Abstract
We previously reported the analysis of genome-wide expression profiles and various diabetes-related traits in a segregating cross between inbred mouse strains C57BL/6J (B6) and DBA/2J (DBA). By considering transcript levels as quantitative traits, we identified several thousand expression quantitative trait loci (eQTL) with LOD score >4.3. We now experimentally address the problem of multiple comparisons by estimating the fraction of false-positive eQTL that are under cis-acting regulation. For this, we have utilized a classic cis–trans test with (B6 × DBA)F1 mice to determine the relative levels of transcripts from the B6 and DBA alleles. The results suggest that at least 64% of cis-acting eQTL with LOD >4.3 are true positives, while the remaining 36% could not be confirmed as truly cis-acting. Moreover, we find that >96% of apparent cis-acting eQTL occur in regions that do not share SNP haplotypes. Cis-acting eQTL serve as an important new resource for the identification of positional candidates in QTL studies in mice. Also, we use the analysis of the correlation structures between genotypes, gene expression traits, and phenotypic traits to further characterize genes expressed in liver that are under cis-acting control, and highlight the advantages and disadvantages of integrating genetics and gene expression data in segregating populations.
Variations in the DNA that lead to variability in the transcript levels of a gene can be expected to exist in any heterogeneous population, given that all genes are under the control of cis-acting elements (e.g., promoter elements, TATA box, etc.). Associations between variations in the DNA of such genes and varying levels of expression can be detected in populations in which the DNA variations are cosegregating with the expression traits (e.g., experimental crosses, human families, etc.). Variability in the expression level or functional efficacy of one gene can, in turn, result in a broader set of expression changes in other genes that do not themselves differ genetically. This results in these secondary genes ultimately seen as being under the control of DNA variations in the primary gene. Thus, the individual variation in gene expression consists of two varieties. The first, termed cis-acting, results from DNA variations of a gene that directly influence transcript levels of that gene. The second variety, termed trans-acting, does not involve DNA variations of the gene in question, but rather, is secondary to alterations of other genetic variations. For example, cis-acting variations in a gene encoding a transcription factor or a splicing factor might be expected to influence the expression of other genes in trans. These upstream genetic perturbations may affect transcript levels or could affect protein function, but, in both cases, the end result is an effect on transcription in trans on a downstream target gene. Genome-wide expression array analyses have revealed thousands of differences in transcript levels between pathologic conditions (e.g., cancer) or between different strains of experimental organisms or cells from different humans (Karp et al. 2000; Jin et al. 2001; Brem et al. 2002; Schadt et al. 2003; Yvert et al. 2003; Monks et al. 2004; Morley et al. 2004). However, until recently, the degree of cis-acting versus trans-acting variations has been unknown. Cowles et al. (2002) examined this issue by systematically testing for cis-acting regulator variation in 69 randomly selected genes across four inbred strains of mice. For this, they used a classic cis–trans test involving the quantitation of transcripts from each allele in heterozygous animals. They demonstrated that of 69 genes, four genes exhibited evidence of cis-acting variation. Another approach to the problem is to measure transcript levels in genetic crosses or human families and map the loci controlling transcript levels using classical linkage analysis. In the case of cis-acting variation, the levels of a transcript would map to the structural gene, producing the transcript, whereas in the case of trans-acting variation, the levels of a transcript would not be expected to map to the structural gene, except by coincidence. In this approach, the transcript levels are analyzed as quantitative traits, and we have referred to the loci controlling transcript levels as expression quantitative trait loci (eQTL). We and others have recently reported such “genetics of gene expression” studies in yeast (Brem et al. 2002), maize, humans, and mice (Chesler et al. 2003; Schadt et al. 2003; Monks et al. 2004; Morley et al. 2004).
We previously applied the genetics of gene-expression approach to a genetic intercross between two common inbred strains of mice, DBA/2J (DBA) and C57BL/6J (B6), by combining genome-wide genotype data at an average density of 13 cM with expression profiling of liver tissue for all F2 mice. The results revealed a surprising level of genetic variation, resulting in the identification of several thousand loci with LOD scores >4.3 (the threshold of significance in an F2 mouse intercross for achieving a genome-wide P-value < 0.05). Of these, approximately one-third exhibited apparent cis-acting regulatory variation as judged by the mapping of the eQTL to within 10 cM of the structural gene locus. To date, no characterization has been done to assess on a broad scale whether the cis-acting eQTL detected in any segregating population, including our F2 cross, are actually likely due to DNA variations in the genes themselves. To assess the fraction of cis-acting eQTL that may be artifacts of probes overlapping DNA polymorphisms, we used the genomic sequence data from multiple mouse strains and single nucleotide polymorphism (SNP) data derived from these sequence data to establish whether the microarray probes associated with cis-acting eQTL reported by Schadt et al. (2003) were enriched for probes overlapping SNPs. Further, because the genomic regions supporting genes with cis-acting eQTL would not be expected to be identical by descent (IBD) between the two strains used to construct the corresponding cross, we used the genomic sequence and associated SNP data to reconstruct haplotypes and used these haplotypes to assess where the two strains (B6 and DBA) are identical by descent (IBD) genome-wide. We validate a fraction of the cis-acting eQTL using a classic cis–trans test in F1 heterozygous mice. Also, the correlation structures induced by cis-acting eQTL are described, demonstrating the advantages and potential disadvantages that arise in integrating gene expression and genetic data to elucidate complex traits. Finally, we illustrate how these cis-acting eQTL can be used to prioritize candidate genes at a classical trait QTL (cQTL) region of interest.
Results
SNPs and cis-acting eQTL
Cis-acting eQTL summaries from the BXD cross
Of the 23,574 genes represented on the mouse microarray used to profile the BXD cross previously reported by Schadt et al. (2003), 21,744 could be reliably mapped to unique autosomal chromosome locations of the mouse genome (NCBI Build 32). Of these 21,744 mapped genes, 3465 had eQTL with LOD scores >4.3, and 913 had eQTL with LOD scores >7.0. Although some eQTL are false positives, these frequencies are far in excess of the numbers one would expect by chance alone. Approximately 34% (1171) of the mapped genes with eQTL exceeding 4.3 had a physical location coincident with the eQTL position, while 68% of the mapped genes with eQTL exceeding 7.0 had a physical location coincident with its eQTL position. Due to the inexact nature of QTL positioning in this type of experimental cross, we called an eQTL and corresponding gene coincident if the physical location of the gene mapped to within 20 mb (∼10 cM) of the cis eQTL peak. For nearly all of these cis eQTL (98.3%) with lod scores >4.3, the gene falls within the 95% confidence interval of the QTL. When assessing the significance of a cis eQTL, it is not necessary to perform a correction for multiple testing over the entire genome, given the only location of interest is the 20-cM window centered at the physical location of the gene of interest. Therefore, to determine the significance associated with the classic LOD score threshold of 4.3 for cis eQTL, multiple testing corrections only need to take into account tests performed in the 20-cM window centered at the gene of interest. In this case, the significance level associated with a LOD score of 4.3 is 0.00066 (as opposed to a genome-wide significance of 0.05). Given this estimate, we would expect only 14 of the 1171 genes detected with cis eQTL to be false-positive eQTL.
Most cis-acting eQTL fall in regions that are not IBD
Genes with true cis eQTL in the BXD cross require polymorphisms in the DNA sequence that differ between the B6 and DBA strains and that ultimately lead to significant differences in transcript levels between these strains. Therefore, we would expect most probe sequences associated with cis-acting eQTL to fall in regions that are not identical by descent (IBD). To assess the proportion of genes with cis eQTL that fall in non-IBD regions, we first estimated the fraction of the B6 and DBA genome that was IBD. We mapped all SNP sequences represented in the Celera Mouse Genome Database to NCBI Build 32 of the public mouse genome assembly. Of these SNPs, 1,064,677 could be reliably mapped to the autosomal chromosomes and were polymorphic between the B6 and DBA strains. By examining the moving average of SNP frequencies along the autosomal genome, we determined that regions with fewer than five informative SNPs per 25-kb window were indicative of regions IBD between B6 and DBA (Fig. 1A). Averaged over the entire genome, we estimated that 57% of the genome between B6 and DBA is IBD. The extent of genome-wide IBD has been investigated before among several strains of mice (Wade et al. 2002; Wiltshire et al. 2003). Our observed estimate for B6 versus DBA is higher than previous estimates of ∼35% (Wiltshire et al. 2003); however, the SNP data used to make this estimate is much more comprehensive than that in prior studies.
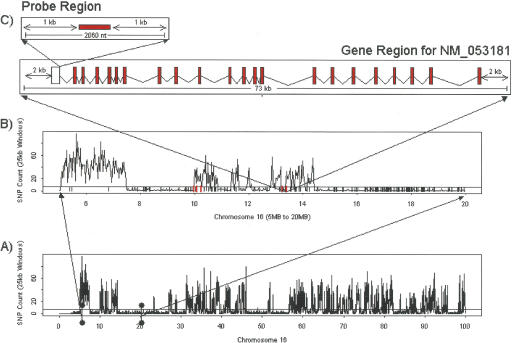
Figure 1.
Informative SNP frequency across chromosome 16 between B6 and DBA. The horizontal line in graphs A and B represents the threshold of five SNP/25 kb. Regions with five or less SNPs in a 25-kb interval were designated IBD between B6 and DBA. (A) The entire chromosome 16; (B) a closer look at an ∼15-Mb region on proximal chromosome 16, where the small vertical red lines at the base of the figure represent genes with cis eQTL in this region. (C) further zooms in on NM_053181, depicting the boundaries for the various regions considered in the SNP overlapping probe analysis.
Next, we defined gene regions for each of the 21,744 genes that mapped to the autosomal genome, where the gene regions were defined as the region between the transcription start and stop sites for the gene, plus 2000 nucleotides flanking the transcription start and stop positions (Fig. 1C). Of these regions, 9932 (∼45%) were overlapping non-IBD regions, close to the overall percentage of the genome we identified as non-IBD between the B6 and DBA strains. In contrast, for the gene regions corresponding to the 1171 genes with strong cis eQTL, a striking 1132 (nearly 97%) were overlapping non-IBD regions. This increase in genes with cis eQTL overlapping non-IBD regions is statistically very significant (Fisher Exact Test P-value <1.0 × 10–300), a result that strongly supports the cis-acting nature of the eQTL in this set. We would expect roughly 1% (14) of the genes detected with cis eQTL to be false-positive events, where the gene is actually under the control of a neighboring gene, so that the cis versus trans control cannot be distinguished by the QTL data alone. While this likely explains a majority of these cases, the data do support that some of the genes detected with cis eQTL in IBD regions as potentially true positives. First, the median distance between these 39 genes and the closest non-IBD regions was only 200 kb, so that cis regulatory elements located further away from the gene-promoter region may explain the cis effects observed for some of the genes. Previous reports have reported functional cis elements as far as 1 Mb away from the gene under study (Qin et al. 2004). Second, even in IBD regions, there are polymorphisms occurring at a low frequency (Fig. 1), and these polymorphisms are most likely due to sequencing errors or spontaneous mutations, where the latter could reasonably be expected to affect transcript abundances in some cases.
Does probe/SNP overlap lead to spurious cis-eQTL?
Even though genes with cis eQTL in the BXD cross overwhelmingly fall in regions that are not IBD, spurious associations between DNA variations in a given gene and variations in transcript abundances for the given gene could arise if the probes used to monitor transcript abundances directly overlap SNPs, thereby influencing transcript abundance measures, even subtly. Therefore, it was of interest to estimate the percentage of cis-eQTL that may be due to probes overlapping SNPs. To obtain reliable estimates using the SNP data represented in the Celera Mouse Genome Database derived from multiple sequenced strains of mice, we first established the extent of strain-specific coverage for each probe region for each probe represented on the microarray used in this study. This step was necessary because it is not possible to conclude that a probe and/or probe region does not overlap any SNPs that are polymorphic between the B6 and DBA strains, unless we know there is genomic sequence supporting the regions from each strain.
Given the computational complexity involved in mapping each of the raw sequence reads represented in the Celera Mouse Genome Database to the corresponding assembled genomic sequence, we instead chose to estimate the percent coverage by examining the SNPs in each of the probe regions for each probe that could be mapped to the genomic sequence, where probe regions in this instance were defined as the 1000 nucleotides flanking the left and right of each 60-nucleotide probe sequence (Fig. 1C). If SNPs were detected in this 2060-nucleotide region, and these SNPs included alleles identified from the B6 and DBA strains, then we concluded that the probe sequence was, in fact, supported by B6 and DBA genomic sequence. Of these 21,744 probe regions that were mapped to the mouse genome, we detected at least one SNP with alleles from the B6 and DBA strains in 14,101 of the probe regions. This provides an estimate of 65% for the percentage of probes on the microarray that were supported by B6 and DBA sequence.
The test for determining whether SNPs have a significant impact on cis-acting eQTL is based on a comparison between the frequency of probes overlapping SNPs over all probes that could be mapped to the mouse genome, and the frequency of probes overlapping SNPs where the probes give rise to a significant cis-acting eQTL. If the SNPs have no effect on the hybridization intensities, then the frequencies of probes overlapping SNPs should be the same between the two groups (the null hypothesis). The overall frequency for probes overlapping one or more SNPs is given by the percentage of probes in the set of 14,101 genes described above that overlap at least one SNP in the probe region. We identified 585 (4.1%) probes that overlapped one or more SNPs. The mean number of SNPs within these 585 probes is 1.19. Of the 585 probes overlapping at least one SNP, 121 represent genes with cis eQTL. Interestingly, the mean amount of SNPs in this group of 121 probes is 1.22, which does not differ significantly from the full set of 585 probes. However, the fact that 21% (121/585) of the probes overlapping SNPs were in genes with cis eQTL is a significant bias. This enrichment could result from the fact that cis eQTL are overwhelmingly found in non-IBD regions of the genome, where there is a higher probability of incurring a SNP within the probe sequence. Therefore, to assess further the potential bias introduced by probes overlapping SNPs, we restricted attention to those probe regions containing exactly one SNP.
Of all the probe regions that mapped to the mouse genome, 3107 contained exactly one SNP. We chose this set as our baseline set of probe regions to consider for more precisely testing whether a bias exists. The baseline frequency for probes overlapping a single SNP is then given by the percentage of probes in the set of 3107 that overlap the one SNP identified in the probe region. In this case, we found 96 probes of 3107 that overlapped the single SNP (3.1%). Next, we restricted attention to those 238 probe regions in the set of 3107 probe regions for which the probes gave rise to cis eQTL with an associated LOD score >4.3. Of these 238 probe regions, 14 had probes that overlapped the single SNP (5.9%). Application of the Fisher Exact Test in this case to determine the probability that this bias could occur by chance resulted in a P-value of 0.013, indicating that the percentage of probe regions with strong cis-acting eQTL that overlap a single SNP is marginally significantly higher than the percentage of probes overlapping a single SNP over all genes considered. Given the low frequency of probes overlapping SNPs, the net impact of this effect is minor with respect to considering a putative cis eQTL as a true or false positive. Also, the potential impact is further mitigated by the observation presented earlier that probes with cis-acting eQTL overwhelmingly fall in regions that are not IBD, compared with probes in general.
Testing whether additive QTL effects for cis eQTL are evenly distributed about zero
Given the slight bias noted above in probes overlapping SNPs for genes with cis eQTL, and given that we do not have a reliable estimate on the false-negative rate for SNPs identified as polymorphic between B6 and DBA in the Celera SNP data, we further investigated the extent to which probes overlapping SNPs may be responsible for cis-acting eQTL by examining whether the additive effects related to the cis-acting eQTL were upwardly biased in the direction of the B6 allele. The biologically intuitive null hypothesis is that the additive effects over all cis eQTL are randomly distributed about 0. Given that an overwhelming majority of the probes on the microarray were designed against B6 sequence, we would expect significantly >50% of the additive effects for cis eQTL to be higher for the B6 allele if probes overlapping SNPs were an issue, since such probes would likely perfectly match the B6 sequence against which it was designed. Of the 1171 genes with cis-acting eQTL considered in this study, 657 had the B6 allele more highly expressed than the DBA allele. That is, just over 56% of the cis-acting eQTL indicated higher expression at the B6 allele compared with the DBA allele. Assuming the probability is 0.5 that we observe such an event for any given gene, the number of observations we make over all genes is binomially distributed, so that the probability we observe at least 657 of 1171 cis eQTL with additive effects biased in the B6 direction is easily computed to be 2.1 × 10–5.
This bias is more significant than the estimates provided above, where only 5.9% of the probes were found to overlap a single SNP, compared with a 12% bias noted here for cis eQTL additive effects. This discrepancy may suggest an appreciable false-negative rate for SNPs identified in the Celera data as polymorphic between the B6 and DBA strains, resulting in a significant number of probes misclassified as not overlapping SNPs. In addition, the SNP/probe overlap statistics discussed above were restricted to probe regions containing exactly one SNP, so that the bias may become more extreme if probe regions containing more than one SNP were considered. In any case, these observed biases speak for the need to provide direct experimental confirmation of the cis eQTL.
Validation of cis-regulated genes
All of the above arguments speaking to the validity of cis eQTL were computational in nature. To provide direct experimental support for the validity of the cis eQTL and to estimate to what extent the previously reported cis-acting eQTL represent type 1 errors, we validated a subset of previously identified cis eQTLs using a classical cis–trans test in which (B6 × DBA)F1 mice were analyzed for the relative levels of transcript from each allele. In a cis-regulated gene, the allelic expression in F1 mice should reflect the ratios observed in the two parental strains, given that all mice examined are exposed to the same environmental perturbations. If the gene is regulated in trans, we expect the allelic ratio of transcripts from each allele to be ∼1:1, because a truly trans-acting regulator should act in a similar manner on both alleles. Of course, a gene could exhibit a combination of trans and cis regulation, although the cis regulatory component should cause the ratio to be different from 1:1, albeit to a lesser extent than might otherwise be the case.
To quantitate transcript levels from each allele in an F1 heterozygote, we selected genes exhibiting the predicted cis-acting eQTL with LOD >4.3 and also exhibiting one or more polymorphisms within the genes-coding sequence (the latter being required to distinguish transcripts from the two alleles). Probes that overlapped SNPs were excluded. This gene set is representative of the 1171 genes with annotated physical location and a cis eQTL with LOD scores >4.3. Although the genes tested were randomly chosen from this pool, it is worth noting that they are biased toward higher LOD scores, with a median LOD score of 11.8. We explored several methods for quantitative genotyping of the transcripts, including allele quantitation using primer extension, quantitative sequence analysis, and restriction enzyme digestion of the RT–PCR product, followed by separation of the labeled product by gel electrophoresis (data not shown). Previous studies have shown comparable results and accurate allelic quantification using primer extension by Pyrosequencing, single-base extention, and RFLP analysis (Shifman et al. 2002). We observed comparable results for the different methods used, but for ease of analysis and flexibility (for example, only a fraction of the SNPs resulted in an RFLP), we chose to use primer extension using Pyrosequencing and sequencing of RT–PCR products across the SNPs. Pyrosequencing is a primer extension-based method that utilizes the release of pyrophosphate during nucleotide incorporation to fuel a cascade of enzymatic reactions, resulting in a quantitative fluorescence emission. The different methods were validated as described in the Methods. In cases where more than one coding SNP was available in the amplicon, the result was consistent throughout all SNPs in the region (Table 1, NM_026981 and AF168680). Moreover, multiple amplicons were used for one gene, and in this case, the two different amplicons yielded similar B6:DBA ratios (Table 1, NM_130447). Finally, where possible, both forward and reverse primers were used with consistent results (data not shown). One gene that exhibited strictly trans regulation was assayed as a negative control (Table 1, NM_028333). The allelic peak areas were determined, and the ratio of B6:DBA allelic transcripts were calculated for each F1. The average of these values over several F1 mice (n ranges from 2 to 8, see Table 1) were then calculated and used as the ratio value. For most analysis, three to five F1 female mice were studied. The difference in the number of animals for the various genes reflects the failure of some assays. These data are summarized in Table 1 and Figure 2. In order to determine the sensitivity of the assay to detect a 1:1 (B6:DBA) ratio, we assayed F1 genomic DNA for a subset of genes. These genes were limited to those in which the entire amplicon analyzed resided within one exon. A total of four genes were analyzed with DNA from two different F1s with multiple replicates. In all, 18 of these 1:1 ratio controls yielded results (four different genes). The 95% confidence interval of the distribution of these results (.96, 1.08) was used as the 95% confidence interval for the detection of a trans-regulated transcript. When this interval was overlapping with the 95% confidence interval of the B6:DBA ratio of any of the genes being tested, the gene was considered under possible trans control.
Table 1.
Results of cis-trans test on putative cis eQTLs
| #a | GenBank accessionb | Peak LOD scorec | SNP positiond | Array data ratio (B6/DBA)e | Sequencing observed ratio (B6/DBA)f | P-valueg | CI95h |
|---|---|---|---|---|---|---|---|
| 1 | NM_010381 | 24.2 | 553 | 0.10 | <0.10 (4) | N/A | N/A |
| 2 | NM_026621 | 21.0 | 353 | 0.20 | <0.10 (4) | N/A | N/A |
| 3 | NM_130447 | 35.6 | 1834 | 0.30 | 0.67 ± .02 (6) | <10-4 | .59, .75 |
| 4 | NM_130447 | 35.6 | 1898 | 0.30 | 0.88 ± .01 (6) | 0.005 | .81, .94 |
| 5 | BC019407 | 36.7 | 879 | 0.30 | 0.19 ± .02 (3) | <10-4 | .14, .24 |
| 6 | NM_008514 | 28.2 | 1035 | 0.30 | 0.57 ± .03 (3) | 0.01 | .35, .76 |
| 7 | NM_008620 | 11.8 | 1557 | 0.30 | 0.49 ± .07 (4) | 0.007 | .25, .74 |
| 8 | AK017526 | 10.7 | 1087 | 0.35 | 0.28 ± .02 (4) | <10-4 | .14, .44 |
| 9 | NM_009252 | 9.2 | 282 | 0.37 | 0.72 ± .03 (4) | 0.03 | .49, .94 |
| 10 | BC023898 | 11.8 | 1299 | 0.40 | 0.17 ± .05 (4) | <10-4 | .06, .28 |
| 11 | NM_011424 | 11.5 | 652 | 0.40 | 1.10 ± .04 (4) | 0.35 | .72, 1.48 |
| 12 | NM_026981 | 31.4 | 164 | 0.40 | 0.76 ± .02 (3) | 0.01 | .63, .90 |
| 13 | NM_026981 | 31.4 | 173 | 0.40 | 0.75 ± .02 (3) | 0.008 | .66, .85 |
| 14 | AK003222 | 5.1 | 767 | 0.40 | 0.40 ± .01 (6) | <10-7 | .36, .43 |
| 15 | AK011491 | 16.5 | 716 | 0.40 | 0.95 ± .03 (7) | 0.39 | .85, 1.06 |
| 16 | AV119440 | 6.6 | 610 | 0.40 | 0.57 ± .05 (4) | 0.002 | .41, .71 |
| 17 | AK007567 | 7.5 | 147 | 0.44 | 0.44 ± .02 (4) | 0.002 | .25, .63 |
| 18 | NM_028333 | N/A | 646 | N/A | 0.98 ± .03 (4) | 0.53 | .82, 1.14 |
| 19 | AK011928 | 7.5 | 93 | 1.50 | 0.64 ± .02 (4) | <10-4 | .56, .71 |
| 20 | AK014150 | 13.1 | 1083 | 1.50 | 1.05 ± .02 (8) | 0.13 | .97, 1.15 |
| 21 | NM_008663 | 10.3 | 1430 | 2.31 | DBA absent | N/A | N/A |
| 22 | NM_027134 | 10.5 | 157 | 2.38 | 1.19 ± .06 (4) | 0.06 | .97, 1.37 |
| 23 | NM_025892 | 10.0 | 131 | 2.46 | 0.87 ± .03 (4) | 0.14 | .67, 1.08 |
| 24 | NM_009735 | 15.8 | 104 | 2.50 | 0.62 ± .01 (4) | <10-5 | .58, .65 |
| 25 | NM_031376 | 11.7 | 1898 | 2.60 | 1.00 ± .06 (3) | 0.97 | .51, 1.51 |
| 26 | AF168680 | 11.5 | 4100 | 2.60 | 2.37 ± .12 (4) | 0.001 | 1.94, 2.79 |
| 27 | AF168680 | 11.5 | 4102 | 2.60 | 2.37 ± .13 (4) | 0.003 | 1.90, 2.83 |
| 28 | AK018666 | 11.5 | 2980 | 2.60 | 2.62 ± .15 (4) | 0.001 | 2.12, 3.11 |
| 29 | NM_025938 | 21.9 | 972 | 2.70 | 0.87 ± .02 (3) | 0.47 | .72, 1.03 |
| 30 | NM_146126 | 13.2 | 1203 | 3.60 | 2.24 ± .09 (3) | 0.004 | 1.74, 2.75 |
| 31 | NM_009104 | 13.9 | 1604 | 3.90 | 2.14 ± .04 (3) | 0.003 | 1.83, 2.45 |
| 32 | NM_053181 | 79.8 | 730 | 0.10 | 3.81 ± .08 (4) | <10-4 | 3.43, 4.20 |
Allele-specific transcription was assessed in (B6 × DBA)F1 mice.
The numbers in the first column refer to those in Figure 2.
GenBank accession number for each gene analyzed in column one.
The peak cis LOD score for each gene observed in microarray analysis.
`SNP position' refers to nucleotide number in the transcript.
The results of the microarray experiment.
The mean ratio of transcripts ± SE in F1 mice determined using classic cis-trans test. In column 6, the number of F1s used for each assay is given in parenthesis.
Each cis-trans result was given a t-test against the null hypothesis that the mean equals 1.0, and the resulting P-value is listed.
CI95 refers to the 95% confidence interval for each B6:DBA ratio.
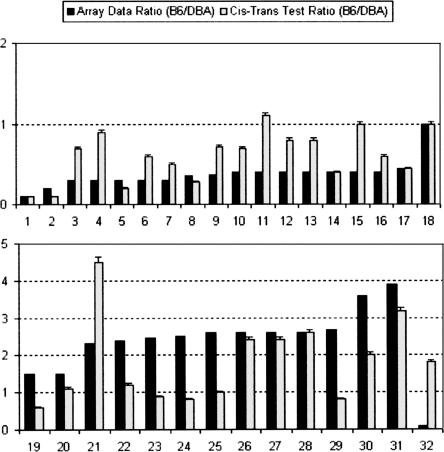
Figure 2.
Results of cis–trans test on a subset of cis-acting eQTLs. See Table 1 for GenBank accession numbers corresponding to gene numbers. Black bars indicate B6:DBA ratio observed in previously described microarray data (Schadt et al. 2003). Light gray bars indicate the observed ratio from the cis–trans test and are displayed with standard error bars. A total of 19 of 29 genes confirm the mode of regulation observed in the microarray data according to our definition described in the text.
Of the 28 predicted cis-acting eQTL studied, 18 confirmed cis regulation as shown by the cis–trans test. Of the remaining 10 genes, seven tested as trans according to the above definition. The remaining three genes resulted in ratios that were in the opposite direction to those observed in the microarray data. There are several possible explanations for the failure to confirm the remaining genes tested. They may be false positives that arose due to spurious associations of genotype and gene-expression level. However, given the significance of most of the cis-acting eQTL that did not validate (median LOD score equal to 13.1), we believe this is the least likely explanation, a more plausible explanation being that some of the genes were under trans regulation of closely linked genes. The initial criterion for a predicted cis-acting eQTL was that the physical location of the gene was within 20 cM of the eQTL peak marker (Schadt et al. 2003). This is a broad interval, which in many cases would extend past the two LOD score drop interval surrounding the apex of the QTL. However, we chose this conservative measure in order to avoid exclusion of any true cis-acting QTL that may have mapped inaccurately due to genotyping inconsistencies. It is plausible that in some cases a trans-acting element resides within this large interval, and the observed ratio is a result of nearby trans-acting regulation. Another reason for the failed confirmations could be due to the differences in age and environmental influences between the two groups of mice used. The F2 mice utilized for our microarray study were much older (16 mo) than the F1 mice (10–12 wk), and the F2 mice had been on a high-fat, atherogenic diet for 16 wk prior to profiling the livers compared with the F1 mice that were on a standard chow diet. These differences could conceivably have an effect on gene expression (Han et al. 2000; Jaenisch and Bird 2003; Sharman et al. 2004), invoking trans effects in the F1 mice, which in some cases override the cis effects of the locus of interest, thereby bringing the allelic expression ratio closer to 1:1.
Correlation patterns for genes with strong cis-acting eQTL and their experimental value
Strong cis-acting eQTL can obscure the true relationship between genes. Figure 3 highlights correlation distributions for genes with strong cis-acting eQTL and genes without strong cis-acting eQTL. What is clear in this figure is the shift to more significant correlations in genes that do not have strong cis-acting eQTL. This result seems counterintuitive, given the cis eQTL considered for this figure all had LOD scores >15. Such strong perturbations explain at least 46% of the variation in the expression trait, and if these perturbations in turn affected the expression levels of other genes, then we would expect those genes so affected to be significantly correlated with the gene, giving rise to that variation and to have eQTL that colocalize with the cis eQTL. However, the distributional patterns shown in Figure 3 indicate that genes whose expression are strongly perturbed by these cis events are actually much less related, expression-wise, to other expression traits, compared with genes without strong cis eQTL. To understand the significantly reduced number of genes whose expression is correlated with the expression of genes with strong cis-acting eQTL, we highlight an extreme example that illustrates relationships among genes with strong cis-acting eQTL that significantly impact the correlation structure between the genes.
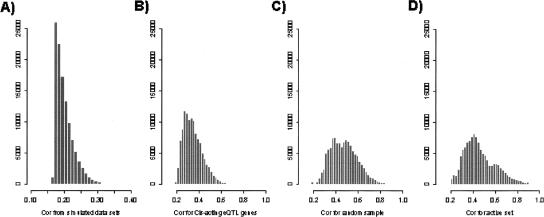
Figure 3.
Significant differences in the distribution of Pearson correlation coefficients involving genes with strong cis-acting eQTL versus genes with weak or no cis-acting eQTL. For each gene considered, the Pearson correlation coefficient was computed between the ml ratio vector for the gene from the BXD population and the ml ratio vectors for every other gene monitored in the BXD population (23,472 genes), resulting in a correlation vector for each gene. Histograms for the correlation coefficients whose significances fell in the ninth percentile were plotted in A–D for different sets of genes. (A) Distribution of correlations for 100 simulated mlratio vectors, representing the random distribution expected if 100 genes with independent mlratio vectors are compared with 23,472 other mlratio vectors assumed to be independent of the 100 simulated mlratio vectors. (B) Correlation distribution for the 100 genes with the strongest cis-acting eQTL in the BXD data set; all cis-acting eQTL in this set had LOD scores >15. (C) Correlation coefficient distribution for 100 genes randomly chosen from the set of 23,500 genes represented on the microarray in the BXD set. (D) Correlation coefficient distribution for 100 genes randomly chosen from the set of most transcriptionally active genes in the BXD set.
Figure 4 highlights two genes, JTP and Pik4cb, with strong cis-acting eQTL that are within 3 cM of each other. The LOD scores associated with the JTP and Pik4cb cis-acting eQTL are 30 and 37, respectively. What is clear from this figure is the significant negative correlation between the expression values of these two genes overall, in contrast to the significant positive correlation within each of the genotype groups. The significant negative correlation overall can be explained in terms of the closely linked cis-acting eQTL for these two genes, as described in Figure 5. Computing the correlation between these two genes conditional on the genotypes at the given locus results in a strong positive correlation between these two genes, as shown in Figure 6. That is, what is likely the biologically relevant component of the correlation between these two genes was completely obscured by the strong cis-acting eQTL that drove a strong negative correlation. This strong negative correlation appears to be an artifact of the closely linked cis-acting eQTL. This pattern of correlation observed here follows a well-studied property of correlations known as Simpon's Paradox (or the Yule-Simpson effect) (Yule 1903; Simpson 1951). Consistent with the correlation patterns shown in Figure 3, the top 10 genes most correlated with Pik4cb and JTP transcript levels (without conditioning on the genotype) all reside on chromosome 3 and all exhibit strong cis eQTL. As expected by Simpon's Paradox, recomputing the correlations between these genes and all of the other expression traits conditional on the genotypes at the Pik4cb/JTP locus resulted in the loss of correlation between the original 10 genes (the correlation having been explained by the closely linked eQTL), while simultaneously giving rise to significant correlations with a completely different set of genes.
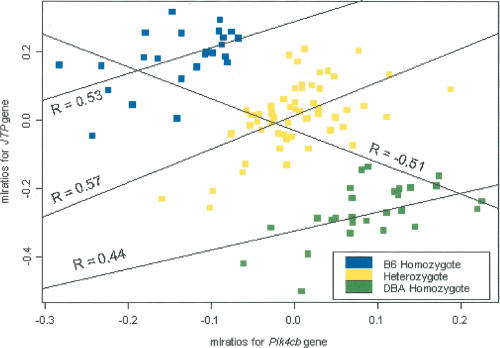
Figure 4.
Scatter plot of the mlratios for the jumping translocation point (JTP) and phosphatidylinositol 4-kinase, catalytic, β polypeptide (Pik4cb) genes in the BXD set. Color coding of each point is given with respect to the genotypes at a locus coincident with the physical location of the JTP and Pik4cb genes (the grouping indicates strong cis-acting eQTL for each gene). The overall correlation is computed to be –0.51, a statistically significant correlation (P = 7.8 × 10–9). The correlation within each genotype group is seen to be statistically significant as well, with a correlation of 0.53 in the B6 group, a correlation of 0.57 in the heterozygote group, and a correlation of 0.44 in the DBA group. Interestingly, the overall correlation is opposite in sign to each of the within-group correlations.
Figure 5.
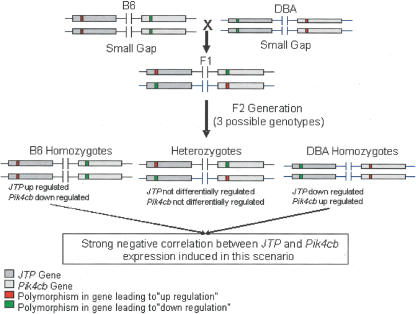
Diagram explaining the overall negative correlation observed in Figure 3.
Figure 6.
Highlighting what may be the biologically relevant component of the correlation between the JTP and Pik4cb gene-expression traits. This scatter plot represents the correlation between the JTP and Pik4cb gene-expression traits conditional on the locus at the JTP and Pik4cb gene locations. The overall correlation in this instance is now significantly positive, almost identical to the overall correlation given in Figure 4, but opposite in sign. Using the overall correlation statistic between these two traits as shown in Figure 4 completely obscures this relationship.
While this is an extreme example, it is of note that most genes with strong cis-acting eQTL tend to be most correlated with other genes that have closely linked, strong cis-acting eQTL. This observation suggests that the distributional shifts noted in Figure 3 are likely the result of the type of confounding effect shown in Figure 4, although in most cases, this effect will not be as extreme. Further, despite this type of confounding of gene–gene interactions, there are many examples where the expression perturbation described by a strong cis-acting eQTL gives rise to a meaningful expression response from other genes, as highlighted below.
Utilization of the information in the data available from the genetics of gene expression approach
As previously demonstrated (Schadt et al. 2003), cis eQTL data from segregating populations can be used to prioritize candidate genes in QTL mapping studies. This concept is illustrated in Figure 7, showing the cis eQTL at a locus controlling plasma total cholesterol levels in the (B6 × DBA)F2 cross. The rate-limiting step in the identification of genes in QTL studies is usually the screening for positional candidates (Flint and Mott 2001). Clearly, not all functional variation will be reflected in differences in transcript levels, but a survey of the genes underlying QTL suggests that a large fraction will exhibit altered expression (Abiola et al. 2003). In order to investigate the correlation structures of these genes with total cholesterol levels, the Pearson's correlation of each transcript level with total cholesterol levels was calculated. These results are reported in Table 2 (COR1). Three genes of the proposed 11 candidate genes with cis eQTL have significant correlations (P < 0.05). Because this correlation could be due to a biological relationship between gene and trait or one simply due to the proximity of the given genes such as that demonstrated above, we then calculated the correlation of all genes with cis-acting eQTL conditional on genotype at the cQTL peak marker (Table 2, COR2). This relationship represents that of environmental and trans-acting effects on the gene of interest and the trait; the logic being that if a gene is indeed related to a trait, these `outside' effects should affect both the gene and trait in a similar manner, and a significant relationship should still exist between the two. After removing the genetic effects of the chromosome 3 locus from the correlation, only one gene remained significantly correlated (BC005709). Although causal or reactant relationships cannot be inferred from this type of analysis, we argue that these data support BC005709 as being the most closely related gene to cholesterol levels, and therefore, the top candidate gene at this locus.
Figure 7.
Utilization of genetics of gene expression data in order to prioritize candidate genes underlying a locus of interest. A cQTL for chow diet total cholesterol levels was previously described on chromosome 3 at 24 cM at the peak marker D3mit241 (Drake et al. 2001; Colinayo et al. 2003). Cis-acting eQTL were identified as transcripts that mapped to the cQTL peak marker or either of the markers flanking this marker with a LOD score >4.3, and physically reside within 20 cM of the peak marker. This resulted in a candidate gene list of 11 genes. The LOD score curve for cholesterol (blue) is shown in relation to those for each of 11 candidate cis eQTLs (black). GenBank accession numbers for the candidate genes are indicated in the corner of each plot.
Table 2.
Candidate gene analysis for chromosome 3 cholesterol QTL
| Genea | LODb | Mbc | COR1d | COR1 P-value | COR2e | COR2 P-value |
|---|---|---|---|---|---|---|
| BC005709 | 6.7 | 87.5 | 0.25 | <.01* | 0.23 | 0.01* |
| AK006728 | 4.5 | 98.9 | 0.2 | 0.03* | 0.18 | 0.06 |
| AK018343 | 5.3 | 90.8 | 0.18 | 0.05* | 0.14 | 0.15 |
| 0610007D09Rik | 17.1 | 71.1 | -0.17 | 0.06 | 0.03 | 0.75 |
| 5430417P09Rik | 19.2 | 69.0 | 0.16 | 0.08 | 0.13 | 0.20 |
| AK018125 | 9.7 | 69.0 | -0.16 | 0.09 | -0.02 | 0.76 |
| AV361415 | 24.5 | 97.7 | -0.14 | 0.13 | -0.01 | 0.91 |
| NM_019410 | 11.9 | 59.2 | -0.14 | 0.14 | 0.01 | 0.92 |
| AK018323 | 13.1 | 60.7 | 0.09 | 0.29 | -0.01 | 0.84 |
| AK017836 | 5.2 | 97.2 | 0.07 | 0.46 | 0.01 | 0.86 |
| AA959924 | 4.8 | 35.0 | -0.05 | 0.59 | 0.04 | 0.70 |
GenBank accession number for each candidate gene.
Peak LOD score for each gene eQTL.
Physical location in megabases of each gene.
Pearson's correlation coefficient for each gene versus cholesterol across F2 mice and associated P-value.
Pearson's correlation coefficient for each gene versus cholesterol conditioned on genotype at the peak marker, D3Mit241, and the associated P-value. Significant correlations are indicated with an asterisk.
Strong cis-acting eQTL can lead to a significant expression response
It is tempting to conclude from the type of correlation structures exhibited in Figures 3 and 4 that many of the cis-acting eQTL give rise to expression perturbations that do not significantly impact the transcriptional network. That is, one can imagine that mutations in genes that lead to consistent expression differences among any number of strains of mice are simply tolerated by these complex systems as the body is well buffered with respect to changes in expression for many genes (i.e., the system tolerates large variations in expression—10 vs. 100 RNA copies per cell—with no effect on function). Therefore, selecting strains of mice to construct a cross from the population of all mouse strains may lead to consistent differences in some sets of genes (i.e., those in which cis-acting eQTL are detected) that have polymorphisms that affect transcription, where the polymorphisms have been tolerated (i.e., not selected against) because they do not functionally perturb the system.
However, another view would be that the response to genes that have strong cis-acting eQTL is context dependent, requiring the right tissue and right environmental conditions to realize differences in a phenotype of interest. Stressing a system in a particular way may lead to a response in the transcriptional network that is associated with phenotypes of interest (Karp et al. 2000). Further, there are a number of genes with strong cis-acting eQTL that do lead to a significant expression response. One such example is given in Figure 8. Here, we see a Riken cDNA clone, 1810073K19Rik, whose expression is strongly controlled by a chromosome 7 eQTL that is coincident with the gene's physical location. However, if we examine those genes most highly correlated with this particular gene, we get a significant number of genes with transcript abundance measures that are highly correlated with 1810073K19Rik, but which physically reside on chromosomes other than 7. Further, the expression of these genes are strongly controlled by the same chromosome 7 QTL controlling for 1810073K19Rik. In such a case, the question naturally arises whether the group of genes that are significantly correlated and linked to the same QTL are actually controlled by one of the genes in the group (e.g., the one with the cis-acting eQTL). In this case, we can apply the type of test described by Zhu et al. (2004) and E.E. Schadt, J. Lamb, X. Yang, J. Zhu, S. Edwards, D. GuhaThukarta, S.K. Sieberts, S. Monks, M. Reitman, C. Zhang, et al. (in prep.) to assess whether the group of genes controlled by the same locus are reacting to the gene with the cis-acting eQTL or whether they are driven independently by a given QTL or multiple closely linked QTL. In the present case, the correlation structures observed between the gene-expression traits and between the genotypes and gene-expression traits at the QTL position are consistent with 1810073K19Rik as the “causal” gene, giving rise to the expression response. This type of pathway analysis was initially introduced by Wright (1921) more than 80 yr ago as a way to quantify the causal relationships between variables once their true relationship had been established by other means. Since this original introduction, inferring causal relationships between correlated variables has been the subject of intense debate. However, in our present application, we know a priori that it is changes in DNA that lead to cis regulation of the expression traits, so that we begin with stronger prior information relating to how genes may be causally associated. The idea of resolving whether two or more genes driven by the same QTL are related in a causal/reactive way or driven independently by the same QTL are more fully explored by us elsewhere (E.E. Schadt, J. Lamb, X. Yang, J. Zhu, S. Edwards, D. GuhaThukarta, S.K. Sieberts, S. Monks, M. Reitman, C. Zhang, et al., in prep.).
Figure 8.
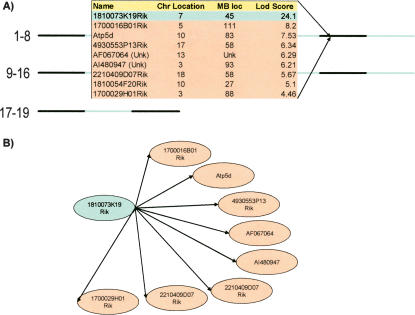
Genes with eQTL that are coincident with the physical location of the 1810073K19Rik gene test as reactive to the strong cis-acting 1810073K19Rik gene-expression perturbation, providing direct support that this cluster of eQTL obtains in response to the 1810073K19Rik perturbation. (A) The list of genes linking to the chromosome 7 locus. Only 1810073K19Rik has its physical location coincident with this locus. (B) Graph supported by examining the correlation structures between the gene-expression traits and locus using the causality test (E.E. Schadt, J. Lamb, X. Yang, J. Zhu, S. Edwards, D. GuhaThukarta, S.K. Sieberts, S. Monks, M. Reitman, C. Zhang, et al., in prep.; Zhu et al. 2004).
Discussion
We have presented the first general characterization of gene-expression traits for which cis-acting eQTL have been detected in segregating mouse populations. We have provided multiple lines of evidence that indicate most of the cis-acting eQTL detected represent true cis gene-expression effects. Only a relatively small number of cis-acting eQTL can be attributed to probes overlapping SNPs, and genes with cis-acting eQTL have been found to reside in regions that are overwhelmingly not IBD and actually increased for single nucleotide polymorphisms. We also tested several cis eQTL using direct experimental validation, confirming that a minimum of 64% of the cis-acting eQTL events detected are true positives. One way to view our results in a more conservative fashion is by making a cutoff for a confirmatory result at a twofold expression difference. Using this interpretation, those genes that resulted in a greater than twofold ratio (B6:DBA) in the microarray experiment, and in the cis–trans test (with consistant directionality), can be considered confirmed. Consistently, those that show a difference less than twofold should confirm in the cis–trans test with a difference less than twofold. Using this seemingly stringent criterion for confirmation, the same 64% of the cis eQTL remain confirmed. Because cis-acting eQTL afford one way to rapidly identify candidate genes underlying clinical trait QTL, it is important to understand the nature of the cis-acting eQTL. To that extent, we have highlighted cases where the correlation structure between two genes with closely linked eQTL can actually obscure the true relationship between the two genes, and where the cis-acting eQTL appears to give rise to a significant expression response in other genes. The potential exists to discriminate cases where the gene expression traits are related in a causal/reactive fashion, or whether they are driven by a single or two or more closely linked QTL. These characterizations represent the first steps that are necessary in more broadly interpreting expression-based QTL in the context of other complex phenotypes. This early work supports the continued investigation of the genetics of gene expression to elucidate complex disease traits and to more generally reconstruct gene networks associated with these diseases, as we have discussed elsewhere (Zhu et al. 2004).
Methods
Animal husbandry and diets
C57BL/6J and DBA/2J mice were purchased from The Jackson Laboratory. (B6 × DBA)F1 mice were reared from these parental mice. All mice were housed under conditions meeting Association for Assessment and Accreditation of Laboratory Animal Care and were housed in groups of four or less animals per cage and maintained on a 12-h light/12-h dark cycle at an ambient temperature of 23°C. They were allowed ad libitum access to water and rodent chow containing 6% fat (Diet No. 8604, Harlan Teklad). All animal care and experimental protocols were approved by the UCLA Animal Research Committee.
Experimental samples and expression data
We have previously described an experiment in which livers from a population of female F2 mice, constructed from a B6 × DBA cross, were profiled using a standard murine microarray consisting of 23,574 genes. The full details of the mouse expression data discussed here are provided by Schadt et al. (2003). This experiment is referred to as the BXD cross in the present study.
Identifying SNPs in probe regions
All of the SNP sequences that were informative between B6 and DBA in the Celera Mouse Genome Database were mapped to NCBI Build 32 of the mouse genome. The probe sequences for each of the genes represented on the microarray described above were then mapped against this same build of the mouse genome using BLAST (Altschul et al.1990). The designated probe and gene regions were defined and uploaded into a relational database. Through a series of SQL queries, the SNP data were intersected with the probe and probe region data, and the SNP frequencies were subsequently summarized after applying various filters as described in the main text.
Reconstructing haplotypes and identifying IBD regions
Once the Celera SNP data were loaded into a relational database, we reconstructed haplotypes for the B6 and DBA strains. To reconstruct the haplotypes, we processed the genomic sequence in 25-kb steps. For each 25-kb window, all SNPs polymorphic between the B6 and DBA strains were identified. The SNP counts in each window were smoothed by averaging with neighboring windows. Smoothed 25-kb regions containing five or fewer SNPs polymorphic between B6 and DBA were considered IBD between the two strains.
Allele-specific transcript quantification
Allele-specific transcript quantification was performed using quantitative sequence analysis and primer extension-based Pyrosequencing. All sequences and SNP data used were obtained from the Celera Discovery System. Transcript sequences were analyzed using Prophet 5.0 (BBN Systems and Technologies, A division of Bolt Beranek and Newman Inc.). (B6 × DBA)F1 mice were sacrificed between the ages of 10 and 12 wk, and the liver immediately frozen in liquid nitrogen. Total RNA was isolated using Qiagen Rneasy kit, with a protocol optimized for liver tissue. For Pyrosequencing analysis, (B6 × DBA)F1 cDNA was produced using Invitrogen Thermoscript reverse transcriptase. cDNA was then amplified using transcript-specific primers flanking an SNP. Extension primers were developed using Pyrosequencing SNP primer design software v1.0.1 (http://www.pyrosequencing.com). Reactions were performed on the Biotage PSQ 96MA, and data analysis was done using PSQMA 2.1. Subtraction of background signal and normalization for nucleotide incorporation efficiency is sequence dependent. Methods are described at http://www.pyrosequencing.com. For quantitative sequencing analysis, RT–PCR amplification of sequences containing the SNP was performed using the Invitrogen Thermo-script two-step procedure. All primer sequences used are supplied in Supplemental Table 1. Excess reactants were removed from the PCR products using the Qiaquick PCR purification kit from Qiagen. Quantitative genotyping of the products was performed using semiquantitative sequence analysis. Sequence analysis was performed using an ABI 3700 (Applied Biosystems Inc.) sequencer, and sequence chromatograms were analyzed using the Chromas 2.23 software (Technelysium Pty. Ltd.). This method of analysis was validated by PCR amplification of fragments of cDNA from each strain, and mixing these products in various ratios, followed by sequencing analysis. Supplemental Figure 1 shows results from one such comparison. In this example, the relationship between the B6:DBA RT–PCR product ratio and the B6:DBA peak area corresponds to a Pearson's correlation coefficient of 0.95 indicating peak area is relative to the quantity of product sequenced. In addition, the concordance of the results from the two methods was tested by assaying eight genes with both methods. These eight genes all resulted in consistent results between the two methods used. The signal strength will vary in sequence analysis depending on the surrounding sequences. To correct for such differences, we amplified parental cDNA and normalized using surrounding sequences shared by the two strains. The ratio of one SNP allele's incorporation efficiency to the other was then obtained. This ratio reflects the amount of differential nucleotide incorporation at that position and was used to normalize the ratios observed in the F1 animals.
Statistical analysis
Statistical analysis was conducted in R–A language and environment version 1.9.1 and in Microsoft Excel. Cis–trans test results were normalized according to the surrounding sequence (each normalization is unique), and detailed methods are available at http://www.pyrosequencing.com. A t-test was then done on each set of resulting B6:DBA ratios to the null hypothesis that μ = 1.0. For the correlation analysis, the Pearson correlations were computed in each case, and the mean log ratio data used in these calculations was assumed to follow a normal distribution, a valid assumption as described by He et al. (2003). The data generated for Figure 3A were simulated such that the mean log ratios for each gene followed a normal distribution with the same mean and variance as the 100 genes used to generate Figure 3B. Further, for all gene-expression traits giving rise to significant cis eQTL (LOD >4.3 as reported in the main text), the residuals were examined after fitting the expression traits to the eQTL genotypes, and almost all were found to be approximately normally distributed.
Acknowledgments
This work was supported by NIH grants HL30568 and HL28481.
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3216905. Article published online ahead of print in April 2005.
References
- Abiola, O., Angel, J.M., Avner, P., Bachmanov, A.A., Belknap, J.K., Bennett, B., Blankenhorn, E.P., Blizard, D.A., Bolivar, V., Brockmann, G.A., et al. 2003. The nature and identification of quantitative trait loci: A community's view. Nat. Rev. Genet. 4: 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Brem, R.B., Yvert, G., Clinton, R., and Kruglyak, L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Chesler, E.J., Wang, J., Lu, L., Qu, Y., Manly, K.F., and Williams, R.W. 2003. Genetic correlates of gene expression in recombinant inbred strains: A relational model system to explore neurobehavioral phenotypes. Neuroinformatics 1: 343–357. [DOI] [PubMed] [Google Scholar]
- Colinayo, V.V., Qiao, J.H., Wang, X., Krass, K.L., Schadt, E., Lusis, A.J., and Drake, T.A. 2003. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mammalian Genome 14: 464–471. [DOI] [PubMed] [Google Scholar]
- Cowles, C.R., Hirschhorn, J.N., Altshuler, D., and Lander, E.S. 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Drake, T.A., Schadt, E., Hannani, K., Kabo, J.M., Krass, K., Colinayo, V., Greaser III, L.E., Goldin, J., and Lusis, A.J. 2001. Genetic loci determining bone density in mice with diet-induced atherosclerosis. Physiol. Genomics 5: 205–215. [DOI] [PubMed] [Google Scholar]
- Flint, J. and Mott, R. 2001. Finding the molecular basis of quantitative traits: Successes and pitfalls. Nat. Rev. Genet. 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Han, E., Hilsenbeck, S.G., Richardson, A., and Nelson, J.F. 2000. cDNA expression arrays reveal incomplete reversal of age-related changes in gene expression by calorie restriction. Mech. Ageing Dev. 115: 157–174. [DOI] [PubMed] [Google Scholar]
- He, Y.D., Dai, H., Schadt, E.E., Cavet, G., Edwards, S.W., Stepaniants, S.B., Duenwald, S., Kleinhanz, R., Jones, A.R., Shoemaker, D.D., et al. 2003. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics 19: 956–965. [DOI] [PubMed] [Google Scholar]
- Jaenisch, R. and Bird, A. 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33: 245–254. [DOI] [PubMed] [Google Scholar]
- Jin, W., Riley, R.M., Wolfinger, R.D., White, K.P., Passador-Gurgel, G., and Gibson, G. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nature Genetics 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Karp, C.L., Grupe, A., Schadt, E., Ewart, S.L., Keane-Moore, M., Cuomo, P.J., Kohl, J., Wahl, L., Kuperman, D., Germer, S., et al. 2000. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 1: 221–226. [DOI] [PubMed] [Google Scholar]
- Monks, S.A., Leonardson, A., Zhu, H., Cundiff, P., Pietrusiak, P., Edwards, S., Phillips, J.W., Sachs, A., and Schadt, E.E. 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M., Molony, C.M., Weber, T.M., Devlin, J.L., Ewens, K.G., Spielman, R.S., and Cheung, V.G. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y., Kong, L.K., Poirier, C., Truong, C., Overbeek, P.A., and Bishop, C.E. 2004. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum. Mol. Genet. 13: 1213–1218. [DOI] [PubMed] [Google Scholar]
- Schadt, E.E., Monks, S.A., Drake, T.A., Lusis, A.J., Che, N., Colinayo, V., Ruff, T.G., Milligan, S.B., Lamb, J.R., Cavet, G., et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Sharman, E.H., Sharman, K.G., Ge, Y.W., Lahiri, D.K., and Bondy, S.C. 2004. Age-related changes in murine CNS mRNA gene expression are modulated by dietary melatonin. J. Pineal. Res. 36: 165–170. [DOI] [PubMed] [Google Scholar]
- Shifman, S., Pisante-Shalom, A., Yakir, B., and Darvasi, A. 2002. Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol. Cell Probes 16: 429–434. [DOI] [PubMed] [Google Scholar]
- Simpson, E.H. 1951. The interpretation of interaction in contingency tables. J. Royal Stat. Soc. 13: 238–241. [Google Scholar]
- Wade, C.M., Kulbokas III, E.J., Kirby, A.W., Zody, M.C., Mullikin, J.C., Lander, E.S., Lindblad-Toh, K., and Daly, M.J. 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574–578. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., Pletcher, M.T., Batalov, S., Barnes, S.W., Tarantino, L.M., Cooke, M.P., Wu, H., Smylie, K., Santrosyan, A., Copeland, N.G., et al. 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. 100: 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. 1921. Correlation and causation. J. Agr. Res. 20: 557–585. [Google Scholar]
- Yule, G.H. 1903. Notes on the theory of association of attributes in Statistics. Biometrika 2: 121–134. [Google Scholar]
- Yvert, G., Brem, R.B., Whittle, J., Akey, J.M., Foss, E., Smith, E.N., Mackelprang, R., and Kruglyak, L. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nature Genetics 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Lum, P.Y., Lamb, J., GuhaThakurta, D., Edwards, S.W., Thieringer, R., Berger, J.P., Wu, M.S., Thompson, J., Sachs, A.B., et al. 2004. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogen. Genome Res. 105: 363–374. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.pyrosequencing.com; Pyrosequencing SNP primer design software v1.0.1.