Abstract
The retrainable, comparative gene predictor N-SCAN integrates multigenome modeling and 5′ untranslated region (5′ UTR) modeling. In this article, we evaluate N-SCAN's transcription-start site (TSS) and first exon predictions both computationally and experimentally. The computational results indicate that N-SCAN is more accurate than any of the other tools we tested at predicting the TSS and the complete first exon. It is the only one of these tools that can predict complete gene structures together with 5′ UTRs. Experimental evaluation shows that N-SCAN can be used to validate novel UTR introns in human gene predictions that do not overlap any RefSeq gene and even to correct RefSeq mRNAs by adding validated UTR exons that are missing from RefSeq.
The accuracy with which the protein coding regions of genes can be predicted is increasing steadily, but state-of-the-art gene prediction systems make no attempt to accurately predict 5′ untranslated regions (UTRs) (Brent 2002; Brent and Guigó 2004). Several programs predict only the transcription start site (TSS), including PromoterInspector (Scherf et al. 2000), CpG-promoter (Isohikhes and Zhang 2000), Eponine (Down and Hubbard 2002), and Dragon Gene Start Finder (DGSF) (Bajic and Seah 2003). FirstEF goes one step further, predicting both boundaries of the first exon (Davuluri et al. 2001; Brent 2002), which is either completely or partially noncoding. DOUBLESCAN (Meyer and Durbin 2002) predicts 5′ UTRs, including noncoding exons, as a part of its overall gene prediction, but no evaluation of the accuracy of its 5′ UTR predictions has been published. In this article we describe how N-SCAN (Gross and Brent 2005), the latest in the series of TWINSCAN programs, predicts 5′ UTRs as part of an integrated gene prediction process.
Accurately predicting UTRs is important because transcriptional regulatory signals are often located adjacent to the TSS, and post-transcriptional regulatory sites can often be found in the 5′ UTR. The 5′ UTR can also serve a variety of more specialized functions. For example, huntingtin, the gene whose expanded (CAG) repeats are responsible for Huntington's disease, contains a second open reading frame (ORF) upstream of the one for the huntingtin protein. The second ORF encodes a peptide that inhibits expression of the huntingtin mRNA (Lee et al. 2002). There is also growing evidence that many translational regulatory signals reside in 5′ UTRs, including signals that govern cap-independent translation initiation (Miskimins et al. 2001) and mRNA stability (Chen et al. 1998).
Accurately modeling 5′ UTRs can also be expected to improve prediction accuracy in the protein coding region. For example, ∼39% of known human genes contain spliced 5′ UTRs (Davuluri et al. 2001). Except for DOUBLESCAN, current gene prediction programs do not predict spliced UTRs, so they are forced to choose between incorporating the UTR splice sites into false coding exons and ignoring them entirely.
Finally, accurate prediction of 5′ UTRs would be useful for designing RT-PCR experiments to verify predicted ORFs. Without accurate 5′ UTR predictions, designing a primer in the 5′ UTR is difficult, because the distance from the start codon to the TSS or first upstream splice site can vary from zero to hundreds of nucleotides.
The gene-prediction system used in this article is N-SCAN. N-SCAN is a version of TWINSCAN that uses multigenome alignments to inform gene prediction, rather than using alignments between the target genome and one informant genome. The multigenome alignments are exploited via a tree-structured HMM that integrates a phylogenetic tree model with the hidden Markov model used for gene finding (Gross and Brent 2005). This method is related to other tree HMM methods (Holmes and Bruno 2001; McAuliffe et al. 2004; Siepel and Haussler 2004a,b), but it differs from them in a number of important details, including the designation of a single target genome in which predictions are to be made. As a result of these differences, NSCAN includes earlier TWINSCAN models (see Korf et al. 2001) as special cases. In this article, we report results on the human genome that were created by using MULTIZ (Blanchette et al. 2004) alignments to the genome sequences of mouse, rat, and chicken, as well as results on the Drosophila melanogaster genome that were created using MULTIZ alignments to Drosophila yakuba, Drosophila pseudoobscura, and Anopheles gambiae (a mosquito). The results include a comparison between our TSS predictions to those of Eponine, DGSF, and FirstEF, which do not make use of genome comparisons or ORF models. We also compare our first exon predictions to those of FirstEF. In addition, two categories of human, spliced 5′ UTRs predicted by N-SCAN are evaluated by RT-PCR and sequencing: those for which the entire gene is missing from the current RefSeq mRNA collection, and those for which the current annotation contains an unspliced 5′ UTR, contradicting our prediction.
Results
Model structure
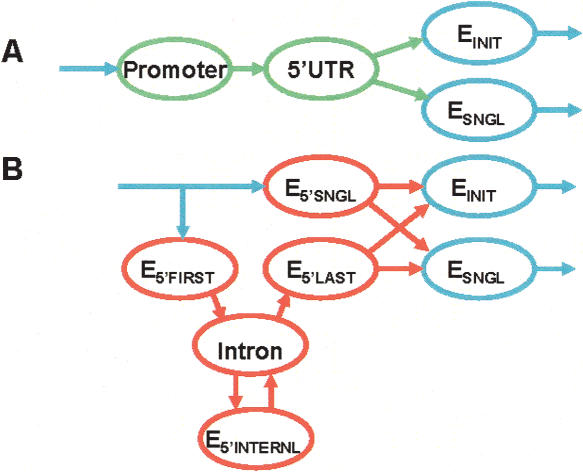
N-SCAN is derived from TWINSCAN 2.0, in which the contiguous region from the predicted promoter to the predicted start codon is loosely labeled as 5′ UTR. The probability model for this region uses a geometric length distribution and the same hexamer distribution as intronic and intergenic regions (Korf et al. 2001). In contrast to TWINSCAN 2.0, N-SCAN uses four generalized states to model the 5′ UTR exonic regions and one geometric state for UTR introns (Fig. 1). N-SCAN's generalized states are not required to have geometric length distributions. One of these states models unspliced UTRs from the TSS to the start codon. The other three model initial noncoding exons (from the TSS to the splice donor), internal noncoding exons (from acceptor to donor), and the noncoding segment of the exon containing the start codon (Table 1). N-SCAN models the DNA sequences of these regions with fourth-order Markov chains. Separate Markov chain parameters are used for the states that fall adjacent to the TSS (E5′SNGL, E5′FIRST) and the states that are separated from the TSS by one or more introns (E5′INTERNL, E5′LAST), because the TSS region has distinctive pentamer frequencies. Each of the four UTR states also has its own evolutionary substitution parameters, and 5′ UTR splice models (which include three bases of the exon) have a substitution model that is distinct from that of the coding splice sites.
Figure 1.
A comparison of state-diagram components that change from TWINSCAN 2.0 to N-SCAN. Blue states are common to TWINSCAN 2.0 and N-SCAN. Green states in TWINSCAN 2.0 are replaced by red states in N-SCAN. (A) Promoter and 5′UTR states in TWINSCAN 2.0. (B) 5′ UTR and intron states in N-SCAN.
Table 1.
Generalized state types by region and bounding features
| Type | Region | 5′ Boundary | 3′ Boundary |
|---|---|---|---|
| ESNGL | CDS | Start codon | Stop codon |
| EINIT | CDS | Start codon | Donor |
| EINT | CDS | Acceptor | Donor |
| ETERM | CDS | Acceptor | Stop codon |
| E5′ SNGL | 5′ UTR | TSS | Start codon |
| E5′ FIRST | 5′ UTR | TSS | Donor |
| E5′ INTERNL | 5′ UTR | Acceptor | Donor |
| E5′ LAST | 5′ UTR | Acceptor | Start codon |
Computational evaluation
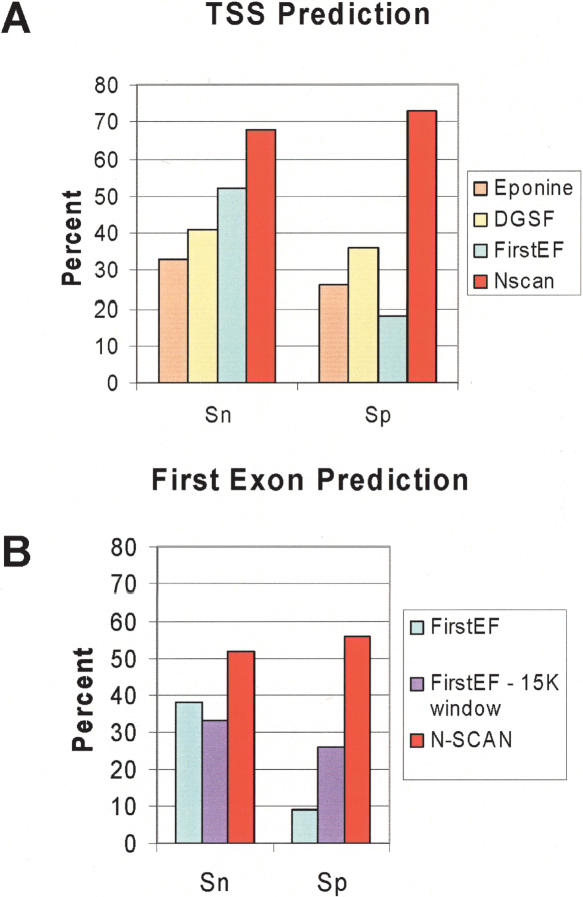
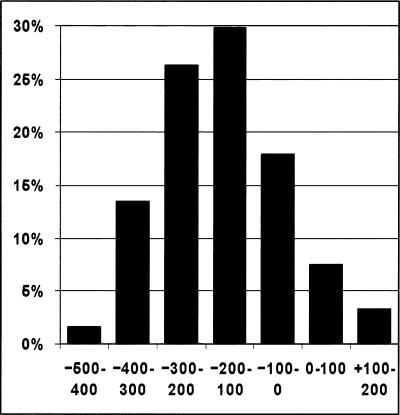
For TSS evaluation we count as a true positive any prediction that falls within a window from –500 bp to +200 bp around an annotated TSS but does not fall in the coding region. TSS annotations were obtained from the Database of Human Transcription Start Sites (DBTSS), a collection of human cDNA sequences from oligo-capped libraries (see Methods). TSS predictions from N-SCAN are compared to strand-specific TSS predictions from Eponine, DGSF, and FirstEF on the human genome. As can be seen from Figure 2A, N-SCAN's sensitivity is at least 15% greater than that of any other prediction method shown. The specificity comparison is even more dramatic—N-SCAN's specificity is double that of DGSF, triple that Eponine, and quadruple that of FirstEF. Within the 700-bp window from –500 to +200, 88% of N-SCAN's TSS predictions fall in the 400 bp upstream of the true TSS (Fig. 3). However, the distribution of distances of N-SCAN predictions from the true TSS peaks ∼100–200 bp upstream of the true TSS, revealing a slight bias in the model. While N-SCAN has a greater concentration of predictions near the TSS than any other system, EPONINE's predictions are less biased, peaking 0–100 bp upstream of the true TSS (Down and Hubbard 2002), while FirstEF's broader distribution peaks 0–100 bp downstream of the true TSS (data not shown).
Figure 2.
Sensitivity (Sn) and specificity (Sp) for 5′ UTR predictions in the human genome by comparison to aligned sequences from DBTSS. All Sp values are scaled assuming 25,000 genes in human genome and assuming the annotation set is a random sample of the full set of human genes. (A) TSS predictions for Eponine (tan), DGSF (light yellow), FirstEF (light blue), and N-SCAN (red). (B) First exon predictions for FirstEF (light blue) and N-SCAN (red) and from FirstEF evaluated in 15-kb windows upstream of annotated start codons (light purple).
Figure 3.
Distribution of the distance between N-SCAN TSS predictions from TSS annotations.
First exon predictions are considered matches if their TSSs are true positives and their splice donors are exactly correct (single exon genes are not considered first exons). First exons predicted by FirstEF are evaluated by using two approaches. The first approach compares predictions against annotations across the entire genome, while the second follows Davuluri's recommendation (Davuluri et al. 2001) that the optimal way to employ FirstEF is in combination with a good gene predictor. Specifically, Davuluri et al. (2001) suggest using FirstEF predictions only if they fall within a 15-kb window upstream of a predicted start codon. This approach reduces sensitivity slightly because some good predictions fall outside the 15-kb windows, but it increases specificity greatly because many false positives lie outside the 15-kb windows (the UCSC FirstEF track records 100,059 FirstEF predictions). To obtain an upper limit on how well this approach can perform, we use known genes to simulate a gene predictor with 100% start codon sensitivity. We evaluate first exon predictions with the whole-genome and 15-kb window approaches and compare with N-SCAN results in Figure 2B. N-SCAN significantly outperforms FirstEF in both sensitivity and specificity—N-SCAN's specificity is about twice that of the best specificity result from FirstEF. One of N-SCAN's advantages is the small number of false positives it predicts; N-SCAN makes 22,242 first exon predictions on the genome (as well as 3508 gene predictions with no splices). If N-SCAN predictions are employed as the gene predictor for FirstEF instead of known genes, the results in Figure 2B are not significantly changed. We find that the main factor determining first exon accuracy is the accuracy of the splice donor prediction; if the donor is predicted correctly, then the first exon is usually correct.
Table 2 shows the match and overlap sensitivity and specificity for noncoding first exons (E5′FIRST, from the TSS to the splice donor) and for the final spliced 5′ UTR segment (E5′LAST, from the splice acceptor to the start codon). These are particularly useful for estimating the success rate for RT-PCR primer placement. The match specificity gives a rough lower bound on successful, first-exon primer placement for spliced 5′ UTRs. If the predicted feature matches the annotation, then a primer placed in the predicted feature is very likely to fall in the true feature. The overlap specificity gives a rough upper bound on successful primer placement. If the predicted feature does not overlap the true feature, then a primer placed in the predicted feature is very unlikely to fall in the true feature. Finally, it is interesting to note that sensitivity and specificity values for E5′LAST are significantly greater than the corresponding values for E5′FIRST. For these features, we observe that the N-SCAN prediction accuracy tends to decrease as distance from the coding region increases.
Table 2.
Sensitivity and specificity for the first and last segments of spliced 5′ UTRs (E5′FIRST and E5′LAST) as well as unspliced 5′ UTRs (E5′SNGL), evaluated by comparison to aligned sequences from DBTSS.
|
Match
|
Overlap
|
||||
|---|---|---|---|---|---|
| Feature | 5′ UTR Type | Sn (%) | Sp (%) | Sn (%) | Sp (%) |
| E5′FIRST | Spliced | 25 | 25 | 43 | 43 |
| E5′LAST | Spliced | 42 | 43 | 54 | 54 |
| E5′SNGL | Unspliced | 66 | 68 | 67 | 70 |
The match (both boundaries are true positives) and overlap (the predicted feature overlaps the annotation by at least one base) values are displayed. Matches are a special case of overlap and hence they contribute to the overlap total. All specificity values are scaled assuming that there are 25,000 genes in the human genome and that the annotation set is a random sample of them. Rounded to the nearest percentage, sensitivity and specificity values are equal for match and overlap for both features.
Splice existence is a measure of how accurately N-SCAN predicts whether a 5′ UTR is spliced. Table 2 displays splice existence results for predictions with correct start codons. N-SCAN identifies unspliced 5′ UTRs as being unspliced more accurately than it identifies spliced 5′ UTRs as being spliced. More generally, N-SCAN is most accurate when predicting features near the coding region (Table 3). Combining results for spliced and unspliced UTRs produces a success rate of 86% (sensitivity must equal specificity in this case). In locating the TSS region from a known start codon, knowledge of whether a 5′UTR is spliced or unspliced can make a significant positional difference. In the unspliced case, the median distance between start codon and TSS is 80 bp, while for spliced 5′ UTRs the median distance is 4600 bp. Therefore, the ability to accurately predict splice existence alone can be a great advantage when searching for the TSS.
Table 3.
Given a correctly predicted start codon, the 5′ UTR splice existence sensitivity (Sn) and specificity (Sp) on the human genome, by comparison to DBTSS
| Sn (%) | Sp (%) | |
|---|---|---|
| N-SCAN Spliced 5′ UTR | 80 | 79 |
| N-SCAN Unspliced 5′ UTR | 89 | 90 |
| N-SCAN Total | 86 | 86 |
To test the applicability of this method outside the vertebrates, we retrained N-SCAN for D. melanogaster by using MULTIZ alignments with D. yakuba, D. pseudoobscura, and A. gambiae. By comparing with the RefSeq annotation set and assuming a total of 13,500 genes (Yandell et al. 2005), we found the TSS sensitivity was 64% and scaled specificity was 65%—comparable to, but slightly lower than, the numbers for human.
Experimental validation of predicted 5′ UTR introns
We tested introns in predicted 5′ UTRs by performing RT-PCR with one primer designed in the first exon and one in the coding region, sequencing the product, and aligning the experimental sequences back to the genome. We tested three groups of genes. Group DBTSS_Control consists of randomly chosen entries from the DBTSS with spliced 5′ UTRs (Suzuki et al. 2002). To construct group No_Overlap, we attempted to design primers for all 870 spliced predictions that do not overlap any RefSeq gene and then selected 74 targets at random from those for which satisfactory primers could be designed. Group Correct_ATG consists of 43 out of 53 cases of spliced predictions that satisfy the following conditions: (1) the predicted start codon matches a RefSeq start codon, (2) the matching RefSeq does not have a spliced 5′UTR, (3) there are no human mRNA entries in GenBank that indicate a spliced 5′ UTR, and (4) there is no corresponding spliced DBTSS entry (for 10 out of 53 cases, satisfactory primers were not found). Table 4 displays both raw and scaled success rates for all three groups. The computational and experimental results are compared in the Discussion.
Table 4.
RT-PCR results
| Group | Trials (count) | Spliced products | Scaled spliced products |
|---|---|---|---|
| DBTSS_Control | 75 | 53 (71%) | 75 (100%) |
| No_Overlap | 74 | 5 (7%) | 7.1 (10%) |
| Correct_ATG | 43 | 6 (14%) | 8.5 (20%) |
The second column shows the observed results. The third column shows scaled results assuming that all DBTSS introns are real, that the observed negative results for DBTSS introns are due to unspecified experimental problems, and that these experimental problems are not biased with respect to any of the three experimental groups.
Discussion
Accurately predicting 5′ UTRs is much harder than is predicting coding regions, since 5′ UTRs do not display codon bias and generally do not contain long ORFs. The results presented above represent a significant advance in predicting complete 5′ UTRs. This advance was achieved by using an integrated probabilistic model of 5′ UTRs and coding regions. The overall model contains submodels for both the DNA sequence and the pattern of cross-species conservation in splice sites, start and stop codons, and the interiors of exons. Distinct submodels were used within coding exons, noncoding regions near the TSS, and noncoding regions that are separated from the TSS by one or more introns. These models reflect differences such as the greater frequency of CpG dinucleotides near the TSSs of many genes and the fact that UTRs are, on average, more conserved than are nonexonic regions but less conserved than are coding regions. These differences, together with the start codon submodel and the absence of in-frame stop codons in coding regions, enable the system to distinguish between coding and noncoding regions of the mRNA.
Because the probability model is integrated, accurate 5′ UTR prediction also resulted in improved coding region accuracy. Such improvements were also reported for DOUBLESCAN, the only previously published integrated model for predicting 5′ UTRs and coding regions (Meyer and Durbin 2002). In TWINSCAN 2.0, which uses only two genomes, the accuracy of start codon prediction is moderately enhanced by the addition of the 5′ UTR model. In N-SCAN, which uses multigenome alignments to detect patterns characteristic of various features within a gene, the 5′ UTR model is critical for the accuracy of coding region predictions. Because 5′ UTR exons are often located adjacent or relatively close to coding sequence, have a relatively high level of conservation, and have high GC content, N-SCAN often mistakes them for coding exons when the 5′ UTR model is removed. In the future it may be possible to use a similar approach for 3′ UTR modeling in order to increase the accuracy with which the terminal exon of the ORF can be predicted (Hajarnavis et al. 2004).
Locating the TSS region is a first step toward identifying transcriptional regulatory signals. N-SCAN's high TSS specificity and sensitivity make it the best available tool for this application. For full ORF gene verification using RT-PCR followed by direct sequencing, it is desirable to place one primer in an exonic region upstream of the start codon. If the primer anneals too close to the start codon, then the region of high-quality sequencing trace may begin after the start codon. Furthermore, verifying the reading frame requires that the high-quality sequence contain an in-frame stop codon upstream of the start codon. However, the further upstream of the start the primer is designed, the greater the risk that it does not anneal to the mature 5′ UTR but rather to a UTR intron or even a genomic sequence upstream of the TSS. That risk can be mitigated by designing primers in the exonic 5′ UTR predicted by N-SCAN. For this application, improved accuracy in 5′ UTR prediction translates into more successful experiments.
Analysis of the distance between the TSS predicted by N-SCAN and the true TSS revealed that more predictions fall upstream of the true TSS than downstream. This is probably because we did not include a specific model for the portion of the CpG island that is upstream of the TSS—the only CpG-rich models occur in the UTR. Thus, the system will tend to place the entire CpG island in the UTR because the UTR model provides the best match for their composition. Adding a state and submodel for the intergenic portion of the CpG island would probably reduce the observed upstream bias in TSS prediction.
By experimentally validating predicted novel UTR introns, we have demonstrated the ability of this system not only to reproduce known spliced 5′ UTRs but to find new ones. Verifying true 5′ UTRs is generally harder than verifying coding exons, since the 5′ ends of mRNAs tend to be degraded first and reverse transcription tends to fail before reaching the 5′ end. Given the relatively advanced state of the human genome annotation, the targets we chose are expected to be particularly difficult. One group of targets consisted of predicted 5′ UTR introns for RefSeq genes where neither the RefSeq itself nor the corresponding DBTSS entry, if any, nor the GenBank mRNAs, indicated such an intron. In effect, we attempted to use a de novo gene prediction program to correct RefSeq. Thus, the scaled success rate of 20% is surprisingly high. Compared with the computational accuracy estimates for spliced first exons (25% exact 43% overlap), the experimental success rate was lower. This is likely due to the fact that these predictions were specifically selected to contradict prior evidence from RefSeq and GenBank mRNAs. The fact that our TSS predictions tend to be a little upstream of the true TSS also hurts the experimental success rate when the primers are upstream of the true TSS. The second group of targets consisted of predicted 5′ UTR introns for genes that do not overlap aligned RefSeq mRNAs. These predicted genes may not exist at all, and if they do, they are likely to be expressed at a much lower level and/or under much more specific conditions than the first group. This is probably responsible for the lower success rate of these predictions. The success rate can be expected to be much higher when testing unannotated 5′-UTR intron predictions in a genome where a higher percentage of true 5′-UTR introns remain unannotated.
We also retrained N-SCAN for D. melanogaster by using multigenome alignments from other insects. This completely automated process yielded TSS accuracies just slightly lower than those achieved on human. We found this remarkable in view of the fact that there is no equivalent of DBTSS for flies and that they appear to lack CpG islands altogether (Hendrich and Tweedie 2003; Aerts et al. 2004), although there are apparently other compositional signals in the vicinity of the TSS (Aerts et al. 2004). The true accuracy of the fly predictions may be even higher, since our estimates had to be made against RefSeq mRNAs, which are less reliable indicators of the true TSS than the sequences in DBTSS.
In an integrated system such as the one presented here, improvements in the coding region model and the UTR models are synergistic. Furthermore, many UTR features, such as splice site signals, are very similar to those in the coding regions. Thus, the accuracy of 5′ UTR prediction is likely to improve in the coming years, along with that of coding region prediction. As a consequence, we expect integrated systems to become the standard for TSS prediction, UTR prediction, and gene prediction.
Methods
Data sets
Build 34 of the human genome was downloaded from http://genome.ucsc.edu. The downloaded files are masked by UCSC using RepeatMasker with default settings for human. We unmasked low complexity and simple repeats. The chromosomes were divided into 1-Mb fragments for processing.
The RefSeq alignments for Build 34 were downloaded from http://genome.ucsc.edu. RefSeq entries without ATG start codons, without TAA, TGA, or TAG stop codons, without GT splice donors, and without AG splice acceptors were removed. If two RefSeq entries overlapped, one was randomly discarded, leaving no overlapping annotations. After processing, 13,311 genes remain in the annotation set.
Release 3.0 of the DBTSS was downloaded (http://dbtss.hgc.jp/index.html). The DBTSS was constructed by sequencing into cDNA libraries constructed by the oligo-capping method (Maruyama and Sugano 1994). These were mapped to RefSeq genes. We aligned each DBTSS sequence to a genomic region around its corresponding RefSeq annotation by using EST_GENOME (http://www.rfcgr.mrc.ac.uk/Registered/Help/est_genome/). We then identified the 5′UTR exon–intron structure and extended the RefSeq annotation. If the RefSeq 5′UTR structure differed from the DBTSS 5′UTR structure, the DBTSS structure was used. There are 6118 genes in the resulting annotation set.
The GenBank human mRNA data set aligned to Build 34 of the genome was downloaded from http://genome.ucsc.edu
Predictions
FirstEF
FirstEF predictions were downloaded from the UCSC track for Build 34 (http://genome.ucsc.edu). Where FirstEF makes two predictions in the same cluster, both were used. The UCSC FirstEF track contained 100,059 first exon predictions.
DGSF
DGSF was downloaded (http://sdmc.lit.org.sg/promoter/dragonGSF1_0/genestart.htm) and TSS predictions generated with default parameters on Build 34 of the human genome. DGSF generated 27,618 TSS predictions.
Eponine
Eponine was downloaded (http://www.sanger.ac.uk/Users/td2/eponine), and predictions were generated with default parameters on Build 34 of the human genome. Eponine generated 58,871 TSS predictions.
Fourfold cross-validation
The RefSeq genes were randomly divided into four sets. DBTSS genes were divided into four sets by assigning them to the same set as their corresponding RefSeq entry. For each set, parameters were trained on the remaining three-fourths of the data and tested on the chosen set. DBTSS genes were used for 5′ UTR feature training. RefSeq genes were used for all other training. All N-SCAN results are fourfold cross-validated.
Evaluation measures
If both boundaries of a UTR segment are evaluated as true positives, then the predicted segment is a correct match. Acceptor, donor, and start-codon boundaries must match exactly for a true positive. A true positive predicted TSS boundary falls within –500 bp and +200 bp of the annotated TSS. Sensitivity is defined as a measure of how well the predictions cover the annotations. The sum of all annotations that are matched by a prediction is divided by the number of annotations to give sensitivity. Specificity is defined as a measure of how well the annotations cover the predictions. The sum of all predictions that are matched by an annotation is divided by the number of predictions to give specificity. There are cases when more than one prediction can be considered as correct (such as TSS predictions in a window) for a particular annotation.
Experimental procedure
We designed PCR primers with Primer3 (Rozen and Skaletsky 2000; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) with default parameters except for PRIMER_MIN_ SIZE = 17, PRIMER_MIN_GC = 30, PRIMER_MAX_GC = 70, PRIMER_OPT_TM = 70, PRIMER_MIN_TM = 65, PRIMER_ MAX_TM = 75, and PRIMER_GC_CLAMP = 2. One primer was placed in the first exon and one in the coding region with a buffer (minimum distance) between the primer and the nearest splice site. The primer design consisted of, at most, three rounds, increasing the amplicon length with each round: a 300–500-bp amplicon and a buffer of 10 bp in round 1, a 500–800-bp amplicon and a buffer of 30 bp in round 2, and an 800–1000-bp amplicon and a buffer of 30 bp in round 3. If a satisfactory primer pair was found, the subsequent rounds were skipped. Primer sequences can be found at http://genes.cse.wustl.edu/brown-2005/.
For all UTR experiments, Poly-A RNA from 20 human tissues obtained from BD Biosciences (http://www.bdbiosciences.com) was pooled. First-strand cDNA was generated by using SuperScript III reverse transcriptase by Oligo-dT priming (Invitrogen). RT was followed by PCR amplification using Phusion High Fidelity Polymerase. PCR products were purified with a QuickStep 2, 96-well PCR Purification Kit from Edge BioSystems (http://www.edgebio.com), and sequenced by using both forward and reverse primers for each predicted gene. Sequencing traces are available on the auxiliary data Web site: http://genes.cse.wustl.edu/brown-2005/ and were submitted to http://www.ncbi.nimh.nih.gov/Traces/.
Acknowledgments
We thank the Baylor College of Medicine Human Genome Sequencing Center, particularly Kim Haeberlen and John McPherson, for RT-PCR reactions and sequencing. We also thank Michael Stevens for designing primers and analyzing sequences. R.H.B. was partially supported by Fellowship HG02635 from the National Human Genome Research Institute. The remainder of the funding for this work was provided by grant HG02278 from the National Human Genome Research Institute to M.R.B.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3696205.
References
- Aerts, S., Thijs, G., Dabrowski, M., Moreau, Y., and De Moor, B. 2004. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics 5: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic, B. and Seah, S. 2003. Dragon Gene Start Finder identifies approximate locations of the 5′ ends of genes. Nucleic Acids Res. 31: 3560–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F., Roskin, R.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent, M.R. 2002. Predicting full-length transcripts. Trends Biotechnol. 20: 273–275. [DOI] [PubMed] [Google Scholar]
- Brent, M.R. and Guigó, R. 2004. Recent advances in gene structure prediction. Curr. Opin. Struct. Biol. 14: 264–272. [DOI] [PubMed] [Google Scholar]
- Chen, C., Del Gatto-Konczak, F., Wu, A., and Karin, M. 1998. Stabilization of interlukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280: 1945–1949. [DOI] [PubMed] [Google Scholar]
- Davuluri, R., Grosse, I., and Zhang, M. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412–417. [DOI] [PubMed] [Google Scholar]
- Down, T. and Hubbard, T. 2002. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 12: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S. and Brent, M. 2005. Using multiple alignments to improve gene prediction. The Ninth International Conference on Research in Computational Molecular Biology (RECOMB). (in press). [DOI] [PubMed]
- Hajarnavis, A., Kork, I., and Durbin, R. 2004. A probabilistic model of 3′ end formation in Caenorhabditis elegans. Nucl. Acids Res. 32: 3392–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich, B. and Tweedie, S. 2003. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19: 269–277. [DOI] [PubMed] [Google Scholar]
- Holmes, I. and Bruno, W.J. 2001. Evolutionary HMMs: A Bayesian approach to multiple alignment. Bioinformatics 17: 803–820. [DOI] [PubMed] [Google Scholar]
- Isohikhes, I. and Zhang, M. 2000. Large-scale human promoter mapping using CpG islands. Nat. Genet. 26: 61–63. [DOI] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17(Suppl 1): S140–S148. [DOI] [PubMed] [Google Scholar]
- Lee, J., Park, E.H., Couture, G., Harvey, I., Garneau, P., and Pelletier, J. 2002. An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res. 30: 5110–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K. and Sugano, S. 1994. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 200: 149–156. [DOI] [PubMed] [Google Scholar]
- McAuliffe, J.D., Pachter, L., and Jordan, M.I. 2004. Multiple-sequence functional annotation and the generalized hidden Markov phylogeny. Bioinformatics 20: 1850–1860. [DOI] [PubMed] [Google Scholar]
- Meyer, I. and Durbin, R. 2002. Comparative ab initio prediction of gene structures using pair HMMs. Bioinformatics 18: 1309–1318. [DOI] [PubMed] [Google Scholar]
- Miskimins, W., Wang, G., Hawkinson, M., and Miskimins, R. 2001. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 21: 4960–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Scherf, M., Klingenhoff, A., and Werner, T. 2000. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: A novel context analysis approach. J. Mol. Biol. 297: 599–606. [DOI] [PubMed] [Google Scholar]
- Siepel, A.C. and Haussler, D. 2004a. Computational identification of evolutionarily conserved exons. In RECOMB. ACM, San Diego, CA.
- ———. 2004b. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21: 468–488. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Nakai, K., and Sugano, S. 2002. DBTSS: Data base of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 30: 328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell, M., Bailey, A.M., Misra, S., Shu, S., Wiel, C., Evans-Holm, M., Celniker, S.E., and Rubin, G.M. 2005. A computational and experimental approach to validating annotations and gene predictions in the Drosophila melanogaster genome. Proc. Natl. Acad. Sci. 102: 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://genome.ucsc.edu; the July 2003 release of Build 34 (hg16) of the human genome, the human mRNA data base, and the FirstEF track of first exon predictions.
- http://www.sanger.ac.uk/Users/td2/eponine; Eponine TSS finder.
- http://sdmc.lit.org.sg/promoter/dragonGSF1_0/genestart.htm; DGSF.
- http://www.rfcgr.mrc.ac.uk/Registered/Help/est_genome/; EST_GENOME.
- http://dbtss.hgc.jp/index.html; Database of Human Transcription Start Sites home.
- http://www.bdbiosciences.com; BD Biosciences home page.
- http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Primer3.
- http://www.ncbi.nimh.nih.gov/Traces
- http://www.edgebio.com; Edge BioSystems home page.
- http://genes.cse.wustl.edu/brown-05-UTR-data/; Supplemental data for this paper