Two model legumes, Medicago truncatula (Mt) and Lotus japonicus (Lj), are currently targets of large-scale genome sequencing projects. As a result, legumes are one of few plant families with extensive genome sequence in multiple species. The prospect of integrating genome information from Mt and Lj together into a reference for legume genomics will provide exciting opportunities for plant biologists. Because the Mt and Lj sequencing efforts are both clone by clone (as opposed to shotgun or filtered genome sequencing strategies), syntenic comparisons between these two genomes and with other plant taxa will be straightforward and highly informative. Already, the Mt and Lj genome sequences offer novel insights into the organization and evolution of legumes, as well as the similarities and differences with genomes of other plant families, such as Arabidopsis (Arabidopsis thaliana; Zhu et al., 2003) and Populus trichocarpa (G. Tuskan, personal communication). A growing number of researchers are using the Mt and Lj genomes to positionally clone genes of biological importance, especially those involved in symbiosis (Schauser et al., 1999; Endre et al., 2002; Krusell et al., 2002; Nishimura et al., 2002; Stracke et al., 2002; Madsen et al., 2003; Ane et al., 2004; Levy et al., 2004). Increasingly, researchers working in broader aspects of plant biology will find the genome sequences of Mt and Lj essential to their research.
In this review, we briefly describe basic features of the Mt and Lj genomes, gleaned from the growing body of genome sequence data. We compare the two genomes through direct sequence comparisons, based on a total of 122 Mb of finished (phase 3) sequence available between the two genomes. These comparisons lay a foundation for integrating knowledge about these two systems and increasing their utility as reference legumes.
WHY M. TRUNCATULA AND L. JAPONICUS?
Since the 1990s, Mt and Lj have played central roles in symbiosis research (Pichon et al., 1992; Crespi et al., 1994; Kapranov et al., 1997; Schauser et al., 1998). Like Arabidopsis before them, Mt and Lj evolved from systems for studying biological questions into models for genomics and genome sequencing. Both exhibit diploid genetics and modest genome sizes (around 500 Mb each), both are tractable to genetic manipulation, and both have strong international research communities. Recent progress in comparative genomics confirms that genomic discoveries made in these two model species can frequently be extended to other legumes, including most members of the large and agriculturally important Papilionoid subfamily.
The decision to focus genome sequencing on reference species played a prominent role in the recent U.S. National Plant Genome Initiative report, published by the U.S. National Academy of Science in 2002 (www.nap.edu/openbook/0309085217/html/). This highly influential report recommended that the plant genomics community concentrate “on a small number of key species for in-depth development of genome-sequence data” (p. 3), and legumes were highlighted for the substantial investment required. The Kazusa DNA Research Institute in Japan had already chosen Lj as its target for legume sequencing with the backing of a large international research community and financial support from the local government of Chiba, Japan. Soon afterward, the University of Oklahoma, with support from the Samuel Roberts Noble Foundation, initiated similar efforts in Mt. This was followed by large-scale support for Mt sequencing by the U.S. National Science Foundation and the European Union 6th Framework Program. At first, sequencing in two model legumes was viewed as a wasteful duplication of effort. Now, it is clear that having two substantially sequenced legume genomes will lead to valuable new discoveries.
Both projects decided to pursue similar strategies in their sequencing efforts. Previous research had indicated that most Mt and Lj genes would be found in euchromatic regions throughout chromosome arms and would be largely absent from the heterochromatin of centromeres and pericentromeres (Kulikova et al., 2001; Pedrosa et al., 2002). Thus, a judicious choice of bacterial artificial chromosome (BAC) and P1 transformation-competent artificial chromosome (TAC) clones (Liu et al., 1999) rich in genes would be expected to uncover the majority of genes in these two genomes. These large insert clones could be anchored to extensive map resources available in Mt (mtgenome.ucdavis.edu) and Lj (www.kazusa.or.jp/lotus/), efficiently creating contiguous or near-contiguous assemblies of genome sequence that comprise most of the genes, the so-called “genespace.” The decision to adopt this anchored, clone-by-clone approach to genome sequencing, rather than a whole-genome shotgun (WGS; Venter et al., 1998) or genome filtering method (Palmer et al., 2003; Whitelaw et al., 2003), was considered essential for making the Mt and Lj genomes as useful as possible to the broader community of legume and plant biologists.
As of January 2005, approximately 134 Mb of the genome sequence in Mt (77 Mb finished, 57 Mb draft) and 165 Mb of the genome sequence in Lj (45 Mb finished, 120 Mb draft) were publicly available. Analysis of these genome sequences demonstrates the wisdom in adopting a clone-by-clone strategy. Gene density is reasonably high in sequenced clones: 149 genes/Mb (1 gene every 6.7 kb) in Mt and 158 genes/Mb (1 gene per 6.3 kb) in Lj. (These estimates are based on Fgenesh predictions [Salamov and Solovyev, 2000] using a Mt-trained matrix, retaining peptides with a BLASTP match at 10e−4 to the UniProt NREF100 database of peptides [Apweiler et al., 2004]. This estimate for Lj differs from the published value of 1 gene per 10.1 kb [Asamizu et al., 2003a] due to the use here of the Fgenesh gene-calling algorithm so Mt and Lj could be compared directly.) Based on these estimates, gene densities in Mt and Lj are lower than in Arabidopsis by a factor of approximately 1.5 (Arabidopsis Genome Initiative, 2000) but still quite high. While there are repetitive elements on most sequenced clones, there are no cases of BACs or TACs from euchromatic regions without genes or consisting entirely of repeats. There also is no indication that Mt or Lj genes are found in islands separated by extensive stretches of retroelements, as observed in maize (Zea mays; SanMiguel et al., 1996), although the longest contiguous stretches examined so far are just 1 Mb. In the case of Mt, for example, 77 Mb of phase 3 (finished) BACs contains approximately 11,500 genes. Assuming there are 35,000 to 40,000 genes overall, then sequencing a total of 230 to 270 Mb will discover essentially all. A growing body of data suggests that other legume species, even those with a much larger genome, such as soybean (Glycine max), may actually comprise genespaces on par with Mt and Lj (Mudge et al., 2004).
GENOME SEQUENCING IN M. TRUNCATULA
Mt (2n = 16) is an annual diploid in the tribe Trifolieae, cultivated as a forage crop and closely related to tetraploid alfalfa (Medicago sativa). In the past few years, more than 190,000 Mt expressed sequence tags (ESTs) have been produced (www.medicago.org/MtDB2/ and www.tigr.org/tdb/tgi/plant.shtml), with corresponding microarray and DNA chips now available. There are also 155,000 sequenced BAC ends (ftp.tigr.org/pub/data/m_truncatula) plus detailed physical and genetic maps (mtgenome.ucdavis.edu). Gene knockout systems involving T-DNA and Tnt1 (Scholte et al., 2002; d'Erfurth et al., 2003), RNA interference (Limpens et al., 2004), and gene TILLING (D. Cook, personal communication) are under development. Combined with the rapidly emerging sequence of its genespace, Mt provides an impressive array of genomic tools to legume biologists.
Fluorescent in situ hybridization (FISH) has been especially influential in guiding the Mt sequencing effort (Kulikova et al., 2001; Choi et al., 2004a; Kulikova et al., 2004). The pachytene chromosomes of Mt are relatively easy to visualize, and all eight chromosomes can be identified by appearance. Significantly, they all show distinct euchromatic arms and heterochromatic centromeres/pericentromeres, although chromosome 6 is substantially more heterochromatic than the rest. Kulikova et al. (2004) found that each of 20 gene-rich BAC clones hybridized exclusively to euchromatic arms, and subsequent FISH analysis confirmed and extended this initial observation, with 80 gene-rich BACs all localizing to euchromatin (Choi et al., 2004a; R. Guerts, personal communication). This analysis also uncovered a translocation involving chromosomes 4 and 8 between the parents of a widely studied Mt mapping population: A20 and Jemalong, the genotype now being sequenced. In summary, cytogenetic and FISH results provided critical support for the BAC-by-BAC strategy adopted for sequencing. It demonstrated that if BAC clones could be efficiently identified as gene rich (initially through the use of EST-based overgoes), then most of the Mt genespace would be uncovered in the course of BAC-by-BAC sequencing.
Genome sequencing began in earnest in 2002 through a collaboration between Bruce Roe at the University of Oklahoma and Doug Cook and Dongjin Kim at the University of California, Davis. This was expanded significantly in 2003 with grants from the National Science Foundation and the European Union (see www.medicago.org/genome/people.php for a complete list of participants). Sequencing is coordinated by an international steering committee, with most of the genespace sequencing scheduled for completion by the end of 2006. Altogether, slightly more than 2,000 BAC clones will be sequenced in the course of the project by the four centers performing the work (Bruce Roe et al., Oklahoma; Chris Town et al., The Institute for Genomic Research [TIGR]; Jane Rogers et al., Sanger Centre; Francis Quétier et al., Genoscope). The most important product of this initiative will be 16 chromosome arm-length sequences, called pseudomolecules after the model of Arabidopsis and rice (Oryza sativa), comprising the complete sequence of each chromosome arm. Realistically, every one of these molecules will still contain gaps, but the gaps will be sized through FISH. The pseudomolecules will extend approximately from telomeres to pericentromeres, and annotation in the form of computer-based predictions of genes and other genomic features will be performed. An international committee known as the International Medicago Genome Annotation Group is coordinating the annotation process and utilizing training sets of Mt gene models fully supported by EST sequence data to train gene prediction algorithms.
As of January 2005, sequencing of 1,165 BAC clones, constituting approximately 133 Mb of the Mt genome, was complete or in progress. After accounting for overlap, this represents about 118 Mb of nonredundant sequence. As noted earlier, approximately 11,500 genes have been predicted among finished BAC clones so far. Most Mt BAC clones are anchored to chromosomal locations through the use of microsatellite and other BAC-based markers or by BAC sequence overlap. In this way, 820 of the sequenced BAC clones have been assigned to a specific chromosome and genetic map location.
Information about the Mt genome sequence can be accessed through a variety of Web sites. Because of the project's international and collaborative nature, data production, storage, and visualization tools are broadly distributed. These resources include the primary Mt genome sequence portal, www.medicago.org/genome at the University of Minnesota, as well as related sites at the University of Oklahoma (www.genome.ou.edu), TIGR (www.tigr.org/tdb/e2k1/mta1/), and the Munich Information Center for Protein Sequences (mips.gsf.de/proj/plant/jsf/medi/index.jsp). Along with the finger-print-contig-based physical and genetic map Web site (mtgenome.ucdavis.edu), the Mt genome sites provide query and visualization tools for BAC-based sequence assemblies, marker-BAC associations, BAC-sequence browsers showing tentative gene calls, and FTP downloads of large genome sequence datasets.
GENOME SEQUENCING IN L. JAPONICUS
Lj (2n = 12) is a diploid self-fertile perennial pasture legume. Several mutants in symbiosis and nitrogen fixation have previously been isolated and the underlying genes identified. Insertional mutagenesis and TILLING systems are available (Schauser et al., 1999; Webb et al., 2000; Perry et al., 2003), as are 110,000 ESTs derived from a variety of different organs (Szczyglowski et al., 1997; Endo et al., 2000; Asamizu et al., 2003b). High-density molecular marker maps plus TAC and BAC genomic libraries facilitate gene identification, map-based cloning, and genome sequencing (Hayashi et al., 2001; Sato et al., 2001).
Cytogenetic analysis of Lj distinguished all six chromosomes based on patterns of heterochromatic regions (Ito et al., 2000; Hayashi et al., 2001; Pedrosa et al., 2002). FISH analysis integrated the genetic and cytogenetic maps of Lj with BAC and plasmid clones from 32 genome regions (Pedrosa et al., 2002), a process that continues with new seed clones from the genome sequencing project (N. Ohmido, personal communication). Like Mt, a difference in chromosome morphology between the two accessions used for genetic mapping (Gifu B-129 and Miyakojima MG-20) revealed a reciprocal translocation between chromosomes 1 and 2. FISH analysis pinpointed the borders and map location of this translocation using sequenced seed clones as probes.
Large-scale genome sequencing of Lj began in 2000 using genotype Miyakojima MG-20. Seed points were chosen along the entire genome based on sequences of ESTs, cDNAs, and gene segments from Lj and other legumes, and corresponding TAC clones were selected for sequencing by PCR. TAC clones were sequenced by shotgun and standard finishing methods, and then gene annotation was performed by a combination of semiautomatic and manual methods. Microsatellite and single nucleotide polymorphism markers generated from genome sequence localized TACs onto the genetic linkage map.
As of October 2004, a total of 1,659 clones had been selected for sequencing in Lj and a total of 162 Mb had been sequenced, including clones still in draft (phase 1) stage. In the 44.9 Mb of finished sequence, 4,089 potential protein-encoding genes are predicted (Sato et al., 2001; Nakamura et al., 2002; Asamizu et al., 2003a; Kaneko et al., 2003; Kato et al., 2003). (This estimate increases to 6,500 when Lj genes are predicted by the Mt-trained Fgenesh algorithm described earlier.) Altogether, 1,310 TACs have been placed on the genetic map using 691 microsatellite and 80 cleaved amplified polymorphic sequence markers and by overlaps among sequenced clones. These TAC-based markers and associated sequence information provide enormous value in gene mapping and map-based cloning in Lj and other legumes.
A Web-based database (www.kazusa.or.jp/lotus/) supports easy access to Lotus genome information generated through the sequencing project. One can retrieve information on DNA markers, genetic linkage maps, recombinant inbred lines, nucleotide sequences of the chloroplast and TAC clones, annotation of predicted genes, and results of similarity searches. Legume Base (www.shigen.nig.ac.jp/legume/legumebase/) is a materials resource database for Lj and soybean, supported by the Japan National Bioresource Project. Resources such as seeds, recombinant inbred lines, and TAC genomic libraries can be obtained through this Web site.
REPEAT SEQUENCES OF M. TRUNCATULA AND L. JAPONICUS
An important feature of the Mt and Lj genomes that can be examined with existing sequence data is the diversity and organization of repeat elements. Of course, both sequencing projects have sought to avoid the highly repetitive sequences found in centromeres and pericentromeres, as this is the rationale for the underlying gene-rich BAC/TAC sequencing strategy. Still, a combination of random and clone-by-clone sequencing plus FISH analysis reveals a great deal about the repeat space of these two legume genomes.
To survey the Mt genome for repeat sequences, Roe and colleagues carried out a pilot WGS of 25,000 reads early in the genome sequencing effort (Roe and Kupfer, 2004). In addition to assembling the entire Mt chloroplast genome sequence, the WGS displayed several novel Mt-specific repeat families. Altogether, 23% of nonchloroplast reads were repetitive and 25% of these clustered into groups of 50 or more, strongly suggesting they were high copy. Four centromere-associated, short-tandem repeat families were examined in detailed. Altogether, these repeats were found to comprise nearly 10% of the Mt genome. Three of these high copy repeats, MtR1, MtR2, and MtR3, were subsequently characterized by FISH (Kulikova et al., 2004). MtR3 is found in stretches 450 kb to 1 Mb in length within centromeres, whereas MtR1 and MtR2 occupy distinct and diagnostic regions within pericentromeric heterochromatin.
In a similar fashion, 37,000 random TAC-end sequences from Lj were characterized and clustered by sequence similarity. Approximately 47% of the TAC ends could be clustered, with 25% of this fraction clustering into high copy repeats. Analyzing consensus sequences for each of these groups revealed five different short tandem repeats, two retroelements, and nine unclassified repeats, including a previously characterized centromere-associated repeat, Ljcen1 (GenBank accession no. AF390569; Pedrosa et al., 2002). Many of these repeats demonstrated characteristic patterns of distribution when examined by FISH. For example, LjRE2 was present only in pericentromeric heterochromatin, LjTR1 was found in chromosome knobs, and LjRE1 was found along the entire lengths of chromosome arms (N. Ohmido, personal communication).
While the WGS and random TAC-end approaches enable comparisons of high copy tandem repeats, full-length BAC and TAC sequences provide opportunities to compare intergenic retrotransposons and DNA transposons of Mt and Lj. With this in mind, we carried out a preliminary RepeatMasker (www.repeatmasker.org) analysis of sequenced BACs and TACs to investigate the interspersed repeats in the available genomic sequence. In contrast with the WGS and random TAC-end results described earlier, just 4.7% of the BAC-by-BAC Mt sequence and 5.7% of the TAC-by-TAC Lj sequence could be classified as repetitive in this analysis (though some repeat classes that had previously been observed in Mt and Lj, including SIRE [Laten and Morris, 1993] and MIRE1 [GenBank accession no. AY196987], were not yet represented in the underlying RepBase database [www.girinst.org/]). Since the BAC and TAC clones sequenced so far were chosen because they were gene rich, this low percentage is not surprising. Indeed, just four sequenced Mt BACs and none of the Lj TACs contain centromeric repeats, and one of these Mt BACs had been chosen expressly so that the centromeric element MtR1 could be examined in detail. Based on this preliminary analysis, the Mt and Lj genespaces appear to be quite similar in their retrotransposon and DNA transposon composition. Both contain large numbers of the same family of LINEs (L1/CIN4), the same families of retrotransposons (Ty1/Copia and Gypsy), as well as similar families of DNA transposons. It is also interesting that the distribution of different repeat families is nonuniform among sequenced clones in both Mt and Lj. For example, LjRE1, a Ty1/Copia element, is found on 27% of sequenced Lj TACs, whereas LjRE2, a Gypsy element, is found on just 0.5% of clones, even though LjRE1 is only 4 times more abundant than LjRE2.
COMPARATIVE GENOMICS WITH OTHER LEGUMES
The most compelling rationale for sequencing genomes of model plant species is the opportunity to extend this information to important crops. A growing number of studies demonstrate macro- and microsynteny among legumes (Menancio-Hautea et al., 1993; Boutin et al., 1995; Simon and Muehlbauer, 1997; Lee et al., 2001; Brauner et al., 2002; Gualtieri et al., 2002; Cannon et al., 2003; Yan et al., 2003, 2004; Choi et al., 2004a, 2004b; Kalo et al., 2004). In a recent study that spanned multiple legume species, macrosynteny between Mt, Lj, and four other legume species was examined in detail (Choi et al., 2004b). The results indicate extensive genome-wide synteny between Mt and Galegoid legumes (such as alfalfa, pea [Pisum sativum], chickpea [Cicer arietinum], and Lj). The high level of macrosynteny between Mt and pea is notable, as the pea genome is roughly 10 times larger than Mt. Long tracts of macrosynteny were also observed between Mt and the Phaseoleae species soybean and mungbean (Vigna radiata), though at levels lower than with Galegoid legumes. These comparisons enabled the construction of a genome-wide picture of legume synteny in the form of concentric circles of corresponding chromosomes anchored by Mt (Choi et al., 2004b), similar to the model previously developed for rice and other grasses (Gale and Devos, 1998).
These macrosynteny results complement a growing number of microsynteny studies that describe similarities at the scale of individual BAC clones or clone contigs between legume genomes. Of course, microsynteny between Mt or Lj and crop species like alfalfa and pea has already enabled the positional cloning of symbiosis genes (Endre et al., 2002; Stracke et al., 2002). However, microsynteny with Mt also extends to the more evolutionarily distant soybean. Specifically, two sequenced regions of soybean totaling 1 Mb in length have been compared to the Mt genome sequence, and the extent of colinearity is impressive (Choi et al., 2004b; J. Mudge, personal communication). Altogether, the syntenic regions comprised nearly 500 predicted genes, with 75% of soybean genes colinear with their Mt homologs, including one segment where 33 of 35 soybean genes with hits to the GenBank nonredundant database were colinear with Mt. In fact, microsynteny between Mt and soybean appears to be widespread throughout the genomes. One study compared 50 pairs of Mt and soybean BAC contigs, comprising nearly 10 Mb in each species, and found that 35% of contigs compared through cross-hybridization and physical mapping exhibited substantial microsynteny, with another 20% exhibiting more limited levels of microsynteny (Yan et al., 2003).
INTEGRATING THE M. TRUNCATULA AND L. JAPONICUS GENOMES
With the growing body of genome sequence for both Mt and Lj, it is clear that detailed comparisons between these two genomes (and also with Arabidopsis and poplar) will reveal exciting new aspects of plant genome organization and evolution. More importantly, detailed comparisons between Mt and Lj will provide a foundation for researchers in other systems to mine these model genomes in a systematic and integrated fashion.
Marker-based comparisons between Mt and Lj have already demonstrated substantial macrosynteny (Choi et al., 2004b). This macrosynteny is punctuated by multiple rearrangements involving translocation of chromosomes arms, as the two species have different chromosome numbers (Mt with eight; Lj with six). Individual BAC- and TAC-based comparisons between Mt and Lj also revealed substantial levels of microsynteny (Cannon et al., 2003; Choi et al., 2004b). For example, 10 BAC/TAC clone pairs broadly spaced throughout the two genomes were compared at the sequence level (Choi et al., 2004b). Within these regions, 91 and 84 genes were identified in Mt and Lj, respectively, and 82% of the Mt genes were conserved between the genomes. With just four exceptions, homologs were present in conserved order and orientation.
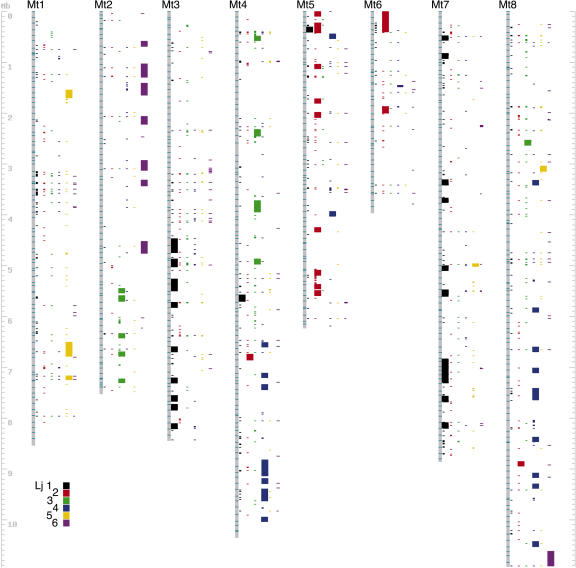
When all currently available phase 2 and 3 Mt and Lj genome sequences are compared, striking large-scale similarities become apparent (Fig. 1). These results significantly expand the scope of earlier comparative mapping studies, where macrosynteny was based on segregation analysis of conserved DNA markers and microsynteny was examined one BAC or TAC clone at a time. Since long stretches of anchored genome sequences can now be compared directly, microsynteny can be integrated into the larger picture of macrosynteny, and commonalities in genome organization can be inferred genome wide. This is illustrated in Figure 1, where 71 Mb of anchored Mt sequence is compared to 34 Mb of anchored Lj sequence. In the figure, all top hits of the Lj genome to Mt are shown. When four or more Lj top hits are colinear on the same Mt BAC, a wide colored block running the length of the Mt BAC is shown, with each Lj chromosome assigned a different color. For example, the bottom of Mt chromosome 3 has 11 clustered BACs, each with blocks of 4 or more colinear homologs on Lj chromosome 1. Altogether, 101 Mt BACs spanning approximately 10 Mb were microsyntenic with a comparable portion of the Lj genome in this analysis.
Figure 1.
Sequence-based synteny between Mt and Lj. Provisional assemblies for all eight of Mt's chromosomes are displayed along side regions of sequence homology in Lj. Mt chromosome assemblies were constructed from sequenced phase 2 and 3 BACs, with order inferred from a combination of genetic map anchors, finger print contigs, and overlapping BAC sequence data. For each BAC along the assembly, the top reciprocal BLAST hits to predicted Mt genes are shown as colored bars for each of the six Lj chromosomes. Broader bars indicate regions of predicted synteny, corresponding to four or more genes with conserved order and orientation in both genomes. The heights of broader bars also indicate the length along Mt chromosome assemblies where synteny is observed.
The many genome segments that fail to exhibit conservation in this study might represent nonsyntenic regions. However, segments that appear to lack synteny are more likely to be cases where corresponding genome regions have not yet been sequenced in one or the other genome sequencing project. Even with all the sequencing that has been accomplished so far, finished and anchored BACs cover just 28% of the Mt genespace, whereas finished and anchored TACs cover just 13% of Lj. For this reason, sequence-based comparisons would be expected to discover just 4% of all potential overlap at this level of genome coverage, assuming unbiased distribution of sequences across the two genespaces.
Even with the relatively limited genome sequence available, it is clear that Mt chromosome 1 shows modest synteny with Lj chromosome 5 (gold); Mt-2 is largely syntenic with Lj chromosomes 3 and 6 (green and purple, respectively); Mt-3 with Lj-1; Mt-4 with Lj-3 and Lj-4; Mt-5 with Lj-2; Mt-6 with Lj-2; Mt-7 with Lj-1; and Mt-8 with Lj-4. Mt chromosome 6, which exhibits the lowest synteny with Lj, is unusual in its high proportion of heterochromatin (Kulikova et al., 2004), lack of marker-based synteny with pea (Choi et al., 2004b), and abundance of nucleotide-binding site-Leu-rich repeat genes (Zhu et al., 2002). If all putative cases of synteny are considered (observed syntenic blocks plus nonsyntenic regions flanked on both sides by syntenic blocks), then more than 75% of the Mt and Lj genespaces are conserved. Not surprisingly, these genome sequence-based relationships mirror and extend marker-based synteny predictions made previously (Choi et al., 2004b).
PERSPECTIVES
In the future, genome-scale comparisons will become increasingly informative as genome sequencing of Mt and Lj nears completion. These comparisons will reveal the detailed processes that shaped the evolution of these two legume genomes and provide increasingly detailed insights into plant genome evolution and organization. Moreover, by viewing genome information from Mt and Lj in an integrated manner, researchers working in other species will find a much richer resource than would have been available with just one. Genome sequencing in other legumes will be better informed by the genomes of these two model legumes, especially the construction of sequence assemblies and scaffolds. Even now, efforts to discover important regulatory elements and novel legume genes, as well as positionally clone mutants and quantitative trait loci, can take advantage of the powerful combination of model genomes provided by Mt and Lj.
Acknowledgments
We thank the outstanding efforts of the many scientists involved in the Mt and Lj genome sequencing initiatives. Complete lists of these individuals can be found at www.medicago.org/genome/people.php and at www.kazusa.or.jp/lotus/people/.
The U.S. component of the Medicago truncatula sequencing effort was initially supported by a grant from the Samuel Roberts Noble Foundation to B.A.R. Current support comes from National Science Foundation Plant Genome Research Program (grant no. 0110206 to D.R.C., D.K., C.D.T., and N.D.Y., and grant no. 0321460 to N.D.Y., B.A.R., and C.D.T.). Funding for Lotus japonicus sequencing comes from the Kazusa DNA Research Institute Foundation.
References
- Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Kato T, Sato S, Nakamura Y, Kaneko T, Tabata S (2003. a) Structural analysis of a Lotus japonicus genome. IV. Sequence features and mapping of seventy-three TAC clones which cover the 7.5 Mb regions of the genome. DNA Res 10: 115–122 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2003. b) Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol Biol 54: 405–414 [DOI] [PubMed] [Google Scholar]
- Boutin SR, Young ND, Olson T, Yu Z-H, Shoemaker R, Vallejos C (1995) Genome conservation among three legume genera detected with DNA markers. Genome 38: 928–937 [DOI] [PubMed] [Google Scholar]
- Brauner S, Murphy RL, Walling JG, Przyborowski J, Weeden NF (2002) STS markers for comparative mapping in legumes. J Am Soc Hortic Sci 127: 616–622 [Google Scholar]
- Cannon SB, McCombie WR, Sato S, Tabata S, Denny R, Palmer L, Katari M, Young ND, Stacey G (2003) Evolution and microsynteny of the apyrase gene family in three legume genomes. Mol Genet Genomics 270: 347–361 [DOI] [PubMed] [Google Scholar]
- Choi HK, Kim D, Uhm T, Limpens E, Lim H, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, et al (2004. a) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166: 1463–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis THN, Doyle J, Kiss GB, et al (2004. b) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101: 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34: 95–106 [DOI] [PubMed] [Google Scholar]
- Endo M, Kokubun T, Takahata Y, Higashitani A, Tabata S, Watanabe M (2000) Analysis of expressed sequence tags of flower buds in Lotus japonicus. DNA Res 7: 213–216 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualtieri G, Kulikova O, Limpens E, Kim DJ, Cook DR, Bisseling T, Geurts R (2002) Microsynteny between pea and Medicago truncatula in the SYM2 region. Plant Mol Biol 50: 225–235 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Miyahara A, Sato S, Kato T, Yoshikawa M, Taketa M, Hayashi M, Pedrosa A, Onda R, Imaizumi-Anraku H, et al (2001) Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res 8: 301–310 [DOI] [PubMed] [Google Scholar]
- Ito M, Miyamoto J, Mori Y, Fujimoto S, Uchiumi T, Abe M, Suzuki A, Tabata S, Fukui K (2000) Genome and chromosome dimensions of Lotus japonicus. J Plant Res 113: 435–442 [Google Scholar]
- Kalo P, Seres A, Taylor SA, Jakab J, Kevei Z, Kereszt A, Endre G, Ellis TH, Kiss GB (2004) Comparative mapping between Medicago sativa and Pisum sativum. Mol Genet Genomics 272: 235–246 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Asamizu E, Kato T, Sato S, Nakamura Y, Tabata S (2003) Structural analysis of a Lotus japonicus genome. III. Sequence features and mapping of sixty-two TAC clones which cover the 6.7 Mb regions of the genome. DNA Res 10: 27–33 [DOI] [PubMed] [Google Scholar]
- Kapranov P, de Bruijn FJ, Szczyglowski K (1997) Novel, highly expressed late nodulin gene (LjNOD16) from Lotus japonicus. Plant Physiol 113: 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (2003) Structural analysis of a Lotus japonicus genome. V. Sequence features and mapping of sixty-four TAC clones which cover the 6.4 Mb regions of the genome. DNA Res 10: 277–285 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Kulikova O, Geurts R, Lamine M, Kim DJ, Cook DR, Leunissen J, de Jong H, Roe BA, Bisseling T (2004) Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. Chromosoma 113: 276–283 [DOI] [PubMed] [Google Scholar]
- Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook D, Huguet T, de Jong JH, Fransz PF, Bisseling T (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27: 49–58 [DOI] [PubMed] [Google Scholar]
- Laten HM, Morris RO (1993) SIRE-1, a long interspersed repetitive DNA element from soybean with weak sequence similarity to retrotransposons: initial characterization and partial sequence. Gene 134: 153–159 [DOI] [PubMed] [Google Scholar]
- Lee JM, Grant D, Vallejos CE, Shoemaker RC (2001) Genome organization in dicots. II. Arabidopsis as a ‘bridging species’ to resolve genome evolution events among legumes. Theor Appl Genet 103: 765–773 [Google Scholar]
- Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Limpens E, Javier R, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96: 6535–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Menancio-Hautea D, Fatokun CA, Kumar L, Danesh D, Young ND (1993) Comparative genome analysis of mungbean (Vigna radiata (L.) Wilczek) and cowpea (V. unguiculata (L.) Walpers) using RFLP mapping data. Theor Appl Genet 86: 797–810 [DOI] [PubMed] [Google Scholar]
- Mudge J, Huihuang Y, Denny RL, Howe DK, Danesh D, Marek LF, Retzel E, Shoemaker RC, Young ND (2004) Soybean BAC contigs anchored with RFLPs: insights into genome duplication and gene clustering. Genome 47: 361–372 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Asamizu E, Kato T, Sato S, Tabata S (2002) Structural analysis of a Lotus japonicus genome. II. Sequence features and mapping of sixty-five TAC clones which cover the 6.5-Mb regions of the genome. DNA Res 9: 63–70 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429 [DOI] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy A, Balija V, Nascimento L, Dike S, de la Bastide M, Martienssen RA, McCombie WR (2003) Maize genome sequencing by methylation filtration. Science 302: 2115–2117 [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161: 1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M (2003) A TILLING reverse genetics tool and a web accessible collection of mutants of the legume Lotus japonicus. Plant Physiol 131: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon M, Journet EP, Dedieu A, de Billy F, Truchet G, Barker DG (1992) Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe BA, Kupfer DM (2004) Sequencing gene rich regions of Medicago truncatula, a model legume. In A Hopkins, ZY Yang, R Mian, M Sledge, RE Barker, eds, Molecular Breeding of Forage and Turf. Kluwer Academic Publishers, Amsterdam, pp 333–344
- Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Sato S, Kaneko T, Nakamura Y, Asamizu E, Kato T, Tabata S (2001) Structural analysis of a Lotus japonicus genome. I. Sequence features and mapping of fifty-six TAC clones which cover the 5.4 Mb regions of the genome. DNA Res 8: 311–318 [DOI] [PubMed] [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259: 414–423 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Scholte M, d'Erfurth I, Rippa S, Mondy S, Jean V, Durand P, Breda C, Trinh H, Rodriguez-Llorente I, Kondorosi E, et al (2002) T-DNA tagging in the model legume Medicago truncatula allows efficient gene discovery. Mol Breed 10: 203–215 [Google Scholar]
- Simon CJ, Muehlbauer FJ (1997) Construction of a chickpea linkage map and its comparison with maps of pea and lentil. J Hered 88: 115–119 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 27: 959–962 [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Hamburger D, Kapranov P, de Bruijn FJ (1997) Construction of a Lotus japonicus late nodulin expressed sequence tag library and identification of novel nodule-specific genes. Plant Physiol 114: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapiller M (1998) Shotgun sequencing of the human genome. Science 280: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Webb KJ, Skot L, Nicholson MN, Jorgensen B, Mizen S (2000) Mesorhizobium loti increases root-specific expression of a calcium-binding protein homologue identified by promoter tagging in Lotus japonicus. Mol Plant Microbe Interact 13: 606–616 [DOI] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120 [DOI] [PubMed] [Google Scholar]
- Yan H, Mudge J, Kim DJ, Shoemaker RC, Cook DR, Young ND (2003) Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor Appl Genet 106: 1256–1265 [DOI] [PubMed] [Google Scholar]
- Yan H, Mudge J, Kim DJ, Shoemaker RC, Cook DR, Young ND (2004) Comparative physical mapping reveals features of microsynteny between the genomes of Glycine max and Medicago truncatula. Genome 47: 141–155 [DOI] [PubMed] [Google Scholar]
- Zhu H, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15: 529–539 [DOI] [PubMed] [Google Scholar]
- Zhu H, Kim DJ, Baek JM, Choi HK, Ellis LC, Kuester H, McCombie WR, Peng HM, Cook DR (2003) Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol 131: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]