Terrestrial autotrophs such as higher plants are confronted with challenges largely unknown to their counterparts in the seas, including extreme patchiness of inorganic nutrients. Evolution of higher plants has yielded interesting and important solutions to the problem of nutrient acquisition on land. Some of the most intriguing of these involve mutually beneficial symbioses. In fact, evolution of a fungal-plant symbiosis around 450 million years ago may have been the key innovation that enabled plants to colonize the land (Pirozynski and Malloch, 1975; Remy et al., 1994). Related mycorrhizal associations continue to exist for more than 90% of land plants, a reflection of their ancient origin and importance. Mycorrhizal associations enhance the ability of plants to scavenge nutrients such as phosphate from the soil, by virtue of the greater volume of soil exploited by the filamentous fungal partner (Smith and Read, 1997; Smith et al., 2003). In return for its services, the fungus is provided with a carbon source, derived from plant photosynthesis, for biosynthesis and energy metabolism. To avoid exploitation, however, plants have evolved a series of checkpoints that help to discern friend from foe, which presumably switch off defense responses against robbers when a friend comes to visit.
A different kind of beneficial plant-microbe interaction that provides a more restricted range of plants with the often-limiting macronutrient nitrogen is symbiotic nitrogen fixation (SNF). This type of symbiosis evolved more recently, some 60 million years ago, and is confined to legumes and a few nonlegumes (Doyle, 1998), which form intracellular symbioses with rhizobia or other nitrogen-fixing bacteria, respectively (Pawlowski and Bisseling, 1996). Once again, the plant provides its beneficial endosymbiont with photosynthate, together with other nutrients, in exchange for valuable fixed nitrogen, in the form of ammonium and amino acids (Udvardi and Day, 1997). Interestingly, some of the signaling pathways that mediate peace between legumes and mycorrhizal fungi also function in the rhizobial symbiosis, and stunning progress has been made recently in identifying a few of the genes involved. Significant progress has also been made in identifying changes in transport and metabolism during symbiosis development, which will contribute to a better understanding of the nature of trade between legumes and their microsymbionts. These breakthroughs largely stem from focused research on two model legumes, Lotus japonicus and Medicago truncatula, and the conjunction of genetics, genomics, and functional genomics. This Update focuses on recent discoveries in the areas of legume-microbe communication and trade, two essential aspects of stable mutualism.
UNDERGROUND PEACE TALKS: RECENT ADVANCES IN LEGUME MICROBE SIGNALING
Symbiotic interactions involve molecular communication between the host plant and its microbial symbiont in the rhizosphere. The legume symbioses, SNF and arbuscular mycorrhizae (AM), share common features in early signaling. Discoveries in the signaling pathways that underpin these symbioses are among the most exciting recent advances in legume research. It is becoming clear that calcium plays a crucial role in the symbiotic signaling and may be a common feature of legume/symbiont peace talks.
NOD FACTOR SIGNALING
The invasion of the bacteria into the plant root occurs through an invagination of the plant cell (termed the infection thread) that initiates at the primary site of the interaction, the root hair cell, but spans the entire root cortex, allowing bacterial invasion into the dividing cells of the nodule primordium. Bacteria are released from the infection thread into membrane bound compartments, where they differentiate into bacteroids. Nod factors, or lipo-chito-oligosaccharide signaling molecules, are central to the initial establishment of the legume-rhizobial symbiosis (Dénarié et al., 1996; Long, 1996; Oldroyd, 2001). Production of this signaling molecule is activated by the release of plant phenolic signals, predominantly flavonoids, into the rhizosphere, where they activate Nod factor production through induction of a set of nod genes in the appropriate rhizobial strain. The nature of both the flavonoid signal and the structure of Nod factor are central to the maintenance of specificity in this interaction, ensuring that the plant only accommodates a friendly rhizobial strain. Nod factors are critical both at the early stages of the interaction and during infection thread development, and may play a role during bacterial release (Ardourel et al., 1994; Downie and Walker, 1999). It is clear that understanding the plant's perception of this signaling molecule is key to understanding this symbiosis. Since the work of Ehrhardt nearly a decade ago (Ehrhardt et al., 1996), it is becoming more apparent that calcium is an important component of Nod factor signaling, and such a calcium-centric viewpoint has been validated by more recent work.
A number of studies using calcium dyes and ion-selective electrodes have indicated significant Nod factor-induced calcium changes in root hair cells (Cardenas et al., 2000). This work can be summarized into two main calcium events: an initial calcium flux that occurs at the tip of the root hair and repetitive cytosolic oscillations of calcium, termed calcium spiking, in the region surrounding the nucleus (Oldroyd and Downie, 2004). These two calcium responses are separated both spatially and temporally, but can also be separated by Nod factor concentrations: 10−12 to 10−9 m Nod factor activates spiking without inducing the flux (Shaw and Long, 2003).
Calcium is a ubiquitous secondary messenger in diverse organisms and can affect a wide variety of cellular events. The Nod factor-induced calcium changes are simply observations and provide little insight into the actual roles that calcium plays during this symbiosis. In animal systems, calcium spiking has been shown to regulate gene expression in response to a signaling molecule (Dolmetsch et al., 1998; Li et al., 1998). A similar signaling role for calcium spiking in Nod factor signal transduction is supported by the fact that a number of genes essential for Nod factor signaling are also required for activation of calcium spiking (Wais et al., 2000; Walker et al., 2000b; Oldroyd and Downie, 2004; Fig. 1).
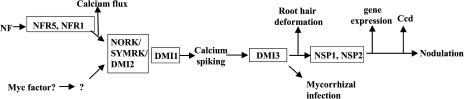
Figure 1.
The Nod factor and mycorrhizal signaling pathways. This signaling pathway has been defined through genetics in the model legumes M. truncatula and L. japonicus, and Medicago sativa. The genes identified are defined in boxes. Mutations in all these genes have been characterized for calcium spiking except SYMRK. In addition, it has been shown that mutations in NFR5 and NFR1 lack the calcium flux, whereas mutations in DMI1 and DMI2 show the first phase of the flux response. Components of the Nod factor (NF) signaling pathway are conserved with mycorrhizal signaling. We presume that genes specific to mycorrhizae must exist at equivalent positions to NFR1 and NFR5 and possibly NSP1 and NSP2.
The molecular identity of the gene products that link Nod factor perception with induction of calcium spiking (Fig. 1) is highly informative of the regulation of calcium signaling in plants. The Nod factor receptor is most likely a heterodimer of two classes of receptor-like kinases that contain LysM domains in the extracellular region (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). LysM domains are present in a number of proteins and have been shown to bind polysaccharides, particularly glucosamine chains, which form the backbone of the Nod factor molecule. Mutations in these LysM receptor-like kinases abolish all Nod factor-induced responses, including calcium spiking and the calcium flux, which supports a role for these proteins at a very early stage of the Nod factor signal transduction pathway (Amor et al., 2003; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003).
Functioning downstream of these LysM receptor-like kinases are DMI1, a putative cation channel, and NORK/SYMRK/DMI2, another receptor-like kinase with Leu-rich repeat domains in the extracellular portion (Endre et al., 2002; Stracke et al., 2002; Ane et al., 2004). Mutations in both of these proteins abolish Nod factor-induced calcium spiking and restrict the calcium flux response to a single rapid calcium increase, rather than a calcium increase that is maintained for a number of minutes as is seen in wild-type plants (Wais et al., 2000; Shaw and Long, 2003). The presence of two receptor-like kinases that most likely make up the Nod factor receptor and a second receptor-like kinase functioning downstream implicates a phosphorylation cascade early in Nod factor signaling, and defining the targets of these kinases is crucial for linking Nod factor perception with the activation of the downstream calcium responses. The putative cation channel DMI1 may have a direct role in calcium spiking, in that it could function as a calcium channel during both the calcium flux and calcium spiking. However, two proteins homologous to DMI1, CASTOR and POLLUX, have recently been identified in L. japonicus that are required for Nod factor signaling and show plastid localization (Imaizumi-Anraku et al., 2005). The plastid is an unlikely internal calcium store for calcium spiking, and, therefore, this localization suggests that this class of putative cation channels is not the calcium channel involved in calcium spiking. Defining the ions transported by these channels is crucial for assessing their role in the Nod factor signaling pathway.
DMI3 of M. truncatula is essential for Nod factor signaling, and mutations in dmi3 are phenotypically identical to dmi1and dmi2 except for the fact that dmi3 mutants can activate calcium spiking (Catoira et al., 2000; Wais et al., 2000). This indicates that DMI3 functions downstream of calcium spiking and is a strong candidate for a protein able to perceive and transduce the calcium spiking signal. This hypothesis is validated by the identity of DMI3: a chimeric calcium/calmodulin-dependent protein kinase (CCaMK; Levy et al., 2004; Mitra et al., 2004a). Clearly, such a calcium activatable kinase has the hallmarks of a protein able to decode the calcium spiking signal. If this is indeed the function of DMI3, the absolute requirement for this activity in Nod factor signaling highlights the central role that calcium spiking plays in transducing the Nod factor signal.
EARLY SIGNALING IN THE AM SYMBIOSIS
The symbiotic organelle of the AM symbiosis, the arbuscule, forms within the root cortex. Entry into the root is achieved through fungal appressoria that develop on the plant epidermal cell surface. While this is the first visible stage of the interaction, there is evidence that the plant and fungus communicate prior to physical contact. AM fungi respond to signals in plant root exudates by altering their respiratory activity and hyphal morphology, while a diffusible signal from the fungus elicits alterations in plant gene expression (Nagahashi and Douds, 1997; Buee et al., 2000; Kosuta et al., 2003; Tamasloukht et al., 2003). The nature of the signaling molecules is not known.
The signaling pathways involved in triggering the cellular events required for development of the association are beginning to be revealed. The DMI genes of M. truncatula and their reciprocal proteins in L. japonicus and pea are not only required for nodulation, but also the early development of the mycorrhizal association (Fig. 1). Mutations in all these genes fail to allow entry of the fungus into the cortex (Sagan et al., 1995; Wegel et al., 1998). This implicates the NORK/SYMRK/DMI2/SYM19 Leu-rich repeat receptor kinase, the DMI1 channel protein, and the DMI3 CCaMK in early mycorrhizal signaling (Endre et al., 2002; Stracke et al., 2002; Ane et al., 2004; Levy et al., 2004; Mitra et al., 2004a). Since the AM symbiosis is the older of the two associations, the legume/rhizobial symbiosis probably co-opted part of a signaling pathway that had been established initially for development of the AM symbiosis. The fact that proteins involved in the induction and perception of calcium spiking in Nod factor signaling are also involved in mycorrhizal signaling implicates calcium as a secondary messenger in the mycorrhizal pathway. Nod factor signaling contains nodulation-specific proteins both upstream and downstream of the conserved pathway, and, similarly, one would expect mycorrhiza-specific signaling proteins. Genetic screens for these mycorrhiza-specific mutants are currently under way in a number of laboratories.
Recently, transcriptional profiling has identified genes whose expression is differentially regulated in M. truncatula in response to appressoria formation and the early stages of development of the symbiosis (Liu et al., 2003; Brechenmacher et al., 2004). Defense gene transcripts are among those that show a transient increase in the early stages of the AM symbiosis, followed by a decrease coincident with proliferation of the fungus within the roots. While similar defense gene expression patterns had been noted earlier, the genome-scale analyses have identified coregulated signal transduction proteins that may be involved in regulating defense responses in the symbiosis (Liu et al., 2003). It would appear that the plant initially reacts in a defensive manner, but, following communications with the fungus, peace ensues and the plant reduces its defenses.
SPECIALIZATION AND TRADE: TWO KEYS TO MUTUALISM
Effective and sustained communication is necessary for any long-term relationship, but it is not sufficient. The evolutionary success of legume-rhizobia and mycorrhizal associations derives from the trade of goods (metabolites) of value to each. The division of labor that underpins such exchanges is achieved by metabolic specialization/differentiation in each symbiont during development of the symbiosis. The following sections describe recent advances in our understanding of these processes.
DIFFERENTIATION OF LEGUMES AND RHIZOBIA DURING SNF
Development of functional nodules requires differentiation of both plant and bacterial cells, the latter being converted to a distinct nitrogen-fixing form called the bacteroid. Transcriptomics and proteomics are beginning to reveal the true extent of plant and bacterial differentiation during nodule development. Hundreds of novel plant genes that are either induced or repressed during nodule development have now been identified using cDNA arrays (Colebatch et al., 2002, 2004; El Yahyaoui et al., 2004; Kouchi et al., 2004; Kuster et al., 2004; Lee et al., 2004), oligonucleotide microarrays (Mitra et al., 2004b), and bioinformatic approaches (Fedorova et al., 2002; Journet et al., 2002). Many of these are involved in metabolism and transport (Colebatch et al., 2004; El Yahyaoui et al., 2004; Kouchi et al., 2004). Coordinate up-regulation of plant genes involved in glycolysis, carbon fixation, and amino acid biosynthesis highlight the importance of these processes in carbon supply for bacteroid nitrogen fixation and plant ammonium assimilation. While compatible solutes may not be a currency of trade between legumes and rhizobia, induction of plant genes involved in polyamine, polyol, and Pro synthesis indicate that nodule cells may have to work overtime for osmotic homeostasis (Colebatch et al., 2004). Of more interest from the point of view of trade between host and microsymbionts is the growing list of nodule-induced plant genes encoding transporters (Colebatch et al., 2004; El Yahyaoui et al., 2004; Kouchi et al., 2004), some of which appear to be located on the symbiosome membrane, based on proteomics data (Saalbach et al., 2002; Wienkoop and Saalbach, 2003; Catalano et al., 2004). The symbiosome membrane is the specialized plant membrane that separates bacteroids from the host cell cytoplasm and controls the traffic of nutrients between the two. Among the most interesting of the nodule-induced transporters identified by transcriptomics are homologs of AgDCAT1, a dicarboxylate transporter of the nonlegume Alnus, which probably delivers carbon substrates to nitrogen-fixing Frankia in Alnus nodules (Jeong et al., 2004). The legume counterparts of AgDCAT1 may play a crucial role in SNF, as dicarboxylic acids are believed to be the primary source of carbon for bacteroid metabolism. Other nodule-induced transporters include putative amino acid transporters, which may provide a missing link in intriguing models of amino acid cycling between legumes and rhizobia (Lodwig and Poole, 2003).
Transcriptome analysis is also making important in-roads in rhizobium biology. DNA arrays containing essentially all the genes of Mesorhizobium loti (Uchiumi et al., 2004) and Sinorhizobium meliloti (Becker et al., 2004) have now been produced and used to obtain the first global view of gene expression in symbiotic rhizobia. Nodule development is accompanied by declining levels of free oxygen, which is a prerequisite for activity of oxygen-labile nitrogenase in rhizobia. Oxygen levels control the expression of many genes in rhizobia (Batut and Boistard, 1994), and low oxygen is believed to be an important trigger for differentiation of rhizobia into bacteroids in nodules. Transcript profiling of oxygen-limited free-living bacteria indicated that up to 5% of S. meliloti genes may be oxygen regulated (Becker et al., 2004). However, microoxic and bacteroid transcriptomes overlapped only partially, indicating that low oxygen in nodules can account for only a fraction of the changes in gene expression observed during symbiotic development. Clearly, other physiological or biochemical factors in nodules are important for bacteroid differentiation, and it will be interesting to learn what these are in the future.
Proteomic analysis has also contributed to knowledge about bacterial and plant cell differentiation during nodule development. The most comprehensive work has been done on free-living and symbiotic forms of S. meliloti isolated from Melilotus alba or M. truncatula (Natera et al., 2000; Djordjevic et al., 2003, 2004). Of 170 bacteroid proteins identified, 27 appear to be symbiosis specific, including nif and fix gene products involved directly or indirectly in nitrogen fixation. Also in this list are a raft of transporters that are presumably involved in nutrient transfer between host and microsymbiont (Djordjevic et al., 2003, 2004). Among the proteins that were reduced or absent in bacteroids compared to cultured cells were several involved in nitrogen regulation and nitrogen assimilation, which is consistent with past observations that ammonium assimilation is repressed in bacteroids. Not all of the differences in the proteomes of free-living and bacteroid forms of S. meliloti are mirrored by corresponding changes in gene transcript levels (Becker et al., 2004). Such discrepancies may reflect different levels of regulation (i.e. transcriptional versus posttranscriptional regulation), which is an interesting area for future research.
DIFFERENTIATION DURING ARBUSCULE FORMATION
Unlike the rhizobium-legume symbiosis, the AM symbiosis does not culminate in the formation of a new plant organ. Instead, there are major rearrangements within the root cortex where terminally differentiated hyphae, termed arbuscules, form extensive dichotomous branching within cortical cells, enveloped within the plant derived periarbuscular membrane (Bonfante-Fasolo, 1984). Transcriptional profiling has allowed the identification of several novel plant genes whose expression is activated coincident with arbuscule development (Liu et al., 2003; Wulf et al., 2003). Further spatial expression analyses revealed at least two distinct gene expression patterns: genes whose expression occurs only in cells with arbuscules and genes whose expression is activated more broadly throughout the cortex in both colonized and noncolonized cells. In addition to identifying candidate genes implicated in development of the biotrophic interface, these analyses point to the existence of both cell autonomous and systemic signaling pathways operating in the AM symbiosis. Current information about the genomes of AM fungi is limited, but analyses of one species, Glomus intraradices, suggest a genome size of 15 Mb (Hijiri and Sanders, 2004). Genome sequencing is in progress and promises to provide the first insights into the genome of a broad-host-range, obligate symbiont.
Recent physiological and molecular data suggest that when plants form an AM symbiosis, they alter their phosphate acquisition pathways significantly. Phosphate transporters operating at the root-soil interface are down-regulated, and the plant relies largely on phosphate delivered by the fungal symbiont (Liu et al., 1998; Chiou et al., 2001; Smith et al., 2003). In mycorrhizal roots, phosphate is acquired by the extraradical fungal hyphae and is then transferred to the arbuscules, where it is released from the fungus and transported across the periarbuscular membrane into the cortical cell. In the past few years, there has been progress in understanding the molecular basis of phosphate transport in the symbiosis, and, most recently, plant phosphate transporters implicated in the uptake of phosphate released from the arbuscule have been reported (Rausch et al., 2001; Harrison et al., 2002; Paszkowski et al., 2002). In potato (Solanum tuberosum), expression of a high-affinity phosphate transporter, StPT3, is induced in mycorrhizal roots, particularly in cells containing arbuscules, making it a strong candidate for involvement in symbiotic phosphate transport (Rausch et al., 2001). Bioinformatic analyses of M. truncatula expressed sequence tag collections and the complete rice (Oryza sativa) genome enabled the identification of the mycorrhiza-specific phosphate transporters MtPT4 and OsPT11. These two transporters share a high level of sequence identity but are less closely related to StPT3 (Harrison et al., 2002; Paszkowski et al., 2002). They also differ from StPT3 in that they are expressed exclusively in cells containing arbuscules, with the protein located on the periarbuscular membrane (Harrison et al., 2002). Finally, in contrast to StPT3, MtPT4 mediates low-affinity phosphate transport in yeast (Harrison et al., 2002). The relative contribution of these transporters to phosphate transport across the periarbuscular membrane remains to be determined. In the meantime, the differences between the phosphate transporters identified in these different plant species are intriguing, particularly because the phosphate transporters operating in nonmycorrhizal roots are relatively well conserved across species. Have these plant species evolved different styles of phosphate transporters to mediate symbiotic phosphate transport, or are there additional transporters, as yet unidentified, in each species?
SUMMARY
The development of model legume systems has revolutionized our understanding of plant symbioses. These models have provided the genetic and genomic platforms essential for dissecting the biological components that underpin these fascinating interactions. The recent isolation of a number of genes essential for both rhizobial and mycorrhizal signal perception provides significant insights into symbiotic signaling pathways and the communication that is required to establish peaceful relationships between these organisms. However, gene isolation is just the beginning, and defining the function of these proteins, in particular how they link Nod factor perception with activation and recognition of calcium signals, is a major challenge for the future. Although impressive in terms of the amount of data that has been generated, the output of the various “omics” technologies has so far been largely descriptive. Ultimately, we must work toward a better understanding of the genes/proteins and processes that are central to both SNF and AM and the integration of data from molecular, cellular, and physiological levels into seamless models of the whole system.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut J, Boistard P (1994) Oxygen control in Rhizobium. Antonie Van Leeuwenhoek 66: 129–150 [DOI] [PubMed] [Google Scholar]
- Becker A, Berges H, Krol E, Bruand C, Ruberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, et al (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact 17: 292–303 [DOI] [PubMed] [Google Scholar]
- Bonfante-Fasolo P (1984) Anatomy and morphology of VA mycorrhizae. In DJ Bagyaraj, ed, VA Mycorrhizae. CRC Press, Boca Raton, FL, pp 5–33
- Brechenmacher L, Weidmann S, van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V (2004) Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14: 253–262 [DOI] [PubMed] [Google Scholar]
- Buee M, Rossignol M, Jauneau A, Ranjeva R, Becard G (2000) The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol Plant Microbe Interact 13: 693–698 [DOI] [PubMed] [Google Scholar]
- Cardenas L, Holdawa-Clarke TL, Sanchez F, Quinto C, Feijo JA, Kunkel JG, Hepler PK (2000) Ion changes in legume root hairs responding to Nod factors. Plant Physiol 123: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano CM, Lane WS, Sherrier DJ (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25: 519–531 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet E, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Liu H, Harrison MJ (2001) The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J 25: 1–15 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15: 411–420 [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debelle F, Prome J-C (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA (2004) Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4: 1859–1872 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Chen HC, Natera S, Van Noorden G, Menzel C, Taylor S, Renard C, Geiger O, Weiller GF (2003) A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol Plant Microbe Interact 16: 508–524 [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS (1998) Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392: 933–936 [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA (1999) Plant responses to nodulation factors. Curr Opin Plant Biol 2: 483–489 [DOI] [PubMed] [Google Scholar]
- Doyle JJ (1998) Phylogenetic perspectives on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci 3: 473–478 [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136: 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijiri M, Sanders IR (2004) The arbuscular mycorrhizal fungus Glomus intraradices is haploid and has a small genome size in the lower limit of eukaryotes. Fungal Genet Biol 41: 253–261 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature (in press) [DOI] [PubMed]
- Jeong J, Suh S, Guan C, Tsay YF, Moran N, Oh CJ, An CS, Demchenko KN, Pawlowski K, Lee Y (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol 134: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Denarie J, Barker DG, Becard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu G-J, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11: 263–274 [DOI] [PubMed] [Google Scholar]
- Kuster H, Hohnjec N, Krajinski F, El Yahyaoui F, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al (2004) Construction and validation of cDNA-based Mt6k-RIT macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108: 95–113 [DOI] [PubMed] [Google Scholar]
- Lee H, Hur CG, Oh CJ, Kim HB, Park SY, An CS (2004) Analysis of the root nodule-enhanced transcriptome in soybean. Mol Cells 18: 53–62 [PubMed] [Google Scholar]
- Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li W, Llopis J, Whitney M, Zlokarnik G, Tsien RY (1998) Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature 392: 936–941 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock L, Endre G, Cho J, Town CD, VandenBosch K, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of the arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig E, Poole P (2003) Metabolism of Rhizobium bacteroids. Crit Rev Plant Sci 22: 37–78 [Google Scholar]
- Long SR (1996) Rhizobium symbiosis: Nod factors in perspective. Plant Cell 8: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004. a) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004. b) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD Jr (1997) Appressorium formation by AM fungi on isolated cell walls of carrot roots. New Phytol 136: 299–304 [Google Scholar]
- Natera SHA, Guerreiro N, Djordjevic MA (2000) Proteome analysis of differentially displayed proteins as a tool for the investigation of symbiosis. Mol Plant Microbe Interact 13: 995–1009 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED (2001) Dissecting symbiosis: developments in Nod factor signal transduction. Ann Bot (Lond) 87: 709–718 [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T (1996) Rhizobial and actinorhizal symbioses: What are the shared features? Plant Cell 8: 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozynski KA, Malloch DW (1975) The origin of land plants: a matter of mycotrophism. Biosystems 6: 153–164 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M (2001) A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–466 [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalbach G, Erik P, Wienkoop S (2002) Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2: 325–337 [DOI] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Sci 111: 63–71 [Google Scholar]
- Shaw SL, Long SR (2003) Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol 131: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ, editors (1997) Mycorrhizal Symbiosis. Academic Press, San Diego
- Smith SE, Smith AF, Jakobsen I (2003) Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol 133: 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Tamasloukht MB, Sejalon-Delmas N, Kluever A, Jauneau A, Roux C, Becard G, Franken P (2003) Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol 131: 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Ohwada T, Itakura M, Mitsui H, Nukui N, Dawadi P, Kaneko T, Tabata S, Yokoyama T, Tejima K, et al (2004) Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J Bacteriol 186: 2439–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarie J, Long SR (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie JA (2000. b) Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc Natl Acad Sci USA 97: 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Schauser L, Sandal N, Stougaard J, Parniske M (1998) Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol Plant Microbe Interact 11: 933–936 [Google Scholar]
- Wienkoop S, Saalbach G (2003) Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Kuster H, Krajinski F (2003) Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact 16: 306–314 [DOI] [PubMed] [Google Scholar]