On December 14 to 15, 2004, some 50 legume researchers and funding agency representatives (the latter as observers) met in Santa Fe, New Mexico, to develop a plan for cross-legume genomics research. This conference was one of the outcomes of the Legume Crops Genome Initiative (LCGI), an organization bringing together the major U.S. legume commodity associations and their respective research communities. The commodities include alfalfa (Medicago sativa), common bean (Phaseolus vulgaris), the cool-season food legumes (pea [Pisum sativum], lentil [Lens culinaris Med.], and chickpea [Cicer arietinum]), peanut (Arachis hypogaea), and soybean (Glycine max L. Merr.). In recent years, legume genomics has been focused primarily on the development of resources and information of two species considered to be model legumes (Medicago truncatula Gaertner and Lotus japonicus [Regel] K. Larsen) and soybean, the legume of principal economic importance in the United States (VandenBosch and Stacey, 2003). The development of similar genomics research in other legumes has lagged, which has limited the overall impact of genomics of the three reference species. Conversely, these three reference species could benefit from the sharing of biological information that already exists in other legume crops.
Therefore, the main goal of this conference was to forge a common plan with specific objectives for cross-legume genomics research. The specific objectives of the conference were to: (1) identify a unifying goal for a cross-legume genome project; (2) identify cross-cutting themes to help integrate the different legume crop genomics programs, including a unified legume genomics information system, nutritional and health-related aspects of legumes, and detailed synteny and comparative genomics of legumes; and (3) outline specific components and milestones for this initiative. The conference was funded by the National Science Foundation (NSF) Plant Genome Research and the U.S. Department of Agriculture (USDA)/Cooperative State Research, Education, and Extension Service/National Research Initiative Plant Genome programs and organized by the authors of this report. A more detailed white paper resulting from this meeting will be posted on the meeting Web site (http://catg.ucdavis.edu).
THE MULTIPLE ROLES OF LEGUMES
An overview of the societal importance of legumes (Leguminosae or Fabaceae) and some of their most salient biological features provides ample justification for a significant investment in genomics of this botanical family. It also helps orient and prioritize this investment according to goals, species, and tools. With some 20,000 species, the legumes are the third largest family of higher plants. In comparison with other families with model species, the Gramineae have only some 10,000 species and the Brassicaceae some 3,500 species. This situation represents a challenge for comparative genomics, the identification of model species, and the determination of synteny.
The Leguminosae are second to cereal crops in agricultural importance based on area harvested and total production. In 2004, more than 300 million metric tons of grain legumes were produced on 190 million ha (or about 13% of total land under cultivation, including arable land and land under permanent crops; http://faostat.fao.org/faostat/collections?subset=agriculture). The diverse roles of legume plants are often overlooked. Grain legumes provide about one-third of all dietary protein nitrogen and one-third of processed vegetable oil for human consumption (Graham and Vance, 2003). Seeds of grain legumes contain at least 20% to 40% of protein. In many places of the world, legumes complement cereals or root crops, the primary source of carbohydrates, in terms of amino acid composition. Whereas cereal seed proteins are deficient in Lys, legume seed proteins are deficient in sulfur-containing amino acids and Trp (Wang et al., 2003). This situation may explain why in most centers of crop domestication, legumes and cereals have been domesticated together (Gepts, 2004). Legumes are also important forages in temperate (e.g. alfalfa, clover [Trifolium spp.]) and tropical (Stylosanthes sp., Desmodium sp.) regions. Of note are tropical legume trees, which play a particularly important role as forage in arid areas (National Academy of Sciences, 1979) and as abundant timber in humid areas (Doyle and Luckow, 2003).
Legumes also provide essential minerals required by humans (Grusak, 2002a) and produce health-promoting secondary compounds that can protect against human cancers (Grusak, 2002b; Madar and Stark, 2002) and protect the plant against the onslaught of pathogens and pests (Dixon et al., 2002; Ndakidemi and Dakora, 2003). In addition to their blood cholesterol-reducing effect (e.g. Andersen et al., 1984), grain legumes generally also have a hypoglycemic effect, reducing the increase in blood Glc after a meal and, hence, blood insulin. Legumes are, therefore, included in the diet of insulin-dependent diabetics (Jenkins et al., 2003). Certain legumes, however, produce antinutritional factors, such as trypsin inhibitors and phytohemagglutinins (Gupta, 1987) and allergens, the latter being a severe problem in peanut (Spergel and Fiedler, 2001). Genomics approaches, including metabolomics and proteomics, are essential to understanding the metabolic pathways that produce these antinutritional compounds and to eliminating these factors from the plant.
The molecular signaling taking place between legumes and rhizobial symbionts, pollinators, and herbivores and carnivores suggests that legumes are an excellent model for the study of molecular signaling among organisms. Legume crops are of great significance because they produce substantial amounts of organic nitrogen fertilizer resulting from a symbiosis between the plant and bacterial symbionts (e.g. Jensen and Hauggaard-Nielsen, 2003; Hirsch, 2004). Rapid progress is being made in unraveling the molecular basis of this nitrogen-fixing symbiotic relationship, principally in the two model legumes M. truncatula and L. japonicus (Oldroyd et al., 2005). Translating this success to other legume species can be realized with genomic approaches (Stougaard, 2001; Broughton et al., 2003). In contrast with other botanical families, wind-pollinated species are extremely rare in the legumes, which are largely insect-pollinated or self-fertilized. Although not unique to the legumes, insect pollination is accompanied by adaptations in the plant host such as the development of specific morphological traits and the production of volatile attractants. Morphological traits include specific inflorescence types such as racemes and pseudoracemes and a zygomorphic (bilateral) flower symmetry (Tucker, 2003). Floral volatiles have been studied in several legumes, including, and not limited to, alfalfa and clover (Henning et al., 1992; Tava and Pecetti, 1997). Interestingly, leaf volatiles also play an important role in communications with insects, particularly as a defense mechanism to attract predators of herbivores (Pichersky and Gershenzon, 2002) in lima bean (Arimura et al., 2000) and L. japonicus (Ozawa et al., 2000).
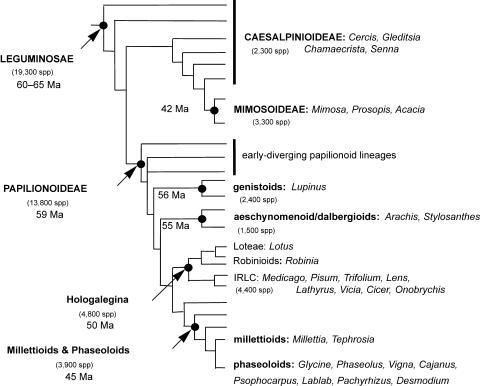
Traditionally, the legume family has been divided into three subfamilies: Caesalpinieae, Mimosoideae, and Papilionoideae. The grain legumes are included in the latter subfamily. Within the Papilionoideae, there are four important clades, which group most of the economically important food and feed legumes (Fig. 1; Doyle and Luckow, 2003). The genistoid clade includes the genus Lupinus and the aeschynomenoid/dalbergioid clade, the peanut. The third and fourth clades have a common ancestor. The Hologalegina clade is split into two subclades, one that includes the Loteae (L. japonicus) and the other that includes species with a chloroplast DNA characterized by the loss of one copy of the inverted repeat found in most angiosperm plants (hence called inverted repeat loss clade). The inverted repeat loss clade includes many species with temperate adaptation (cool-season legumes), such as alfalfa (and M. truncatula), chickpea, faba bean (Vicia faba), lentil, and pea. The fourth clade, the phaseoloid/millettioid clade, includes several legumes that are better adapted to more tropical climates (warm-season legumes), such as common bean, cowpea (Vigna unguiculata L. Walp.), pigeon pea (Cajanus cajan L. Millsp.), and soybean (Doyle and Luckow, 2003).
Figure 1.
Simplified schematic tree of legume family (modified from Doyle and Luckow, 2003). The three subfamilies (Caesalpinioideae, Mimosoideae, and Papilionoideae) and major subclades identified by recent molecular phylogenetic studies are shown in boldface (Kajita et al., 2001; Wojciechowski et al., 2004) and their positions are indicated by black circles (estimated number of taxa from Lewis et al., 2005, and ages [in millions of years] from Lavin et al., 2005).
Phylogenetic relationships within the legume family (Wojciechowski et al., 2004) are reflected in relatively high similarity or synteny at the genome level among the cool-season legumes, including Medicago sp. and pea (Kalo et al., 2004) or between the warm-season legumes common bean and soybean (Lee et al., 2001), but limited synteny is present among other legumes (for example, between cool-season and warm-season legumes; Choi et al., 2004; Zhu et al., 2005). Through a comprehensive assessment of synteny, comparative genomics to assess synteny can facilitate back-and-forth use of genomics resources between different legume species, making the research cost-effective and efficient. In addition, it can speed up gene identification in species that are less tractable because they have a large genome or are less easily transformed. Despite these studies assessing overall levels of synteny, no systematic determination of (micro- and macro-) syntenic relationships among legume species has been attempted as has been accomplished in cereals. Thus, a detailed determination of these relationships is a critical need to allow translation of genomic information among the legume reference and crop species.
Bringing the genomic and biological knowledge in reference legumes to bear on other food and feed legumes of major economic importance, including cool-season pulses (e.g. pea, lentil, and chickpea), warm-season food legumes (e.g. peanut and common bean), and forage legumes (e.g. alfalfa and clover) represents a major scientific opportunity. Each legume presents unique features of economic and scientific interest. Examples include the geotropic peg and pod development of peanut (Pattee et al., 1998), drought tolerance and vernalization in chickpea (Abbo et al., 2002), forage nutritive value in alfalfa (Riday et al., 2002), biologically interesting mutants in pea (e.g. DeMason and Villani, 2001; Novak, 2003), evolution of domestication (Koinange et al., 1996), coevolution of host and pathogen/pests in common bean (e.g. Geffroy et al., 2000; Aguilar et al., 2004), and differential resistance of legume species to closely related groups of pathogens such as fusarium and rust (Gray et al., 1999; Tenuta, 2004). Examination of these specific features will require a combination of genomic resources from the reference species together with those developed in the crops themselves.
A VISION: LEGUMES AS A MODEL PLANT FAMILY: GENOMICS FOR FOOD AND FEED
The CATG conference participants agreed for the first time on the development of a 10-year prioritized plan for cross-legume genomics focused on the single theme of legume genomics for food and feed. Cross-legume genomics seeks to advance: (1) knowledge about the legume family as a whole; (2) understanding about the evolutionary origin of legume-characteristic features such as rhizobial symbiosis, flower and fruit development, and its nitrogen economy; and (3) pooling of genomic resources across legume species to address issues of scientific, agronomic, environmental, and societal importance. Thus, the CATG initiative seeks to develop the study of the organization and function of a unified legume genome in all its diversity. This implies translation of genomic information and tools developed for the reference legumes to other legumes and, conversely, utilization of the extensive biological and agronomic knowledge accumulated in crop legumes to improve our understanding of the biology of reference legumes. To be fully effective, a genome project across a botanical family like the Leguminosae needs to allow researchers to go back and forth among species and not just in one direction, i.e. from reference or model species to crop species. However, because resources are limited, the development of genomic tools needs to be carefully prioritized.
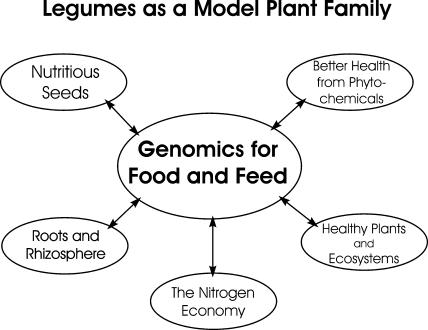
The general theme of improvement of food and feed represents a clear vision for the future for legume genomics, as well as an emphatic statement directed primarily toward the public, who will be the ultimate beneficiary of genomic activities. This unified theme combines several areas of research (Fig. 2). First and foremost, it recognizes the importance of grain legumes (also known as pulses) as essential sources of dietary protein for humans and animals, as well as health-related phytochemicals such as dietary fiber, hormone analogs, and antioxidants. Genomics provides essential tools to fully understand the molecular and metabolic basis of the synthesis of these compounds, to increase their content in seeds and pods, and to better manipulate interactions between the plant's genetic makeup and its environment. A focus on seeds also underscores the importance of the genomics of reproductive biology in the development of higher-yielding, more nutritious legume cultivars.
Figure 2.
Major research areas supporting improved food and feed as a major goal of cross-legume genomics.
One of the signature features of legumes is the association between plants and rhizobial and mycorrhizal symbionts. The application of genomics has led to substantial and rapid advances in our understanding of the molecular basis of the two types of symbioses in M. truncatula and L. japonicus (Oldroyd et al., 2005). Studies of the rhizosphere in legumes are among the most developed of all botanical families and can lead to significant advances in plant health and growth.
The common currency underlying protein-rich seed/forage and rhizobial symbiosis is nitrogen. The nitrogen-rich life style can explain, in part, the success and diversity of the legume family (McKey, 1994) and represents a significant contribution to agricultural and natural ecosystems. Comparative and functional genomics will extend the knowledge gained in the model legumes to crop legumes and should lead to more efficient nitrogen-fixing cultivars.
Finally, the contributions of legumes could not be fully realized without low incidence of diseases and pests that affect the family. Recent advances in our understanding of disease resistance genes will be complemented by the application of genomics to fully understand the mechanisms by which legumes resist or tolerate pathogens and pests. Toward this end, molecular markers developed during genomics projects will assist breeders in developing new, resistant cultivars.
A STRATEGY TOWARD UNIFIED LEGUME GENOMICS
The path to better food and forage legumes requires a detailed knowledge of the different genes involved in the biochemical pathways leading up to key nutritional compounds, including the expression patterns and levels of these genes and their interactions. The tools of genomics, including bioinformatics and synteny analysis, provide the opportunity not only to obtain this comprehensive type of metabolical information, but also to integrate research efforts across different species. At the CATG conference, participants considered information on legume phylogeny and economic importance, among other factors, to establish a cross-legume genomics strategy and priorities for the development of legume genomic resources over the period of the coming 10 years. Specifically, four tiers of investment of genomic tools were recognized in the legumes. These tiers are described below (in decreasing order of priority and investment).
The Reference Legumes M. truncatula, L. japonicus, and Soybean
Cross-legume genomics research will be organized around two major clades that include most of the economically important legumes. These two clades correspond to the Hologalegina (cool-season legumes) and phaseoloid/millettioid (warm-season legumes) clades (Fig. 1; Doyle and Luckow, 2003). Jointly, they represent a large part of the economic legumes, especially for food and feed, and capture approximately 40% of the phenotypic variation among all legumes (J. Doyle, personal communication). The relatively broad taxonomic distance separating the two clades (Doyle and Luckow, 2003; Wojciechowski et al., 2004; Lavin et al., 2005) warrants the development of one or two reference systems within each one. M. truncatula and L. japonicus represent models for the cool-season legumes and soybean for the warm-season legumes. For each of these reference systems, considerable investments have already been made and should be continued in order to develop the full range of genomics resources, including sequencing of the entire genome or at the very least the gene-rich regions and extensive transcriptomics, proteomics, and metabolomics resources.
Extended Genomic Tools for Targeted Species Phaseolus and Arachis
Phaseolus and Arachis will be targeted for the development of a range of extended genomic resources. These include development of a physical map accompanied by bacterial artificial chromosome (BAC)-end sequencing, molecular markers based on BAC-end sequencing, anchoring of physical and genetic maps, expressed sequence tags (ESTs) of the major organs, especially those involved in reproductive development, microarray and DNA chip resources, and ultimately sequencing of the Phaseolus genome and gene-rich regions for Arachis. In addition to their economic importance on a worldwide basis, special arguments support more extensive development of genomic tools in these two crops. The small, diploid genome of Phaseolus is a key to understanding genome structure and expression in the phaseoloid/millettioid group (Fig. 1), which includes soybean. Arachis occupies a phylogenetically more distant position from the two foci proposed here (Fig. 1), providing perspective on the evolution of the foci.
Translational Genomic Tools for Other Major Legume Crops and Targeted Experimental Systems
All legumes belonging to the two foci will benefit from the development of translational tools that enable the sharing of genetic and genomics information among the various species. The translation tools consist primarily of cross-legume markers (extending the preliminary work of Brauner et al., 2002; Choi et al., 2004; and Kalo et al., 2004 by expanding the number of markers and species across the two major foci and peanut), EST libraries from several critical tissues, and BAC libraries with extensive coverage of the genome. In addition, for those species still lacking genetic maps and recombinant inbred populations, development of these resources should be assigned a high priority. The crops recommended for targeting include pea, lentil, chickpea, faba bean, alfalfa, clover, cowpea, pigeon pea, and lupin (Lupinus spp.). With progress in sequencing, some genomes in this group may be sequenced at least in part (gene-rich regions). They include the genomes of pea (to permit more productive use of the extensive biochemical and physiological literature for this species) and chickpea, one of the most drought-tolerant species among legumes of major economic importance.
Develop Additional, Selected Legume Experimental Systems
Some of the fundamental biological questions, such as the origin of legume-characteristic traits, require an evolutionary approach that encompasses the entire legume family. Such traits include reproductive development (especially floral and pod development), the origin of nodulation, and the evolution and importance of polyploidy. The two foci, while providing coverage for most economic legumes involved in food and feed, do not come close to covering the biodiversity included in the Fabaceae. Hence, to address the issue of legume-characteristic traits, other species may have to be considered, including species in the basal clades of the Papilionoideae, the Caesalpinieae (e.g. Chamaecrista sp.), and the Mimosoideae. In these species, ad hoc genomic resources targeted toward evolutionary genomics questions of interest will have to be developed to allow comparisons with reference and other legume species.
TOOLS AND TIMELINE TO ADDRESS STRATEGIC GOALS
The timeline is divided into short-term (years 1–3), medium-term (years 4–6), and long-term (years 7–10) periods.
Structural Genomics, Genome Sequencing, and Synteny Mapping
Genomics resources need to be developed or expanded within the reference legumes as well as for the major crop legumes to enhance the translation of genomics information across species. The CATG consensus, benchmarked over the next decade, envisions the following.
Within 3 Years
A legume synteny project should be completed, in which 500 gene-based markers developed primarily from important biochemical genes will be mapped across major crop legumes in at least one mapping population per species. These markers should be explicitly tied to whole genome sequences of reference species. Additional segregating populations useful for mapping important traits should be developed. Physical mapping should be initiated by end-sequencing BAC libraries currently available in crop species and fingerprinting BAC libraries that have been constructed from parents of mapping populations. A legume-wide and plant-wide workshop specifically addressing potential solutions to the long-term maintenance and conservation of genomic resources is called for.
Within 6 Years
A genome-wide survey of genetic diversity among germplasm accessions of each major legume species should be undertaken, using common markers to facilitate translation of the results. Evaluation of the extent of linkage disequilibrium in exotic and domesticated germplasm should be started. Phenotypic evaluation of multiple populations per species should be conducted so that the locations of quantitative trait loci for important agronomic traits, such as seed composition and yield, can be identified by genetic and association mapping. The accumulation of mapping information will facilitate the exploration of syntenic region across legumes. Physical maps should be constructed for common bean and diploid Arachis. The soybean genome, at least its gene-rich regions, should be sequenced by the fourth or fifth year.
Within 10 Years
The genome sequences of Phaseolus and Arachis and selected BAC sequencing in Pisum and Cicer should be completed. Further genetic mapping in all species will continue.
Bioinformatics
Scientists at the CATG conference made the following recommendations.
Within 3 Years
Develop a virtual, easy-to-navigate one-stop legume information network. By one-stop, we refer by analogy to Google and how it can be seen as a single, yet nonexclusive, information resource. Such a system should be able to semantically respond to queries for both information and services. Current and future legume information resources can be viewed as components to this network. Attribution to the sources of the data will help assure a willingness to register with the network and provide a peer-review mechanism for quality assurance. While it was beyond the scope of this meeting to recommend a particular technological approach to assure these attributes, there was considerable enthusiasm expressed for the utilization of the BioMOBY technology platforms (Schiltz et al., 2004) as a possible solution. Species-level biologist/curator of data and information and cross-species working groups composed of biologist/curators will be put in place.
Within 6 Years
Legume information resources will accommodate proteomic and metabolomic data and information as well as analysis and visualization tools for these emerging sources of information.
Within 10 Years
The legume community should become a leader in the development of new bioinformatics tools. There is an opportunity to do so with development of a one-stop legume information network. There are also opportunities to develop novel analysis and visualization tools for both integrative and comparative research questions. For example, there are needs for: (1) visualization of comparative maps at the level of linkage maps, physical maps (BAC-level), and homologous (orthologous) sequences; and (2) the development of researcher-centric (breeders, geneticists, biochemists, and molecular biologists) interfaces. These tools can be extended to develop common biological and bioinformatic frameworks for all plants and animals to maximize the benefits of comparative biology.
Throughout the 10-Year Period
As new resources become available, workshops, education, and ongoing feedback from user community will be needed for all bioinformatics efforts.
The Increasingly Prominent Role of Functional Genomics (ESTs, Transcriptomics, Proteomics, and Metabolomics)
Breakthroughs in understanding the relationship between genotype and phenotype will come about through proteomics and metabolomics. Legumes are especially appropriate for research in the areas of proteomics and metabolomics. As information on DNA sequences becomes more common in legumes, especially the reference legumes, the next steps will naturally focus increasingly on the expression of DNA sequence. Transcript profiling, proteomics, and metabolomics are essential tools to fully understand the synthesis of compounds that are at the basis of this research community's focus on food and feed. In addition to protein, carbohydrates, and lipids, an understanding of the secondary metabolism and mineral nutrition of legumes is an important goal. The secondary metabolism involves interactions between legumes and pathogens, symbiotic organisms, predators, and pollinators. It is also the basis for many of the nutritional benefits ascribed to legumes. Extensive transcriptomics (e.g. Colebatch et al., 2004), proteomics (Watson et al., 2004), and metabolomics (Sumner et al., 2003) programs are already under way in the model legumes M. truncatula and L. japonicus.
Within 3 Years
These resources need to be further developed in the two model legume species in parallel with the ongoing whole-genome sequencing efforts. The modest EST resources in major legume crops (other than soybean) must be expanded to include a broader range of tissues and environmental conditions.
Within 6 Years
Transcriptomic, proteomic, and metabolomic resources should be extended to soybean. Additional resources to be developed include full-length cDNA libraries from seed, mature leaf, nodule, flower, and pod for common bean and peanut from which DNA microarray and oligonucleotide gene chips can be developed for these two species.
Within 10 Years
Selected resources should be extended to major crop legumes. Protein fragment databases, gel fingerprints for the important tissues of the major legumes, chips, including arrays of legume transcription factors and proteins (or antibodies to the proteins) of the developing flower, seed, and root, should be developed.
Education, Outreach, and Recruitment of Young Scientists
Because of their important role in human nutrition and the environment, legumes offer many opportunities in education and outreach. Ever since Mendel first discovered the fundamental laws of genetics through his studies of garden peas, legumes have helped people to understand biology. Now classroom and laboratory activities can introduce important concepts like genetic inheritance, symbiosis, and seed and flower development to young people. Already, new legume-based projects for K-12 students are being developed, like the “Bacteria and Plants Team Up!” packet created at Indiana University with NSF Plant Genome support. In addition, the role of legumes on a worldwide basis as a source of food, forage, and timber can be highlighted, as well as their role in ecosystem health and the nitrogen cycle. In the process, the benefits of good nutrition can be introduced to young people by making them familiar with the central role of legumes and health-promoting compounds in their diet (e.g. the place of legumes in the USDA food pyramid). Furthermore, research experience in legume genomics and genetics for K-12 teachers can, for example, also be provided through the Research Experience for Teachers (RET) program of NSF. Workshops describing legume genomics and bioinformatics for applied plant scientists and crop breeders need to be developed and expanded.
Bringing in the International Legume Community, Including Researchers Working on Legume Crops of the Developing World
A mechanism will be established to ensure international coordination of legume genomics efforts and involving researchers, funding agencies, farmers, and other stakeholders.
A large proportion of worldwide legume production occurs in developing countries, where legumes fulfill a vital role in human nutrition, as tropical forages and as timber species. For several species, such as common bean and chickpea, strong collaborative links among scientists throughout the world, including those of developing countries, will provide additional opportunities for funding and research and will significantly increase the impact of legume genomics. The Phaseolus genomics initiative (Phaseomics; Broughton et al., 2003), with representatives of over 20 countries, provides an example of current collaboration. The scope of some projects, such as genome sequencing is beyond the capacity of single countries (e.g. the International Medicago truncatula Genome Sequencing Initiative). In addition, legume genomics projects and research centers in other countries provide opportunities for collaborations. These include the European Grain Legumes Integrated Project, which is part of the Framework 6 Food Quality and Safety research program of the European Union (http://www.eugrainlegumes.org), the Australian Centre of Excellence for Integrative Legume Research at the University of Queensland (http://www.cilr.uq.edu.au), and the Generation Challenge Program of the Consultative Group for International Agricultural Research (http://www.generationcp.org/vw/index.php). The introduction of Asian soybean rust (Phakopsora pachyrhizi Sydow) into the United States highlights the potential not only for cross-legume genomics research (e.g. between common bean and soybean) but also for international collaborations given the widespread distribution of the pathogen in Asia, Africa, and the Americas.
CONCLUSION
This white paper marks a major departure from previous legume genomic research, which was focused primarily on the development of genomic tools and biological investigations for individual species. The approach endorsed by CATG participants and described in this paper represents a coordinated effort for the development and research involving genomics across the legume family. To paraphrase Bennetzen and Freeling (1997) when they spoke about cereals, the legume genomics community is now pursuing a unified legume genome in the hope of achieving synergy in synteny.
Acknowledgments
We are grateful to our fellow legume researchers for their enthusiasm and positive spirits, which made the CATG meeting a success. We thank D. Zamir, J. Doyle, A. Hirsch, M. Grusak, and E. Pichersky for exciting and stimulating plenary lectures, and J. Doyle and M. Wojciechowsky for their assistance with Figure 1.
This work was supported by the National Science Foundation Plant Genome Research Program and by the U.S. Department of Agriculture/Cooperative State Research, Education, and Extension Service/National Research Initiative Plant Genome Program.
References
- Abbo S, Lev-Yadun S, Galwey N (2002) Vernalization response of wild chickpea. New Phytol 154: 695–701 [DOI] [PubMed] [Google Scholar]
- Aguilar OM, Riva O, Peltzer E (2004) Analysis of Rhizobium etli and of its symbiosis with wild Phaseolus vulgaris supports coevolution in centers of host diversification. Proc Natl Acad Sci USA 101: 13548–13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JW, Story L, Sieling B, Chen W-JL, Petro MS, Story J (1984) Hypocholesterolemic effects of oat-bran or bean intake for hypercholesterolemic men. Am J Clin Nutr 40: 1146–1155 [DOI] [PubMed] [Google Scholar]
- Arimura G-i, Ozawa R, Shimoda T, Nishioka T, Boland W, Takabayashi J (2000) Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 406: 512–515 [DOI] [PubMed] [Google Scholar]
- Bennetzen J, Freeling M (1997) The unified grass genome: synergy in synteny. Genome Res 7: 301–306 [DOI] [PubMed] [Google Scholar]
- Brauner S, Murphy RL, Walling JG, Przyborowski J, Weeden NF (2002) STS markers for comparative mapping in legumes. J Am Soc Hortic Sci 127: 616–622 [Google Scholar]
- Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.): model food legumes. Plant Soil 252: 55–128 [Google Scholar]
- Choi H-K, Mun J-H, Kim D-J, Zhu H, Baek J-M, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101: 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- DeMason DA, Villani PJ (2001) Genetic control of leaf development in pea (Pisum sativum). Int J Plant Sci 162: 493–511 [Google Scholar]
- Dixon RA, Achnine L, Kota P, Liu C-J, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence: a genomics perspective. Mol Plant Pathol 3: 371–390 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131: 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy V, Sévignac M, De Oliveira J, Fouilloux G, Skroch P, Thoquet P, Gepts P, Langin T, Dron M (2000) Inheritance of partial resistance against Colletotrichum lindemuthianum in Phaseolus vulgaris and co-localization of QTL with genes involved in specific resistance. Mol Plant-Microbe Interact 13: 287–296 [DOI] [PubMed] [Google Scholar]
- Gepts P (2004) Domestication as a long-term selection experiment. Plant Breed Rev 24: 1–44 [Google Scholar]
- Graham PH, Vance CP (2003) Legumes. Importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LE, Achenbach LA, Duff RJ, Lightfoot DA (1999) Pathogenicity of Fusarium solani f. sp. glycines isolates on soybean and green bean plants. J Phytopathol 147: 281–284 [Google Scholar]
- Grusak MA (2002. a) Enhancing mineral content in plant food products. J Am Coll Nutr 21: 178S–183S [DOI] [PubMed] [Google Scholar]
- Grusak MA (2002. b) Phytochemicals in plants: genomics-assisted plant improvement for nutritional and health benefits. Curr Opin Biotechnol 13: 508–511 [DOI] [PubMed] [Google Scholar]
- Gupta YP (1987) Anti-nutritional and toxic factors in food legumes: a review. Qual Plant Plant Foods Hum Nutr 37: 201–228 [DOI] [PubMed] [Google Scholar]
- Henning J, Peng Y, Montague M, Teuber L (1992) Honey bee (Hymenoptera: Apidae) behavioral response to primary alfalfa (Rosales: Fabaceae) floral volatiles. J Econ Entomol 85: 233–239 [Google Scholar]
- Hirsch AM (2004) Plant-microbe symbioses: a continuum from commensalism to parasitism. Symbiosis 37: 345–363 [Google Scholar]
- Jenkins DJA, Kendall CWC, Marchie A, Jenkins AL, Augustin LSA, Ludwig DS, Barnard ND, Anderson JW (2003) Type 2 diabetes and the vegetarian diet. Am J Clin Nutr 78: 610S–616S [DOI] [PubMed] [Google Scholar]
- Jensen ES, Hauggaard-Nielsen H (2003) How can increased use of biological N2 fixation in agriculture benefit the environment? Plant Soil 252: 177–186 [Google Scholar]
- Kajita T, Ohashi H, Tateishi Y, Bailey CD, Doyle JJ (2001) rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst Bot 26: 515–536 [Google Scholar]
- Kalo P, Seres A, Taylor SA, Jakab J, Kevei Z, Kereszt A, Endre G, Ellis THN, Kiss GB (2004) Comparative mapping between Medicago sativa and Pisum sativum. Mol Genet Genom 272: 235–246 [DOI] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P (1996) Genetic control of the domestication syndrome in common-bean. Crop Sci 36: 1037–1045 [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol (in press) [DOI] [PubMed]
- Lee JM, Grant D, Vallejos CE, Shoemaker RC (2001) Genome organization in dicots. II. Arabidopsis as a “bridging species” to resolve genome evolution events among legumes. Theor Appl Genet 103: 765–773 [Google Scholar]
- Lewis GP, Schrire BD, Mackinder BA, Lock M, editors (2005) Legumes of the World. Royal Botanic Garden, Kew, UK (in press)
- Madar Z, Stark AH (2002) New legume sources as therapeutic agents. Br J Nutr 88: S287–S292 [DOI] [PubMed] [Google Scholar]
- McKey D (1994) Legumes and nitrogen: the evolutionary ecology of a nitrogen-demanding lifestyle. Adv Legume Syst 5: 211–228 [Google Scholar]
- National Academy of Sciences (1979) Tropical Legumes: Resources for the Future. National Academy of Sciences, Washington, DC
- Ndakidemi PA, Dakora FD (2003) Review: legume seed flavonoids and nitrogenous metabolites as signals and protectants in early seedling development. Funct Plant Biol 30: 729–745 [DOI] [PubMed] [Google Scholar]
- Novak K (2003) Allelic relationships of pea nodulation mutants. J Hered 94: 191–193 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Udvardi M (2005) Peace talks and trade deals: keys to long-term harmony in legume-microbe symbioses. Plant Physiol 137: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa R, Shimoda T, Kawaguchi M, Arimura G-i, Horiuchi J-i, Nishioka T, Takabayashi J (2000) Lotus japonicus infested with herbivorous mites emits volatile compounds that attract predatory mites. J Plant Res 113: 427–433 [Google Scholar]
- Pattee HE, Stalker HT, Giesbrecht FG (1998) Reproductive efficiency in reciprocal crosses of Arachis monticola with A. hypogaea subspecies. Peanut Sci 25: 7–12 [Google Scholar]
- Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Riday H, Brummer EC, Moore KJ (2002) Heterosis of forage quality in alfalfa. Crop Sci 42: 1088–1093 [Google Scholar]
- Schiltz G, Gessler D, Stein L (2004) Semantic MOBY. Position paper for the W3C workshop on Semantic Web for Life Sciences. http://lists.w3.org/Archives/Public/public-swls-ws/2004Sep/att-0036/smoby-w3c-sw-ls.pdf. (February 4, 2005)
- Spergel JM, Fiedler JM (2001) Natural history of peanut allergy. Curr Opin Pediatr 13: 517–522 [DOI] [PubMed] [Google Scholar]
- Stougaard J (2001) Genetics and genomics of root symbiosis. Curr Opin Plant Biol 4: 328–335 [DOI] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Tava A, Pecetti L (1997) Volatiles from Medicago sativa complex flowers. Phytochemistry 45: 1145–1148 [Google Scholar]
- Tenuta A (2004) Soybean rust infosheet. http://www.gov.on.ca/OMAFRA/english/crops/facts/soybean_rust.htm. (February 4, 2005)
- Tucker SC (2003) Floral development in legumes. Plant Physiol 131: 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Domoney C, Hedley CL, Casey R, Grusak MA (2003) Can we improve the nutritional quality of legume seeds? Plant Physiol 131: 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Lei ZT, Dixon RA, Sumner LW (2004) Proteomics of Medicago sativa cell walls. Phytochemistry 65: 1709–1720 [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analyses of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Zhu H, Choi H-K, Cook DR, Shoemaker RC (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137: 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]