 6, and 10 of adulthood. Day 1 of adult animals is a developmentally defined stage after finishing larval 4 (L4) development. Day 6 in fer-15 and day 10 in daf-2 nematode populations marked the last time points at which all worms of the cultures were alive, healthy, and active. We investigated age-synchronized, healthy, and active worm populations at all time points. Age synchronization of the worm populations is possible due to the sterile (fer-15) genetic background.
RNA preparation
Frozen animals were crushed in liquid nitrogen using a mortar and pestle for 5–10 min. Total RNA was isolated by the guanidinium isothiocyanate: phenol method (Chomczynski and Sacchi 1987). The yields of total RNA from 3.5 mL and 5.0 mL of settled fer-15 worms at days 1 and 6 were 2.7 mg and 4 mg, respectively. For 5 mL, 2.5 mL, and 10 mL of day 1, 6, and 10 of settled daf-2 worms, the total RNA yields were 2.0 mg, 2.7 mg, and 4 mg, respectively.
Generation of SAGE tags and libraries
For removal of contaminating genomic DNA, the total RNA was treated with DNase I (Invitrogen). cDNA synthesis and the subsequent steps of SAGE library preparation were performed using Invitrogen's I-SAGE kit and protocol. NlaIII was used as the anchoring enzyme and BsmFI as the tagging enzyme. In addition, Phase Lock Gel (Eppendorf) was used at every phenol-chloroform extraction step to increase the recovery and purity of DNA (Ye et al. 2000). During library construction, ditags were amplified by 27–29 cycles of PCR.
SAGE library sequencing
The library clones were sequenced on an ABI Prism 3700 DNA analyzer using BigDye primer cycle sequencing reagents (Applied Biosystems). Sequence reads were processed, and their quality was assessed using Phred (Ewing and Green 1998; Ewing et al. 1998). Vector sequences were then removed, and 14 base-pair tags were isolated. Linker sequences and duplicate ditags were also removed, and only tags of ≥95% accuracy (derived from Phred quality scores) were kept for further analysis. Total tag counts in each library were as follows: fer-15 day 1 (62,104 tags), fer-15 day 6 (68,345 tags), daf-2 day 1 (59,940 tags), daf-2 day 6 (63,883 tags), and daf-2 day 10 (66,355 tags). The same quality filters were used for all five SAGE libraries.
Analysis of SAGE tags
For comparison of individual SAGE libraries we used the DiscoverySpace platform (http://www.bcgsc.ca/bioinfo/software/discoveryspace/), a software tool specifically developed for analysis of SAGE data sets (S. Zuyderduyn, S.J. Jones, and M.A. Marra, in prep.). DiscoverySpace allows comparison of two or more SAGE libraries, and returns details regarding respective tag frequencies and probability scores for all common or differential expressed tags. This software links tags directly to a database of predicted C. elegans transcripts, the “virtual transcriptome” (Pleasance et al. 2003; McKay et al. 2004). Pairwise library distances, which were based on differences between normalized tag counts, were calculated as follows:
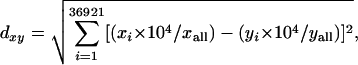 |
where d is the distance between x and y libraries in a multidimensional space in which each tag defines one dimension; xi and yi are the counts of the i tag, and xall and yall are the total number of tags in libraries x and y, respectively.
Another source for comparing tag expression profiles within libraries is provided at http://tock.bcgsc.bc.ca/cgi-bin/sage. This Web-based tool allows searches for tags as well as keywords (e.g., gene names). Tags that matched two or more transcripts and matched genes in the antisense orientation were generally excluded from the analysis.
We analyzed five SAGE libraries of adult nematodes; three daf-2 (days 1, 6, and 10) and two control populations (days 1 and 6). The process of isolating genes important for the increased lifespan of daf-2 worms included comparison of SAGE libraries, selection of relevant tags by applying stringent inclusion criteria, tag-to-gene mapping, and grouping the identified genes according to their biological function. WormBase release WS107 (http://www.wormbase.org/) was used as a reference to determine tag-to-gene matches and to assess the known or inferred gene functions. The actual tag counts in each library were normalized to 70,000 tags per library to simplify data comparison. The fold increase or decrease was calculated using tag frequencies. Tag frequencies were calculated by dividing individual tag counts by the total tag counts in the library. Spreadsheets summarizing tag-to-gene mapping for all five libraries are posted at: http://elegans.bcgsc.ca/home/sage.html.
Global analysis of metabolism-associated genes
C. elegans genes that are associated with the Gene Ontology term “metabolism” (GO:0008152) were identified through WormBase (http://www.wormbase.org/); 4647 genes are associated with this GO term (release WS 119), and 1860 of these had unambiguous matches with tags in daf-2 and control SAGE libraries. Tags were sorted according to their abundance within the libraries. We considered only 900 of the most abundant tags to be meaningful, as the remainder had very low expression levels. We compared expression of these 900 genes between daf-2 and control libraries on a global scale. Fold differences in expression levels were calculated by dividing the number of tags that were significantly increased in one library from the number of tags that were significantly increased in another library.
Statistical analysis
P-values for all data comparisons were calculated with DiscoverySpace (S. Zuyderduyn, S.J. Jones, and M.A. Marra, in prep.), using the Audic and Claverie (1997) test statistic. P-values < 0.05 for differential expression were considered significant.
Acknowledgments
This work was funded by a New Emerging Team Grant from the Canadian Institute of Health Research (CIHR) to M.A.M., A.B.W., and others, and NIH grant #AG12689 to D.L.R. and M.A.M. M.A.M. and S.J.M.J. are Scholars of the Michael Smith Foundation for Health Research. J.H.W. is supported by an Erwin-Schrödinger Fellowship from the Austrian Science Foundation (FWF). We thank Jennifer Asano, Susanna Chan, and Pawan Pandoh for their contributions to SAGE library construction, Allen Delaney for bioinformatics assistance, Patrice Albert and Amy Cox for the survival studies, numerous members of the Genome Sciences Centre sequencing group for superlative technical assistance, and Joseph Connors and Nhu Le for critically reading the manuscript.
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3274805. Article published online ahead of print in April 2005.
References
- Ailion, M., Inoue, T., Weaver, C.I., Holdcraft, R.W., and Thomas, J.H. 1999. Neurosecretory control of aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 96: 7394–7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi, A., Yeh, W.H., Tait, D., Hedgecock, E.M., and Riddle, D.L. 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes & Dev. 14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira, N., Berman, J.R., and Kenyon, C. 2003. Healthy animals with extreme longevity. Science 302: 611. [DOI] [PubMed] [Google Scholar]
- Audic, S. and Claverie, J.M. 1997. The significance of digital gene expression profiles. Genome Res. 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Bargmann, C.I. and Horvitz, H.R. 1991. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251: 1243–1246. [DOI] [PubMed] [Google Scholar]
- Braeckman, B.P., Houthoofd, K., De Vreese, A., and Vanfleteren, J.R. 1999. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr. Biol. 9: 493–496. [DOI] [PubMed] [Google Scholar]
- Braeckman, B.P., Houthoofd, K., and Vanfleteren, J.R. 2002a. Assessing metabolic activity in aging Caenorhabditis elegans: Concepts and controversies. Aging Cell 1: 82–88; 102–103. [DOI] [PubMed] [Google Scholar]
- Braeckman, B.P., Houthoofd, K., De Vreese, A., and Vanfleteren, J.R. 2002b. Assaying metabolic activity in ageing Caenorhabditis elegans. Mech. Ageing Dev. 123: 105–119. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini, B.D., Barlera, S., Franzosi, M.G., Beceiro, V.L., Introna, M., and Tognoni, G. 2003. APO B gene polymorphisms and coronary artery disease: A meta-analysis. Atherosclerosis 167: 355–366. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Cohen, P., Miyazaki, M., Socci, N.D., Hagge-Greenberg, A., Liedtke, W., Soukas, A.A., Sharma, R., Hudgins, L.C., Ntambi, J.M., and Friedman, J.M. 2002. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science 297: 240–243. [DOI] [PubMed] [Google Scholar]
- Corder, E.H., Saunders, A.M., Strittmatter, W.J., Schmechel, D.E., Gaskell, P.C., Small, G.W., Roses, A.D., Haines, J.L., and Pericak-Vance, M.A. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261: 921–923. [DOI] [PubMed] [Google Scholar]
- Dorman, J.B., Albinder, B., Shroyer, T., and Kenyon, C. 1995. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141: 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Finch, C.E. and Ruvkun, G. 2001. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2: 435–462. [DOI] [PubMed] [Google Scholar]
- Gems, D. and Riddle, D.L. 2000. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics 154: 1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems, D., Sutton, A.J., Sundermeyer, M.L., Albert, P.S., King, K.V., Edgley, M.L., Larsen, P.L., and Riddle, D.L. 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150: 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, J.W. and Riddle, D.L. 1984. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev. Biol. 102: 368–378. [DOI] [PubMed] [Google Scholar]
- Harman, D. 1956. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 11: 298–300. [DOI] [PubMed] [Google Scholar]
- Harris, T.W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. 2004. WormBase: A multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32: D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, S.T. and Johnson, T.E. 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11: 1975–1980. [DOI] [PubMed] [Google Scholar]
- Holzenberger, M., Dupont, J., Ducos, B., Leneuve, P., Geloen, A., Even, P.C., Cervera, P., and Le Bouc, Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421: 182–187. [DOI] [PubMed] [Google Scholar]
- Hsu, A.L., Murphy, C.T., and Kenyon, C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Johnson, T.E., de Castro, E., Hegi de Castro, S., Cypser, J., Henderson, S., and Tedesco, P. 2001. Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. Exp. Gerontol. 36: 1609–1617. [DOI] [PubMed] [Google Scholar]
- Jones, S.J., Riddle, D.L., Pouzyrev, A.T., Velculescu, V.E., Hillier, L., Eddy, S.R., Stricklin, S.L., Baillie, D.L., Waterston, R., and Marra, M.A. 2001. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 11: 1346–1352. [DOI] [PubMed] [Google Scholar]
- Keidar, S., Etzioni, A., Brook, J.G., Gershoni-Baruch, R., and Aviram, M. 1990. Compound heterozygosity for abetalipoproteinaemia and familial hypobetalipoproteinaemia. J. Med. Genet. 27: 133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C., Chang, J., Gensch, E., Rudner, A., and Tabtiang, R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kimura, K.D., Tissenbaum, H.A., Liu, Y., and Ruvkun, G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Klass, M. and Hirsh, D. 1976. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260: 523–525. [DOI] [PubMed] [Google Scholar]
- Koubova, J. and Guarente, L. 2003. How does calorie restriction work? Genes & Dev. 17: 313–321. [DOI] [PubMed] [Google Scholar]
- Larsen, P.L., Albert, P.S., and Riddle, D.L. 1995. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139: 1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K., Dorman, J.B., Rodan, A., and Kenyon, C. 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. [DOI] [PubMed] [Google Scholar]
- Link, C.D., Taft, A., Kapulkin, V., Duke, K., Kim, S., Fei, Q., Wood, D.E., and Sahagan, B.G. 2003. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer's disease model. Neurobiol. Aging 24: 397–413. [DOI] [PubMed] [Google Scholar]
- Lund, J., Tedesco, P., Duke, K., Wang, J., Kim, S.K., and Johnson, T.E. 2002. Transcriptional profile of aging in C. elegans. Curr. Biol. 12: 1566–1573. [DOI] [PubMed] [Google Scholar]
- McElwee, J., Bubb, K., and Thomas, J.H. 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121. [DOI] [PubMed] [Google Scholar]
- McKay, S.J., Johnson, R., Khattra, J., Asano, J., Baillie, D.L., Chan, S., Dube, N., Fang, L., Goszczynski, B., and Ha, E. 2004. Gene expression profiling of cells, tissues and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68: 159–169. [DOI] [PubMed] [Google Scholar]
- Murphy, C.T., McCarroll, S.A., Bargmann, C.I., Fraser, A., Kamath, R.S., Ahringer, J., Li, H., and Kenyon, C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G.I., Lee, L., Tissenbaum, H.A., and Ruvkun, G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- Paradis, S., Ailion, M., Toker, A., Thomas, J.H., and Ruvkun, G. 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes & Dev. 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance, E.D., Marra, M.A., and Jones, S.J. 2003. Assessment of SAGE in transcript identification. Genome Res. 13: 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, P., Lim, C.S., Johnsen, R., Albert, P.S., Pilgrim, D., and Riddle, D.L. 1996. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science 274: 1389–1391. [DOI] [PubMed] [Google Scholar]
- Rubner, M. 1908. Das Problem der Lebensdauer. Oldenburg, Munich.
- Sohal, R.S. and Weindruch, R. 1996. Oxidative stress, caloric restriction, and aging. Science 273: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman, J.R., Talbot, D.A., Selman, C., Snart, S., McLaren, J.S., Redman, P., Krol, E., Jackson, D.M., Johnson, M.S., and Brand, M.D. 2004. Uncoupled and surviving: Individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell 3: 87–95. [DOI] [PubMed] [Google Scholar]
- Tatar, M., Kopelman, A., Epstein, D., Tu, M.-P., Yin, C.-M., and Garofalo, R.S. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110. [DOI] [PubMed] [Google Scholar]
- Tatar, M., Bartke, A., and Antebi, A. 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351. [DOI] [PubMed] [Google Scholar]
- Van Voorhies, W.A. 2002. The influence of metabolic rate on longevity in the nematode Caenorhabditis elegans. Aging Cell 1: 91–101. [DOI] [PubMed] [Google Scholar]
- Van Voorhies, W.A. 2003. Is life span extension in single gene long-lived Caenorhabditis elegans mutants due to hypometabolism? Exp. Gerontol. 38: 615–618. [DOI] [PubMed] [Google Scholar]
- Van Voorhies, W.A. and Ward, S. 1999. Genetic and environmental conditions that increase longevity in Caenorhabditis elegans decrease metabolic rate. Proc. Natl. Acad. Sci. 96: 11399–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren, J.R. 1993. Oxidative stress and ageing in Caenorhabditis elegans. Biochem. J. 292: 605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren, J.R. and De Vreese, A. 1996. Rate of aerobic metabolism and superoxide production rate potential in the nematode Caenorhabditis elegans. J. Exp. Zool. 274: 93–100. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. 1995. Serial analysis of gene expression. Science 270: 484–487. [DOI] [PubMed] [Google Scholar]
- Walker, G.A. and Lithgow, G.J. 2003. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2: 131–139. [DOI] [PubMed] [Google Scholar]
- Weinert, B.T. and Timiras, P.S. 2003. Invited review: Theories of aging. J. Appl. Physiol. 95: 1706–1716. [DOI] [PubMed] [Google Scholar]
- Ye, S.Q., Zhang, L.Q., Zheng, F., Virgil, D., and Kwiterovich, P.O. 2000. miniSAGE: Gene expression profiling using serial analysis of gene expression from 1 microg total RNA. Anal. Biochem. 287: 144–152. [DOI] [PubMed] [Google Scholar]
- Zeeberg, B.R., Feng, W., Wang, G., Wang, M.D., Fojo, A.T., Sunshine, M., Narasimhan, S., Kane, D.W., Reinhold, W.C., Lababidi, S., et al. 2003. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol. 4: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.wormbase.org/; WormBase, an annotated database for C. elegans genes, proteins, and genomic sequences.
- http://tock.bcgsc.bc.ca/cgi-bin/sage; Genome Sciences Centre, BC Cancer Agency, C. elegans SAGE analysis tool (multisage).
- http://elegans.bcgsc.ca/home/sage.html; Spreadsheets summarizing tag-to-gene mapping for all five libraries.
- http://www.bcgsc.ca/bioinfo/software/discoveryspace/; Discovery Space Software.