Abstract
The obligate intracellular bacterial pathogen Chlamydophila abortus strain S26/3 (formerly the abortion subtype of Chlamydia psittaci) is an important cause of late gestation abortions in ruminants and pigs. Furthermore, although relatively rare, zoonotic infection can result in acute illness and miscarriage in pregnant women. The complete genome sequence was determined and shows a high level of conservation in both sequence and overall gene content in comparison to other Chlamydiaceae. The 1,144,377-bp genome contains 961 predicted coding sequences, 842 of which are conserved with those of Chlamydophila caviae and Chlamydophila pneumoniae. Within this conserved Cp. abortus core genome we have identified the major regions of variation and have focused our analysis on these loci, several of which were found to encode highly variable protein families, such as TMH/Inc and Pmp families, which are strong candidates for the source of diversity in host tropism and disease causation in this group of organisms. Significantly, Cp. abortus lacks any toxin genes, and also lacks genes involved in tryptophan metabolism and nucleotide salvaging (guaB is present as a pseudogene), suggesting that the genetic basis of niche adaptation of this species is distinct from those previously proposed for other chlamydial species.
The Chlamydiaceae is a phylogenetically distinct Gram-negative bacterial family, encompassing two genera (Chlamydia and Chlamydophila), which are subdivided into three (Chlamydia muridarum, Chlamydia suis, and Chlamydia trachomatis) and six (Chlamydophila pneumoniae, Chlamydophila abortus, Chlamydophila caviae, Chlamydophila felis, Chlamydophila pecorum, and Chlamydophila psittaci) defined species, respectively (Everett et al. 1999). Chlamydiaceae undergo a unique biphasic developmental cycle switching from an infectious, but metabolically inactive, form called the elementary body (EB) to the noninfectious, metabolically active cell type known as the reticulate body (RB). The infectious process is initiated by the attachment of EBs to susceptible host cells, particularly mucosal epithelial cells. Upon entry the EB differentiates into the metabolically active form, RB, which multiplies by binary fission within specialized intracellular vacuoles called inclusions. These inclusions evade fusion with lysosomes, thus avoiding the host endocytic pathway, but instead intercept the exocytic pathway, appearing as secretory vacuoles to the host cell (Hackstadt et al. 1997). RBs transform back into EBs 2–3 d after infection (depending on species) and are then released by lysis or exocytosis to complete the infectious process. EBs can be spread by aerosols, through ingestion, and by direct physical contact.
Although the developmental cycle is common to all Chlamydiaceae, the disease outcomes and associated sequelae, host range, and tissue tropisms vary markedly. C. trachomatis is the world's leading cause of preventable blindness (trachoma), and is the most common sexually transmitted pathogen in the United Kingdom and USA, giving rise to such conditions as pelvic inflammatory disease and epididymitis. Chlamydophila pneumoniae (formerly Chlamydia pneumoniae), which is associated with hosts as diverse as humans, horses, frogs, and marsupials, causes acute respiratory infections and is also implicated in chronic conditions such as adult-onset asthma and atherosclerosis (Everett et al. 1999; Saikku 1999). In contrast, Cp. caviae is host-restricted and causes inclusion conjunctivitis (GPIC) and genital tract infections in guinea pigs. Several of the animal-infective Chlamydophila, namely, Cp. abortus (ruminant abortion; formerly Chlamydia psittaci serotype 1), Cp. psittaci (causative agent of avian chlamydiosis), and Cp. felis (feline conjunctivitis), can also cause acute zoonotic infections in humans (for review, see Longbottom and Coulter 2003).
We chose to sequence a low passage strain of Cp. abortus (S26/3), which is a pathogen of significant economic importance and represents the first genome from a zoonotic chlamydial pathogen to be determined. Cp. abortus is principally a pathogen of ruminants and pigs. In the United Kingdom, and countries of Northern Europe, it is the most common cause of abortion in sheep (ovine enzootic abortion, or OEA) and goats, as a result of its ability to colonize the placenta (Rodolakis and Souriau 1989; Longbottom and Coulter 2003). Cp. abortus also represents a threat to human health because it can cause zoonotic infections; pregnant women who are exposed to infected animals are also at risk of abortion and life-threatening illness (Longbottom and Coulter 2003).
The obligate intracellular nature and lack of genetic tools have hampered the molecular genetic investigation of chlamydiae. However, their obvious importance as pathogens of humans and animals has made them attractive targets for genome sequencing, and representative genomes from four different chlamydial species have been published (Stephens et al. 1998; Kalman et al. 1999; Read et al. 2000, 2003; Shirai et al. 2000). Thus far all of the published Chlamydiaceae genomes belong to important pathogens of humans or animals. While human chlamydial research relies on the use of comparative animal model systems, the principal advantage for those working in animal chlamydial research is that the cellular and molecular pathogenesis studies, and vaccine studies, can be performed in the natural host.
Although the clinical manifestations of chlamydial infections are protean, whole-genome sequencing has shown that their genomes are remarkably conserved, with only a small proportion of the genes being species-specific. Unlike many human and animal pathogens (Parkhill et al. 2001a; Deng et al. 2002), there is little evidence of recent horizontal acquisition of DNA. This is consistent with the closeted lifestyle of the metabolically active RB developmental form, which is principally found within an intracellular environment. An exception to this is the presence of phage-related genes in Cp. caviae and Cp. pneumoniae (Read et al. 2000, 2003). Like the genomes of other sequenced obligate intracellular pathogens, such as the Rickettsiales (Andersson et al. 1998; Zomorodipour and Andersson 1999; Ogata et al. 2001), chlamydial genomes are small in size and are thought to be undergoing a process of reductive evolution. Consistent with this, many genes for steps in metabolic pathways such as de novo amino acid, nucleotide, and co-factor synthesis are often absent, suggesting a general reliance on host-derived intermediates (Stephens et al. 1998; Read et al. 2000).
The availability of several taxonomically distinct Chlamydiaceae genomes provides a unique opportunity to investigate the microevolutionary events that generate genetic plasticity and contribute to the speciation in this important family of pathogens. A comparative genomic approach has been used to identify the genetic diversity of this ovine pathogen, and investigate possible implications for host niche adaptation.
Results
General features of the genome
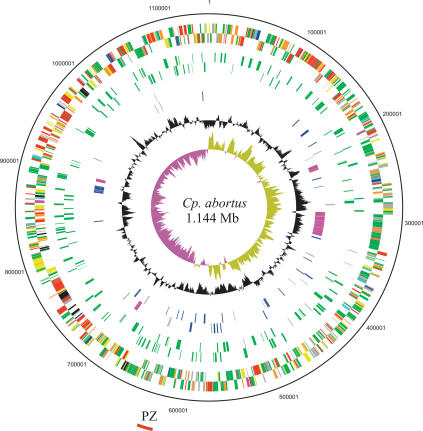
The general features of the Cp. abortus genome are shown in Figure 1 and summarized in Table 1. The genome is composed of a 1,144,377-bp circular chromosome with an overall G+C content of 39.87%. The origin of replication was assigned based on the GC deviation of the genome as previously described (Parkhill et al. 2001a). Characteristically for the Chlamydophila, Cp. abortus possesses only single copies of the 23S, 16S, and 5S rRNA genes, in contrast to Chlamydia species, which possess two copies (Everett et al. 1999). The annotation identified 961 predicted coding sequences (CDS), representing a coding density of 88%. Of the predicted CDS, 746 have been given functional assignments based on previous experimental evidence or database similarity and motif matches, and for those with no functional assignment (215 CDS), 110 were only significantly similar to proteins from other members of the Chlamydiaceae, and 15 predicted CDS returned no significant database hits. In total, 38 tRNAs were identified, which corresponded to all the amino acids except selenocysteine.
Figure 1.
Circular representation of the Cp. abortus chromosome. The outer scale shows the size in base pairs. From the outside in, circles 1 and 2 show the position of genes transcribed in a clockwise and anticlockwise direction, respectively (for color codes, see below); circles 3 and 4 CDS encoding all membrane proteins (green) minus the Pmp and TMH/Inc-family proteins in the clockwise and anticlockwise directions, respectively; circles 5 and 6 show members of the Pmp (purple) and TMH/Inc protein families (blue) in the clockwise and anticlockwise directions, respectively. Circle 7 shows a plot of G+C content (in a 10-kb window); circle 8 shows a plot of GC skew ([G–C]/[G+C]; in a 10-kb window). Genes in circles 1 and 2 are color coded according to the function of their gene products: (dark green) membrane or surface structures; (yellow) central or intermediary metabolism; (cyan) degradation of macromolecules; (red) information transfer/cell division; (purple) degradation of small molecules; (pale blue) regulators; (dark blue) pathogenicity or adaptation; (black) energy metabolism; (orange) conserved hypothetical; (pale green) unknown; (brown) pseudogenes. The position of the plasticity zone (PZ) is shown as a red arc outside of the scale ring.
Table 1.
Summary of the Cp. abortus genome features compared to representatives of the other sequenced chlamydial genomes
| Cp. abortus (S26/3) | Cp. caviae (GPIC) | C. trachomatis (serovar D) | C. muridarum (Nigg) | Cp. pneumoniae (AR39) | |
|---|---|---|---|---|---|
| Genome size (bp) | 1,144,377 | 1,173,390 | 1,042,519 | 1,072,950 | 1,229,858 |
| % GC of genome | 39.87 | 39.22 | 41.31 | 40.34 | 40.57 |
| % GC of CDSa | 40.5 | 38.82 | 41.66 | 40.69 | 41.29 |
| % coding | 88.2 | 89.4 | 90.1 | 90.0 | 89.0 |
| No. of CDSa | 961 | 1009 | 894 | 921 | 1130 |
| Avg. aa % ID to Cp. abortus orthologs | — | 85 | 65 | 66 | 69 |
| No. of Pmp proteins | 18 | 18b | 9 | 9 | 21 |
| No. of tRNA | 38 | 38 | 37 | 37 | 38 |
| No. of rRNA operons | 1 | 1 | 2 | 2 | 1 |
CDS, coding sequences.
Seventeen reported in the original publication (Read et al. 2003) lacked CCA00285.
There is no evidence of recent horizontal gene transfer in Cp. abortus, including a complete lack of any phage genes that have been seen in other sequenced Chlamydophila species (Read et al. 2000, 2003).
Whole-genome comparisons with members of the Chlamydiaceae
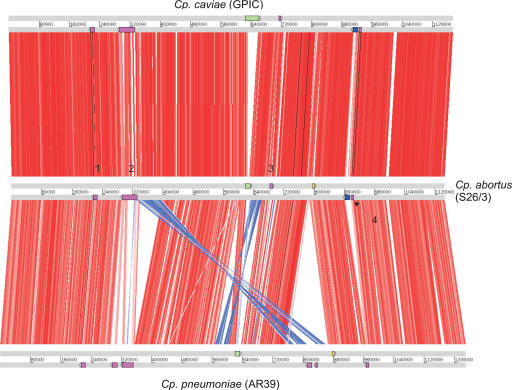
Whole-genome comparisons of Cp. abortus with the other published Chlamydiaceae genomes show that there is a high level of conservation. The sequenced Chlamydophila genomes (Fig. 2) are essentially collinear with the majority of the sequence, or the “core” sequence, being conserved between all three genomes. The most striking rearrangement is the reciprocal recombination event seen in the replication terminus of Cp. pneumoniae (as previously described; Read et al. 2000, 2003). Disruptions to this apparent uniformity in Cp. abortus (we refer to these as variable regions; summarized in Fig. 2) are confined to the plasticity zone (PZ) or replication termination region terminus, the two major clusters of polymorphic membrane protein (Pmp) genes, the novel transmembrane head (TMH)/Inc protein gene cluster, and the locus encoding the biotin biosynthetic operon (discussed in more detail below).
Figure 2.
Global comparison between Cp. caviae (GPIC), Cp. abortus (S26/3), and Cp. pneumoniae (AR39). The figure shows ortholog matches (see Methods; displayed using ACT http://www.sanger.ac.uk/Software/ACT/) of the three sequenced Chlamydophila genomes. Gray bars represent the forward and reverse strands of DNA with the scale marked in base pairs. The red bars between the DNA lines represent individual forward ortholog matches, with inverted matches colored blue. The positions of the PZ region (green), pmp clusters (purple; numbered 1–4), tmh cluster (blue), and biotin biosynthetic operon (yellow) are shown as colored boxes on the DNA lines. All the genomes have been oriented at the origin of replication to start at the hemB gene.
More detailed analysis of the Cp. abortus core genome regions showed, by clustering orthologous groups, that it encoded 840 CDS. Functional analysis of these core CDS showed that not only were the general metabolic capabilities of Cp. abortus the same as previously described for other Chlamydiaceae (Stephens et al. 1998; Read et al. 2000, 2003), apart from those mentioned below, but that Cp. abortus also possesses intact copies of all the CDS thought to encode the chlamydial type three secretion system (CDS CAB034–CAB041, CAB442–CAB446, and CAB904– CAB924)—further circumstantial evidence that this system is essential for both human- and animal-infective Chlamydiaceae.
CDS encoded within the non-core “variable” regions were divided into 25 CDS that were unique to Cp. abortus (including CAB598, a pmp gene), 15 CDS found in Cp. abortus and Cp. pneumoniae strain AR39 only (including the biotin biosynthetic operon genes; CAB685–CAB689), and 52 CDS shared between Cp. abortus and Cp. caviae but absent from Cp. pneumoniae strain AR39 (including thiE and thiM [CAB198 and CAB199] involved in thiamine biosynthesis; CAB600–CAB602, which are predicted to encode an ABC-type membrane transport system; and recF [CAB430]) (see Supplemental Table 1).
Small-scale gene variation: Pseudogenes
Of the 961 CDS predicted for Cp. abortus, 29 were pseudogenes (∼3% of the total CDS). The majority of these encode membrane or exported proteins (16 CDS), of which four encode Pmp-family proteins (CAB270, CAB273, CAB279, and CAB596; discussed below) and nine encode conserved hypothetical proteins. The remaining pseudogenes included argR, encoding the arginine repressor (CAB529), and guaB (CAB551), involved in purine nucleotide biosynthesis.
The frameshift mutations of seven of the 29 pseudogenes occurred within homopolymeric tracts (CAB279, CAB356, CAB383A, CAB516, CAB543, CAB596, and CAB820). By comparing the sequence of homopolymeric tracts from orthologous genes in the other chlamydial genomes it was evident there was significant variation in the length of these sequences. For example, the Cp. abortus pmp-family CDS CAB820 carries a T(×8) tract, whereas the ortholog of this CDS in Cp. caviae (CCA00855) possesses a T(×11) tract, and all the Cp. pneumoniae CAB820 orthologs a T(×9) tract (CP0952 [strain AR39], CPn0914 [strain CML029], CpB0946 [strainTW183], and CPj0914 [strain J138]). Moreover, CCA00855 appears to be intact, but the CAB820 orthologs in the four independently sequenced Cp. pneumoniae genomes are in the frameshifted state.
Furthermore, whole-genome assembly data showed that the homopolymeric tracts found within CAB279, CAB596, and CAB598 (all encoding members of the Pmp family), as well as a CDS encoding a protein of unknown function, CAB853, varied in length such that intact and frameshifted variants were represented in the sequence data, suggesting that they could be subject to phasevariable expression by slip-strand pairing (Viratyosin et al. 2002).
It is notable that some of the CDS that are intact in Cp. abortus are pseudogenes in the other Chlamydophila including rluC (CAB379), which encodes 23S rRNA pseudouridine synthase C and has a functional ortholog in Cp. caviae (CCA00391; originally annotated as a pseudogene) (Read et al. 2003) but is disrupted by a frameshift mutation in Cp. pneumoniae (e.g., CP0352 in AR39). Several other CDS involved in pseudouridine synthesis are annotated as pseudogenes in Cp. caviae: truA, truB, rluB, and rluD. However, on closer inspection all these genes appear to be intact, consistent with them being essential for the formation of two universally conserved ribosomal RNA pseudouridine residues (Raychaudhuri et al. 1998; Ofengand 2002).
Other gene variations include gatA, encoding glutamyl-tRNA amidotransferase subunit A, which is intact in both Cp. abortus (CAB286) and Cp. pneumoniae (CP0772) but is a pseudogene in Cp. caviae (CCA00288).
Large-scale gene variation
Many of the gene differences of the Chlamydiaceae are clustered into discrete regions. It is likely that these regions hold important genetic clues to explain the wide ranging niche adaptation of this bacterial family. These regions include the PZ, which has been shown to vary markedly both in sequence and gene content in the other fully sequenced chlamydial genomes (Read et al. 2000, 2003). The PZ in Cp. caviae (∼35 kb), bounded by accB and guaB, carries genes involved in tryptophan biosynthesis, purine nucleotide interconversion (guaBA–add cluster), and a large toxin gene that is similar to the EHEC adherence factor (CCA00558) (Read et al. 2003). In comparison, the Cp. abortus PZ is considerably smaller than that of Cp. caviae, being ∼12 kb and encoding 11 genes (compared to 22 in Cp. caviae). The Cp. abortus PZ lacks most of the genes that are found in the PZ of the other chlamydiae, including the trp gene cluster (trpABFCDR, kynU, prsA), guaA, add, and those for any recognizable toxin genes. Genes in common with the other PZ regions include accB (CAB539), accC (CAB540), and CAB548 (conserved hypothetical protein). This region also carries a guaB pseudogene, suggesting that the purine nucleotide interconversion operon once present at this locus is now in the process of being lost from Cp. abortus.
Several other genes within the Cp. abortus PZ are similar to hypothetical proteins from Cp. caviae, including CAB545 (similar to CCA00557) and the two pseudogenes CAB541 (similar to CCA00555) and CAB543 (similar to CCA00556). The Cp. pneumoniae strain AR39 PZ is also much reduced (∼12 kb) compared to Cp. caviae, although it retains the purine nucleotide interconversion genes add, guaB, and guaA. Both the Cp. abortus and all of the sequenced Cp. pneumoniae guaB genes are pseudogenes, although they have distinct mutations (Read et al. 2000; this study).
Because of the variability of the PZ regions, we designed PCR primers to amplify the PZ region of six Cp. abortus strains to see if there was also intraspecies variation. Bovine (LV350/93), ovine (A22), and caprine (LLG and FAS) abortion strains, in addition to vaccine strains (A22 and 1B), isolated from different geographical areas in Europe and Africa, were chosen for this analysis (see Methods). PCR products of predicted size were amplified from all six strains, and these were digested with EcoRI, HindIII, PstI, and XbaI for restriction fragment length polymorphism analysis (RFLP). None exhibited a restriction digestion pattern significantly different from Cp. abortus strain S26/3 (data not shown), although strain LLG showed some variation in the sizes of fragments. Sequencing of the LLG PCR fragment revealed no difference in gene content, but highlighted differences in the pseudogene content; CAB543 (conserved inner membrane protein) and CAB551 (guaB) were both intact in strain LLG. In addition, there was an insertion of 130 nt at the 3′-end of the pseudogene CAB541 (data not shown).
The biotin gene cluster (bioBFDA) is a region that shows limited distribution in the Chlamydiaceae (data not shown); the cluster has been deleted from Cp. caviae, C. trachomatis, and C. muridarum but remains intact in both Cp. abortus and all of the sequenced Cp. pneumoniae strains.
Another significant region of variability in the Cp. abortus genome is located immediately upstream of the 5S rRNA gene. In Cp. caviae this region carries a pseudogene (CCA00886) with similarity to the virulence-associated invasion/intimin-family of outer membrane proteins from Gram-negative bacteria (Read et al. 2003). The genome sequence of Cp. abortus encodes two CDS (CAB852 and CAB853), in place of the intimin-family gene: one (CAB852) encoding a conserved membrane protein, which is similar to CP0984 in Cp. pneumoniae AR39; and the other (CAB853) a hypothetical protein, which is unique to Cp. abortus. The analogous region in Cp. pneumoniae AR39 is occupied by genes encoding proteins of unknown function, including CP0984.
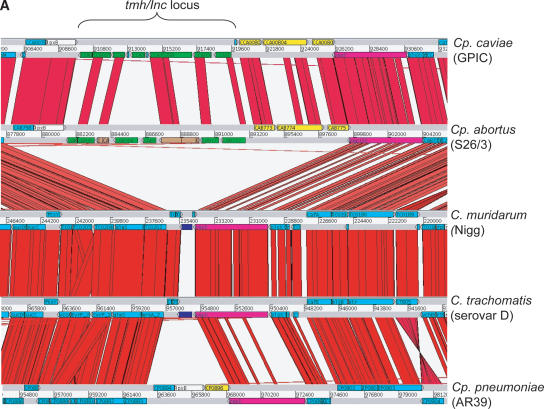
The Cp. abortus transmembrane head (TMH) protein family
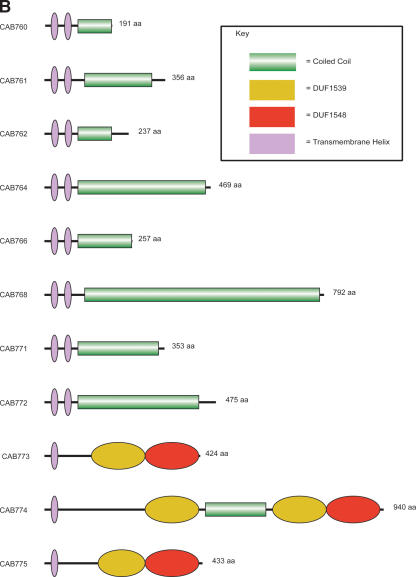
Another region showing limited distribution between chlamydial species encodes 11 CDS (CAB760–CAB775; CAB762 and CAB768 are pseudogenes) (Fig. 3A). CAB764 and CAB766 appear to have arisen following a duplication event. Eight of the CDS (CAB760–CAB762, CAB764, CAB766, CAB768, CAB771, and CAB772) encode products with paired N-terminal transmembrane (TM) domains followed by α-helical coiled-coil domains of varying lengths (Fig. 3B). The amino acid composition of these proteins is rich in leucine, glutamate, and serine residues. Because of the presence of the paired N-terminal TM domains, we termed these proteins transmembrane head proteins (TMH).
Figure 3.
(A) Comparison of the tmh locus of Cp. caviae (GPIC), Cp. abortus (S26/3), C. muridarum (Nigg), C. trachomatis (serovar D), and Cp. pneumoniae (AR39). ACT (see Fig. 2) comparison of amino acid matches between the complete six-frame translations (computed using TBLASTX) of representatives of the five sequenced Chlamydiaceae species. The red bars spanning between the genomes represent individual TBLASTX matches. CDS are marked as colored boxes positioned on the gray DNA lines: tmh family (green for intact genes, brown for pseudogenes), pmpD (pink), those carrying both DUF1539 and DUF1548 motifs (yellow), CDS encoding products with paired N-terminal TM domains (dark blue), lipid A biosynthesis (white), and all others (light blue). The scale is marked in base pairs. The region shown for Cp. pneumoniae strain AR39 is identical in all of the other sequenced Cp. pneumoniae strains (data not shown). (B) Architecture of the proteins encoded at the tmh locus of Cp. abortus. The amino acid sequences of the two pseudogenes CAB762 and CAB768 have been artificially reconstructed for the purpose of this analysis. See key for color codes. (DUF) Domain of unknown function.
The remaining three CDS within this region (CAB773–CAB775) encode proteins with a single N-terminal TM domain followed by two further conserved domains that are currently of unknown function (DUF1539 and DUF1548; DUF, domain of unknown function) (Fig. 3B).
Although the TMH locus is also present in Cp. caviae, a comparison of protein orthologs encoded in the Cp. abortus and Cp. caviae TMH loci showed that the level of amino acid sequence similarity was significantly lower than the genome average of 85% (SD 11), ranging between 32% and 60%. Comparison of the analogous regions in C. trachomatis, C. muridarum, and Cp. pneumoniae strain AR39 revealed significant levels of variation in gene content, although some of the encoded proteins possess paired N-terminal TM domains (CT813, TC199, and CP0896, respectively) and CP0896 possesses both DUF1539 and DUF1548 domains (Fig. 3A). When the DUF1539 and DUF1548 protein profiles were used to search the wider protein database (UNIPROT), they were found to be unique to Chlamydophila.
The presence of the paired N-terminal TM helices in the TMH-family proteins suggests that they may belong to a much larger family of proteins, the Inc-protein family. The gene products of paralogous inc genes have been shown to share little or no significant similarity in their primary sequence, but all possess unique paired hydrophobic domains of 50–80 amino acids in either the N-terminal (IncA) or C-terminal (IncB and IncC) regions. The lack of signal sequences (other than for CAB766) and the presence of paired hydrophobic domains and coiledcoil regions makes CAB760–CAB772 candidate Inc-effector proteins.
To extend these observations, using the criterion employed to identify Inc-family proteins in previous studies (Bannantine et al. 1998a,b, 2000; Bannantine and Stabel 1999), we identified 57 intact genes and six pseudogenes that are predicted to be c-inc genes (this figure includes the Cp. abortus orthologs of IncA [CAB536], IncB [CAB477], and IncC [CAB476] and the eight TMH-family protein-encoding genes) (see Supplemental Table 2). Almost half of the c-inc genes were clustered, with 29 CDS found in 10 loci, the largest of which was the tmh locus (CAB760–CAB772). Interestingly, most of the inc CDS were located toward the genome terminus compared to the overall distribution of other membrane protein genes as a whole (Fig. 1).
Polymorphic membrane proteins
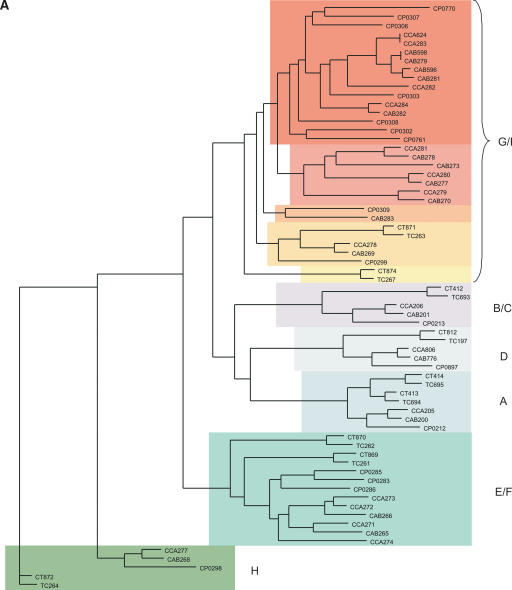
Cp. abortus carries several important putative outer membrane protein (Pomp) genes, including pomp98A (CAB282) and the two adjacent CDS, pomp90A (CAB279) and pomp91A (CAB281) (Longbottom et al. 1996, 1998b). Duplicates of pomp90A and pomp91A had also been identified and we have shown them to be located on the other replichore, ∼360 kb away: pomp90B (CAB598) and pomp91B (CAB596) (Fig. 2, pmp clusters 2 and 3). The products of these genes are part of a larger family of proteins known as the polymorphic membrane proteins (Pmp) that are represented in all of the sequenced Chlamydiaceae genomes (Vretou et al. 2003): 9, 21, and 17 pmp genes have been reported within the genome sequences of C. trachomatis/C. muridarum, Cp. pneumoniae, and Cp. caviae, respectively (Stephens et al. 1998; Kalman et al. 1999; Read et al. 2003).
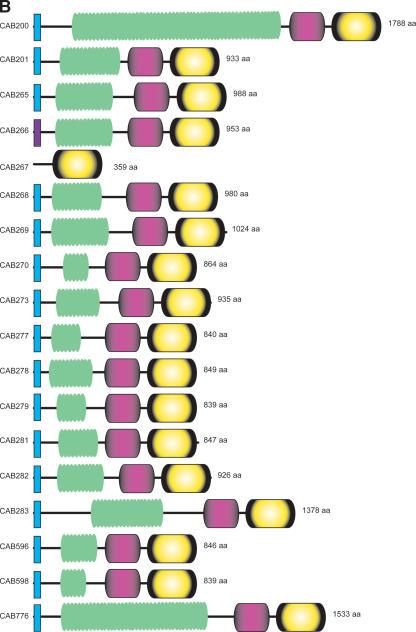
Protein clustering analysis, using TribeMCL (see Methods) showed that the Cp. abortus pmp gene family is made up of 18 CDS located in four loci (including the pomp genes) (Figs. 1 and 2) composed of a singleton (CAB776), two pairs of genes (CAB200, CAB201 and CAB596, CAB598), and a large cluster of 13 genes (CAB265–CAB270, CAB273, CAB277–CAB279, CAB281–CAB283).
The products of these pmp genes range in size from 40 to 189.5 kDa, and all terminate in a phenylalanine residue. All of the Cp. abortus Pmps, with the exception of CAB266 (which has a membrane-spanning domain but one that is not predicted to be a signal sequence) and CAB267 (which is a likely gene remnant), possess N-terminal signal sequences with potential signal peptidase 1 or signal peptidase II cleavage sites, which suggests that most are targeted to the outer membrane and is consistent with the localization of some to the EB surface (Longbottom et al. 1998a; Pedersen et al. 2001; Tanzer et al. 2001).
Two of the pmp genes carry multiple frameshifts and/or a premature stop codon (CAB270 and CAB273) and are likely to be pseudogenes. However, CAB279 (pomp90A) and CAB596 (pomp91B) have single frameshifts located within homopolymeric tracts of G(×17) and G(×16), respectively, which as mentioned above were seen to vary in length in a minority of library clones used to generate the whole-genome sequence. Homopolymeric nucleotide tracts are also found in other pmp-family members that appeared intact in the final genome assembly: CAB266, CAB281, and CAB598, which possess G(×8), G(×10), and G(×15) tracts, respectively. Of these three CDS the homopolymeric tract of CAB598 was also seen to vary in length, composed of between 15 and 18 G residues.
The Pmp family of proteins have a conserved domain architecture consisting of an N-terminal repeat region (defined by the motif GG[A/L/V/I][I/L/V/Y] and FXXN) and a C-terminal domain with similarities to the passenger domain of autotransporter proteins (Everett and Hatch 1995; Grimwood and Stephens 1999; Grimwood et al. 2001; Henderson and Lam 2001; Subtil et al. 2001). We investigated the domain structure of all the Pmp proteins from Cp. abortus and found that the number of N-terminal tandem repeats varies from three to 27 (Table 2; Fig. 4B). These repeats were found to be short in length and highly variable (investigated using NJ trees; see Methods).
Table 2.
Location of Pmp conserved repeats, middle (PMP_M), and autotransporter domains
| Name | No. of repeatsa | Amino acid range of repeatsd | PMP_M domaind | Autotransporter domaind |
|---|---|---|---|---|
| CAB200 | 27 | 140-1200 | 1266-1451 | 1497-1780 |
| CAB201 | 9 | 120-390 | 466-615 | 660-924 |
| CAB265 | 9 | 50-350 | 453-673 | 711-980 |
| CAB266 | 9 | 110-380 | 456-639 | 677-940 |
| CAB267 | 0 | — | — | 164-359 |
| CAB268 | 6 | 180-360 | 478-646 | 690-972 |
| CAB269 | 9 | 130-420 | 506-706 | 745-1016 |
| CAB270b | 3 | 150-240 | 353-522 | 563-856 |
| CAB273b | 4 | 120-300 | 428-591 | 632-927 |
| CAB277 | 4 | 90-230 | 341-499 | 541-832 |
| CAB278 | 4 | 100-300 | 351-508 | 549-841 |
| CAB279c | 4 | 150-270 | 348-500 | 539-826 |
| CAB281 | 3 | 150-240 | 353-509 | 548-839 |
| CAB282 | 6 | 150-330 | 440-591 | 631-918 |
| CAB283 | 6 | 290-620 | 839-1046 | 1085-1370 |
| CAB596c | 4 | 150-310 | 354-508 | 547-838 |
| CAB598 | 3 | 150-270 | 348-500 | 539-826 |
| CAB776 | 19 | 180-900 | 1017-1207 | 1251-1520 |
The number of repeats was estimated using both visual and computational evidence.
Pseudogenes for which the sequence has been reconstructed in silico.
Phase-variable CDS for which the sequence has been reconstructed in silico.
Represents amino acid positions.
Figure 4.
(A) Clustering of the Pmp-family proteins of sequenced chlamydial species. Phylogenetic tree of the Pmp-family proteins from Cp. abortus (S26/3), Cp. caviae (GPIC), Cp. pneumoniae (AR39), C. muridarum (Nigg), and C. trachomatis (serovar D). The Pmp protein families (A–I) are marked as previously assigned (Grimwood and Stephens 1999). (B) Architecture of the Pmp-family proteins of Cp. abortus. Conserved domains are color coded: signal sequence (light blue), transmembrane domain (dark purple), Pmp-repeat region (green), central PMP_M region (light purple), and autotransporter-like domain (yellow). The amino acid sequences of the pseudogenes CAB270 and CAB273 and the phase-variable genes CAB279 and CAB596 have been reconstructed in silico for the purpose of this analysis. CAB267 is a gene remnant.
The protein profiles of the Cp. abortus Pmp-family proteins also revealed the presence of a novel conserved middle domain, denoted PMP_M, which is present in all of the Pmp-family proteins (Table 2). This PMP_M domain has been submitted to the PFAM database and includes several motifs and residues that are highly conserved among members of this protein family (see Supplemental Fig. 1).
Previous phylogenetic analysis of all of the Pmp proteins identified six Pmp families: A, B/C, D, E/F, G/I, and H (Fig. 4A; Grimwood and Stephens 1999). To reduce the possible interference by the variable repeats present in the N terminus, phylogenetic trees of the Pmp-family proteins were based on an alignment of the conserved central PMP_M and C-terminal autotransporter domains. It is apparent from the Cp. abortus genome that, as in Cp. pneumoniae and Cp. caviae, while there are similar numbers of pmpA-F-family CDS, there is a significant expansion of pmpG-family genes, when compared to C. trachomatis (Fig. 4A). Moreover, with the additional Pmp proteins from sequenced genomes there appear to be five subfamilies within the G/I grouping (Fig. 4A).
Of the non-chlamydial proteins currently in UNIPROT, there are several bacterial genera that carry the Pmp-like repeat domains, but only Escherichia coli encodes gene products (K12-b2233 [YfaL; previously reported (Grimwood and Stephens 1999)], CFT073-c2775, 0157:H7 [RIMD] - Ecs3116, 0157:H7 [EDL933] - z3487) with more extensive similarity to the Pmp proteins, possessing five to six copies of the N-terminal repeats and a C-terminal autotransporter domain. The conserved chlamydial PMP_M domain is missing from these E. coli proteins. In place of PMP_M there is a motif that is similar to a domain found in the Bordetella pertussis protein pertactin, a filamentous hemagglutinin thought to promote adhesion to target mammalian cells (Emsley et al. 1996).
Discussion
Whole-genome comparison of Cp. abortus with other members of the Chlamydiaceae showed a remarkable level of conservation in both sequence and gene content. This is in striking contrast to the significant differences in host range, tissue tropisms, and disease outcomes observed for this group of human and animal pathogens. It was not possible, within the scope of this study, to consider the contribution of amino acid substitutions and differences in, for example, the timing and level of gene expression, although this will be extremely important to our understanding of this important group of pathogens. Instead, we have identified the major regions of variation within this highly conserved core genome that distinguish the sequenced members of the Chlamydophila and have focused our analysis on them.
The PZ has been shown to be the site of the most extensive gene differences between the Chlamydiaceae. Compared to the other chlamydial genomes, the PZ of both Cp. abortus and Cp. pneumoniae are smaller and have considerably fewer CDS. To confirm that the Cp. abortus strain S26/3 PZ was representative of the species, we amplified the PZ loci from six additional Cp. abortus isolates. The PZ regions from these isolates were shown by RFLP analysis to be identical to strain S26/3 with only the PZ region of variant strain LLG, which displays differences in inclusion morphology and antigenic diversity compared to other Cp. abortus strains (Vretou et al. 1996), showing any minor sequence variation. Consequently, it appears that the PZ region of Cp. abortus isolates is relatively stable at least in the context of this relatively small sample size.
The reduced size of the Cp. abortus PZ genome is due in part to the loss of the tryptophan biosynthetic operon (trp). This is significant because one of the primary host immune responses to chlamydial infection involves the production of the proinflammatory cytokine interferon-γ (IFN-γ). IFN-γ induces expression of indoleamine-2,3-dioxygenase (IDO), which degrades host tryptophan (Trp) to kynurenine, thus depriving the chlamydiae of Trp for growth and multiplication (Entrican 2002). Furthermore, in human and mouse placentae at least (there is no information for the ruminant placenta), Trp is also degraded through the constitutive expression of IDO in trophoblast cells (Entrican 2002). However, despite the obvious requirement for Trp, Cp. abortus does succeed in growing in the placenta. It is not clear how this occurs, but a possible explanation is that Cp. abortus is able to scavenge enough Trp from nutrients being passed from the mother to the developing fetus. Other explanations could include temporal differences in IDO expression and complex immunological and physiological interactions during pregnancy, which have yet to be elucidated (Entrican 2004).
The loss of the trp genes is not peculiar to Cp. abortus; all the Cp. pneumoniae strains and C. muridarum lack all of the trp biosynthesis genes, and so appear to be auxotrophic for host Trp. These differences suggest that, as with Cp. abortus, Trp synthesis is not required in these species for transmission or survival. In contrast, Cp. caviae has the most complete set of trp genes seen in the Chlamydiaceae, which should allow for the production of Trp from anthranilate (Read et al. 2003). It is possible that such gene loss has driven the differing tissue tropisms of the chlamydiae, although we feel this is unlikely and that gene loss probably followed niche adaptation. Either way, with little evidence of lateral gene transfer in the Chlamydiaceae the loss of the trp cluster may have put certain species in an evolutionarily vulnerable position.
The Cp. caviae, C. muridarum, and C. trachomatis PZ regions also encode toxin genes, which are notably absent from the same region of Cp. abortus. Elsewhere on the genome Cp. caviae encodes an intimin/invasin-like gene (CCA00886), remnants of which can be found in C. muridarum (Read et al. 2003); an intact version of this gene is present in the swine pathogen C. suis, and it is absent from Cp. abortus, Cp. pneumoniae, Cp. felis, Cp. psittaci, and Cp. pecorum (Liu et al. 2004). Since Cp. abortus is a recent clinical isolate that is unlikely to have lost these genes through prolonged laboratory passage, and none of these invasion/colonization factors are present in many other chlamydial species, this may underscore their importance in generating the niche differences observed for the Chlamydiaceae. However, it is worth noting that several of these genes are present as pseudogenes, including CCA00886, and although this may be a laboratory artifact, it might suggest that they are in the process of being lost and so call into question their actual importance.
The sporadic distribution of the biotin biosynthetic genes, present in Cp. abortus (CAB685–688; bioBFDA, respectively) but absent from Cp. caviae, may also reflect niche differences of this group of pathogens. There is an increased requirement for biotin during pregnancy in women (Mock et al. 2002), with many women becoming biotin-depleted and having to take supplements. Perhaps a similar situation arises in sheep during pregnancy, resulting in a lack of availability of biotin for this placental pathogen, and hence this operon would be important for colonization of this niche.
In addition to differences between chlamydial genomes that have arisen because of gene loss, there were also regions within the core sequence that appear to have undergone accelerated levels of variation and perhaps gene expansion by gene duplication. Such regions were found to encode TMH/Inc and Pmp protein families. The TMH-family proteins possess paired hydrophobic N-terminal domains followed by extensive coiled-coil regions. It is likely that the TMH proteins are part of a larger family of chlamydial proteins, termed the Inc family, noted for their targeting to the host inclusion membrane, and are suggested to be involved in inclusion development and avoidance of lysosomal targeting (Rockey et al. 2000).
In addition to the TMH proteins, we identified 55 other candidate Inc proteins (C-Inc) encoded by Cp. abortus, 14 of which showed a similar domain structure to the TMH proteins, composed of the paired hydrophobic regions accompanied by coiled-coil domains (this includes IncA, IncB, and IncC, although the relative order of the two domains was reversed in the Cp. abortus IncB and IncC protein sequences compared to the TMH protein structure). Coiled-coil regions are generally associated with protein–protein interactions and are present as functional domains in a variety of proteins including the protective M proteins of Streptococci, hemagglutinins of H. influenzae and transcriptional regulators (for review, see Lupas 1996), and this is perhaps consistent with their proposed role in the Chlamydiaceae.
While polymorphic membrane proteins constitute only a minor component of the chlamydial outer membrane, antibodies raised to them have significantly reduced the infectivity of EBs in vitro. Their high immunogenicity is also likely to be significant in the protective immunity conferred by chlamydial outer membrane preparations (Tan et al. 1990; Cevenini et al. 1991; Longbottom et al. 1998b; Vretou et al. 2003; Wehrl et al. 2004). In total, Cp. abortus carries 18 pmp genes. All of the Cp. abortus pmp gene products (with the exception of CAB267) possess characteristic N-terminal repeats present in a highly variable number (Table 2). The short length of these repeats suggests that gene variation occurs by frequent strand slippage (Lovett 2004). Unlike other repetitive proteins such as the Tropheryma whipplei WiSP proteins (Bentley et al. 2003), there is no obvious conservation with sequences outside of these repeats. The rapid evolution of the Pmp proteins has occurred through frequent duplication and deletion events, and therefore suggests that localized mutagenesis is the dominant force introducing variation within these proteins.
The central region of the Pmp proteins was also found to carry a conserved motif we designated PMP_M. The function of PMP_M remains unknown. However, Wehrl et al. (2004) have shown that once the Cp. pneumoniae PmpD protein is exported to the outer membrane and the N-terminal signal sequence is cleaved, two additional cleavage products are observed representing the N-terminal 660 amino acids and the central region (amino acids 661–1146). It is possible that PMP_M occludes the “sticky” N-terminal repetitive regions, which are thought to be involved in cell–cell attachment, until safely localized to the outside of the cell, when this central region may be cleaved to expose the N-terminal binding region. However, it is equally possible that PMP_M may be involved in localization/maintenance on the outer surface of the cell.
Two of the pmp genes contain frameshift mutations located within homopolymeric tracts that varied in length, presumably by the process of slip-strand mispairing making the expression of their protein products phase-variable. Similarly, the expression of the apparently intact pmp-family gene CAB598 was also found to be phase-variable. This is consistent with previous observations relating to the expression of pmp genes in other chlamydiae (Pedersen et al. 2001).
The presence of mutations was not restricted to the pmp genes. Overall Cp. abortus carries a similar proportion of pseudogenes to other niche-adapted bacterial pathogens such as Yersinia pestis and Salmonella enterica serovar Typhi. However, in these pathogens the presence of a high number of pseudogenes has been associated with a recent and dramatic change in lifestyle (Parkhill et al. 2001a,b). This appears not to be the case for the chlamydiae. The recent genome sequence of the related environmental Parachlamydiaceae, Acanthamoeba sp UWE25, has shown that the Chlamydiales had developed the characteristic biphasic developmental cycle and were adapted for an intracellular lifestyle very early on in their evolution (as much as 700 million years ago) (Horn et al. 2004). Consequently, it seems unlikely that the significant numbers of pseudogenes found in Cp. abortus are vestiges of such an ancient event. It is therefore of note that the proportion of pseudogenes in the two extended gene families of Cp. abortus: the tmh/inc genes, 9.5%, and the pmp-family genes, 17%, is significantly higher than the genome average of 2.8%. It is therefore tempting to speculate that both the pmp and tmh/inc genes are subject to greater selective pressures, which has resulted in the significant intra and interspecies variations within these protein families. Consequently, like the pmp genes, the tmh genes are obvious candidates for genes involved in more recent niche adaptations.
Although the chlamydial genomes are very well conserved, multiple whole-genome sequencing has undoubtedly had a dramatic effect on chlamydial research. We have shown that Cp. abortus conforms to the common theme observed from the analysis of other Chlamydiaceae, in that among the highly conserved core genome there are significant species variations that may account for the observed differences in tissue tropism, clinical sequelae, and disease outcomes. We have shown that the genetic differences of Cp. abortus compared to other chlamydial genomes include the lack of the trp operon and variation in the full complement of extended gene families such as the pmp family and perhaps the tmh family. Other differences may be associated with the presence or absence of single genes such as the toxins/invasins and the biotin operon, all of which are likely to have a significant bearing on the niche adaptation of this bacterium.
Methods
Bacterial strains, culture, and preparation of genomic DNA
The strain chosen for sequencing was S26/3, which was isolated in Scotland in 1979 from a vaccinated ewe that aborted (McClenaghan et al. 1984). The genomic DNA used for sequencing was derived from the fourth passage of an original isolate, propagated in fertile hens eggs, and represents a relatively fresh clinical isolate rather than a laboratory-adapted strain.
The Cp. abortus S26/3 strain was propagated in McCoy cells. Chlamydial EBs were harvested and purified from 10 225-cm2 flasks of infected cells by homogenization using a ground-glass homogenizer, followed by extraction with 1% N-lauroylsarcosine in 20 mM Tris-HCl (pH 7.5), 150 mM KCl (TKCl), and centrifugation through 15% sucrose in TKCl, as described previously (McClenaghan et al. 1984). Genomic DNA was prepared by lysis of the EBs at 37°C for 30 min with 0.5% SDS in 0.1 M Tris-HCl (pH 8.0), 0.1 M EDTA, 150 mM NaCl, and 100 μg/mL DNase-free RNase, followed by incubation with 150 μg/mL proteinase K at 50°C for 2 h. DNA was extracted twice with Tris-buffered (pH 8.0) phenol/chloroform/isoamyl alcohol (25:24:1), once with chloroform/isoamyl alcohol (24:1), and precipitated by adding 0.1 vol of 3 M sodium acetate (pH 5.2) and 2 vol of absolute ethanol. Spooled DNA was solubilized in 10 mM Tris-HCl (pH 8.0), and the concentration and purity were determined using a GenQuant Pro spectrophotometer (Amersham Biosciences).
Preparation of genomic DNA libraries, sequencing, and annotation
The DNA was fragmented by sonication, and several libraries were generated in pUC18 using size fractions ranging from 1.4 to 4 kb. The whole genome sequence was obtained from 20,394 end sequences (giving 9.39× coverage) derived from these libraries using dye terminator chemistry on ABI3700 automated sequencers. End sequences from larger insert plasmid (pMAQ1b; SmaI; 4–5 kb insert size) and BAC (pBACe3.6; BamHI; 23–48 kb insert size) libraries were used as a scaffold. Manual finishing ensures that every base is covered by at least two clones of high-quality sequence in each direction, or alternatively with complementary sequencing chemistries. Every consensus base must have a Phred quality score (Ewing and Green 1998) of >30 (i.e., <1/1000 chance of error). The measured accuracy statistics indicate that the chance of an incorrect base in the full genome sequence is 1 in 101,330,904 bp (a likely number of errors in the Cp. abortus S26/3 sequence being 0.01).
The sequence was assembled, finished, and annotated as described previously (Parkhill et al. 2000), using the program Artemis (http://www.sanger.ac.uk/Software/Artemis/; Rutherford et al. 2000) to collate data and facilitate annotation.
In silico analysis and database submission
The genome sequences of C. trachomatis (serovar D), C. muridarum (formerly C. trachomatis mouse pneumonitis [MoPn] strain Nigg), Cp. caviae (GPIC), and the Cp. pneumoniae (strains AR39, CWL029, TW183, J138) isolates were compared pairwise using the Artemis Comparison Tool (ACT) (http://www.sanger.ac.uk/Software/ACT/). Pseudogenes had one or more mutations that would ablate expression; each of the inactivating mutations was subsequently checked against the original sequencing data. Proteins were clustered into related family groups by the Markov Clustering method TribeMCL (Enright et al. 2002). Signal sequences were predicted using the SignalP 2.0 Server, Center for Biological Sequence Analysis, Technical University of Denmark (Nielsen et al. 1997).
Orthologous gene sets were identified by reciprocal FASTA searches, wherein homologous CDS were identified by the highest scoring hit again yielding the original query as highest scoring hit in the reverse search direction. Only those pairs of homologous CDS were retained for further analysis where the predicted amino acid identity was ≥40% (≥30% for the TMH/Inc-family protein orthologs) over 80% of the protein length. This strategy was applied to pairwise comparisons of the genomes of Cp. abortus, Cp. caviae, and Cp. pneumoniae strain AR39. Pseudogenes were not included in this analysis.
Conserved protein domains were identified from multiple protein alignments using MAFFT (Katoh et al. 2002) or ClustalX (Thompson et al. 1997). These conserved domains were used as seeds sequences for iterative searches by use of HMMER2 (http://hmmer.wustl.edu/) against protein databases made from the other sequenced chlamydial gene sets and/or the UNIPROT database (Apweiler et al. 2004). Alignment and analysis of the Pmp protein repeats were performed using neighbor-joining tree (NJ-tree) analysis (Saitou and Nei 1987) using the Belvu software (http://www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html). Trees were drawn using PHYML (Guindon and Gascuel 2003) based on a Maximum Likelihood approach (Hasegawa et al. 1991) and using the JTT substitution model (Jones et al. 1992).
All novel protein motifs defined in this study have been submitted to the PFAM database (http://www.sanger.ac.uk/Software/Pfam/tsearch.shtml; Bateman et al. 2004). The nucleotide sequence of the whole genome of Cp. abortus S26/3 was submitted to EMBL and assigned accession number CR848038.
PCR amplification of the plasticity zone of different Cp. abortus strains
Primer pairs were designed to the Cp. abortus S26/3 genome sequence in an attempt to amplify the plasticity zone (PZ) or replication termination region (RTR) of other Cp. abortus strains. The strains chosen were the ovine abortion vaccine strain A22 (isolated in Scotland in the early 1950s); the temperature-sensitive mutant vaccine strain 1B (Intervet UK) (Rodolakis and Souriau 1983); caprine abortion strain 15 (isolated in Tunisia; from H. Krauss, Giessen, Germany); bovine abortion strain LV350/93 (bovine abortion isolate; from S. Magnino, Pavia, Italy); and caprine abortion strains LLG and FAG (from O. Papadopoulos, Thessaloniki, Greece) (Vretou et al. 1996). Primer pairs annealed within CDS CAB538 (5′-CCGTTTCTGCCTTGGTTTATGAT-3′) and CAB553 (5′-TCACGATGAATATAAAGACGCTCCTA-3′), and CAB539 (5′-CGTAATAGCATGAAGCGTTTTGTGA-3′) and CAB552 (5′-GTTATCGACACTGCTCATGGACACTC-3′), and were predicted to amplify PCR products of 12.4 kb and 11.4 kb, respectively. The PCR reactions were performed according to manufacturers' instructions (Expand Long Template PCR System; Roche, UK) using 250–500 ng of DNA per reaction and the following conditions: denature at 94°C for 2 min (1 cycle); denature at 94°C for 10 sec, anneal at 50°C for 30 sec, and elongate at 68°C for 10 min (10 cycles); denature at 94°C for 10 sec, anneal at 50°C for 30 sec, and elongate at 68°C for 10 min + 20 sec increments (20 cycles); and a final extension cycle at 68°C for 7 min.
Acknowledgments
We thank the core sequencing and informatics teams at the Sanger Institute for their assistance and the Wellcome Trust for its support of the Sanger Institute Pathogen Sequencing Unit. The authors also thank the Scottish Executive Environment and Rural Affairs Department for funding this work.
[Supplemental material is available online at www.genome.org. The genome sequence data from this study have been submitted to EMBL under the accession number CR848038. The following individuals kindly provided DNA samples as indicated in the paper: H. Krauss, S. Magnino, and O. Papadopoulos.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3684805. Article published online ahead of print in April 2005.
References
- Andersson, S.G.E., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C.M., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140. [DOI] [PubMed] [Google Scholar]
- Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2004. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine, J.P. and Stabel, J.R. 1999. Identification of Mycobacterium paratuberculosis antigens present within infected macrophages. Mol. Biol. Cell 10: 1047. [Google Scholar]
- Bannantine, J.P., Rockey, D.D., and Hackstadt, T. 1998a. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 28: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Bannantine, J.P., Stamm, W.E., Suchland, R.J., and Rockey, D.D. 1998b. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 66: 6017–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine, J.P., Griffiths, R.S., Viratyosin, W., Brown, W.J., and Rockey, D.D. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2: 35–47. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S.D., Maiwald, M., Murphy, L.D., Pallen, M.J., Yeats, C.A., Dover, L.G., Norbertczak, H.T., Besra, G.S., Quail, M.A., Harris, D.E., et al. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361: 637–644. [DOI] [PubMed] [Google Scholar]
- Cevenini, R., Donati, M., Brocchi, E., Desimone, F., and Laplaca, M. 1991. Partial characterization of an 89-kDa highly immunoreactive protein from Chlamydia psittaci A/22 causing ovine abortion. FEMS Microbiol. Lett. 81: 111–116. [DOI] [PubMed] [Google Scholar]
- Deng, W., Burland, V., Plunkett, G., Boutin, A., Mayhew, G.F., Liss, P., Perna, N.T., Rose, D.J., Mau, B., Zhou, S.G., et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184: 4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Charles, I.G., Fairweather, N.F., and Isaacs, N.W. 1996. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381: 90–92. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Van Dongen, S., and Ouzounis, C.A. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entrican, G. 2002. Immune regulation during pregnancy and host–pathogen interactions in infectious abortion. J. Comp. Pathol. 126: 79–94. [DOI] [PubMed] [Google Scholar]
- ———. 2004. IDO: A crossroads of immunology and physiology? J. Reprod. Immunol. 61: 63–65. [DOI] [PubMed] [Google Scholar]
- Everett, K.D.E. and Hatch, T.P. 1995. Architecture of the cell-envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177: 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett, K.D.E., Bush, R.M., and Andersen, A.A. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. System. Bacteriol. 49: 415–440. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Grimwood, J. and Stephens, R.S. 1999. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4: 187–201. [DOI] [PubMed] [Google Scholar]
- Grimwood, J., Olinger, L., and Stephens, R.S. 2001. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun. 69: 2383–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. and Gascuel, O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hackstadt, T., Fischer, E.R., Scidmore, M.A., Rockey, D.D., and Heinzen, R.A. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5: 288–293. [DOI] [PubMed] [Google Scholar]
- Hasegawa, M., Kishino, H., and Saitou, N. 1991. On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol. 32: 443–445. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R. and Lam, A.C. 2001. Polymorphic proteins of Chlamydia spp.—Autotransporters beyond the Proteobacteria. Trends Microbiol. 9: 573–578. [DOI] [PubMed] [Google Scholar]
- Horn, M., Collingro, A., Schmitz-Esser, S., Beier, C.L., Purkhold, U., Fartmann, B., Brandt, P., Nyakatura, G.J., Droege, M., Frishman, D., et al. 2004. Illuminating the evolutionary history of chlamydiae. Science 304: 728–730. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kalman, S., Mitchell, W., Marathe, R., Lammel, C., Fan, L., Hyman, R.W., Olinger, L., Grimwood, L., Davis, R.W., and Stephens, R.S. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21: 385–389. [DOI] [PubMed] [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K., and Miyata, T. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Plaut, R., Kaltenboek, B., Rank, R., Magnino, S., Dean, D., and Bavoil, P. 2004. Genetic variation in the 5S rRNA–nqrF intergenic segment in Chlamydia spp. In Proceedings of the Fifth Meeting of the European Society for Chlamydia Research. Pauker Nyomdaipari Kft, Budapest.
- Longbottom, D. and Coulter, L.J. 2003. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 128: 217–244. [DOI] [PubMed] [Google Scholar]
- Longbottom, D., Russell, M., Jones, G.E., Lainson, F.A., and Herring, A.J. 1996. Identification of a multigene family coding for the 90 kDa proteins of the ovine abortion subtype of Chlamydia psittaci. FEMS Microbiol. Lett. 142: 277–281. [DOI] [PubMed] [Google Scholar]
- Longbottom, D., Findlay, J., Vretou, E., and Dunbar, S.M. 1998a. Immunoelectron microscopic localisation of the OMP90 family on the outer membrane surface of Chlamydia psittaci. FEMS Microbiol. Lett. 164: 111–117. [DOI] [PubMed] [Google Scholar]
- Longbottom, D., Russell, M., Dunbar, S.M., Jones, G.E., and Herring, A.J. 1998b. Molecular cloning and characterization of the genes coding for the highly immunogenic cluster of 90-kilodalton envelope proteins from the Chlamydia psittaci subtype that causes abortion in sheep. Infect. Immun. 66: 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, S.T. 2004. Encoded errors: Mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253. [DOI] [PubMed] [Google Scholar]
- Lupas, A. 1996. Coiled coils: New structures and new functions. Trends Biochem. Sci. 21: 375–382. [PubMed] [Google Scholar]
- McClenaghan, M., Herring, A.J., and Aitken, I.D. 1984. Comparison of Chlamydia-psittaci isolates by DNA restriction endonuclease analysis. Infect. Immun. 45: 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock, D.M., Quirk, J.G., and Mock, N.I. 2002. Marginal biotin deficiency during normal pregnancy. Am. J. Clin. Nutr. 75: 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural. Syst. 8: 581–599. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514: 17–25. [DOI] [PubMed] [Google Scholar]
- Ogata, H., Audic, S., Renesto-Audiffren, P., Fournier, P.E., Barbe, V., Samson, D., Roux, V., Cossart, P., Weissenbach, J., Claverie, J.M., et al. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293: 2093–2098. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Achtman, M., James, K.D., Bentley, S.D., Churcher, C., Klee, S.R., Morelli, G., Basham, D., Brown, D., Chillingworth, T., et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404: 502–506. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Dougan, G., James, K.D., Thomson, N.R., Pickard, D., Wain, J., Churcher, C., Mungall, K.L., Bentley, S.D., Holden, M.T.G., et al. 2001a. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413: 848–852. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Thomson, N.R., Titball, R.W., Holden, M.T.G., Prentice, M.B., Sebaihia, M., James, K.D., Churcher, C., Mungall, K.L., et al. 2001b. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413: 523–527. [DOI] [PubMed] [Google Scholar]
- Pedersen, A.S., Christiansen, G., and Birkelund, S. 2001. Differential expression of Pmp10 in cell culture infected with Chlamydia pneumoniae CWL029. FEMS Microbiol. Lett. 203: 153–159. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S., Conrad, J., Hall, B.G., and Ofengand, J. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, T.D., Brunham, R.C., Shen, C., Gill, S.R., Heidelberg, J.F., White, O., Hickey, E.K., Peterson, J., Utterback, T., Berry, K., et al. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, T.D., Myers, G.S.A., Brunham, R.C., Nelson, W.C., Paulsen, I.T., Heidelberg, J., Holtzapple, E., Khouri, H., Federova, N.B., Carty, H.A., et al. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): Examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31: 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey, D.D., Lenart, J., and Stephens, R.S. 2000. Genome sequencing and our understanding of chlamydiae. Infect. Immun. 68: 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis, A. and Souriau, A. 1983. Response of ewes to temperature-sensitive mutants of Chlamydia psittaci (Var-Ovis) obtained by NTG mutagenesis. Ann. Rech. Vet. 14: 155–161. [PubMed] [Google Scholar]
- ———. 1989. Variations in the virulence of strains of Chlamydia psittaci for pregnant ewes. Vet. Rec. 125: 87–90. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.A., and Barrell, B. 2000. Artemis: Sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Saikku, P. 1999. Epidemiology of Chlamydia pneumoniae in atherosclerosis. Am. Heart J. 138: S500–S503. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Shirai, M., Hirakawa, H., Kimoto, M., Tabuchi, M., Kishi, F., Ouchi, K., Shiba, T., Ishii, K., Hattori, M., Kuhara, S., et al. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28: 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R.S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R.L., Zhao, Q.X., et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282: 754–759. [DOI] [PubMed] [Google Scholar]
- Subtil, A., Parsot, C., and Dautry-Varsat, A. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39: 792–800. [DOI] [PubMed] [Google Scholar]
- Tan, T.W., Herring, A.J., Anderson, I.E., and Jones, G.E. 1990. Protection of sheep against Chlamydia psittaci infection with a subcellular vaccine containing the major outer-membrane protein. Infect. Immun. 58: 3101–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer, R.J., Longbottom, D., and Hatch, T.P. 2001. Identification of polymorphic outer membrane proteins of Chlamydia psittaci 6BC. Infect. Immun. 69: 2428–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viratyosin, W., Campbell, L.A., Kuo, C.C., and Rockey, D.D. 2002. Intrastrain and interstrain genetic variation within a paralogous gene family in Chlamydia pneumoniae. BMC Microbiol. 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vretou, E., Loutrari, H., Mariani, L., Costelidou, K., Eliades, P., Conidou, G., Karamanou, S., Mangana, O., Siarkou, V., and Papadopoulos, O. 1996. Diversity among abortion strains of Chlamydia psittaci demonstrated by inclusion morphology, polypeptide profiles and monoclonal antibodies. Vet. Microbiol. 51: 275–289. [DOI] [PubMed] [Google Scholar]
- Vretou, E., Giannikopoulou, P., Longbottom, D., and Psarrou, E. 2003. Antigenic organization of the N-terminal part of the polymorphic outer membrane proteins 90, 91A, and 91B of Chlamydophila abortus. Infect. Immun. 71: 3240–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrl, W., Brinkmann, V., Jungblut, P.R., Meyer, T.F., and Szczepek, A.J. 2004. From the inside out—Processing of the Chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51: 319–334. [DOI] [PubMed] [Google Scholar]
- Zomorodipour, A. and Andersson, S.G.E. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett. 452: 11–15. [DOI] [PubMed] [Google Scholar]
Web site references
- http://hmmer.wustl.edu/; profile hidden Markov models for biological sequence analysis.
- http://www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html; Belvu software for Multiple sequence alignment.
- http://www.sanger.ac.uk/Software/ACT/; ACT comparative sequence analysis tool.
- http://www.sanger.ac.uk/Software/Artemis/; Artemis sequence annotation tool.
- http://www.sanger.ac.uk/Software/Pfam/tsearch.shtml;; PFAM protein family database.