Abstract
Homeotic (Hox) genes are usually clustered and arranged in the same order as they are expressed along the anteroposterior body axis of metazoans. The mechanistic explanation for this colinearity has been elusive, and it may well be that a single and universal cause does not exist. The Hox-gene complex (HOM-C) has been rearranged differently in several Drosophila species, producing a striking diversity of Hox gene organizations. We investigated the genomic and functional consequences of the two HOM-C splits present in Drosophila buzzatii. Firstly, we sequenced two regions of the D. buzzatii genome, one containing the genes labial and abdominal A, and another one including proboscipedia, and compared their organization with that of D. melanogaster and D. pseudoobscura in order to map precisely the two splits. Then, a plethora of conserved noncoding sequences, which are putative enhancers, were identified around the three Hox genes closer to the splits. The position and order of these enhancers are conserved, with minor exceptions, between the three Drosophila species. Finally, we analyzed the expression patterns of the same three genes in embryos and imaginal discs of four Drosophila species with different Hox-gene organizations. The results show that their expression patterns are conserved despite the HOM-C splits. We conclude that, in Drosophila, Hox-gene clustering is not an absolute requirement for proper function. Rather, the organization of Hox genes is modular, and their clustering seems the result of phylogenetic inertia more than functional necessity.
Homeotic (Hox) genes were discovered in Drosophila melanogaster as mutations that transform one body part into another. Lewis (1978) and Kaufman et al. (1980) found that these genes are clustered and arranged in the chromosome in the same order as their domains of action in the body of flies. Homologous Hox genes were subsequently found in many other animals and their arrangement in complexes (HOM-C) shown to be the general rule (McGinnis and Krumlauf 1992; Ruddle et al. 1994). Hox genes encode transcription factors involved in the determination of segment identity along the anteroposterior body axis, and thus, play a fundamental role in animal development. The conserved colinearity between Hox gene chromosomal arrangement and expression domain is a basic notion of developmental biology, yet this is an enigmatic phenomenon for which no single satisfactory explanation exists (Kmita and Duboule 2003). Furthermore, HOM-C splits have been observed in Drosophila (Von Allmen et al. 1996; Lewis et al. 2003; Negre et al. 2003), Bombyx (Yasukochi et al. 2004), nematodes (Aboobaker and Blaxter 2003), and tunicates (Ikuta et al. 2004; Seo et al. 2004).
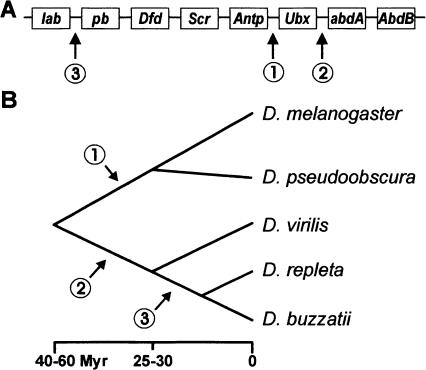
Ten genes arranged in a single complex comprised the ancestral HOM-C of arthropods (Cook et al. 2001; Hughes and Kaufman 2002; Hughes et al. 2004). In winged insects, including Drosophila, the genes Hox3 and fushi tarazu (ftz) lost their homeotic function, and thus, only eight truly homeotic genes remain. Three different splits of the ancestral HOM-C have been found so far in the Drosophila genus (Fig. 1A). In D. melanogaster, the complex is split between the genes Antennapedia (Antp) and Ultrabithorax (Ubx), leaving two separate gene clusters as follows: the Antennapedia complex, ANT-C (Kaufman et al. 1990) that specifies the identity of the mouth parts and anterior thorax, and the Bithorax complex, BX-C (Duncan 1987; Martin et al. 1995) involved in the development of the posterior thorax and abdomen. In D. pseudoobscura, the HOM-C is also similarly divided in the ANT-C and BX-C complexes (Lewis et al. 2003). A different split between Ubx and abdominal A (abdA) occurs in D. virilis (Von Allmen et al. 1996), D. repleta (Ranz et al. 2001), D. buzzatii, and other species of the Drosophila subgenus (Negre et al. 2003; Fig. 1B). Finally, an additional split, between labial (lab) and proboscipedia (pb), is present in D. buzzatii and other species of the repleta group (Negre et al. 2003). This third split separated the gene lab far from pb and the anterior genes of the Hox complex and relocated it near the posterior genes abdA and Abdominal B (AbdB) in a flagrant violation of the colinearity rule. The functional consequences of these splits are unknown.
Figure 1.
Genomic (A) and phylogenetic (B) localization of the three Hox gene complex splits observed in the Drosophila genus. (A) Ancestral arrangement of the eight Hox genes within the insects is as follows: labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp), Ultrabithorax (Ubx), abdominal A (abdA), and Abdominal B (AbdB). (B) Phylogenetic relationships and divergence times for the five Drosophila species included in this study. D. melanogaster and D. pseudoobscura belong to the Sophophora subgenus. D. repleta and D. buzzatii (both in the repleta species group) and D. virilis (virilis species group) belong to the Drosophila subgenus (see Negre et al. 2003 for details).
In order to ascertain the consequences of Drosophila HOM-C splits, we have carried out a genomic and functional characterization of the two splits present in D. buzzatii. We isolated and sequenced two BAC clones containing the lab-abdA and pb chromosomal regions of D. buzzatii. The gene organization in these regions is compared with that of the homologous regions in D. melanogaster and D. pseudoobscura to map the precise site of the two splits. None of the two splits has altered the coding regions of Hox genes. We then searched for Conserved Noncoding Sequences (CNS), which are putative regulatory sequences, around the genes lab, pb, and abdA, to find out whether the splits removed or altered any Hox-gene enhancer. The position of CNS around Hox genes is compared with experimentally identified Hox-gene enhancers, and the arrangement of CNS is compared between Hox and non-Hox genes. Finally, we analyzed the expression patterns of three Hox genes, lab, pb, and abdA, in four Drosophila species with different Hox-gene organizations (with and without the splits) in whole-mount embryos and imaginal discs. The results show that, in Drosophila species, Hox genes, as well as their regulatory regions and expression patterns, are conserved, despite the Hox complex breaks. Thus, the functional significance of the Hox-gene clustering in Drosophila is questionable.
Results
Molecular characterization of Hox-gene complex breakpoints
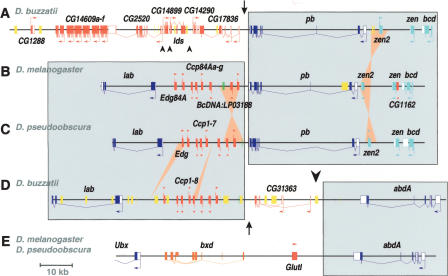
To characterize the two HOM-C splits present in D. buzzatii, we isolated and sequenced two BAC clones, one (5H14, 124,024 bp) containing the lab-abdA region, and another (40C11, 132,938 bp) including the pb region (see Methods). The organization of the two regions of D. buzzatii chromosome 2 is shown in Figure 2 along with the homologous regions of D. melanogaster and D. pseudoobscura for comparison. D. melanogaster and D. pseudoobscura are homosequential in the analyzed regions, except where indicated. The sequenced pb region (Fig. 2A) contains 16 ORFs including Dbuz\pb, Dbuz\zerknüllt (Dbuz\zen), Dbuz\zerknüllt-related (Dbuz\zen2), and Dbuz\bicoid (Dbuz\bcd). These four genes are present in the ANT-C of D. melanogaster and also in the homologous region of D. pseudoobscura (Fig. 2B,C). The orientation of Dbuz\zen2 is the same as that of Dpse\zen2, but inverted with regard to Dmel\zen2. The remaining 12 genes in this region are orthologous to D. melanogaster genes from four different regions (84D1–2, 89D2, 84E5, and 91D4–5) of chromosomal arm 3R. One of the genes, CG14609, is represented by six copies, in contrast to the single copy present in D. melanogaster or D. pseudoobscura. A total of four breakpoints are fixed in this region between D. buzzatii and D. melanogaster beside the zen2 microrearrangement. That corresponding to the lab-pb split is located in the ∼3-kb intergenic segment between Dbuz\pb and Dbuz\CG17836 (Fig. 2A).
Figure 2.
Gene organization of the lab-abdA and pb genomic regions of D. buzzatii compared with the homologous regions of D. melanogaster and D. pseudoobscura. The localization of the lab-pb split (arrow) and the Ubx-abdA split (large arrowhead) are indicated. (A) Sequence of D. buzzatii BAC 40C11 containing the pb region. (B) Organization of the lab-pb region in D. melanogaster. (C) Idem in D. pseudoobscura. (D) Sequence of D. buzzatii BAC 5H14 containing the lab-abdA region. (E) Organization of the abdA region in D. melanogaster and D. pseudoobscura. Genes are represented as open (UTRs) and filled boxes (coding sequences) with arrows indicating the sense of transcription. Hox genes are colored in dark blue, Hox-derived genes in light blue, non-Hox genes in red, noncoding RNA genes in orange, and the BcDNA:LP03188 and orthologous sequences in green. Transposable element insertions (usually ISBu elements, see Negre et al. 2003) are shown as yellow boxes. Large shaded rectangles include homologous Hox-gene regions in different species. Ochre triangles denote small inversions and insertions or deletions. Small arrowheads show breakpoints between D. buzzatii and D. melanogaster in non-Hox regions.
The sequenced lab-abdA region contains 11 ORFs, including Dbuz\lab, the cuticular cluster genes (Dbuz\Ccp), and Dbuz\abdA (Fig. 2D). The number of Ccp copies (including the gene Edg) is eight in the three species, but there is a small inversion encompassing two copies (plus the cDNA BcDNA:LP03188) in D. melanogaster in comparison to D. buzzatii or D. pseudoobscura, as well as one gain and one loss (Fig. 2B–D). These 11 genes come from three different regions (84A2–5, 86E11–13, and 89E2) of D. melanogaster chromosomal arm 3R, which means two fixed breakpoints between D. buzzatii and D. melanogaster, beside the small inversion of Ccp genes. One breakpoint corresponds to the lab-pb split and is found ∼40 kb upstream of Dbuz\lab, in the 5-kb between the sequence similar to BcDNA:LP03188 and the gene Dbuz\CG31363. The second breakpoint is that of the Ubx-abdA split and is located between 11 and 15 kb downstream of Dbuz\abdA. The two breakpoints are separated by a DNA segment of only ∼22 kb encoding a single gene, Dbuz\CG31363 (Fig. 2D).
Conserved noncoding sequences in Hox gene regions
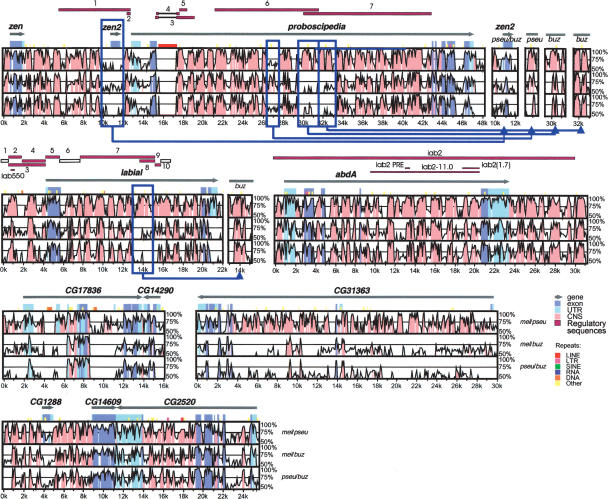
We analyzed the conservation of noncoding sequences around the three Hox genes lab, pb, and abdA by comparing the sequences of the three species D. buzzatii, D. melanogaster, and D. pseudoobscura as done previously by other authors (Bergman and Kreitman 2001; Bergman et al. 2002) (see Methods). Figure 3 shows the VISTA graph, where the conservation between the aligned sequences is plotted (when higher than 50%) and the regions that meet the selected criteria (75% identity in a 25-bp window) are highlighted for both coding and noncoding sequences. A preliminary analysis showed no differences between intergenic and intronic regions, in agreement with previous studies (Bergman and Kreitman 2001). Thus, CNS are defined as intergenic (excluding UTRs) or intronic sequences that meet the above criteria. The characteristics of observed CNS are given in Tables 1 and 2, and the results of statistical analysis are shown in Supplemental Tables S1 and S2.
Figure 3.
Nucleotide sequence conservation in the lab-abdA and pb regions between Drosophila species. The three panels in each VISTA plot represent pairwise comparisons between D. melanogaster and D. pseudoobscura (mel/pse), D. melanogaster and D. buzzatii (mel/buz) and D. pseudoobscura and D. buzzatii (pse/buz). The x-axis represents D. melanogaster coordinates, and y-axis sequence identity (50%–100%). Gray arrows show the location and orientation of genes. Conservation in exons and UTRs is shown in dark and light blue, respectively. Pink regions represent CNS. Experimentally identified regulatory sequences (solid purple bars) or segments with negative results (empty bars) are indicated on top of each plot. Five microinversions detected in the lab or pb regions are enclosed in blue frames, and the VISTA graphs generated with the inverted sequences shown to the right of the main plots. VISTA plots for the CG17836-CG14290, CG31363, and CG1288-CG14609-CG2520 regions (adjacent to Hox genes) are shown at the bottom of the figure for comparison.
Table 1.
Characteristics of conserved noncoding sequences (CNS) detected with mVISTA in comparisons of Hox gene regions between D. melanogaster (mel), D. pseudoobscura (pse), and D. buzzatii (buz)
| Region | Noncoding nucleotides | Species pair | Number of CNS | Densitya(SD) | Mean size (nt) (SD) | Mean nucleotide identity (%) | Sequence in CNS (%) |
|---|---|---|---|---|---|---|---|
| lab | 19,227 | mel/pse | 129 | 6.71 (0.59) | 53.20 (34.03) | 87.69 | 35.69 |
| mel/buz | 73 | 3.80 (0.44) | 46.70 (24.35) | 87.33 | 17.73 | ||
| pse/buz | 84 | 4.37 (0.48) | 44.54 (27.05) | 88.08 | 19.46 | ||
| pb | 42,056 | mel/pse | 265 | 6.30 (0.39) | 55.04 (35.79) | 87.29 | 34.68 |
| mel/buz | 196 | 4.66 (0.33) | 41.88 (22.38) | 87.09 | 19.52 | ||
| pse/buz | 215 | 5.11 (0.35) | 42.92 (24.65) | 86.38 | 21.94 | ||
| abdA | 26,043 | mel/pse | 169 | 6.49 (0.50) | 59.29 (38.43) | 87.44 | 38.45 |
| mel/buz | 126 | 4.84 (0.43) | 45.98 (26.15) | 86.18 | 22.25 | ||
| pse/buz | 141 | 5.41 (0.46) | 46.11 (24.97) | 85.44 | 24.97 | ||
| Total | 87,326 | mel/pse | 563 | 6.45 (0.27) | 55.89 (36.23) | 87.42 | 36.03 |
| Hox gene regions | mel/buz | 395 | 4.52 (0.23) | 44.08 (24.04) | 86.83 | 19.94 | |
| pse/buz | 440 | 5.04 (0.24) | 44.25 (25.53) | 86.39 | 22.30 |
Density = number of CNS per kilobase.
Table 2.
Characteristics of conserved noncoding sequences (CNS) detected with mVISTA in comparisons of non-Hox gene regions between D. melanogaster (mel), D. pseudoobscura (pse), and D. buzzatii (buz)
| Region | Noncoding nucleotides | Species pair | Number of CNS | Densitya(SD) | Mean size (nt) (SD) | Mean nucleotide identity (%) | Sequence in CNS (%) |
|---|---|---|---|---|---|---|---|
| CG1288-CG2520 | 18,333 | mel/pse | 127 | 6.93 (0.61) | 43.48 (28.16) | 86.31 | 30.12 |
| mel/buz | 65 | 3.55 (0.44) | 45.26 (31.51) | 86.30 | 16.05 | ||
| pse/buz | 67 | 3.65 (0.45) | 42.44 (29.40) | 86.81 | 15.59 | ||
| CG17836-CG14290 | 10,921 | mel/pse | 46 | 4.21 (0.62) | 45.02 (33.27) | 82.67 | 18.96 |
| mel/buz | 18 | 1.65 (0.39) | 39.61 (22.67) | 82.88 | 6.53 | ||
| pse/buz | 22 | 2.01 (0.43) | 42.09 (24.34) | 84.34 | 8.48 | ||
| CG31363 | 27,510 | mel/pse | 153 | 5.56 (0.45) | 35.51 (14.95) | 87.17 | 19.75 |
| mel/buz | 22 | 0.80 (0.17) | 26.09 (3.94) | 82.93 | 2.09 | ||
| pse/buz | 23 | 0.84 (0.17) | 28.78 (7.70) | 83.23 | 2.41 | ||
| Total | 56,764 | mel/pse | 326 | 5.74 (0.32) | 39.96 (24.15) | 86.09 | 22.95 |
| non-Hox gene regions | mel/buz | 105 | 1.84 (0.18) | 40.28 (27.50) | 85.27 | 7.45 | |
| pse/buz | 112 | 1.97 (0.19) | 39.59 (25.88) | 85.76 | 7.83 |
Density = number of CNS per kilobase.
When D. buzzatii is compared with D. melanogaster or D. pseudoobscura, 395 and 440 CNS are found, respectively, around the three Hox genes (Table 1). This gives a density of 4.5 and 5 CNS per kilobase, respectively. These conserved blocks show a mean size of 44 bp with 86.5% nucleotide identity and represent 20%–22% of the analyzed noncoding sequence. When D. melanogaster and D. pseudoobscura are compared, 563 CNS are detected (6.5/kb) with a mean size of 55 bp and an average identity of 87.4%. In this comparison, the sequence in CNS represents 36% of noncoding sequence. In all three comparisons, the three regions around the Hox genes lab, pb, and abdA are homogeneous with little variation either in CNS density, size, or nucleotide identity (Supplemental Table S1). It is worth noting that CNS are coincident in all three comparisons (Fig. 3), which means that all CNS detected when comparing D. buzzatii with either D. melanogaster or D. pseudoobscura are also found in the comparison between the latter two species. Although most CNS keep colinearity (relative position and orientation), we could identify four microinversions, around 1–2 kb in size. One is located within the large intron of lab and the other three in introns 2 and 3 of pb (Fig. 3).
D. buzzatii is equally distant phylogenetically from either D. melanogaster or D. pseudoobscura (Fig. 1). The latter two species belong to the same subgenus and are phylogenetically closer. We compared the characteristics of the CNS found in the three pairwise comparisons. As expected, there are no statistical differences between the CNS found when comparing D. buzzatii with either D. melanogaster or D. pseudoobscura (Supplemental Table S2). The CNS density and the proportion of sequence in CNS are significantly higher when comparing the phylogenetically closer species D. melanogaster and D. pseudoobscura. Increasing divergence time does not seem to affect the nucleotide identity of the CNS, although the size of the CNS detected in the Hox-gene regions shows a significant decrease (Supplemental Table S2).
Conserved noncoding sequences in non-Hox gene regions
To find out whether the observed pattern of CNS is a particular feature of Hox genes, we also analyzed the presence of CNS in regions of the sequenced BACs adjacent, but unrelated, to Hox genes. We used the three microsyntenic regions between D. buzzatii, D. melanogaster, and D. pseudoobscura longer than 10 kb, i.e., the CG31363 gene region, between lab and abdA, and the CG17836-CG14290 and CG1288-CG2520 regions, near pb (Fig. 2). These regions include one, two, and three genes, respectively. The pattern of CNS detected is shown in Figure 3 and summarized in Table 2. In the comparisons with D. buzzatii, we found around 100 CNS (∼2/kb), which represents <8% of noncoding sequence. Thus, in these non-Hox regions, a much smaller number of CNS is observed and the proportion of sequence in CNS is also significantly lower than in Hox-gene regions (Supplemental Table S1). In the D. melanogaster–D. pseudoobscura comparison, there are 326 CNS (5.7/kb) which represents a 23% of noncoding sequence. Thus, in this case, the density is similar between Hox and non-Hox-gene regions, but the size of CNS is significantly smaller in the latter regions (Supplemental Table S1). Consequently, the proportion of sequence in CNS is also significantly lower in the non-Hox-gene regions. It should be noted that non-Hox regions show a significant variation for CNS density and also for the proportion of sequence in CNS that is not observed in Hox-gene regions (Supplemental Table S1). The higher variation observed between non-Hox regions is probably due to the heterogeneity of the sample from a functional point of view. There is little information available on the function and expression pattern of the six non-Hox genes analyzed, which probably represent a mixture of genes with different regulatory needs and number of enhancers.
Conservation of known regulatory sequences
Regulatory sequences of the genes lab, pb, and abdA have been experimentally identified in D. melanogaster (Karch et al. 1985; Chouinard and Kaufman 1991; Kapoun and Kaufman 1995; Martin et al. 1995). We compared their position with the pattern of CNS found around Hox genes. As shown in Figure 3, the regulatory sequences identified in D. melanogaster generally contain or correspond to CNS in D. buzzatii. For instance, CNS are found in the sites corresponding to the iab2 PRE and iab2(1.7) enhancers of abdA (Shimell et al. 1994, 2000). Similarly, a prominent conservation peak is observed at the site of the lab550 enhancer, which directs the expression of lab in the embryo midgut (Marty et al. 2001). Also, the inverted segment found in the large intron of the lab gene roughly corresponds to the segment responsible for lab expression in the posterior midgut. Sequence details of the lab550 and iab2(1.7) enhancer and binding site conservation are shown in Supplemental Figure S1. The Homeotic Response Element (HOMRE) of the lab550 enhancer contains four binding sites; all of them are conserved in the three species. In the iab2(1.7) enhancer, there are five Hunchback (HB)-binding sites, three of which are conserved in the three species, whereas the other two vary in position between species. This enhancer also contains a unique Krüppel (KR)-binding site, where point mutations in D. melanogaster cause gain-of-expression mutants (Hab1 and Hab2) (Shimell et al. 1994). This binding site is conserved in all three species (Supplemental Fig. S1). The conservation between D. melanogaster and D. buzzatii around abdA ends 9 kb (in D. melanogaster) and 11 kb (in D. buzzatii) downstream of this gene (Fig. 3). This boundary lies between the iab2 and pbx regulatory sequences, which control the expression of abdA and Ubx, respectively (Karch et al. 1985). We have shown that the iab2 region downstream of abdA is conserved in D. buzzatii. We have not sequenced the Ubx region in D. buzzatii, but we assume that the pbx regulatory sequence will conserve its position upstream of Ubx, i.e., there are no rearrangements between Ubx and its regulatory sequences (see below).
It is worth noting though, that CNS were also found in fragments not experimentally tested or described as with no effect on expression (Fig. 3). This observation suggests that the regulation of these genes may be even more complex than currently envisaged, and that more regulatory modules may be operative in nature than those experimentally identified in the laboratory.
Hox gene expression patterns
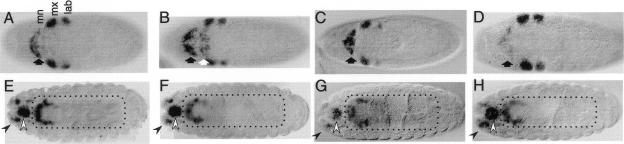
The conservation of regulatory sequences suggests that splits of the HOM-C had no consequences on Hox-gene expression. To test this prediction, we compared the expression patterns of the Hox genes lab, pb, and abdA between D. melanogaster, D. virilis, D. buzzatii, and D. repleta. These four Drosophila species represent three different Hox-gene organizations (Figs. 1,2). D. melanogaster possess the Antp-Ubx split only, whereas D. virilis has the Ubx-abdA split instead. Both D. buzzatii and D. repleta present the Ubx-abdA and lab-pb splits. We used in situ hybridization and antibody staining to whole-mount embryos and to imaginal discs from third instar larvae and prepupae (see Methods). Detailed results are given in Figure 4 and Supplemental Figures S2–S6. The expression patterns of the four species closely follow those described for D. melanogaster (for review, see Hughes and Kaufman 2002). Interspecific variation was detected only in the pb gene, which in D. virilis presents an extra domain in the embryo mesoderm (Fig. 4). As this expression domain is not shared by D. melanogaster, it is seemingly not related with the lab-pb split. Although our analysis is qualitative, and slightly quantitative changes or domain changes of a few cells may remain undetected, it shows that the reorganization of the HOM-C caused no major alterations of the expression patterns of the three Hox genes adjacent to the splits, in good agreement with the conservation of regulatory sequences (see above). Likewise, Bomze and López (1994) found that the expression pattern of Ubx in embryos is conserved between D. melanogaster, D. pseudoobscura, D. virilis, and D. hydei (a species of the repleta group), despite their different Hox-gene organization (Figure 1).
Figure 4.
Expression pattern of pb in embryos. (A–D) stage 11 embryos, (E–H) stage 17 embryos. (A,E) D. melanogaster,(B,F) D. virilis,(C,G) D. buzzatii, and (D,H) D. repleta. (A–D) Expression on the ectoderm of the maxillary and labial lobes. Later in development (E–H) pb is detected in the derivatives of the maxillary (white arrowhead) and labial (black arrowhead) lobes, and in the ventral nervous system (boxed area). (A–D) pb expression is detected in the mesodermal layer of the mandibular segment (black arrow) in all four species. In D. virilis only (B), pb is also expressed in the mesodermal layer of the maxillary segment (white arrow). The mandibular (mn), maxillary (mx), and labial (lab) segments are shown in A.
Discussion
zen2 predates the Drosophila radiation
The zen and bcd genes come from a duplication of Hox3 in the ancestor of Cyclorraphan flies (Stauber et al. 2002). A second duplication of zen gave birth to zen2, which was thought to be a recent event in D. melanogaster (Randazzo et al. 1993), where it has no discernible function. However, the existence of Dpse\zen2 and Dbuz\zen2 shows that the zen–zen2 duplication must predate the divergence of the Sophophora and Drosophila subgenus, and that this gene has been kept during at least 40–60 Myr of evolution. Whether this gene is also present in other flies outside of the Drosophila genus is still unknown.
Patterns of conserved noncoding sequence evolution
Cis-Regulatory Modules (CRM) are transcription regulatory DNA segments (from a few hundred base pair to 1 kb in size) that control gene expression in higher eukaryotes (Wray et al. 2003). CRM have a complex structure still not fully understood. They contain one or several binding sites for different transcription factors, which act cooperatively to activate or repress transcription of the target gene. As CRM are functionally constrained to maintain the expression of the target gene, they evolve slower than nonfunctional sequences. Therefore, the conservation of noncoding sequences between phylogenetically distant species may be used as a guide for identification of regulatory sequences. Several recent studies (Bergman and Kreitman 2001; Bergman et al. 2002; Cooper and Sidow 2003; Nobrega et al. 2003; Santini et al. 2003) support the use of comparative sequence analysis and characterization of CNS as a useful approach to detect putative CRM in Drosophila and other organisms. The clustering of previously characterized transcription-factor binding sites may be also used for detection of CRM (Berman et al. 2004). However, the absence of high-quality binding data for most Drosophila transcription factors represent a great current limitation in the widespread application of this method.
We exhaustively searched for CNS around lab, pb, and abdA and around adjacent non-Hox genes by comparing three species pairs. A plethora of highly conserved blocks was found surrounding the three Hox genes in the comparison between the phylogenetically distant species D. buzzatii and D. melanogaster or D. pseudoobscura (Fig. 1). The proportion of noncoding sequence included in CNS was 20%–22%. In most cases, these CNS keep their relative position and colinearity, although a few microrearrangements were found. The interpretation of these CNS as regulatory sequences is supported by the high neutral substitution rate (Moriyama and Gojobori 1992) and intrinsic rate of DNA loss (Petrov et al. 1996; Singh and Petrov 2004) in Drosophila. Noncoding sequences are not expected to be conserved between such distantly related species unless they are functionally constrained. The coincidence between CNS and known enhancers such as iab2 PRE or lab550 (Supplemental Fig. S1) further supports this interpretation.
A lower CNS density was observed around non-Hox genes. This result fits well with previous observations showing that genes with complex developmentally regulated expression show a higher degree of conservation in noncoding regions than more simple genes with metabolic or housekeeping functions (Bergman and Kreitman 2001; Bergman et al. 2002; Halligan et al. 2004). Moreover, Hox genes are associated with larger noncoding regions. Hox genes harbor some of the longest introns of any Drosophila gene (Moriyama et al. 1998) and mean intron size is significantly greater in the Hox than in the non-Hox genes analyzed here (F = 4.69, df =1, P < 0.05). This observation also fits with the notion that the amount of noncoding DNA must be larger in those genes with complex developmental functions in order to harbor the required CRM (Nelson et al. 2004).
HOM-C evolution in Drosophila
In Drosophila, Hox genes are arranged in the same 5′→3′ orientation (with only one exception, the Deformed gene in D. melanogaster). Their regulatory sequences are usually located upstream of each gene and in the introns. If we look at the three HOM-C splits known in Drosophila, a common pattern arises. As can be seen in Figure 2, the lab–pb split took place close to the 3′ end of pb and far from the lab 5′ end. Likewise, the split between the genes Ubx and abdA took place near the abdA 3′ end and far from the Ubx 5′ end, in the short space between their respective regulatory sequences pbx and iab2. This is approximately the same position where an experimental break that does not affect development has been observed (Struhl 1984), although the deficiencies used in the complementation tests both carry a fraction of the pbx and iab2 regions. Finally, sequence comparison between D. melanogaster and D. virilis (Lewis et al. 2003) show that both the insertion of the CG31217 gene and the Antp–Ubx split took place close to the Ubx 3′ end, and far from the Antp 5′ end (results not shown). Thus, all three splits seem to have occurred far from the 5′ end of one gene and much closer to the 3′ end of the next one, in such a way as to keep in place the regulatory sequences of both genes. In this way, rearrangements did not alter any of the known regulatory sequences of these Hox genes; this would explain the absence of gene expression changes.
In the repleta group species, the anterior gene lab is located near the posterior genes abdA and AbdB. The sequence analysis shows that lab and abdA are only 75 kb apart and show the same orientation. The breakpoint of the lab–pb split occurred at ∼22 kb from that of the Ubx–abdA split. None of those splits seem to have affected the regulatory regions of the Hox genes, because the expression patterns of lab and abdA are unaffected. Although it is intriguing, the proximity between these genes in the D. buzzatii genome seems purely accidental and lacking any functional significance.
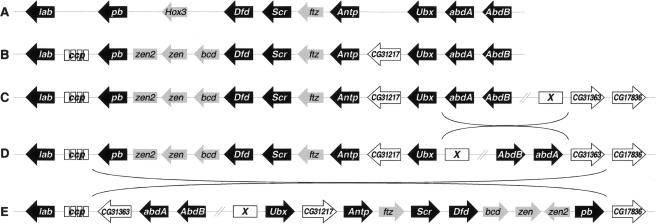
The most likely mechanisms for the generation of the HOM-C splits are paracentric inversions (Ranz et al. 2001; Gonzalez et al. 2002). A plausible reconstruction of HOM-C evolution in the Drosophila subgenus that accounts for the current organization of Hox genes in D. buzzatii is shown in Figure 5. In lower Dipterans, such as Anopheles gambiae, the eight Hox genes, plus Hox3 and ftz, are arranged as a single cluster (Powers et al. 2000). Before the radiation of the Drosophila genus, two transpositions occurred as follows: the Ccp gene cluster between lab and pb, and the gene CG31217 between Antp and Ubx (Lewis et al. 2003). Also, zen, zen2, and bcd evolved from the Hox3 gene (see above). In the lineage of the Drosophila subgenus, an inversion took place with one breakpoint between Ubx and abdA (split 2 in Fig. 1) and the other one between CG31363 and an unknown ORF (X). This HOM-C structure is now present in species of the Drosophila subgenus outside the repleta group, such as D. virilis (see Fig. 1). A second inversion, in the ancestor of the repleta group, split the HOM-C between lab and pb (split 3 in Fig. 1). This inversion, which relocated lab close to abdA, had one breakpoint between pb and the Ccp cluster genes and the second breakpoint between CG31363 and CG17836. These two genes are not adjacent in the D. melanogaster genome, but we infer that they were so in the ancestor of the Drosophila subgenus.
Figure 5.
Reconstruction of the Hox gene complex evolution in the Drosophila subgenus. Genes are shown as arrows when the orientation 5′→3′ is known, and as rectangles otherwise. Hox genes are in black, Hox-related genes in gray, and non-Hox genes in white. (A) Lower Dipterans. (B) Before the radiation of the Drosophila genus. (C) Drosophila subgenus after its separation from that of the Sophophora subgenus. (D) Ancestor of the repleta group. (E) Present arrangement of Hox genes in Drosophila buzzatii (cf. Fig. 2A,D).
Do flies have a Hox gene complex?
Despite the striking conservation of Hox-gene clustering in metazoans, if we compare two of the most deeply studied organisms, Drosophila and vertebrates, important differences arise (Ferrier and Minguillon 2003; Santini et al. 2003; Wagner et al. 2003). Drosophila Hox-gene regions (1) are much larger than those of vertebrates, e.g., the human HoxA cluster is only 110 kb long, whereas the D. melanogaster HOM-C spans 665 kb; (2) contain transposable element insertions, which are remarkably absent in those of vertebrates; (3) contain also non-Hox genes that are inserted between the Hox genes, and tandem duplications within the complex, such as those of the zen-related genes; (4) allow for small inversions of Hox genes, such as Dfd (Randazzo et al. 1993), and non-Hox genes, such as zen2 (Fig. 2); and (5) are split in three ways in different lineages, apparently without consequences on gene expression. These observations suggest a highly dynamic evolution in Drosophila that contrasts with the compact structure seen in vertebrates. Thus, the splits of HOM-C in Drosophila indicate a release of functional requirements present in other metazoan.
Moreover, Drosophila is not the only organism known to have a split HOM-C. Split Hox-gene complexes were also known in nematodes, and recently have been described in Bombyx and tunicates. What do those organisms have in common in addition to the split HOM-C? Vertebrate development follows a rostral-to-caudal temporal progression, and the colinearity of Hox genes is not only spatial, but also temporal (the Hox clock) (Kmita and Duboule 2003). In the tunicate Oikopleura, Hox gene expression still evokes spatial colinearity but not temporal (Seo et al. 2004), which favors the argument that the constraining force of HOM-C structure conservation is temporal colinearity (Ferrier and Minguillon 2003). In nematodes, the pattern of Hox-gene evolution seems indicative of the move to a deterministic developmental mode (Aboobaker and Blaxter 2003). Bombyx embryogenesis, which is difficult to assign to a short or a long germ insect, is characterized by a quick development (Davis and Patel 2002). Drosophila is a long germ insect, where all Hox genes are activated almost simultaneously during the cellular blastoderm stage. Thus, none of these organisms seems to show temporal colinearity. A common feature between all organisms shown so far to have a split Hox complex seems to be a derived mode of embryogenesis characterized by a fast early development.
The loss of temporal progression in the activation of Hox genes in a very rapid mode of embryogenesis could be the ultimate cause for the modular organization of those Hox “clusters,” where modules can be taken apart without loss of function. Given the high rate of chromosomal rearrangement in the genus Drosophila (Ranz et al. 2001; Gonzalez et al. 2002), we anticipate that an even greater variety of Hox-gene organizations will be discovered when more species are investigated. It is ironical that Hox-gene colinearity was discovered in Drosophila, an organism with a partially disassembled complex, which may be the by-product of phylogenetic inertia more than that of functional necessity.
Methods
Flies
D. buzzatii stock st-1 was used for construction of a genomic BAC library (González et al. 2005). The following species and stocks were used for gene expression experiments: D. buzzatii (j19), D. repleta (1611.2), D. virilis (Tokyo-Japan), and D. melanogaster (Canton S and Oregon R).
BAC sequencing
The genomic BAC library was screened with probes from the lab, pb, and abdA genes (González et al. 2005). Positive clones were used to build physical maps for the lab-abdA and pb chromosomal regions, and one BAC clone from each region was chosen for sequencing. Shotgun sublibraries were constructed for each BAC using the vector TOPO, and enough plasmid clones were sequenced by both ends to reach an ∼6× redundancy. Reads were assembled with the PHRED-PHRAD-CONSED software (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998) and sequences finished with one round of AUTOFINISH (Gordon et al. 2001), followed by PCR to bridge the remaining gaps. A continuous high-quality sequence (PHRED score >40) was obtained for BAC clones 5H14 (124,024 bp), and 40C11 (132,938 bp). Statistic details of the sequencing process are given in Supplemental Table S3.
Sequence annotation
Nucleotide sequences were annotated with the aid of GENESCRIPT (Hudek et al. 2003) and ARTEMIS (Berriman and Rutherford 2003). Predicted ORFs were corroborated with GOFIGURE (Khan et al. 2003) for automatic Gene Ontology (Harris et al. 2004) annotation, and BLAST (McGinnis and Madden 2004) for similarity searches. D. buzzatii sequences were compared with those of homologous regions in D. melanogaster (Celniker et al. 2002) and D. pseudoobscura (Richards et al. 2005) genomes. D. melanogaster sequences used were as follows: AE001572 (ANT-C), DMU31961 (BX-C), and AE003692, AE003672, AE003713, AE003676, and AE003724 (other regions). D. pseudoobscura contigs AADE01000437 (lab), AADE01000149 (pb), AADE01000036 (abdA), and AADE01000014, AADE000175, AADE01002495, AADE01000322 (non-Hox genes) were identified with Genome VISTA (Dubchak et al. 2000) and the regions of interest annotated.
Analysis of regulatory sequences
Pairwise alignments of six homologous genomic regions between D. buzzatii, D. melanogaster, and D. pseudoobscura were performed with the AVID global-alignment tool using default parameters (Bray et al. 2003). CNS were identified in the alignments with mVISTA (Mayor et al. 2000) using a window size of 25 bp and a minimum identity of 75%. Statistical tests were carried out to compare the characteristics of the CNS found in the different regions. Comparisons of CNS size distributions, which depart significantly from normality, were conducted using the G-test (Sokal and Rohlf 1995). The number of CNS and the proportion of sequence within CNS was scored for 1-kb windows along the analyzed regions (masking out exons). The resulting variables (density and percent sequence in CNS) as well as the nucleotide identity (per CNS) were tested using ANOVA (Sokal and Rohlf 1995). A complete list of CNS detected is provided in Supplemental Table S5.
Gene-expression experiments
In situ hybridizations and antibody staining were performed to whole-mount embryos and to imaginal discs from third-instar larvae and prepupae as described (Alonso and Akam 2003; Suzanne et al. 2003). cDNA clones were obtained for lab from the four species, pb from D. buzzatii and D. melanogaster and abdA from D. buzzatii, D. repleta, and D. virilis as described (Negre et al. 2003) (for primers see Supplemental Table S4). Sense and antisense RNA probes were produced as described (Suzanne et al. 2003). When no species-specific probe was available, at least two different ones were used in independent experiments, and the results were always consistent. Specific antibodies against the protein were used for abdA (Macias et al. 1990).
Acknowledgments
We thank Claudio Alonso, Joaquin Ariño, Anna Barceló, James Castelli-Gair, Jose Felix de Celis, Susan Celniker, Ariel Chipman, Josefa González, Roger Hoskins, Barret Pfeiffer, Theresa Ren, Armand Sánchez, Chung Li Shu, and three anonymous referees for help or comments on the manuscript. This work was supported by grants BMC2002-01708 awarded to A.R. and BMC2002-00300 to E.S.-H. from the Dirección General de Enseñanza Superior e Investigación Científica (MEC, Spain), an Institutional Grant from the Fundación Ramón Areces, a doctoral FI/DGR fellowship from the Generalitat de Catalunya awarded to B.N., and a doctoral FPI fellowship from the Ministerio de Ciencia y Tecnología (BES-2003–0416) awarded to S.C.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY900631–AY900632 and AY897430–AY897434.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3468605.
References
- Aboobaker, A. and Blaxter, M. 2003. Hox gene evolution in nematodes: Novelty conserved. Curr. Opin. Genet. Dev. 13: 593–598. [DOI] [PubMed] [Google Scholar]
- Alonso, C.R. and Akam, M. 2003. A Hox gene mutation that triggers nonsense-mediated RNA decay and affects alternative splicing during Drosophila development. Nucleic Acids Res. 31: 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, C.M. and Kreitman, M. 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Bergman, C., Pfeiffer, B., Rincon-Limas, D., Hoskins, R., Gnirke, A., Mungall, C., Wang, A., Kronmiller, B., Pacleb, J., Park, S., et al. 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3: research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, B.P., Pfeiffer, B.D., Laverty, T.R., Salzberg, S.L., Rubin, G.M., Eisen, M.B., and Celniker, S.E. 2004. Computational identification of developmental enhancers: Conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman, M. and Rutherford, K. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform. 4: 124–132. [DOI] [PubMed] [Google Scholar]
- Bomze, H.M. and López, A.J. 1994. Evolutionary conservation of the structure and expression of alternatively spliced Ultrabithorax isoforms from Drosophila. Genetics 136: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N., Dubchak, I., and Pachter, L. 2003. AVID: A global alignment program. Genome Res. 13: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard, S. and Kaufman, T.C. 1991. Control of expression of the homeotic labial (lab) locus of Drosophila melanogaster: Evidence for both positive and negative autogenous regulation. Development 113: 1267–1280. [DOI] [PubMed] [Google Scholar]
- Cook, C.E., Smith, M.L., Telford, M.J., Bastianello, A., and Akam, M. 2001. Hox genes and the phylogeny of the arthropods. Curr. Biol. 11: 759–763. [DOI] [PubMed] [Google Scholar]
- Cooper, G.M. and Sidow, A. 2003. Genomic regulatory regions: Insights from comparative sequence analysis. Curr. Opin. Genet. Dev. 13: 604–610. [DOI] [PubMed] [Google Scholar]
- Davis, G.K. and Patel, N.H. 2002. Short, long, and beyond: Molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47: 669–699. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10: 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, I. 1987. The bithorax complex. Annu. Rev. Genet. 21: 285–319. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Ferrier, D.E. and Minguillon, C. 2003. Evolution of the Hox/ParaHox gene clusters. Int. J. Dev. Biol. 47: 605–611. [PubMed] [Google Scholar]
- González, J., Ranz, J.M., and Ruiz, A. 2002. Chromosomal elements evolve at different rates in the Drosophila genome. Genetics 161: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J., Nefedov, M., Bosdet, I., Casals, F., Calvete, O., Delprat, A., Shin, H., Chiu, R., Mathewson, C., Wye, N., et al. 2005. A BAC-based physical map of the Drosophila buzzatii genome. Genome Res. (in press). [DOI] [PMC free article] [PubMed]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., and Green, P. 2001. Automated finishing with autofinish. Genome Res. 11: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D.L., Eyre-Walker, A., Andolfatto, P., and Keightley, P.D. 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32: D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudek, A.K., Cheung, J., Boright, A.P., and Scherer, S.W. 2003. Genescript: DNA sequence annotation pipeline. Bioinformatics 19: 1177–1178. [DOI] [PubMed] [Google Scholar]
- Hughes, C.L. and Kaufman, T.C. 2002. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 4: 459–499. [DOI] [PubMed] [Google Scholar]
- Hughes, C.L., Liu, P.Z., and Kaufman, T.C. 2004. Expression patterns of the rogue Hox genes Hox3/zen and fushi tarazu in the apterygote insect Thermobia domestica. Evol. Dev. 6: 393–401. [DOI] [PubMed] [Google Scholar]
- Ikuta, T., Yoshida, N., Satoh, N., and Saiga, H. 2004. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. 101: 15118–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoun, A.M. and Kaufman, T.C. 1995. A functional analysis of 5′, intronic and promoter regions of the homeotic gene proboscipedia in Drosophila melanogaster. Development 121: 2127–2141. [DOI] [PubMed] [Google Scholar]
- Karch, F., Weiffenbach, B., Peifer, M., Bender, W., Duncan, I., Celniker, S., Crosby, M., and Lewis, E.B. 1985. The abdominal region of the bithorax complex. Cell 43: 81–96. [DOI] [PubMed] [Google Scholar]
- Kaufman, T.C., Lewis, R., and Wakimoto, B. 1980. Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: The homoeotic gene complex in polytene chromosome interval 84A-B. Genetics 94: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, T.C., Seeger, M.A., and Olsen, G. 1990. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv. Genet. 27: 309–362. [DOI] [PubMed] [Google Scholar]
- Khan, S., Situ, G., Decker, K., and Schmidt, C.J. 2003. GoFigure: Automated Gene Ontology annotation. Bioinformatics 19: 2484–2485. [DOI] [PubMed] [Google Scholar]
- Kmita, M. and Duboule, D. 2003. Organizing axes in time and space; 25 years of colinear tinkering. Science 301: 331–333. [DOI] [PubMed] [Google Scholar]
- Lewis, E.B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Lewis, E.B., Pfeiffer, B.D., Mathog, D.R., and Celniker, S.E. 2003. Evolution of the homeobox complex in the Diptera. Curr. Biol. 13: R587–R588. [DOI] [PubMed] [Google Scholar]
- Macias, A., Casanova, J., and Morata, G. 1990. Expression and regulation of the abd-A gene of Drosophila. Development 110: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Martin, C.H., Mayeda, C.A., Davis, C.A., Ericsson, C.L., Knafels, J.D., Mathog, D.R., Celniker, S.E., Lewis, E.B., and Palazzolo, M.J. 1995. Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. 92: 8398–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty, T., Vigano, M.A., Ribeiro, C., Nussbaumer, U., Grieder, N.C., and Affolter, M. 2001. A Hox complex, a repressor element and a 50 bp sequence confer regional specificity to a DPP-responsive enhancer. Development 128: 2833–2845. [DOI] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- McGinnis, W. and Krumlauf, R. 1992. Homeobox genes and axial patterning. Cell 68: 283–302. [DOI] [PubMed] [Google Scholar]
- McGinnis, S. and Madden, T.L. 2004. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32: W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E.N. and Gojobori, T. 1992. Rates of synonymous substitution and base composition of nuclear genes in Drosophila. Genetics 130: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E.N., Petrov, D.A., and Hartl, D.L. 1998. Genome size and intron size in Drosophila. Mol. Biol. Evol. 15: 770–773. [DOI] [PubMed] [Google Scholar]
- Negre, B., Ranz, J.M., Casals, F., Cáceres, M., and Ruiz, A. 2003. A new split of the Hox gene complex in Drosophila: Relocation and evolution of the gene labial. Mol. Biol. Evol. 20: 2042–2054. [DOI] [PubMed] [Google Scholar]
- Nelson, C.E., Hersh, B.M., and Carroll, S.B. 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 5: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega, M.A., Ovcharenko, I., Afzal, V., and Rubin, E.M. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Petrov, D.A., Lozovskaya, E.R., and Hartl, D.L. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Powers, T.P., Hogan, J., Ke, Z., Dymbrowski, K., Wang, X., Collins, F.H., and Kaufman, T.C. 2000. Characterization of the Hox cluster from the mosquito Anopheles gambiae (Diptera: Culicidae). Evol. Dev. 2: 311–325. [DOI] [PubMed] [Google Scholar]
- Randazzo, F.M., Seeger, M.A., Huss, C.A., Sweeney, M.A., Cecil, J.K., and Kaufman, T.C. 1993. Structural changes in the antennapedia complex of Drosophila pseudoobscura. Genetics 134: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J.M., Casals, F., and Ruiz, A. 2001. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 11: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S., Liu, Y., Bettencourt, B.R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M.J., Chen, R., Meisel, R.P., et al. 2005. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddle, F.H., Bartels, J.L., Bentley, K.L., Kappen, C., Murtha, M.T., and Pendleton, J.W. 1994. Evolution of Hox genes. Annu. Rev. Genet. 28: 423–442. [DOI] [PubMed] [Google Scholar]
- Santini, S., Boore, J.L., and Meyer, A. 2003. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 13: 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.C., Edvardsen, R.B., Maeland, A.D., Bjordal, M., Jensen, M.F., Hansen, A., Flaat, M., Weissenbach, J., Lehrach, H., Wincker, P., et al. 2004. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431: 67–71. [DOI] [PubMed] [Google Scholar]
- Shimell, M.J., Simon, J., Bender, W., and O'Connor, M.B. 1994. Enhancer point mutation results in a homeotic transformation in Drosophila. Science 264: 968–971. [DOI] [PubMed] [Google Scholar]
- Shimell, M.J., Peterson, A.J., Burr, J., Simon, J.A., and O'Connor, M.B. 2000. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 218: 38–52. [DOI] [PubMed] [Google Scholar]
- Singh, N.D. and Petrov, D.A. 2004. Rapid sequence turnover at an intergenic locus in Drosophila. Mol. Biol. Evol. 21: 670–680. [DOI] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1995. Biometry: The principles and practice of statistics in biological research. W.H. Freeman and Co., New York.
- Stauber, M., Prell, A., and Schmidt-Ott, U. 2002. A single Hox3 gene with composite bicoid and zerknullt expression characteristics in non-Cyclorrhaphan flies. Proc. Natl. Acad. Sci. 99: 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, G. 1984. Splitting the bithorax complex of Drosophila. Nature 308: 454–457. [Google Scholar]
- Suzanne, M., Estella, C., Calleja, M., and Sánchez-Herrero, E. 2003. The hernandez and fernandez genes of Drosophila specify eye and antenna. Dev. Biol. 260: 465–483. [DOI] [PubMed] [Google Scholar]
- Von Allmen, G., Hogga, I., Spierer, A., Karch, F., Bender, W., Gyurkovics, H., and Lewis, E. 1996. Splits in fruitfly Hox gene complexes. Nature 380: 116. [DOI] [PubMed] [Google Scholar]
- Wagner, G.P., Amemiya, C., and Ruddle, F. 2003. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl. Acad. Sci. 100: 14603–14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G.A., Hahn, M.W., Abouheif, E., Balhoff, J.P., Pizer, M., Rockman, M.V., and Romano, L.A. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Yasukochi, Y., Ashakumary, L., Wu, C., Yoshido, A., Nohata, J., Mita, K., and Sahara, K. 2004. Organization of the Hox gene cluster of the silkworm, Bombyx mori: A split of the Hox cluster in a non-Drosophila insect. Dev. Genes Evol. 214: 606–614. [DOI] [PubMed] [Google Scholar]