Abstract
The Medicago truncatula line 2HA has a 500-fold greater capacity to regenerate plants in culture by somatic embryogenesis than wild-type Jemalong. We have compared proteomes of tissue cultures from leaf explants of these two lines. Both 2HA and Jemalong explants were grown on media containing the auxin 1-naphthaleneacetic acid and the cytokinin 6-benzylaminopurine. Proteins were extracted from the cultures at different time points (2, 5, and 8 weeks), separated by two-dimensional gel electrophoresis, and detected by silver staining. More than 2,000 proteins could be reproducibly resolved and detected on each gel. Statistical analysis showed that 54 protein spots were significantly (P < 0.05) changed in expression (accumulation) during the 8 weeks of culture, and most of these spots were extracted from colloidal Coomassie-stained two-dimensional gel electrophoresis gels and were subjected to matrix-assisted laser desorption ionization time-of-flight mass spectrometry or liquid chromatography-tandem mass spectrometry analysis. Using a publicly available expressed sequence tag database and the Mascot search engine, we were able to identify 16 differentially expressed proteins. More than 60% of the differentially expressed protein spots had very different patterns of gene expression between 2HA and Jemalong during the 8 weeks of culture.
Plants must coordinate the growth of root and shoot meristems to maintain an appropriate balance of root and shoot organs and to respond and adapt to various environmental conditions. This balance is achieved by an intermeristem coordination of growth and development of the plant and involves the interplay of several long-range signals (Wopereis et al., 2000; Jiang and Gresshoff, 2002; Searle et al., 2003). Somatic (or asexual) embryogenesis (SE) is the process whereby somatic cells differentiate into embryos and ultimately into plants via a series of characteristic morphological stages, particularly the later stages, which resemble the zygotic stages of development (Zimmerman, 1993; Schmidt et al., 1997). SE is the developmental restructuring of somatic cells toward the embryogenic pathway and forms the basis of cellular totipotency in higher plants (Nolan et al., 2003; Imin et al., 2004). Two experimental approaches are available to examine this process in detail: (1) leaf cells grown in culture from protoplasts to form calli and, subsequently, the generation of embryos and then the development of plants (Rose and Nolan, 1995); and (2) leaf explants, which form calli in culture, and the concomitant production of embryos and vascular tissues. Depending on the plant system, auxin and/or cytokinin are required to enable embryogenesis to occur in culture (Schmidt et al., 1997; Somleva et al., 2000; Baudino et al., 2001; Hecht et al., 2001). In Medicago truncatula (Australian barrel medic), Nolan et al. (2003) found that embryogenesis required both auxin and cytokinin addition, although some embryos could form on cytokinin alone.
The first appearance of embryos from mesophyll protoplasts occurs between 8 to 9 weeks in M. truncatula and has a reasonable degree of synchrony, thus enabling a developmental study of the molecular changes taking place in the dividing cells. This meristematic system has ideal attributes: the regenerative capacity of the mutant line 2HA, which is 500-fold more embryogenic than its isogenic line Jemalong, and access to a developing genome sequence. We are using this system to investigate meristematic growth and differentiation in culture and have identified differentially expressed proteins during the first developmental stages of SE (Imin et al., 2004). The second experimental system, that of leaf explant tissue culture, has the advantage of being able to manipulate the type of differentiating cells observed by changing the phytohormones added to the culturing media (Schmidt et al., 1997; Nolan et al., 2003; Thomas et al., 2004), and embryos are initiated more rapidly in 4 to 6 weeks (Nolan et al., 1989).
When both M. truncatula cv Jemalong and 2HA explant tissues are cultured in medium with addition of auxin alone, they produce numerous roots but no embryos (Nolan et al., 2003). On media containing both auxin and cytokinin, the 2HA explants form embryos. Generally, cultivar Jemalong does not form embryos but does produce early vascularization in the calli. The pasture legume M. truncatula is one of the model systems for the analysis of the unique biological and fundamental processes governing legume biology. Recent genomic tools, advanced DNA sequencing programs, and expressed sequence tag (EST) libraries have been developed for this legume, and a number of researchers have taken advantage of these resources and initiated the establishment of proteome reference maps for M. truncatula tissues, roots, leaves, flowers, seeds, cell cultures, and SE cultures (Mathesius et al., 2001; Bestel-Corre et al., 2002, 2004; Gallardo et al., 2003; Watson et al., 2003; Imin et al., 2004; Valot et al., 2004). Most of these studies used a combination of two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption ionization time-of-flight mass spectroscopy (MALDI-TOF-MS). Collectively, these maps provide a basis for future comparative investigations of plant-microbe interactions and developmental biology in M. truncatula. In the study by Mathesius et al. (2001), proteins from root extracts were separated on 2-DE gels, yielding reproducibly 2,500 proteins per gels. More than 200 root (Mathesius et al., 2001) and 100 leaf proteins have been identified by peptide mass fingerprinting using the M. truncatula EST or tentative cluster (TC) databases for reliable identification of the proteins. A database of identified proteins has been compiled (http://semele.anu.edu.au/2d/2d.html). This proteome reference map was used to test against the proteomes of related legumes (Mathesius et al., 2002) and to evaluate proteome reference maps for somatic embryonic tissues of M. truncatula (Imin et al., 2004).
Thus, analyses of gene expression during SE can provide information about the earlier stages of plant development (Zimmerman, 1993). Two-dimensional gel electrophoresis-based proteomic approaches have been applied to investigate molecular changes during SE in several species (Choi and Sung, 1984; Gianazza et al., 1992; Reinbothe et al., 1992; Boyer et al., 1993; Pedroso et al., 1995; Yoshida et al., 1995; Dodeman and Ducreux, 1996; Giroux and Pauls, 1996; Dodeman et al., 1998; Sallandrouze et al., 1999) with limited success. Large-scale transcription analyses of embryogenesis have also been reported in several species (Giroux and Pauls, 1997; van Zyl et al., 2002, 2003; Stasolla et al., 2003; Thibaud-Nissen et al., 2003; Stasolla et al., 2004).
Numerous genes have been identified as specifically expressed during SE (Mordhorst et al., 1997; Chugh and Khurana, 2002; Fehér et al., 2003). These genes include hormone-responsive genes such as auxin-inducible genes (Walker and Key, 1982), late embryo abundant genes (Yang et al., 1997), calmodulin (Overvoorde and Grimes, 1994), calcium-dependent/calmodulin-independent protein kinases (Lindzen and Choi, 1995), calmodulin-like protein kinases (Davletova et al., 2001), SERK genes (Schmidt et al., 1997; Nolan et al., 2003; Thomas et al., 2004), homeobox-containing genes (Kawahara et al., 1995; Meijer et al., 1997), chitinases (De Jong et al., 1992), arabinogalactans (Baldwin et al., 2001), lipid transfer proteins (Sterk et al., 1991), WUSCHEL (Zuo et al., 2002), BABY BOOM (Boutilier et al., 2002), and LEAFY COTYLEDON genes (Meinke, 1992; Meinke et al., 1994), to name a few. As yet, little is known about the induction and maintenance process of the genes involved in the SE processes.
In this study, we have used leaf explant tissues of 2HA and Jemalong to investigate the protein profiles and their changes during embryo induction and formation.
RESULTS
Changes in Protein Expression Profiles during the Course of Tissue Culture and Embryo Formation
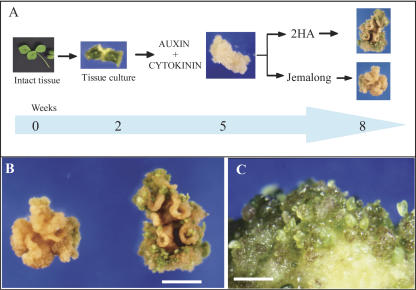
Leaf explants of the highly embryogenic M. truncatula seed line 2HA and its near isogenic line Jemalong were cultured on media containing 1-naphthaleneacetic acid (NAA) and 6-benzylaminopurine (BAP) for 2, 5, and 8 weeks. A schematic diagram of tissue culturing and sampling for proteomic analysis is shown in Figure 1A. At 2 weeks, there were no visible differences between the 2HA and Jemalong. After 5 weeks in culture, embryos began to emerge from 2HA tissue. At 8 weeks, there were numerous embryos (Fig. 1, B and C) in the highly embryogenic line 2HA. By contrast, the Jemalong explants rarely formed embryos (Fig. 1, A and B).
Figure 1.
Explant tissue culturing in M. truncatula. A, Schematic diagram of the experimental design for the proteomic analysis of M. truncatula explant cultures grown on auxin and cytokinin. Samples used for proteomic analysis were collected at 2, 5, and 8 weeks. B, Calli from leaf explants of the highly regenerable seed line 2HA and its near isogenic line Jemalong. Line 2HA shows the formation of numerous embryos. Bar = 1 cm. C, Enlargement of 2HA culture showing green embryos. Bar = 5 mm.
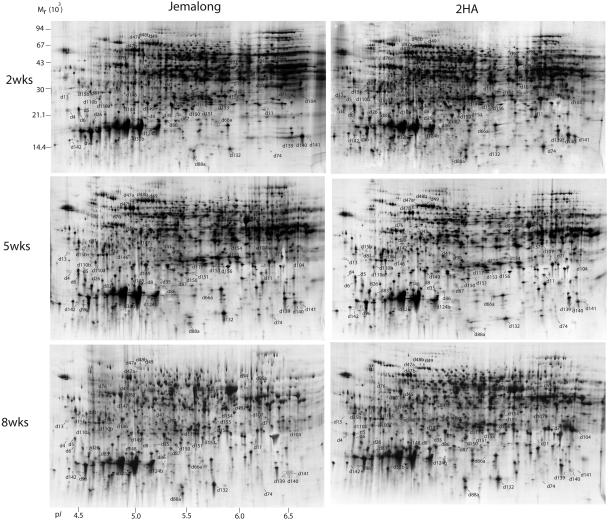
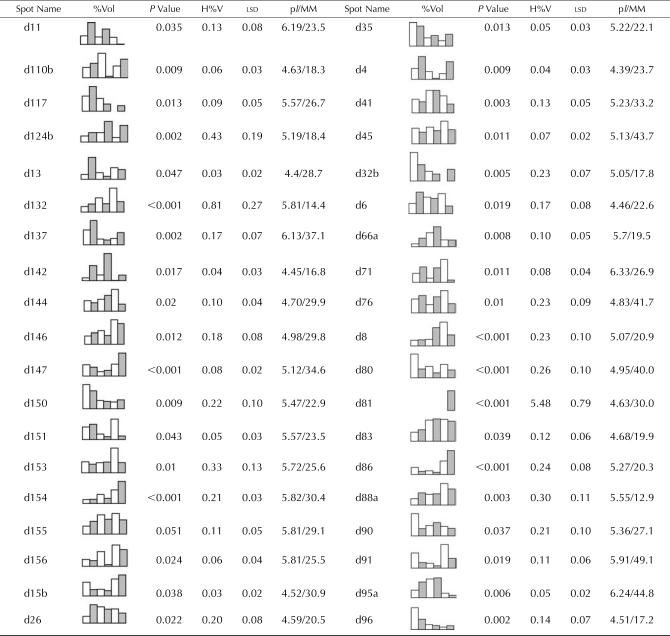
Total proteins were extracted from leaf explants of both 2HA and Jemalong at 2, 5, and 8 weeks and analyzed by 2-DE (Fig. 2). Melanie 4 image analysis of the gels revealed that more than 2,000 proteins were reproducibly resolved in silver-stained gels over a pH range of 4 to 7 and a size range of 8 to 120 kD. Silver-stained gels were used for the comparisons and quantitative analysis. To compare proteome patterns of Jemalong and 2HA at three time points (2, 5, and 8 weeks), the gels were compared to one another, and the protein spots that showed changes were recorded. Selected parts of the gels are highlighted in Figure 3 to show the comparisons of the protein spots. While the majority of the protein spots (more than 90% of the total spots) did not show any change, we determined by inspection more than 200 spots as changed between 2HA and Jemalong at three time points, and these spots were selected for further quantitative analysis. Statistical analysis showed that only 54 protein spots were changed significantly (P < 0.05). Representative graphs for the mean percentage volumes (%Vol) of each of these spots are given in Tables I and II. Some proteins (d147, d154, d41, d96, d139, d140, and d141) had a similar trend of expression between Jemalong and 2HA during 2 to 8 weeks of tissue culture, but the level of expression of these proteins was significantly different. By contrast, many proteins (60%) had a different pattern of expression between Jemalong and 2HA. Some proteins (d11, d149, and d110a) changed little in Jemalong during culture but changed significantly in 2HA. For example, d11 was expressed at a minimum level in Jemalong throughout the 8 weeks of culture. However, it was highly expressed at 2 weeks in 2HA, decreased to almost one-half at 5 weeks, and became undetectable at 8 weeks. d104 had similar expression in Jemalong. By contrast, d104 had low levels of expression at 2 weeks, gradually increased at 5 weeks, and accumulated to a maximum at 8 weeks in 2HA. Some proteins (d132, d153, d45, and d80) showed little change in 2HA, but either increased (d132, d153, and d45) or decreased (d80) in Jemalong. Expression of some proteins (d110a, d6, and d26) was reversed in Jemalong and 2HA. For example, d6 and d26 were increased in Jemalong but decreased in 2HA during culture. d110b expression was increased at 5 weeks and then decreased at 8 weeks in Jemalong, whereas its expression decreased at 5 weeks and then increased at 8 weeks in 2HA. Some proteins (d81 and d148) only expressed in 2HA but were not detected in Jemalong. Due to the complexity of the protein expression in culture, it was difficult to classify these expressions in a small number of subgroups in respect to a Jemalong and 2HA comparison.
Figure 2.
Two-dimensional gel electrophoresis proteome maps of M. truncatula explant cultures. Protein spots were assigned arbitrary identifiers as shown in Tables I and II. First-dimension focusing used 24-cm IPG strips with a linear pH gradient 4 to 7 loaded with 150 μg of total proteins for each strip. In the second dimension, 12–14%T SDS-PAGE gels were used. Proteins were visualized by silver staining. Two regions are enlarged in Figure 3.
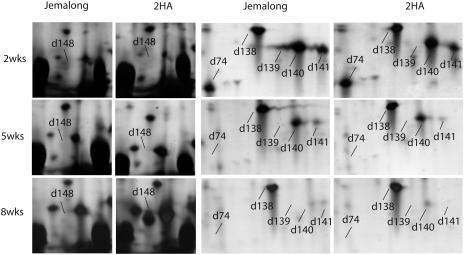
Figure 3.
Enlargement of selected regions in Figure 2 to highlight some of the differentially expressed protein spots. Note the differentially expressed protein spot d148 is on the left panel, and spots d74, d139, d140, and d141 are on the right panel. These images were captured from the annotated gels in Melanie 4.
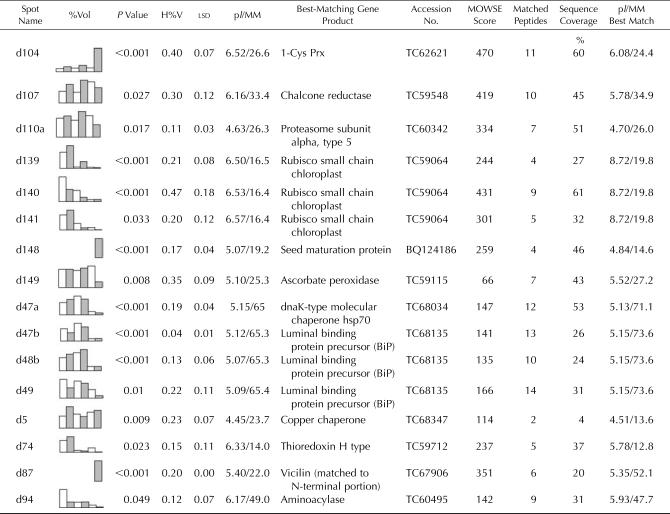
Table I.
Identified M. truncatula proteins whose expression levels changed between Jemalong and 2HA during 2, 5, and 8 weeks of tissue culture
Each entry contains an identifier/spot name, which corresponds to the protein spots marked in Figures 2 and 3. Graphical representation of relative expression (%Vol) is given for each protein spot that showed significant differences between treatments. The columns in the graphs from left to right are: Jemalong 2 weeks (white), 2HA 2 weeks (shaded), Jemalong 5 weeks (white), 2HA 5 weeks (shaded), Jemalong 8 weeks (white), and 2HA 8 weeks (shaded). For each comparison, the P value, the highest %Vol of the six treatments (H%V), and the lsd are given. The P value is from the significant difference between treatments. The pI and relative molecular mass (MM; kD) were calculated from the position on the 2-DE gel. Each entry also contains the best-matching TC identifier (www.tigr.org), the MOWSE score, the number of matched peptides, the percentage of sequence coverage of the match, and the predicted pI and calculated molecular mass (MM; kD) of the protein encoded by the best-matching gene. See “Materials and Methods” for details.
Table II.
Unidentified M. truncatula proteins whose expression levels changed between Jemalong and 2HA during 2, 5, and 8 weeks of tissue culture
Each entry contains an identifier/spot name, which corresponds to the protein spots marked in Figure 2. Graphical representation of relative expression (%Vol) is given for each protein spot that showed significant differences. The columns in the graphs from left to right are: Jemalong 2 weeks (white), 2HA 2 weeks (shaded), Jemalong 5 weeks (white), 2HA 5 weeks (shaded), Jemalong 8 weeks (white), and 2HA 8 weeks (shaded). For each comparison, the P value, the highest %Vol of the six treatments (H%V), and the lsd are given. The pI and relative molecular mass (MM; kD) were calculated from the position on the 2-DE gel.
Many of these proteins (85%) were analyzed by MALDI-TOF-MS, or by tandem MS (MS/MS) if they could not be matched to the identified proteins in the proteome reference map of M. truncatula embryogenic cultures (Imin et al., 2004), and 16 proteins were identified with high confidence as shown in Table I. As expected, Rubisco small chain proteins (spots d139, d140, and d141; Fig. 3) were gradually decreased in both Jemalong and 2HA during explant cultures, which originated from the young leaves. A similar decreasing trend also was observed for an aminocylase (spot d94) and a thioredoxin H (Trx H)-type protein (d74; Fig. 3). Also, as expected, proteins known to be involved in seed formation were expressed only the 8-week-old cultures of the highly embryogenic 2HA line but not in Jemalong. These included a seed maturation protein (d148; Fig. 3) and a vicilin protein (d87). Chaperone proteins (dnaK-type hsp70, spot d47a, and luminal binding proteins d47b, d48b, and d49) showed a decrease in the 8-week cultures in both Jemalong and 2HA, although their expression levels were different. Spot d104 (Fig. 3) was identified as a 1-Cys peroxiredoxin (Prx), which increased to a high level at 8 weeks in 2HA. Interestingly, all the matching M. truncatula ESTs to 1-Cys Prx were only found in the developing reproductive tissues and in the late-stage developing seeds.
DISCUSSION
Comparison of Jemalong and 2HA Responses to Phytohormones
The mutant line Jemalong 2HA was originally isolated from cultivar Jemalong as a highly regenerable seed line that formed embryos at 500-fold more than its near isogenic parent line Jemalong (Nolan et al., 1989; Rose and Nolan, 1995). The resultant plants breed true for this greatly increased embryo formation in culture and, thus, the line 2HA was a valuable resource for legume regeneration (Rose et al., 1999). The regeneration of plants provides a unique stem cell system for investigating meristem formation in embryogenesis research and plant totipotency. Two experimental approaches are available for these studies: one involving the formation of protoplasts from leaf mesophyll cells and the other using leaf explant tissue cultures. In either case, if the calli are maintained on auxin, then rooting and vascularity occur for both cultivar Jemalong and 2HA cultures. If cultures are maintained on cytokinin alone, then only 2HA will form a few embryos, whereas Jemalong calli make no embryos and slowly turn brown. On media containing both auxin and cytokinin, 2HA forms numerous embryos Jemalong generally does not.
M. truncatula cv Jemalong and its embryogenic mutant 2HA leaf explants showed no morphological differences during the early stages of callus development. However, with continued culturing, 2HA formed embryos and vascular tissue, whereas Jemalong formed only vascular tissues. Thus, there are two clear potential pathways of differentiation that can be investigated with the explant system. During this process, there was a differential gene expression resulting in the synthesis of new mRNAs and proteins, which in turn elicited diverse cellular and physiological responses (Chugh and Khurana, 2002). SE studies have sought to describe the array of genes that are activated or differentially expressed during the process. Although numerous genes have been identified as specifically expressed during SE (Mordhorst et al., 1997; Chugh and Khurana, 2002), attempts to identify early embryo control genes have been disappointing (Boutilier et al., 2002). In our system, both NAA and BAP are needed for embryogenic callus formation and maximum embryogenesis; NAA or BAP alone is not sufficient. In the presence of NAA, root differentiation occurs from the proliferating calli. In the presence of auxin and cytokinin, 2HA proliferating calli differentiate into many somatic embryos. As a result of these interactions, developmentally regulated and differentially expressed proteins have been detected and identified during the formation of embryos by applying the 2-DE-based proteomics approach.
Comparison of Proteomes of Protoplast and Leaf Explant Cultures
Some comparisons can be made between the proteome at the globular phase of mesophyll protoplasts grown in culture (Imin et al., 2004) and our results with leaf explant culture. The 2, 5, and 8 weeks of the leaf explant cultures approximate to the 40, 60, and 80 d of the protoplast-derived cultures. The 60-d-old cultures were largely at the globular stage of SE. Interestingly, very early in both experimental systems, the PR10 group of proteins appeared to be the most abundant. While these proteins were first described as pathogenesis-related proteins, they probably have key roles in many plant developmental programs and biotic and abiotic stress responses (Hashimoto et al., 2004). Alternatively, they play a role in phytohormone homeostasis, as they were found to bind cytokinin and brassinosteroids and might thus provide a cellular mechanism to regulate the active cytokinin pool in the cell (Bujacz et al., 2003; Markovic-Housley et al., 2003). Since PR10-1 also is inducible by auxin (Poupard et al., 2001) and both auxin and cytokinin are involved in triggering embryo formation, PR10-1 might be a good candidate protein that mediates phytohormone responses in the cell.
Proteomic Analysis Identified Differentially Expressed Proteins
Proteomic analysis of explant cultures in Jemalong and 2HA identified 54 proteins that are differentially expressed. Although the majority of these differentially displayed proteins were analyzed by MS or by gel matching to the proteome reference map of M. truncatula embryogenic culture (Imin et al., 2004), only 16 proteins were identified with high confidence (Table I). The low percentage of identification was due to either the low abundant nature of the proteins, the poor signal generated from the MS analysis, the possible heavy posttranslational modifications of MS-suitable peptides, or the limitation of the databases used for matching. The major database used for searches was the The Institute for Genomic Research (TIGR) M. truncatula gene index, containing 17,610 TCs and 19,376 singletons (http://www.tigr.org/), which is estimated to represent less than 40% of the expressed genome. It is quite likely that some of the proteins analyzed may not be in the public domain yet. We are establishing a proteome database of M. truncatula SE, and any further identification will be deposited into the publicly available database at http://semele.anu.edu.au/2d/2d.html as part of our ongoing effort. Among the identified proteins, Rubisco small chain proteins (spots d139, d140, and d141; Fig. 3) were gradually decreased in both Jemalong and 2HA during explant cultures, which originated from the young leaves. As such, the decreased trend of Rubisco small chain proteins can be used as a marker for the dedifferentiation and proliferation of the mesophyll tissues. Two of the most abundant proteins were an abscisic acid (ABA)-responsive protein with homology to the pathogenesis-related protein PR10-1 and PR10-1 itself (data not shown) in both Jemalong and 2HA. They were not detected in the young leaves from which the explant cultures originated. Interestingly, they changed little throughout the 8 weeks of culture, suggesting a general role for ABA-responsive proteins and PR10 proteins in cell maintenance or cell defense. Chaperone proteins (dnaK-type hsp70, spot d47a, and luminal binding proteins d47b, d48b, d49), in general, showed a decrease in the 8-week-old cultures in both Jemalong and 2HA, although their expression levels were different. This may imply that a higher level of expression of the chaperones is required for the maintenance of cells during early culture. We also identified proteins involved in seed formation as expressed only in the highly embryogenic 2HA of 8-week-old cultures. These include a seed maturation protein (d148; Fig. 3) and a vicilin (d87). A recent study in M. truncatula showed an increase in accumulation of seed storage proteins, including vicilins, during seed development (Gallardo et al., 2003). SE closely resembles the zygotic embryogenesis, and our approach of inducing embryogenesis in 2HA is a valid comparison of the two systems.
Thioredoxin H Expresses Early in Development
One of the most interesting proteins identified in both studies of proteomic analysis of SE using mesophyll protoplasts (Imin et al., 2004) and explant cultures was Trx H (spot d74; Fig. 3). It appeared early in both protoplast and explant cultures and became undetectable at later stages of cell proliferation. In contrast with Rubisco small chain proteins, which expressed in leaf tissues, the expression of Trx H was not detected in leaf tissue or isolated protoplasts (data not shown), suggesting it is induced only after putting the tissues in culture. There was a higher expression in 2HA than in Jemalong in this early induction process. This MtTrx H protein is highly homologous (71% identical) to the Arabidopsis (Arabidopsis thaliana) ah1 protein, which belongs to the subgroup 1 of the plant Trx H family. The members of the Trx H group are encoded by a multigenic family of eight genes in Arabidopsis (Reichheld et al., 2002). The Trx H group is ubiquitous proteins, which regulate a myriad of posttranscriptional biological functions in eukaryotic cells and are involved in reserve breakdown that sustains early seedling growth (Gelhaye et al., 2004). The reduction of the first subgroup of Trx H is mediated by NADPH-thioredoxin reductase, and many of these reactions take place in specific cells and play a role in the redox regulation of components of the vascular tissues (Gelhaye et al., 2004). The Trx H group of proteins is involved in a wide scope of biological functions, acting as cofactors, transcription regulators, protein binding regulators, protein folding catalysts, growth factors, and antioxidants. In SE formation from explant cultures, MtTrx H was reduced in expression at 5 weeks and could not be detected in the 8-week-old cultures in 2HA. Similarly, it was present in the vegetatively growing cultures from protoplasts but could not be detected in late globular phase cultures (Imin et al., 2004; N. Imin, F. de Jong, U. Mathesius, R.J. Ray, and B.R. Rolfe, unpublished data). These results suggest that MtTrx H plays an important role during early stages of commitment from the vegetative stage to a pathway of cellular differentiation and proliferation.
Expression Pattern of 1-Cys Peroxiredoxin Implies a Role in Embryogenesis
Another interesting protein found in this study is the 1-Cys Prx. As described in “Results,” the expression of 1-Cys Prx (d104; Fig. 3) remained at a minimum level in Jemalong during 8 weeks of culture. By contrast, it showed a slight increase at 5 weeks before reaching a much higher level of expression at 8 weeks in the highly embryogenic line 2HA. All the matching M. truncatula ESTs to 1-Cys Prx (d104) were only found in cDNA libraries of developing reproductive tissues and late-stage developing seeds. Prx are thiol-dependent antioxidants containing one (1-Cys) or two (2-Cys) conserved Cys residues that protect lipids, enzymes, and DNA against reactive oxygen species (Lim et al., 1993; Chae et al., 1994; Netto and Stadtman, 1996; Baier and Dietz, 1997). In plants, antioxidant activity has been demonstrated for the 2-Cys BAS1 (Baier and Dietz, 1997) and the 1-Cys PER1 (Stacy et al., 1996) of barley (Hordeum vulgare). Transgenic plants overexpressing the rice (Oryza sativa) 1-Cys Prx (R1C-Prx) exhibit higher resistance to oxidative stress, suggesting antioxidant activity (Lee et al., 2000). The plant 1-Cys Prx genes pBS128 from brome grass (Goldmark et al., 1992), Per1 in barley (Aalen et al., 1994), AtPER1 in Arabidopsis (Haslekas et al., 1998), and FePer1 in buckwheat (Fagopyrum esculentum; Lewis et al., 2000) are expressed in developing seeds. Recently, Haslekas et al. (2003b) reported that the seed 1-Cys Prx antioxidant AtPER1 is not involved in dormancy but contribute to inhibition of germination during stress.
In mature barley embryos, Per1 transcript levels increase upon addition of exogenous ABA (Aalen et al., 1994). Per1 also is responsive to salt stress in an ABA-dependent manner but to mannitol in an ABA-independent manner (Espelund et al., 1995). In the ABA-insensitive abi3-1 mutant, the AtPER1 transcript level is strongly reduced, suggesting ABI3, a transcription activator, to be a prime regulator of AtPER1 (Haslekas et al., 2003a). An AtPER1::β-glucuronidase expression study showed the AtPER1 promoter drives stage- and tissue-specific expression during embryo and endosperm development, with the earliest expression detected at the late globular stage in a region of the chalazal endosperm termed the chalazal cyst, and at a later stage embryonic AtPER1::β-glucuronidase increased greatly (Haslekas et al., 2003a). In our tissue culture study, we transferred both Jemalong and 2HA calli after 3 weeks of culture to the media to which 1 μm ABA was added to improve embryo formation (see “Materials and Methods”). We did not observe any obvious increase in 1-Cys Prx (d104) expression upon addition of exogenous ABA in Jemalong (Table I). However, 2HA responded with a slight increase at 5 weeks (that was 2 weeks after transferring to media that contained 1 μm ABA) and with a significantly high level of expression at 8 weeks. We propose that 1-Cys Prx is mainly associated with embryo development rather than with a simple response to exogenous ABA.
The genetic mutation(s) of the hyperembryogenic line 2HA is not known, nor are the molecular processes that lead to the SE. We have applied proteomic analyses successfully to dissect molecular processes of an unknown genotype with a known phenotype and detected 53 proteins spots as differentially displayed between hyperembryogenic M. truncatula line 2HA and its near isogenic line Jemalong during 8 weeks of culture. We identified 16 proteins (e.g. Trx H and 1-Cys Prx) associated with SE. This proteomic data will provide a strong reference for the SE of M. truncatula at the protein level.
MATERIALS AND METHODS
Chemicals
All chemicals used were of the highest obtainable grade and are outlined in Guerreiro et al. (1997) and Imin et al. (2001) unless indicated. Milli-Q quality water (Millipore, Bedford, MA) with resistance of greater than 18 MΩ cm was used throughout.
Plant Materials, Growth, and Tissue Culture
Medicago truncatula cv Jemalong and a highly regenerable seed line 2HA, which was derived from cultivar Jemalong (Nolan et al., 1989; Thomas et al., 1990; Rose et al., 1999), were used for the plant growth explant tissue culture as described (Nolan and Rose, 1998; Nolan et al., 2003). Seeds of M. truncatula cv Jemalong were obtained from the National Medicago Collection (Northfield Research Laboratories, Adelaide, Australia). The basal medium used for the culture was P4, which is based on Gamborg's B5 medium as described by Thomas et al. (1990). In the usual culture procedure, leaf explants were plated onto P4 medium containing 10 μm NAA and 4 μm BAP (P4 10:4) for 3 weeks and then transferred to P4 medium with 10 μm NAA, 4 μm BAP, and 1 μm ABA (P4 10:4:1) with subculture to fresh P4 10:4:1 medium every 4 weeks. Cultures were incubated in the dark. This culture procedure has been optimized to maximize embryo number and quality in 2HA cultures (Nolan and Rose, 1998; Nolan et al., 2003).
Extraction of Explant Culture Proteins
Before extraction, several explant cultures of each of the different time points and hormone treatments were pooled together to make an approximately 400-mg sample for each repeat. The extraction procedure was the same as described previously (Imin et al., 2001, 2004). The protein concentration of the supernatant was determined by the Bradford assay (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard. The supernatants were stored in aliquots at −80°C or directly loaded for isoelectric focusing.
Two-Dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis was carried out in a horizontal electrophoresis system, Multiphor II (APB, Uppsala) as described previously (Imin et al., 2001, 2004).
Silver Staining, Colloidal Coomassie Staining, and Image Analysis
Proteins on analytical and preparative 2-DE gels were visualized by silver and colloidal Coomassie blue staining, respectively, as described (Imin et al., 2001, 2004). Stained gels were scanned at 600-dpi resolution using a UMAX ASTRA-2400S scanner fitted with a UTA-II transparency adapter under Photoshop 7.0 (Adobe Systems, Mountain View, CA). Silver-stained gels were scanned using transparency mode, whereas Coomassie-stained gels were scanned using reflective mode with an opaque white background. Spot detection was performed with Melanie 4 (Swiss Institute for Bioinformatics, Geneva). The spot detection parameters were optimized by checking all different protein spots (big, intense, small, and faint) in certain regions of the gels. To identify differentially expressed (accumulated) proteins, first the gels were compared to one another by eye, and spots that changed were recorded. Then, all these protein spots were quantified, and the %Vol were recorded for each spot using the Melanie 4 program. The %Vol is the volume of a spot divided by the total Vol over the whole gel, where the Vol is the integration of intensity of the spot over the spot area. As such, %Vol can be used to minimize the gel-to-gel variations. The %Vol of protein spots were used for statistical analysis using Genstat 7.0 (VSN International, Herts, UK). For quantitative analysis, the %Vol of each spot was quantified using Melanie 4 and analyzed by Genstat 7.0. At least three biological repeats were done for each sample, and at least three reproducible gels were used for the gel analysis. There were substantial variations between the repeats. This is quite different to other proteome analyses we have conducted previously (Imin et al., 2001, 2004; Kerim et al., 2003), in which we observed less variation between the repeats. We think this is due to the nature of tissue culture, where the time course of dedifferentiation and redifferentiation processes are less tightly controlled. As such, we have focused only on protein spots that are consistently changed between the repeats. A restricted maximum likelihood (REML) model was used to determine if there was a statistically significant difference in spot %Vol between treatments. Restricted maximum likelihood is a linear mixed model that uses fixed (treatment) and random (repeat) data to calculate the chi-squared statistic, and it can also handle zero values and unbalanced data. All treatments were compared and tests with P <0.05 were selected and further analyzed by comparing the difference between treatments with the mean lsd calculated from the mean squared error of the means. Where (treatment 1 − treatment 2) > lsd, it was assumed that the %Vol of the spot varied significantly between treatments.
MALDI-TOF-MS and MS/MS Analysis
Proteins spots were excised from Coomassie-stained polyacrylamide gels, then destained and digested with sequencing-grade modified trypsin (Promega, Madison, WI). MALDI-TOF-MS acquisition was performed by an Omniflex MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). For MS/MS analysis, the selected spots were digested as described above and then analyzed on a Thermo Finnigan ProteomeX Workstation (San Jose, CA).
Database Searching
Searches were mainly done against the M. truncatula gene index database (MtGI release 7; June 2003) that contains approximately 37,000 minimally redundant M. truncatula TC entries downloaded from TIGR Web site (ftp://ftp.tigr.org/pub/data/tgi/Medicago_truncatula/). The search engine Mascot (Matrix Science, London) was used to search the MtGI database, for which scores greater than 66 are significant (P < 0.05) as described (Imin et al., 2004).
Acknowledgments
We thank Charles Hocart and Carolyn McKinlay for the MS/MS analysis and Yoko Nitanai for the preparation of plant materials. We are grateful to Ulrike Mathesius, Giel van Noorden, and Peta Holmes for their help in data analysis. We acknowledge Steve Graham's contribution in this project. We also thank Ulrike Mathesius for her critical reading of the manuscript.
This work was supported by the Australian Research Council (grant no. CEO348212).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055277.
References
- Aalen RB, Opsahl-Ferstad HG, Linnestad C, Olsen OA (1994) Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J 5: 385–396 [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz KJ (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Baldwin TC, Domingo C, Schindler T, Seetharaman G, Stacey N, Roberts K (2001) DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol Biol 45: 421–435 [DOI] [PubMed] [Google Scholar]
- Baudino S, Hansen S, Brettschneider R, Hecht VF, Dresselhaus T, Lorz H, Dumas C, Rogowsky PM (2001) Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 213: 1–10 [DOI] [PubMed] [Google Scholar]
- Bestel-Corre G, Dumas-Gaudot E, Gianinazzi S (2004) Proteomics as a tool to monitor plant-microbe endosymbioses in the rhizosphere. Mycorrhiza 14: 1–10 [DOI] [PubMed] [Google Scholar]
- Bestel-Corre G, Dumas-Gaudot E, Poinsot V, Dieu M, Dierick JF, van TD, Remacle J, Gianinazzi-Pearson V, Gianinazzi S (2002) Proteome analysis and identification of symbiosis-related proteins from Medicago truncatula Gaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis 23: 122–137 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer C, Hilbert JL, Vasseur J (1993) Embryogenesis-related protein synthesis and accumulation during early acquisition of somatic embryogenesis competence in Cichorium. Plant Sci 93: 41–53 [Google Scholar]
- Bujacz GD, Pasternak O, Fujimoto Y, Hashimoto Y, Sikorski MM, Jaskolski M (2003) Crystallization and preliminary crystallographic studies of mung bean cytokinin-specific binding protein. Acta Crystallogr D59: 522–525 [DOI] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670–27678 [PubMed] [Google Scholar]
- Choi HH, Sung ZR (1984) Two-dimensional gel analysis of carrot somatic embryonic proteins. Plant Mol Biol Rep 2: 19–25 [Google Scholar]
- Chugh A, Khurana P (2002) Gene expression during somatic embryogenesis—recent advances. Curr Sci 86: 715–730 [Google Scholar]
- Davletova S, Meszaros T, Miskolczi P, Oberschall A, Torok K, Magyar Z, Dudits D, Deak M (2001) Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J Exp Bot 52: 215–221 [PubMed] [Google Scholar]
- De Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodeman VL, Ducreux G (1996) Total protein pattern expression during induction and development of carrot somatic embryos. Plant Sci 120: 57–69 [Google Scholar]
- Dodeman VL, Le Guilloux M, Ducreux G, de Vienne D (1998) Somatic and zygotic embryos of Daucus carota L. display different protein patterns until conversion to plants. Plant Cell Physiol 39: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Espelund M, Debedout JA, Outlaw WH, Jakobsen KS (1995) Environmental and hormonal-regulation of barley late-embryogenesis-abundant (lea) messenger-RNAs is via different signal-transduction pathways. Plant Cell Environ 18: 943–949 [Google Scholar]
- Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult 74: 201–228 [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133: 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42: 265–271 [DOI] [PubMed] [Google Scholar]
- Gianazza E, De Ponti P, Scienza A, Villa P, Martinelli L (1992) Monitoring by two-dimensional electrophoresis somatic embryogenesis in leaf and petiole explants from Vitis. Electrophoresis 13: 203–209 [DOI] [PubMed] [Google Scholar]
- Giroux RW, Pauls KP (1996) Characterization of embryogenesis-related proteins in alfalfa (Medicago sativa). Physiol Plant 96: 585–592 [Google Scholar]
- Giroux RW, Pauls KP (1997) Characterization of somatic embryogenesis-related cDNAs from alfalfa (Medicago sativa L.). Plant Mol Biol 33: 393–404 [DOI] [PubMed] [Google Scholar]
- Goldmark PJ, Curry J, Morris CF, Walker-Simmons MK (1992) Cloning and expression of an embryo-specific mRNA up-regulated in hydrated dormant seeds. Plant Mol Biol 19: 433–441 [DOI] [PubMed] [Google Scholar]
- Guerreiro N, Redmond JW, Rolfe BG, Djordjevic MA (1997) New Rhizobium leguminosarum flavonoid-induced proteins revealed by proteome analysis of differentially displayed proteins. Mol Plant Microbe Interact 10: 506–516 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T (2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol 45: 550–559 [DOI] [PubMed] [Google Scholar]
- Haslekas C, Grini PE, Nordgard SH, Thorstensen T, Viken MK, Nygaard V, Aalen RB (2003. a) ABI3 mediates expression of the peroxiredoxin antioxidant AtPER1 gene and induction by oxidative stress. Plant Mol Biol 53: 313–326 [DOI] [PubMed] [Google Scholar]
- Haslekas C, Stacy RA, Nygaard V, Culianez-Macia FA, Aalen RB (1998) The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol Biol 36: 833–845 [DOI] [PubMed] [Google Scholar]
- Haslekas C, Viken MK, Grini PE, Nygaard V, Nordgard SH, Meza TJ, Aalen RB (2003. b) Seed 1-cysteine peroxiredoxin antioxidants are not involved in dormancy, but contribute to inhibition of germination during stress. Plant Physiol 133: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Imin N, De Jong F, Mathesius U, van Noorden G, Saeed NA, Wang XD, Rose RJ, Rolfe BG (2004) Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Proteomics 4: 1883–1896 [DOI] [PubMed] [Google Scholar]
- Imin N, Kerim T, Weinman JJ, Rolfe BG (2001) Characterisation of rice anther proteins expressed at the young microspore stage. Proteomics 1: 1149–1161 [DOI] [PubMed] [Google Scholar]
- Jiang Q, Gresshoff PM (2002) Shoot control of hypernodulation and aberrant root formation in the har-1 mutant of Lotus japonicus. Funct Plant Biol 29: 1371–1376 [DOI] [PubMed] [Google Scholar]
- Kawahara R, Komamine A, Fukuda H (1995) Isolation and characterization of homeobox-containing genes of carrot. Plant Mol Biol 27: 155–164 [DOI] [PubMed] [Google Scholar]
- Kerim T, Imin N, Weinmann JJ, Rolfe BG (2003) Proteome analysis of male gametophyte development in rice anthers. Proteomics 3: 738–751 [DOI] [PubMed] [Google Scholar]
- Lee KO, Jang HH, Jung BG, Chi YH, Lee JY, Choi YO, Lee JR, Lim CO, Cho MJ, Lee SY (2000) Rice 1Cys-peroxiredoxin over-expressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett 486: 103–106 [DOI] [PubMed] [Google Scholar]
- Lewis ML, Miki K, Ueda T (2000) FePer 1, a gene encoding an evolutionarily conserved 1-Cys peroxiredoxin in buckwheat (Fagopyrum esculentum Moench), is expressed in a seed-specific manner and induced during seed germination. Gene 246: 81–91 [DOI] [PubMed] [Google Scholar]
- Lim YS, Cha MK, Kim HK, Uhm TB, Park JW, Kim K, Kim IH (1993) Removals of hydrogen peroxide and hydroxyl radical by thiol-specific antioxidant protein as a possible role in vivo. Biochem Biophys Res Commun 192: 273–280 [DOI] [PubMed] [Google Scholar]
- Lindzen E, Choi JH (1995) A carrot cDNA encoding an atypical protein kinase homologous to plant calcium-dependent protein kinases. Plant Mol Biol 28: 785–797 [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H (2003) Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol 325: 123–133 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Imin N, Chen H, Djordjevic MA, Weinman JJ, Natera SH, Morris AC, Kerim T, Paul S, Menzel C, et al (2002) Evaluation of proteome reference maps for cross-species identification of proteins by peptide mass fingerprinting. Proteomics 2: 1288–1303 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Keijzers G, Natera SH, Weinman JJ, Djordjevic MA, Rolfe BG (2001) Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1: 1424–1440 [DOI] [PubMed] [Google Scholar]
- Meijer AH, Scarpella E, van Dijk EL, Qin L, Taal AJ, Rueb S, Harrington SE, McCouch SR, Schilperoort RA, Hoge JH (1997) Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J 11: 263–276 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst AP, Toonen MAJ, Devries SC (1997) Plant embryogenesis. Review. Crit Rev Plant Sci 16: 535–576 [Google Scholar]
- Netto LE, Stadtman ER (1996) The iron-catalyzed oxidation of dithiothreitol is a biphasic process: hydrogen peroxide is involved in the initiation of a free radical chain of reactions. Arch Biochem Biophys 333: 233–242 [DOI] [PubMed] [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133: 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Rose RJ (1998) Plant regeneration from cultured Medicago truncatula with particular references to abscisic acid and light treatments. Aust J Bot 46: 151–160 [Google Scholar]
- Nolan KE, Rose RJ, Gorst JR (1989) Regeneration of Medicago truncatula from tissue culture: increased somatic embryogenesis from regenerated plants. Plant Cell Rep 8: 278–281 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Grimes HD (1994) The role of calcium and calmodulin in carrot somatic embryogenesis. Plant Cell Physiol 35: 135–144 [Google Scholar]
- Pedroso MC, Hilbert JL, Vasseur J, Pais MS (1995) Polypeptides associated with the induction of direct somatic embryogenesis in Camellia japonica leaves. 1. Identification of embryo-specific polypeptides. J Exp Bot 46: 1579–1584 [Google Scholar]
- Poupard P, Brunel N, Leduc N, Viemont JD, Strullu DG, Simoneau P (2001) Expression of a Bet v 1 homologue gene encoding a PR 10 protein in birch roots: induction by auxin and localization of the transcripts by in situ hybridization. Aust J Plant Physiol 28: 57–63 [Google Scholar]
- Reichheld JP, Mestres-Ortega D, Laloi C, Meyer Y (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol Biochem 40: 685–690 [Google Scholar]
- Reinbothe C, Tewes A, Reinbothe S (1992) Altered gene expression during somatic embryogenesis in Nicotiana plumbaginifolia and Digitalis lanata. Plant Sci 82: 47–58 [Google Scholar]
- Rose RJ, Nolan KE (1995) Regeneration of Medicago truncatula from protoplasts isolated from kanamycin-sensitive and kanamycin-resistant plants. Plant Cell Rep 14: 349–353 [DOI] [PubMed] [Google Scholar]
- Rose RJ, Nolan KE, Bicego L (1999) The development of the highly regenerable seed line Jemalong 2 HA for transformation of Medicago truncatula: implications for regenerability via somatic embryogenesis. J Plant Physiol 155: 788–791 [Google Scholar]
- Sallandrouze A, Faurobert M, El Maataoui M, Espagnac H (1999) Two-dimensional electrophoretic analysis of proteins associated with somatic embryogenesis development in Cupressus sempervirens L. Electrophoresis 20: 1109–1119 [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, Devries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Somleva MN, Schmidt EDL, de Vries SC (2000) Embryonic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep 19: 718–726 [DOI] [PubMed] [Google Scholar]
- Stacy RA, Munthe E, Steinum T, Sharma B, Aalen RB (1996) A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol 31: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Bozhkov PV, Chu TM, Van Zyl L, Egertsdotter U, Suarez MF, Craig D, Wolfinger RD, Von Arnold S, Sederoff RR (2004) Variation in transcript abundance during somatic embryogenesis in gymnosperms. Tree Physiol 24: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Stasolla C, van Zyl L, Egertsdotter U, Craig D, Liu W, Sederoff RR (2003) The effects of polyethylene glycol on gene expression of developing white spruce somatic embryos. Plant Physiol 131: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC (1991) Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3: 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132: 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Meyer D, Himber C, Steinmetz A (2004) Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol Biochem 42: 35–42 [DOI] [PubMed] [Google Scholar]
- Thomas MR, Johnson LB, White FF (1990) Selection of interspecific somatic hybrids of Medicago truncatula by using Agrobacterium-transformed tissues. Plant Sci 69: 189–198 [Google Scholar]
- Valot B, Gianinazzi S, Eliane DG (2004) Sub-cellular proteomic analysis of a Medicago truncatula root microsomal fraction. Phytochemistry 65: 1721–1732 [DOI] [PubMed] [Google Scholar]
- van Zyl L, Bozhkov PV, Clapham DH, Sederoff RR, von Arnold S (2003) Up, down and up again is a signature global gene expression pattern at the beginning of gymnosperm embryogenesis. Gene Expr Patterns 3: 83–91 [DOI] [PubMed] [Google Scholar]
- van Zyl L, von Arnold S, Bozhkov P, Chen Y, Egertsdotter U, MacKay J, Sederoff R, Shen J, Zelena L, Clapham D (2002) Heterologous array analysis in Pinaceae: hybridization of high density arrays of Pinus taeda cDNA from needles and embryogenic cultures of P. taeda, P. sylvestris or Picea abies. Comp Funct Genomics 3: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Key JL (1982) Isolation of cloned cDNAs to auxin-responsive poly(A)+ RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci USA 79: 7185–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131: 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yang H, Saitou T, Komeda Y, Harada H, Kamada H (1997) Arabidopsis thaliana ECP63 encoding a LEA protein is located in chromosome 4. Gene 184: 83–88 [PubMed] [Google Scholar]
- Yoshida KT, Mizobuchifukuoka R, Sakata M, Takeda G (1995) Detection of embryogenesis- and organogenesis-specific glycoproteins in rice calli. Breed Sci 45: 493–496 [Google Scholar]
- Zimmerman LJ (1993) Somatic embryogenesis: a model for early development in higher plants. Plant Cell 5: 1411–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30: 349–359 [DOI] [PubMed] [Google Scholar]