Abstract
The genes encoding the small nucleolar RNA (snoRNA) species snR190 and U14 are located close together in the genome of Saccharomyces cerevisiae. Here we report that these two snoRNAs are synthesized by processing of a larger common transcript. In strains mutant for two 5′→3′ exonucleases, Xrn1p and Rat1p, families of 5′-extended forms of snR190 and U14 accumulate; these have 5′ extensions of up to 42 and 55 nucleotides, respectively. We conclude that the 5′ ends of both snR190 and U14 are generated by exonuclease digestion from upstream processing sites. In contrast to snR190 and U14, the snoRNAs U18 and U24 are excised from the introns of pre-mRNAs which encode proteins in their exonic sequences. Analysis of RNA extracted from a dbr1-Δ strain, which lacks intron lariat-debranching activity, shows that U24 can be synthesized only from the debranched lariat. In contrast, a substantial level of U18 can be synthesized in the absence of debranching activity. The 5′ ends of these snoRNAs are also generated by Xrn1p and Rat1p. The same exonucleases are responsible for the degradation of several excised fragments of the pre-rRNA spacer regions, in addition to generating the 5′ end of the 5.8S rRNA. Processing of the pre-rRNA and both intronic and polycistronic snoRNAs therefore involves common components.
Eukaryotic cells contain a large number of small nucleolar RNA (snoRNA) species that play major roles in the processing and modification of the pre-rRNAs (reviewed in references 29 and 41). The mode of synthesis of many of the snoRNA species differs from that of other small RNAs. Rather than being expressed from simple genes, most of the known snoRNAs are encoded in the intronic sequences of genes which also encode mRNAs in the exonic sequences. This was first observed for the mammalian U14 snoRNA (27) and has subsequently been demonstrated for several other species (for reviews, see references 29, 31, and 37). In most cases there is some relationship between the protein product of the host gene and ribosome synthesis or function. Several snoRNAs are located in the introns of genes encoding ribosomal proteins (r-proteins), nucleolar proteins, or translation factors (reviewed in reference 29). It has been suggested that this provides a mechanism for the coregulation of the synthesis of the snoRNAs and other components involved in ribosome synthesis. No small RNA species other than the snoRNAs are known to follow this pathway of biosynthesis.
The mechanism of synthesis of vertebrate snoRNAs has been the subject of considerable interest. The faithful synthesis of snoRNAs in Xenopus oocytes and in vitro has been reported. The best-studied example is the U16 snoRNA, which is encoded within intron III of the L1 r-protein gene (7, 9, 13, 34). Synthesis of U16 is mutually exclusive with pre-mRNA splicing and involves endonucleolytic cleavage within intron III, followed by trimming to generate the 5′ and 3′ ends of the mature snoRNA. A similar pathway of processing has been reported for U18, which is also encoded in introns of the L1 r-protein gene (34). Processing of other snoRNAs, i.e., U15 (44), U17 (10, 21, 22), and U19 (20), appears to be slightly different; for these snoRNAs, no upstream endonuclease cleavage site was identified, and both 5′ and 3′ processing appear to be purely by exonuclease digestion of either the pre-mRNA or the excised intron. The exonucleases that carry out these processing reactions have not been identified.
Two homologous proteins with in vitro 5′→3′ exoribonuclease activity have been purified from yeast (19, 24, 38). These are Xrn1p (also known as Rar5p, Kem1p, Dst2p, and Sep1p) (reviewed in reference 18), which functions in the cytoplasm (17), and Rat1p (also known as Tap1p and Hke1p) (1, 2, 12, 19), which functions in the nucleus (17). The genes encoding each of these proteins have been cloned by a number of different groups by using selection techniques which are not obviously related, although in several cases they potentially involve RNA metabolism. We have previously reported that strains which have the XRN1 gene deleted and carry a temperature-sensitive lethal mutation in RAT1 are impaired in the formation of the 5′ end of the 5.8S rRNA (14). In addition, xrn1 mutant strains accumulate high levels of an excised pre-rRNA spacer fragment containing the 5′ region of internal transcribed spacer 1 (ITS1) between sites D and A2 (Fig. 1B) (39). Other studies have shown that xrn1 mutants are impaired in 5′→3′ degradation of mRNA (15, 32, 33). Homologs of both Rat1p and Xrn1p have been identified in mice (5, 36), and mouse Xrn1p can functionally replace the yeast protein, showing that their functions have been highly conserved in evolution.
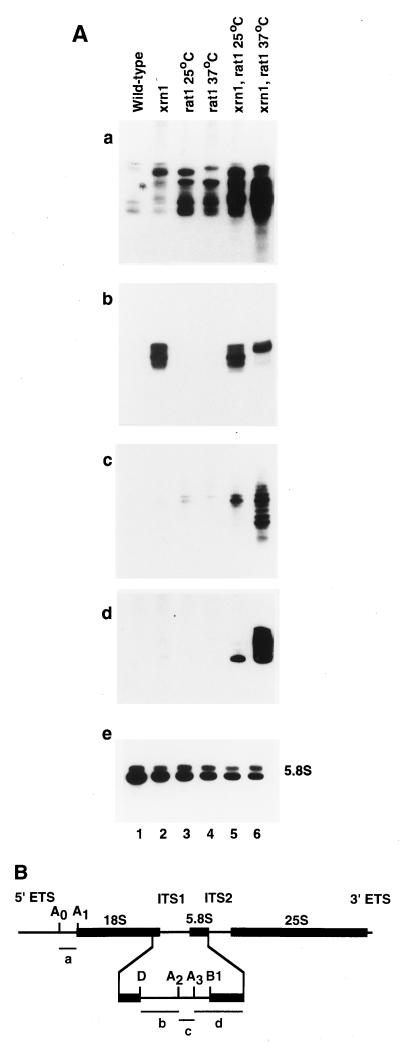
FIG. 1.
Accumulation of excised pre-rRNA fragments in exonuclease mutants. (A) Northern hybridization of RNAs extracted from strains of the indicated genotypes following growth at 25°C or 2 h after transfer to 37°C. Panels: a, Riboprobe specific for the A0-A1 region; b, probe specific for the D-A2 region (oligonucleotide 002); c, riboprobe specific for the A2-A3 region; d, 5′-extended 5.8S detected with the probe for the A3-B1 region (oligonucleotide 003); e, mature 5.8S rRNA (oligonucleotide 017). RNA was separated on a gel containing 8% polyacrylamide. (B) Structure of the pre-rRNA, showing the locations of processing sites above the pre-rRNA. Lines below the pre-rRNA indicate the species detected in panels a to d of panel A. The effects of the mutations on the accumulation of the fragments referred to as a, b, c, and d are shown in panels a, b, c, and d, respectively, of panel A.
The yeast U18 and U24 snoRNAs are intron encoded (23, 29, 35), like their vertebrate homologs (8, 35). Vertebrate U14 is encoded within introns of the hsc70 gene (27, 29), but the genomic arrangement is different in Saccharomyces cerevisiae. The yeast SNR128 gene, which encodes U14, lacks consensus promoter elements and is located just 67 bp 3′ to the end of the gene encoding another snoRNA, snR190 (46). The U14 and snR190 snoRNAs lack the 5′ cap structure characteristic of primary transcripts of RNA polymerase II, and both species show 5′ heterogeneity, consistent with synthesis by processing of larger transcripts (reference 3 and this work). This suggested that these snoRNAs are transcribed from a polycistronic snoRNA precursor, as is also the case for maize U14 (25, 26). Here we analyze the in vivo processing of both intron-encoded and polycistronic snoRNAs in yeast and report that formation of their 5′ ends involves the 5′→3′ exonucleases Rat1p and Xrn1p. We also show that these enzymes process a variety of pre-rRNA spacer fragments, in addition to forming the 5′ end of the 5.8S rRNA.
MATERIALS AND METHODS
Strains and media.
The growth and handling of S. cerevisiae were by standard techniques. Cultures were grown in SD minimal medium containing 2% glucose, 0.67% yeast nitrogen base (Difco), and appropriate supplements. The strains used were as follows: XRN1 strain, MATa trp1-1 ura3-52 his3-11,15 ade2-1 (strain W303-1A; kindly provided by S. Kearsey); xrn1-Δ strain, same as the XRN1 strain except for xrn1::URA3 (strain R934; kindly provided by S. Kearsey) (in the xrn1::URA3 mutation the URA3 gene is inserted at a point corresponding to 97 amino acids from the amino terminus of the protein encoded by the gene; no Xrn1p can be detected on Western blots prepared from this strain [17a]); rat1-1 strain, MATa rat1-1 leu2-Δ1 ura3-52 his3-Δ200 (DAH18; kindly provided by C. Cole) (2); rat1 xrn1 strain, MATa rar5::URA3 rat1-1 (strain 966-1C; kindly provided by S. Kearsey); DBR1 strain MATa trp1-Δ1 his3-Δ200 leu2-Δ1 ura3-167 (strain YH8; kindly provided by J. Boeke) (11); and dbr1-Δ strain, same as the DBR1 strain but with Δdbr1::HIS3 (strain KC99) (11).
RNA extraction, Northern hybridization, and primer extension.
RNA extraction (43) Northern hybridization (40), and primer extension (42) were performed as previously described. For Northern hybridization and primer extension, total RNA equivalent to that of cells at an optical density at 600 nm of 0.1 (approximately 2 × 106 cells; equivalent to 4 μg of RNA for wild-type cells) was used for each sample. Prior to RNA extraction, the rat1-1 and tap1-1 strains were pregrown at 25°C to mid-log phase (optical density at 600 nm = 0.3) and either harvested or transferred to 37°C for 2 or 6 h prior to being harvested.
Hybridization probes. (i) rRNA probes.
For the A0-A1 and A2-A3 fragments, riboprobes overlapping the entire regions were used (28, 45). The hybridization probe for the D-A2 fragment was oligonucleotide 002 (GCTCTTTGCTCTTGCC), the oligonucleotide probe for the A2-A3 region was oligonucleotide 003 (TGTTACCTCTGGGCCC), the probe for the 5′-extended 5.8S rRNA was oligonucleotide 001 (CCAGTTACGAAAATTCTTG), the probe for the 5.8S rRNA was oligonucleotide 017 (GCGTTGTTCATCGATGC), the probe for the 5S rRNA was oligonucleotide 041 (CTACTCGGTCAGGCTC), the probe for the 5′ region of ITS2 was oligonucleotide 013 (GGCCAGCAATTTCAAGTTA), the probe for the 3′ region of ITS2 was oligonucleotide 006 (AGATTAGCCGCAGTTGG), and the probe for the external transcribed spacer (ETS) 5′ to site A0 was oligonucleotide 024 (TCGGGTCTCTCTGCTGC).
(ii) snoRNA probes.
The oligonucleotide complementary to the 5′ flanking sequence of snR190 was CAATCAATTCTTCTTTTCTG (αsnR190+1), the internal snR190 oligonucleotide was CGTCATGGTCGAATCGG (αsnR190), the oligonucleotide complementary to the 5′ flanking sequence of U14 was ATATATTATCTGTCTCCTC (αU14-9), the internal U14 oligonucleotide was TGCGAATGTTAAGGAACC (αU14), the internal U24 oligonucleotide was TCAGAGATCTTGGTGATAAT (αU24), the U24 3′ flanking oligonucleotide was AAACCATTCATCAGAG (U24-3′fl), the U18 internal oligonucleotide was GTCAGATACTGTGATAGTC (αU18), and the U18 3′ flanking oligonucleotide was GCTCTGTGCTATCGTC (U18-3′fl).
RESULTS
Excised pre-rRNA spacer fragments accumulate in 5′→3′ exonuclease mutants.
RNA was extracted from a strain carrying an xrn1::URA3 gene disruption and from an otherwise isogenic wild-type strain following growth at 25°C and from the rat1-1 mutant strain and the rat1-1 xrn1-Δ double-mutant strain following growth at 25°C and after transfer to 37°C for 2 or 6 h. All data shown are for the 2-h time point; the 6-h time point gave similar results. Growth of rat1-1 strains essentially ceased within 2 h of transfer to 37°C (2).
Endonucleolytic cleavage of the pre-rRNA releases a number of discrete fragments from the transcribed spacer regions (see Fig. 1B for the structure of the pre-rRNA). The levels of these excised fragments were assessed by Northern hybridization (Fig. 1A). Strong accumulation of a number of spacer fragments was observed in the exonuclease mutants. Hybridization with a probe specific for the region from site A0, in the 5′ ETS, to site A1, the 5′ end of the 18S rRNA, showed that this fragment (fragment a in Fig. 1B) accumulated significantly in either the xrn1-Δ or rat1-1 single-mutant strain. The accumulation was, however, much stronger in the double mutant at 37°C, the nonpermissive temperature for the rat1-1 strain (Fig. 1A, panel a). This indicates that both exonucleases normally play roles in the degradation of this pre-rRNA region. The largest band visible in the wild-type strain probably corresponds to the full-length A0-A1 fragment (91 nucleotides [nt]) (45), indicating that most of the accumulated spacer fragments in the mutants have undergone some digestion.
Cleavage at sites A0 and A1 is inhibited in strains with the U3 snoRNA depleted (16); to confirm the identification of the hybridizing RNA, the A0-A1 probe was used on a Northern blot of RNA from a strain genetically depleted of U3 by growth of a GAL::U3 strain (16) on glucose medium. As expected, the A0-A1 fragment was lost during U3 depletion (data not shown).
Interestingly, cleavage at site A0 can be detected by primer extension in wild-type strains by using a primer which hybridizes 3′ to site A1, (6, 16, 45), but no effects of the exonuclease mutations were observed with this primer (data not shown). We conclude that while degradation of the excised A0-A1 fragment from A0 very rapidly follows processing at A1, the 5′ exonuclease digestion occurs only after cleavage of A1.
Strains carrying mutations of XRN1 have previously been reported to accumulate the pre-rRNA fragment from site D, the 3′ end of the 18S rRNA, to site A2 in ITS1 (fragment b in Fig. 1B). As expected, the xrn1-Δ strain strongly accumulated this fragment (Fig. 1A, panel b, lane 2). The fragment which accumulated in the rat1-1 xrn1-Δ double-mutant strain at the nonpermissive temperature (Fig. 1A, panel b, lane 6) was, however, significantly longer than that observed in the xrn1-Δ single mutant (Fig. 1A, panel b, lane 2) or in the double mutant grown at the permissive temperature for the rat1-1 strain (Fig. 1A, panel b, lane 5). We conclude that while the degradation of fragment b is largely due to Xrn1p, Rat1p also plays a role in this activity, at least in xrn1 mutants.
A hybridization probe specific for the region between the A2 and A3 cleavage sites in ITS1 (fragment c in Fig. 1B) showed that this fragment was accumulated in the rat1-1 single-mutant strain (Fig. 1A, panel c), with much stronger accumulation in the double-mutant strain, particularly after growth at the nonpermissive temperature for the rat1-1 strain (Fig. 1A, panel c, lane 6). Clear accumulation was not seen in the xrn1-Δ single-mutant strain. The hybridization probe used for Fig. 1A, panel c, is an antisense riboprobe to the A2-A3 region. Hybridization with an oligonucleotide complementary to the sequence immediately 5′ to site A3 gave the same result (data not shown), suggesting that the accumulated fragments have undergone partial digestion from the 5′ end.
We also show the level of the mature 5.8S rRNA (Fig. 1A, panel e) and the accumulation of the 5′-extended form of the 5.8S rRNA (fragment d in Fig. 1B) by using a hybridization probe to the 3′ region of ITS1 (Fig. 1A, panel d). Here, too, accumulation was strongest in the double-mutant strain grown at the nonpermissive temperature (Fig. 1A, panel d, lane 6). As previously reported (14), the major product has undergone partial digestion from the 5′ end to the base of a stable stem structure located between site A3 and the 5′ end of the 5.8S rRNA.
Probes to the 5′ or 3′ region of ITS2 or further upstream in the 5′ ETS did not detect accumulation of pre-rRNA fragments in the mutant strains (data not shown; see Materials and Methods for details of the hybridization probes used). From these data we conclude that both Xrn1p and Rat1p play roles in the turnover of several excised fragments of the pre-rRNA. In each case, the steady-state level of the pre-rRNA fragment was very low in wild-type cells, indicating that degradation very rapidly follows generation of the excised pre-rRNA fragments.
5′-extended forms of snR190 and U14 accumulate in 5′→3′ exonuclease mutants.
Northern hybridization with probes to the mature snR190 (Fig. 2A, panel b) and U14 (Fig. 2A, panel d) sequences revealed a reduction in the levels of the mature snoRNAs compared to 5S rRNA (Fig. 2A, panel e), particularly in the xrn1-Δ rat1-1 double mutant at 37°C. The mature snoRNAs are expected to be stable, so the reduction observed 2 h after transfer to nonpermissive conditions indicates that the synthesis of new snR190 and U14 was strongly inhibited. The rat1-1 strain essentially ceases growth after 2 h at 37°C (reference 2 and data not shown), and further reduction was not observed at later time points (data not shown).
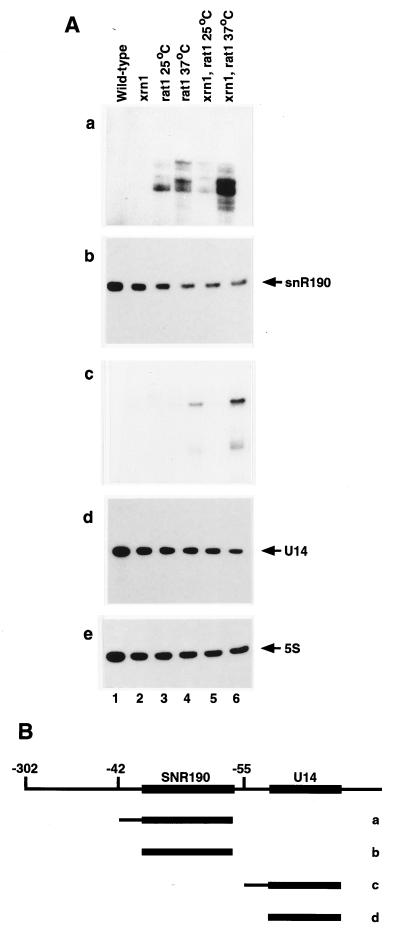
FIG. 2.
Northern analysis of the synthesis of U14 and snR190. (A) Northern hybridization of RNAs extracted from strains of the indicated genotypes following growth at 25°C or 2 h after transfer to 37°C. Panels: a, 5′-extended forms of snR190 (oligonucleotide αsnR190+1); b, mature snR190 (oligonucleotide αsnR190); c, 5′-extended forms of U14 (oligonucleotide αU14-9); d, mature U14 (oligonucleotide αU14); e, mature 5S rRNA (oligonucleotide 041). RNA was separated on a gel containing 8% polyacrylamide. (B) Predicted structure of the precursor to snR190 and U14. Lines below the pre-snoRNA indicate the species detected in panels a to d of panel A.
Probes to the 5′ flanking sequences of snR190 (Fig. 2A, panel a) and U14 (Fig. 2A, panel c) revealed the accumulation of 5′-extended forms of the snoRNAs in the exonuclease mutants. For both snR190 and U14 the 5′-extended RNA was most clearly seen in the double mutant at the nonpermissive temperature (Fig. 2A, panels a and c, lanes 6). A lower accumulation of 5′-extended snR190 was seen in the rat1-1 single-mutant strains. In addition, longer RNAs were weakly detected; RNAs with the same gel mobility were detected by both snR190 and U14 internal and 5′ flanking probes (data not shown), indicating that these represent common transcripts of snR190 and U14 pre-snoRNAs. The 5′-extended snoRNA species accumulated to much lower levels than the mature snoRNAs (ca. 10%), suggesting that other pathways for their turnover exist.
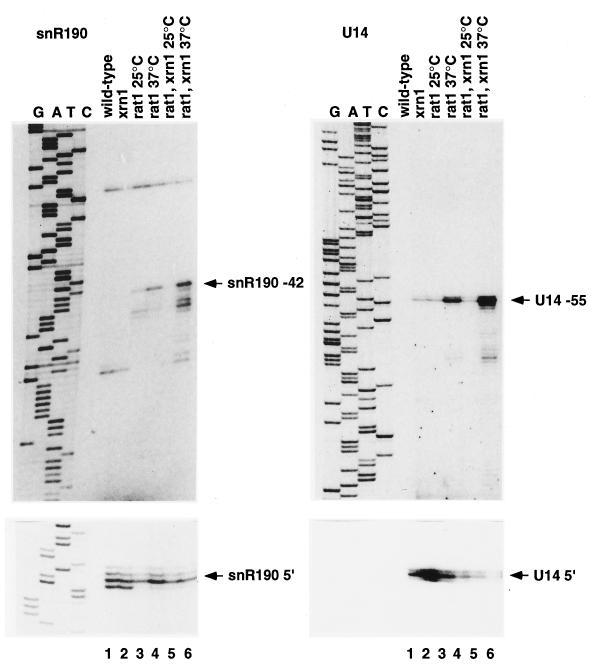
Primer extension was used to better characterize the 5′-extended forms of snR190 and U14 seen in the exonuclease mutant strains. The rat1-1 mutant strains accumulated a ladder of 5′-extended forms of snR190 which extend to a position 42 nt longer than mature snR190, but not beyond. This accumulation was evident in the rat1-1 single mutant (Fig. 3, lanes 3 and 4) but was much stronger in the double-mutant strain at the nonpermissive temperature (Fig. 3, lane 6). Consistent with the results of Northern hybridization, the level of mature U14 fell substantially in the rat1-1 strains, particularly in the rat1-1 xrn1-Δ strain at 37°C (Fig. 3, lane 6). A ladder of 5′-extended forms of U14, which extended to a position 55 nt longer than mature U14 but not beyond, was also observed in the rat1-1 strain at 37°C (Fig. 3, lane 4) and was more strongly accumulated in the xrn1-Δ rat1-1 double-mutant strain (Fig. 3, lane 6).
FIG. 3.
Primer extension analysis of the synthesis of U14 and snR190. 5′-extended forms of snR190 and U14 are detected in 5′→3′ exonuclease mutants. Primer extension was performed with the αU14 or αsnR190 oligonucleotide on RNA extracted from strains of the indicated genotypes following growth at 25°C or 2 h after transfer to 37°C. The lower panels show shorter exposures of the same primer extensions as the corresponding upper panels.
As with pre-rRNA processing (14), the accumulation of the pre-snoRNAs was similar but not identical in strains carrying the tap1-1 mutation (data not shown), which is allelic with rat1-1 (1). From these data we conclude that Rat1p has the major activity in the processing of mature snR190 and U14 from 5′-extended pre-snoRNAs, with Xrn1p also playing a role, at least in rat1-1 mutant strains.
In primer extension analysis, the internal snR190 oligonucleotide also gave a strong stop at a position 302 nt 5′ to the end of the mature snR190 (data not shown). A primer extension stop at a similar position was detected with an internal U14 oligonucleotide, but this product is too long for its end to be accurately identified. In addition, the internal U14 primer gave a stop at the position corresponding to the 5′ end of the mature snR190; as expected, this stop was reduced in the xrn1-Δ rat1-1 double-mutant strain (data not shown). These data are consistent with processing of both U14 and snR190 from a common pre-snoRNA transcript.
Processing of intron-encoded snoRNAs.
In contrast to snR190 and U14, the snoRNAs U18 (4) and U24 (35) are encoded in introns of the genes EFB1 (encoding EF-1 beta) and BEL1(encoding a G-beta-like protein of unknown function), respectively. To determine whether intron debranching is required for their synthesis, RNA was extracted from a strain with DBR1, the gene encoding the intron lariat debranching enzyme (11), deleted. Strains lacking this activity are viable and accumulate the excised intron lariats to high levels (11). Since these are circular molecules, synthesis of intron-encoded snoRNA species that are processed exclusively by exonucleases is expected to be severely inhibited in the dbr1-Δ strain. In contrast, synthesis of snoRNAs that are generated by pathways involving endonuclease cleavage is predicted to be little affected.
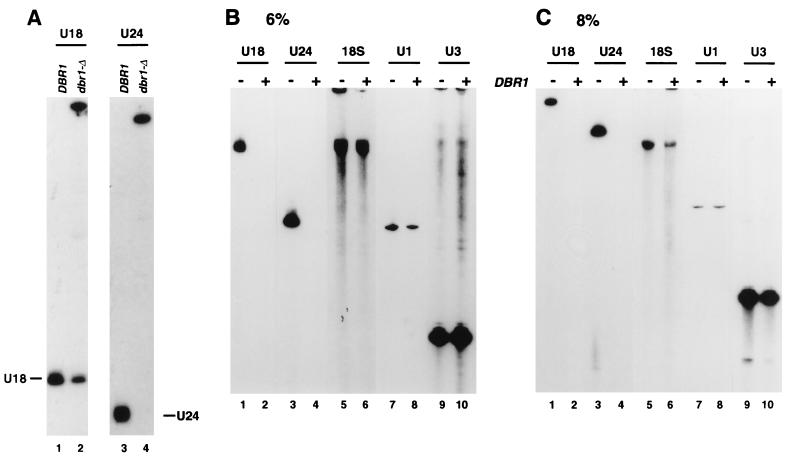
Northern hybridization (Fig. 4A) showed that formation of mature U24 was almost entirely abolished in the dbr1-Δ mutant. This strongly indicates that U24 is synthesized exclusively by exonuclease digestion. Synthesis of U18 was also reduced, but only to ∼30% of the wild-type level. Slow-migrating RNA species were detected with both the U18 and U24 probes in the dbr1-Δ strain. To determine whether these represent the intron lariats, their mobilities on gels containing either 6% (Fig. 4B) or 8% (Fig. 4C) polyacrylamide were compared. The relative mobility of circular RNA species compared to linear RNA is expected to be more retarded in the 8% gel than the 6% gel, and this was observed. Strains carrying dbr1-Δ are reported to accumulate intron lariats which lack the sequence 3′ to the intron branch site (11). This has not been tested for the lariats containing the U24 and U18 sequences but is likely to be the case.
FIG. 4.
Lariat forms of U18 and U24 accumulate in an intron-debranching mutant. (A) In RNA from the dbr1-Δ strain, mature U18 and U24 are underaccumulated and longer forms are detected. (B and C) RNAs extracted from the dbr1-Δ strain and an otherwise isogenic DBR1 strain were separated by long migration on gels containing either 6% (B) or 8% (C) polyacrylamide. Lanes + and −, DBR1 and dbr1-Δ strains, respectively. The slow-migrating forms of U18 and U24 differ in their relative migrations on the gels compared to the linear RNA species U3 (311 nt), U1 (568 nt), and 18S rRNA (1,860 nt). Note that mature U18 and U24 are substantially smaller than the linear RNA species shown and have been lost from the gels in panels B and C.
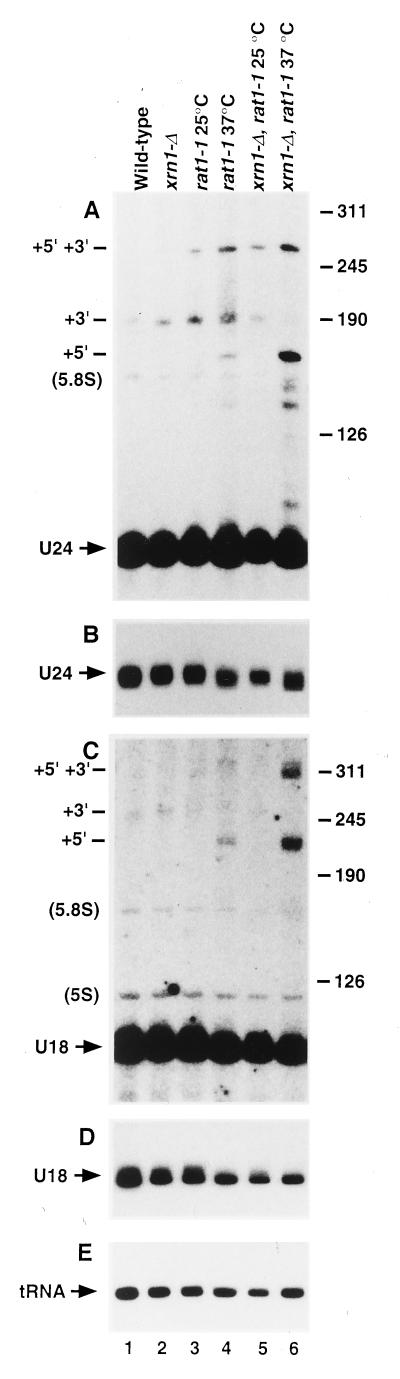
The effects of 5′→3′ exonuclease mutations on the synthesis of U18 and U24 were also assessed. Northern hybridization (Fig. 5) showed some reduction in the levels of mature U24 (Fig. 5B) and U18 (Fig. 5D) relative to tRNA3Leu (Fig. 5E) 2 h after transfer to 37°C, indicating an inhibition of their synthesis. Interestingly, the mobility of U24 was reproducibly shifted downwards, i.e., to slightly shorter forms. Since the location of the 5′ end of U24 is unaltered on primer extension (see Fig. 6), this presumably represents the formation of 3′-shortened species. Larger forms of U18 and U24 were detected in the rat1-1 mutant strain at 37°C (Fig. 5, lane 4) and were more abundant in the xrn1-Δ rat1-1 double mutant (Fig. 5, lane 6). For both snoRNAs two major extended species were detected. The gel mobility of the larger U24 species (+5′ +3′ in Fig. 5A) is appropriate for the entire intron (273 nt). From its mobility, the smaller species (+3′ in Fig. 5A) is predicted to be 3′ matured but 5′ extended to the end of the intron (169 nt). Consistent with this, the U24 +5 +3 band hybridized to the U24-3′ flanking oligonucleotide probe, while the U24 +5′ band did not (data not shown); as the probe lies across the 3′ end of the mature U24, the U24 +5′ species is very likely to be fully 3′ matured. The accumulation of the U24 intron in the exonuclease mutant suggests that some inhibition of 3′ processing also occurs. In other systems, e.g., pre-tRNA processing, 3′ processing is inhibited in the absence of prior 5′ processing. In the wild-type strain an RNA species of ∼190 nt (+5′ in Fig. 5A) is detected with the U24 probe. This is in agreement with the size of a pre-snoRNA which is 5′ mature but 3′ extended to the end of the intron (192 nt). Consistent with this, the 190-nt species is depleted in the double-mutant strain (Fig. 5, lane 6) and is detected with the U24-3′ probe in the same samples (data not shown). From the primer extension data (see below), this species would not, however, be expected in the rat1-1 mutant strain, and we assume that the band at this position in Fig. 5, lane 4, represents an aberrant processing intermediate.
FIG. 5.
Northern analysis of the synthesis of U18 and U24. Longer forms of U18 and U24 are detected in 5′→3′ exonuclease mutants. Northern hybridization of RNAs extracted from strains of the indicated genotypes is shown. For U18, RNA was separated on a gel containing 6% polyacrylamide; for U24, which is smaller, a gel containing 8% polyacrylamide is shown. The positions of migration of U14 (126 nt), snR190 (190 nt), snR10 (245 nt) and U3 (311 nt) determined by subsequent Northern hybridization of the same filters are indicated, as are the positions of migration of mature 5S and 5.8S rRNAs. Species predicted to be 5′- and 3′-extended forms of U24 and U18 are indicated (+5′, +3′, and +5′ +3′).
FIG. 6.
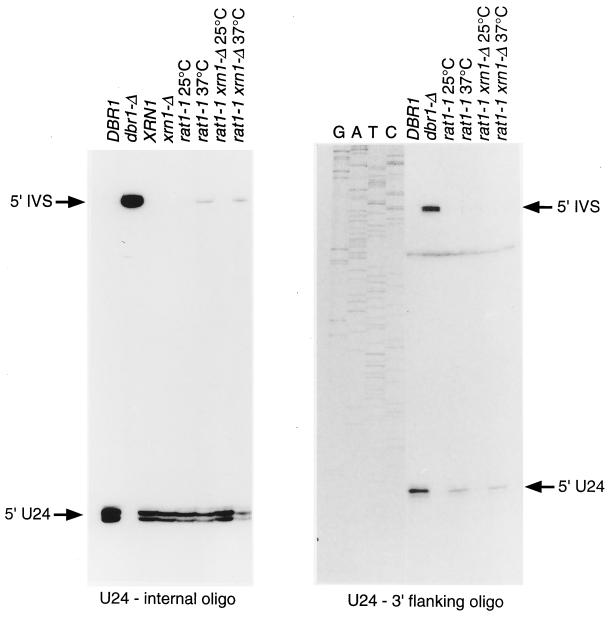
Primer extension analysis of the synthesis of U24. (Left) Primer extension with a probe to mature U24 (αU24). (Right) Primer extension with an oligonucleotide (oligo) spanning the 3′ end of U24 (U24-3′fl). The positions of the mature 5′ end of U24 and the intron 5′ splice site (5′ IVS) are indicated on the DNA sequence ladder.
In the case of U18, the mobility of the U18 +5′ band in Fig. 5C, lane 6, is consistent with a species which is 3′ mature but 5′ extended to the end of the intron (220 nt). The mobility of the larger U18 +5′ +3′ species in Fig. 5C, lane 6, is, however, greater than that predicted for the intact intron (366 nt); the faint band above this species may represent the intact intron. The U18 +5′ +3′ species may therefore be a form of the intron which has undergone partial digestion. A faint band present in the wild-type but not in the double-mutant strain (+3′ in Fig. 5C) shows a gel mobility consistent with U18 that is 5′ mature but 3′ extended to the end of the intron (253 nt)
The synthesis of U24 and U18 was also examined by primer extension. In the dbr1-Δ strain, the mature 5′ end of U24 is lost and a strong primer extension stop at the position corresponding to the 5′ end of the mRNA intron was observed (labeled 5′ IVS in Fig. 6) with either an internal U24 oligonucleotide or an oligonucleotide complementary to the 3′ flanking sequence. With the internal U24 oligonucleotide, an RNA species extended to the 5′ end of the intron was detected in the rat1-1 strains at 37°C (Fig. 6); a reduction in the mature 5′ end of U24 was also observed, particularly in the xrn1-Δ rat1-1 double-mutant strain at 37°C. A striking reduction in the stop corresponding to the mature 5′ end of U24 was observed in the rat1-1 strains at 37°C by using the 3′ flanking oligonucleotide, which detects the 3′-immature pre-snoRNA species. This is consistent with the inhibition of 5′ processing of newly synthesized U24 in the mutant. Similar results were obtained for U18 (data not shown), with the interesting difference that the mature 5′ end of U18 was only weakly detected with the 3′ flanking oligonucleotide even in the wild-type strain. This suggests that 3′ processing of U18 normally precedes 5′ processing.
We conclude that the 5′ ends of U18 and U24 are synthesized by exonuclease digestion requiring Rat1p, with Xrn1p also playing a role, at least in the rat1-1 strains. U24 can be synthesized only from the debranched intron lariat, while U18 can be generated with moderate efficiency in the absence of intron debranching.
DISCUSSION
Results from higher eukaryotic systems indicate that the 5′ ends of many snoRNA species are generated by 5′→3′ exonuclease activities (7, 13, 20, 22, 34, 44). Two 5′→3′ exonucleases, Xrn1p and Rat1p, have been identified in S. cerevisiae, and we therefore tested whether mutants defective in these activities are also defective in the 5′ processing of pre-snoRNA species. The data presented here establish that Xrn1p and Rat1p play roles in the formation of the 5′ ends of the yeast snoRNAs, snR190, U14, U18, and U24. In each case, the rat1-1 mutation led to some accumulation of 5′-extended species at 37°C, but this was stronger in the xrn1-Δ rat1-1 double-mutant strain, whereas the xrn1-Δ single mutation alone had little effect. After 2 h at the nonpermissive temperature, depletion of the mature snoRNAs was observed in the rat1-1 xrn1-Δ strains, indicating that the major biosynthetic pathway was inhibited. As rat1-1 strains rapidly cease growth at 37°C (2), stronger depletion of the mature snoRNAs was not observed at later time points. The pre-rRNA spacer fragments and the 5′-extended snoRNA and 5.8S rRNA species were, however, present at relatively low levels compared to the mature rRNA and snoRNAs. The xrn1-Δ allele is a gene disruption construct (14), whereas the rat1-1 allele is a temperature-sensitive point mutation (2). It is not clear whether Rat1p retains some residual processing activity in the rat1-1 strain or whether the accumulated RNAs were degraded by another pathway. In the rat1-1 mutant strain, the nuclear poly(A) signal is lost after prolonged incubation at the nonpermissive temperature (2), suggesting that there is some residual activity in the mutant. A complex of 3′→5′ exonucleases processes the 3′ end of the 5.8S rRNA (30) and degrades the excised pre-rRNA spacer 5′ to site A0 (11a). This complex may also contribute to turnover of the RNAs that accumulate in the 5′→3′ exonuclease mutants.
Northern and primer extension data indicate that snR190 and U14 are synthesized from a common, dicistronic transcript. In the rat1-1 and xrn1-Δ rat1-1 strains, ladders of 5′-extended snoRNAs that extend to positions −42 for snR190 and −55 for U14 were detected. These sites do not have any obvious homology to each other or to consensus snoRNA promoter sequences. Moreover, U14 position −55 lies only 12 nt 3′ to the mature snR190 region, making it unlikely to be a transcription start site. We propose that snR190 position −42 and U14 position −55 represent intermediate sites in the processing of larger pre-snoRNA species. These could be the products of either endonuclease or 5′→3′ exonuclease digestion. However, exonuclease digestion would presumably have to involve an exonuclease(s) other than Xrn1p and Rat1p, and other 5′→3′ exonuclease have not been identified in yeast extracts (19, 24, 38). Moreover, the 5′ cap structure would be expected to confer protection against exonucleases, and an endonuclease activity is more probable. The furthest-upstream primer extension stop that we detected lies 302 nt 5′ to snR190. It has not been established whether this represents the transcription start site or is a further upstream processing site; however, the 5′ end of the next open reading frame lies only 190 nt upstream of this site, making this likely to be the start site.
U18 and U24 are synthesized from the introns of host genes that also encode mRNAs. U24 can be synthesized only from the debranched intron lariat; the snoRNA was found almost entirely in circular form in a mutant which lacks intron-debranching activity. This strongly indicates that both 5′ and 3′ processing of the pre-snoRNA are exclusively exonucleolytic. Moreover, little if any processing of U24 can occur on the unspliced pre-mRNA. In the case of U18, synthesis of the mature snoRNA was reduced to ∼30% of the wild-type level in the debranching mutant, indicating that the major processing pathway is also via exonuclease digestion. Residual processing might be due to endonuclease cleavage of the intron lariat or exonuclease digestion of the unspliced pre-mRNA. For both U18 and U24, pre-snoRNAs that were 5′ extended to the intron 5′ splice site in the rat1-1 and xrn1-Δ rat1-1 strains accumulated, indicating that these are the 5′→3′ exonucleases responsible for processing the pre-snoRNAs. Primer extension specifically on pre-U24, using a 3′-flanking oligonucleotide, failed to detect the mature 5′ end of the snoRNA in the rat1-1 strains at 37°C, demonstrating the inhibition of 5′ processing.
In each case the accumulation of 5′-extended snoRNA species was much stronger in the rat1-1 strain than in the xrn1-Δ strain, indicating that Rat1p is the major pre-snoRNA-processing activity in wild-type cells. Since Rat1p functions in the nucleus, the presumed site of pre-snoRNA processing, while Xrn1p functions in the cytoplasm (17), it may be that the processing activity normally resides only in Rat1p, with Xrn1p functioning to process the accumulated pre-snoRNAs in the rat1-1 mutant strains.
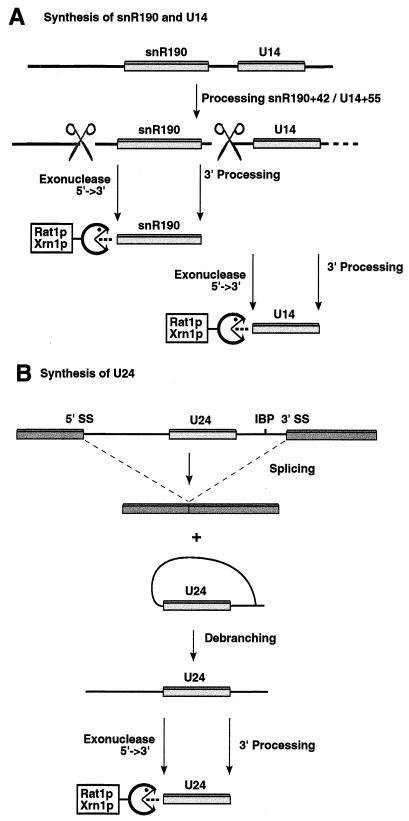
Together, the data suggest the models shown in Fig. 7. We envisage that snR190 and U14 are synthesized from a dicistronic pre-snoRNA species which extends from a position 302 nt 5′ to snR190 to beyond the 3′ end of U14 (Fig. 7A). This is processed, probably endonucleolytically, at positions 42 nt 5′ to snR190 and 55 nt 5′ to U14, within the intergenic spacer region. These processing reactions are followed by 5′ and 3′ trimming reactions which generate the mature snoRNAs. In contrast, U24 (Fig. 7B) is processed from the excised pre-mRNA intron. In wild-type cells processing is probably exonucleolytic from the debranched intron lariat.
FIG. 7.
(A) Model for the processing of pre-snR190 and pre-U14. The coding sequences of snR190 and U14 lie in the same orientation in the genome and are separated by only 67 nt (46). We propose that they are synthesized from a common precursor which extends from 302 nt 5′ to snR190 to beyond the 3′ end of U14. Cleavage of the pre-snoRNA at snR190 position −42 and U14 position −55 is envisaged to be followed by exonuclease digestion by Rat1p to the 5′ ends of the snoRNAs and 3′ trimming. (B) Model for the processing of pre-U24. U24 is encoded in the intron of the BEL1 gene (23, 35) and is generated from the excised intron lariat. Following intron debranching, processing is envisaged to consist of exonuclease digestion by Rat1p to the 5′ end of the snoRNA and 3′ trimming. IBP, intron branch point.
In general, the host genes for vertebrate snoRNAs encode protein products that have some relationship to ribosome synthesis or function. This coexpression may facilitate the coordinated synthesis of the protein and snoRNA products. The data reported here extend the interaction between the synthesis of the snoRNAs and the function of the nucleolus by demonstrating that the snoRNAs and pre-rRNAs are processed by common components. It is possible that as the snoRNAs developed, they simply made use of whatever processing machinery was available. Alternatively, the use of common components might have been selected because of the obvious possibilities that it offered for coregulation of the synthesis of the rRNAs and snoRNAs. Such coregulation might indeed be the reason that so many snoRNAs, but no other known small RNA species, are synthesized by such excision mechanisms.
All studies on the in vitro processing of vertebrate snoRNAs have implicated 5′→3′ exonuclease activities in formation of the 5′ ends of the snoRNAs (7, 13, 22, 34, 44; reviewed in references 20 and 29). In no case have the nucleases yet been identified, but we strongly predict that, at least in some cases, these activities will involve the vertebrate homologs of Rat1p and Xrn1p (5, 36).
ACKNOWLEDGMENTS
We thank Charles Cole and David Amberg for the rat1-1 strain, Ben Hall for the tap1-1 strain, Jeff Boeke for the dbr1-Δ strain, Steve Kearsey and Lydia Jane Brimage for all the xrn1::URA3 strains, and Phil Mitchell for critical reading of the manuscript. We particularly thank Bertran Séraphin for his helpful advice.
This work was partially supported by the Wellcome Trust.
REFERENCES
- 1.Aldrich T L, Di Segni G, McConaughy B L, Keen N J, Whelen S, Hall B D. Structure of the yeast TAP1 protein: dependence of transcription activation on the DNA context of the target gene. Mol Cell Biol. 1993;13:3434–3444. doi: 10.1128/mcb.13.6.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 3.Balakin A G, Lempicki R A, Huang G M, Fournier M J. Saccharomyces cerevisiae U14 small nuclear RNA has little secondary structure and appears to be produced by post-transcriptional processing. J Biol Chem. 1994;269:739–746. [PubMed] [Google Scholar]
- 4.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;85:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Bashkirov V I, Scherthan H, Solinger J A, Buerstedde J M, Heyer W D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffarelli E, De Gregorio E, Fatica A, Prislei S, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reactions and the stability of the mature snoRNAs. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 9.Caffarelli E, Maggi L, Fatica A, Jiricny J, Bozzoni I. A novel Mn++-dependent ribonuclease that functions in U16 snoRNA processing in X. laevis. Biochem Biophys Res Commun. 1997;233:514–517. doi: 10.1006/bbrc.1997.6487. [DOI] [PubMed] [Google Scholar]
- 10.Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman K B, Boeke J D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 11a.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.85 rRNA in Saccharomyces cerevisiae. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 12.Di Segni G, McConaughy B L, Shapiro R A, Aldrich T L, Hall B D. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol Cell Biol. 1993;13:3424–3433. doi: 10.1128/mcb.13.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. A novel small nuclear RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu C L, Stevens A. Yeast cells lacking 5′ 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes J M X, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Kearsey, S. Personal communication.
- 18.Kearsey S, Kipling D. Recombination and RNA processing: a common strand? Trends Cell Biol. 1991;1:110–112. doi: 10.1016/0962-8924(91)90101-e. [DOI] [PubMed] [Google Scholar]
- 19.Kenna M, Stevens A, McCammon M, Douglas M G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T, Bortolini M-L, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 22.Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 24.Larimer F W, Hsu C L, Maupin M K, Stevens A. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 25.Leader D J, Sanders J F, Waugh R, Shaw P, Brown J W S. Molecular characterisation of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as polycistronic U14 transcripts. Nucleic Acids Res. 1994;22:5196–5203. doi: 10.1093/nar/22.24.5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leader D L, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leverette R D, Andrews M T, Maxwell E S. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- 28.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome; a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonuclease activities. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 31.Mount S, Henikoff S. Nested genes take flight. Curr Biol. 1993;3:372–374. doi: 10.1016/0960-9822(93)90205-3. [DOI] [PubMed] [Google Scholar]
- 32.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 33.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prislei S, Michienzi A, Presutti C, Fragapane P, Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993;21:5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu L H, Henry Y, Nicoloso M, Michot B, Azum M C, Renalier M H, Caizergues-Ferrer M, Bachellerie J P. U24, a novel intron-encoded small nucleolar RNA with two 12nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shobuike T, Sugano S, Yamashita T, Ikeda H. Characterization of cDNA encoding mouse homolog of fission yeast dhp1+ gene: structural and functional conservation. Nucleic Acids Res. 1995;23:357–61. doi: 10.1093/nar/23.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993;75:403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- 38.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 39.Stevens A, Hsu C L, Isham K R, Larimer F W. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J Bacteriol. 1991;173:7024–7028. doi: 10.1128/jb.173.21.7024-7028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 42.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tollervey D, Mattaj I W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tycowski K T, Shu M-D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 45.Venema J, Henry Y, Tollervey D. Two distinct recognition signals define the site of endonucleolytic cleavage at the 5′ end of yeast 18S rRNA. EMBO J. 1995;14:4883–4892. doi: 10.1002/j.1460-2075.1995.tb00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zagorski J, Tollervey D, Fournier M J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensable and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]