Abstract
Decades of previous efforts to develop renal-sparing polyene antifungals were misguided by the classic membrane permeabilization model1. Recently, the clinically vital but also highly renal-toxic small-molecule natural product amphotericin B was instead found to kill fungi primarily by forming extramembraneous sponge-like aggregates that extract ergosterol from lipid bilayers2-6. Here we show that rapid and selective extraction of fungal ergosterol can yield potent and renal-sparing polyene antifungals. Cholesterol extraction was found to drive the toxicity of amphotericin B to human renal cells. Our examination of high-resolution structures of amphotericin B sponges in sterol-free and sterol-bound states guided us to a promising structural derivative that does not bind cholesterol and is thus renal sparing. This derivative was also less potent because it extracts ergosterol more slowly. Selective acceleration of ergosterol extraction with a second structural modification yielded a new polyene, AM-2-19, that is renal sparing in mice and primary human renal cells, potent against hundreds of pathogenic fungal strains, resistance evasive following serial passage in vitro and highly efficacious in animal models of invasive fungal infections. Thus, rational tuning of the dynamics of interactions between small molecules may lead to better treatments for fungal infections that still kill millions of people annually7,8 and potentially other resistance-evasive antimicrobials, including those that have recently been shown to operate through supramolecular structures that target specific lipids9.
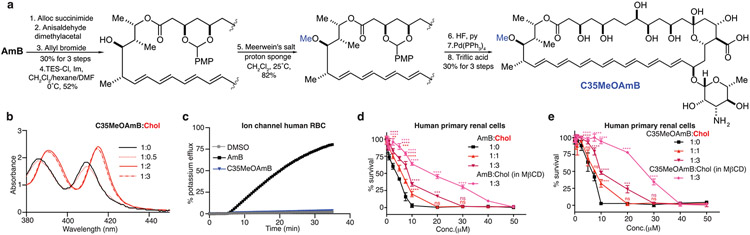
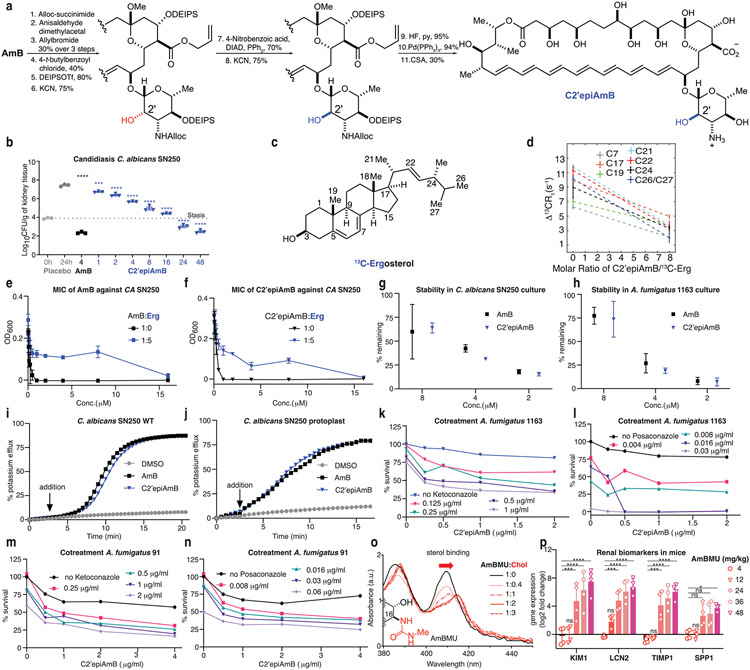
A cholesterol (Chol)-binding but non-membrane-permeabilizing variant of amphotericin B (AmB), C35MeOAmB, retains toxicity to primary human renal cells, and precomplexation of AmB or C35MeOAmB with Chol mitigates this toxicity (Extended Data Fig. 1). Thus, the sterol sponge mechanism2-5,10 also drives renal toxicity, and selective extraction of ergosterol (Erg) over Chol may lead to a renal-sparing polyene antifungal.
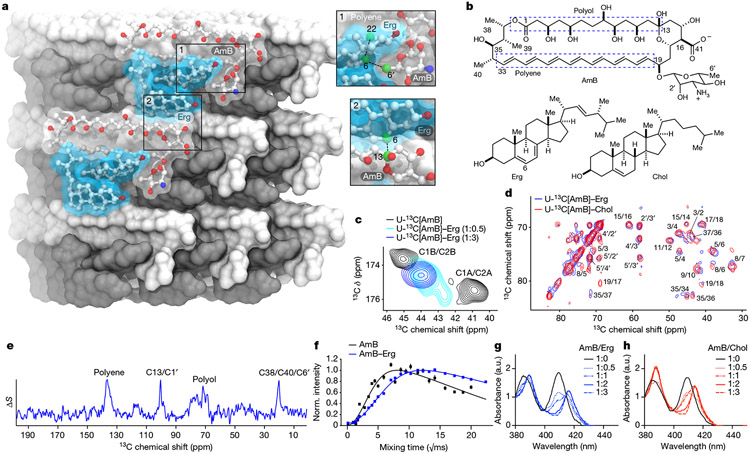
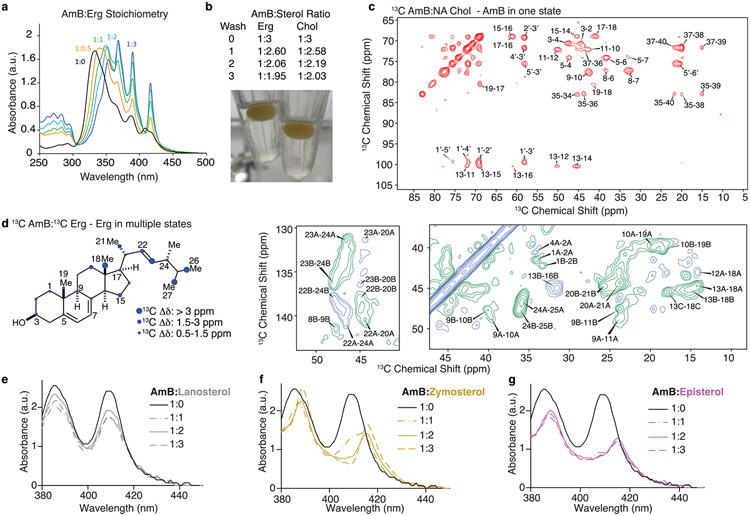
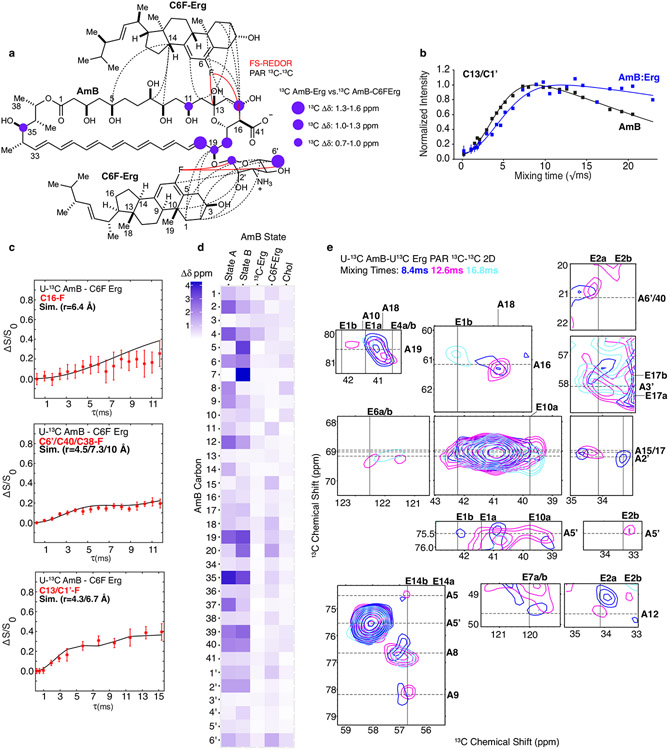
To enable rational pursuit of this objective, we aimed to structurally characterize AmB–Erg and AmB–Chol sponge complexes. We first established a complexation protocol for preparing AmB–sterol complexes with sufficient homogeneity to obtain well-resolved solid-state nuclear magnetic resonance (SSNMR) spectra (Methods). Following Erg titration, 13C–13C two-dimensional (2D) spectra of 13C[AmB] revealed a shift from the two states recently observed in the apo sponge4 to a single state (Fig. 1c and Extended Data Fig. 2c). Changes in the SSNMR spectra as well as changes in the UV–Vis signals following sterol titration both saturate at a 1:2 AmB/sterol ratio, consistent with the same stoichiometry obtained from sterol complexation and washing experiments (Fig. 1c,g and Extended Data Fig. 2a,b). To obtain structures of the sterol-bound complex, Xplor-NIH simulated annealing calculations were carried out using a minimal unit consisting of one AmB and two Erg molecules and 13C–13C and 13C–19F restraints obtained from a suite of high-resolution magic-angle spinning SSNMR studies with 13C[AmB]–13C[Erg] and 13C[AmB]–C6F[Erg] complexes11-17 (Methods, Extended Data Figs. 3 and 5, and Extended Data Table 1). The calculations revealed clathrate-like lattices of AmB molecules encasing Erg guests with sterol A and B rings in close proximity to the C11–C20 region and the mycosamine appendage consistent with broadband and frequency-selective rotational-echo double-resonance (REDOR) experiments, 13C[AmB] chemical shift differences between the 13C[AmB]–Erg and 13C[AmB]–C6F[Erg] complexes, and proton assisted recoupling (PAR) 13C–13C 2D cross-peaks (Fig. 1a,e and Extended Data Fig. 3). Both the C11–C20 region and the mycosamine appendage are highly conserved in the large family of naturally occurring glycosylated polyene macrolides, many examples of which, including natamycin and nystatin, were recently shown to primarily kill fungi through a conserved sterol sponge mechanism of action5. 1H–13C spin-diffusion experiments further revealed dehydration at the polyene and C13/C1′ signals in sterol-saturated samples compared to homogenized AmB consistent with sterol association at these sites (Fig. 1f and Extended Data Fig. 3b). Results from UV–Vis sterol titrations (Fig. 1g,h and Extended Data Fig. 2e-g), sterol-bound 13C–13C spectra (Fig. 1d) and sterol washing experiments carried out on AmB–Chol complexes (Methods and Extended Data Fig. 2b) were very similar to those obtained for AmB bound to Erg, indicating that AmB adopts a highly similar lattice conformation with both sterols.
Fig. 1 ∣. AmB sterol sponge clathrates similarly encapsulate Erg and Chol.
a, SSNMR structure of the AmB–Erg complex, computed with Xplor-NIH simulated annealing, with expansions highlighting interactions between the C6 region of Erg and C13, and polyene and mycosamine regions of AmB observed in 13C{19F} REDOR experiments. b, Molecular structures of AmB, Erg and Chol. c, 13C13C 2D spectra of homogenized 13C[AmB] (black cross-peaks) and 13C[AmB]–Erg complexes prepared with AmB/Erg molar ratios of 1:0.5 (cyan peak) and 1:3 (blue peak) showing the C1–C2 cross-peak collapsing from two peaks to one peak following sterol titration. U-13C, uniformly 13C labelled. d, 13C13C 2D 50-ms dipolar assisted rotational resonance (DARR) overlay of 13C[AmB]–Chol (red) and 13C[AmB]–Erg (blue) showing AmB in the same, single state with each sterol. e,13C{19F} REDOR broadband difference spectrum (ΔS = S0 – S) for U-13C[AmB]–C6F[Erg] at 3.84-ms dephasing showing intensity from 13C[AmB] signals in close proximity to the 19F label at the Erg C6 position. f,13C-detected 1H1H polarization transfers from water to the 13C[AmB] polyene signal for homogenized 13C[AmB] (black line) and the 13C[AmB]–Erg complex (blue lines). Each data point comes from one peak (n = 1). Error bars represent uncertainties from the signal-to-noise ratio of each spectrum. Norm., normalized. g,h, UV–Vis spectra of AmB following titration with increasing molar ratios (0, 0.5, 1, 2 and 3) of Erg (g) and Chol (h). Data in g,h are representative of at least three biological replicates; a.u., arbitrary units.
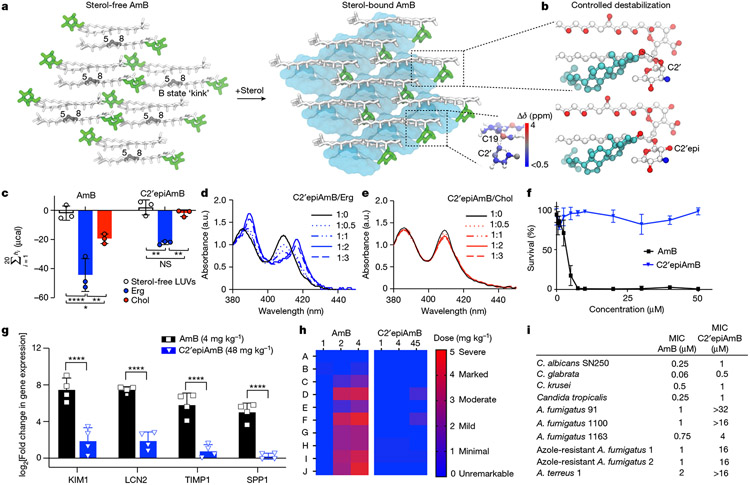
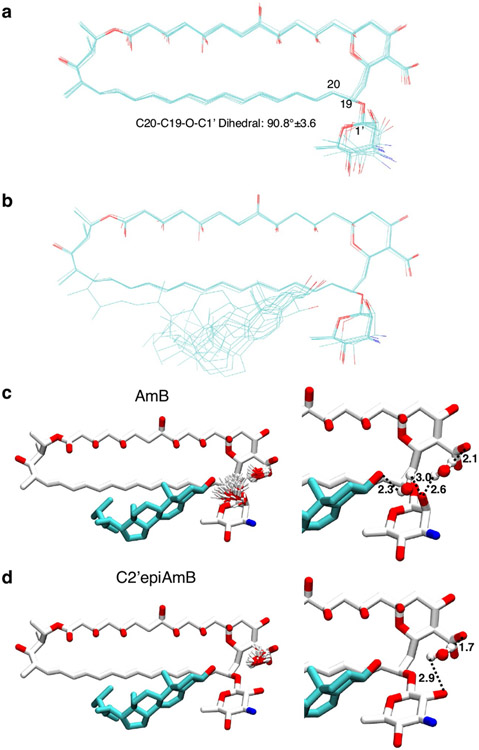
The AmB sponge has higher affinity towards the natural target Erg over Chol, as evidenced by a larger net exotherm following titration of AmB with POPC liposomes containing Erg versus Chol in isothermal calorimetry (ITC) experiments (Fig. 2c). We thus questioned whether controlled destabilization18-22 of these clathrate–guest complexes could eliminate the less favourable binding of Chol while preserving sufficient capacity for binding Erg (Fig. 2b). Understanding how protein conformational changes drive biological function23 can help guide the development of selective interactions between small molecules and proteins24, but this same roadmap has been challenging to achieve for selective interactions between small molecules. To identify structural features to target for selectively disrupting sterol complexation, we compared the AmB sterol-bound complex to the apo-AmB structure (Fig. 2a). The higher-order lattice organizations of the two structures differ with substantially larger AmB nearest-neighbour distances observed in the sterol-bound complex. For six of the ten lowest-energy lattices, including the lowest-energy solution shown in Figs. 1a and 2a,b, a sterol 3β-hydroxyl group, present in both Erg and Chol, is within 4 Å of the C2′-OH of AmB, consistent with direct hydrogen-bonding interactions25. We also noticed close proximity between the AmB polyene and the sterol B-ring (2.8–3.9 Å; Fig. 2b), suggesting an additional π-stacking-like interaction that would be present in the diene-containing Erg but not Chol26-29. At the intramolecular level, the AmB polyene macrolide skeleton shifts from two states (one all staggered and one kinked in the polyol region) to one state (with an all-staggered polyol motif). There is also a large shift in the relative orientation of the mycosamine to the polyene macrolide (Fig. 2a and Extended Data Fig. 4a). Consistent with this conformational change, there is a large chemical shift perturbation at C19 following sterol binding as well as chemical shift perturbations of 1–1.6 ppm at C1′ and C2′ (Fig. 2a, Extended Data Fig. 3d and Supplementary Table 6).
Fig. 2 ∣. Controlled destabilization of AmB–sterol clathrates results in Erg selectivity.
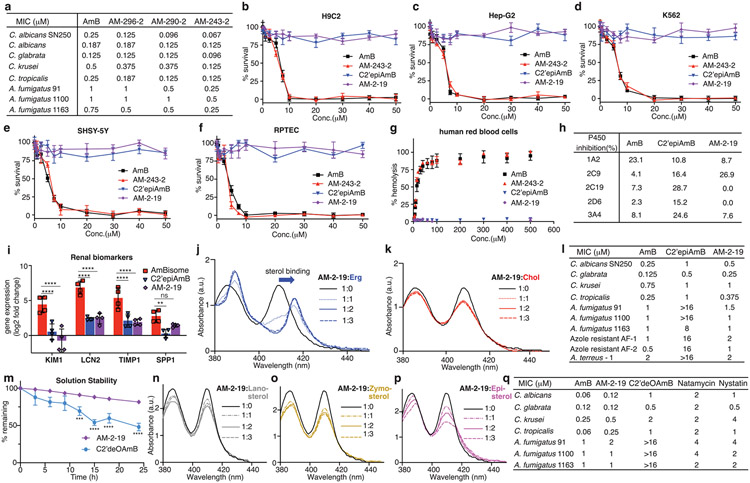
a, Comparison of apo-AmB and sterol-bound AmB clathrate structures highlighting changes in the mycosamine orientation, with the inset showing chemical shift perturbations relative to the apo state A. b, Expansion of the sterol-bound complex showing proximity of the sterol 3β-OH to the C2′OH and disruption of this interaction following in silico epimerization of the C2′centre. c, ITC assays comparing C2′epiAmB and AmB binding affinity to sterol-free, Erg-containing and Chol-containing large unilamellar vesicles (LUVs; n = 3). (AmB–Erg binding data reported in ref. 5.) Pairwise statistical analysis by two-way analysis of variance (ANOVA) with Tukey’s multiple comparison test; *P = 0.0245, **P = 0.0019, ****P < 0.0001, **P = 0.0022, **P = 0.0089. NS, not significant. d,e, UV–Vis spectra of C2′epiAmB following titration with increasing molar ratios (0, 0.5, 1, 2 and 3) of Erg (d) and Chol (e); a.u., arbitrary units. f, C2′epiAmB does not kill human primary renal cells in vitro up to 50 μM concentration (n = 3). g, C2′epiAmB does not elevate kidney damage biomarkers in female CD-1 mice measured 24 h post single IV dosage (n = 4 per group). Pairwise statistical analyses by two-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001. h, Heatmap representation of kidney histopathological scores (A, mixed-cell infiltrate, interstitial; B, tubular basophilia, cortex, medulla; C, Tubular cellular casts, cortex; D, tubular cellular casts, medulla; E, tubular degeneration and necrosis, cortex; F, tubular degeneration and necrosis, medulla; G, tubular dilatation, cortex; H, tubular protein casts, cortex; I, tubular protein casts, medulla; J, vascular congestion, medulla) in mice 24 h post single IV dosage of AmB and C2′epiAmB (n = 4 per group). Both AmB and C2′epiAmB were formulated as 1:2 deoxycholate to improve aqueous solubility. i, Comparison of AmB and C2′epiAmB MICs. Results are means ± s.d. Data presented in d,e are all representative of at least three independent experiments.
Given the compelling evidence—major change in mycosamine orientation following sterol binding, low-energy SSNMR structures consistent with direct hydrogen bonding from the C2′OH to the sterol 3β-OH (Fig. 2b and Extended Data Fig. 4b), further evidence for water-bridging interactions between C2′-OH and the 3β-OH suggested by molecular dynamics simulations on a hydrated sterol-bound AmB structure (Extended Data Fig. 4c), previous evidence for water-bridged hydrogen bonding between the C2′ and C13 hydroxyl groups4,30, and our previous observation that deletion of the C2′-OH group results in an AmB derivative, C2′deoxyAmB, with improved selectivity for Erg over Chol10—we reasoned that hydrogen bonding interactions involving C2′-OH could play an important role in stabilizing the sterol-bound conformations. As is common with 2-deoxysugar appendages that lack an electron-withdrawing group at C2′ and are thus readily hydrolysed through intermediate oxocarbenium ion formation, C2′deOAmB proved to be unstable (Extended Data Fig. 7m) and thus impractical for scalable synthesis10,31. Guided by our new structural understanding of AmB–sterol sponges, we reasoned that stabilizing interactions involving C2′-OH could alternatively be disrupted through epimerization of the C2′ stereogenic centre (C2′epiAmB), which in turn might lead to controlled destabilization of the sterol-bound clathrates while preserving stability of the glycosylated polyene macrolide (Fig. 2b,c). Consistent with this hypothesis, molecular dynamics simulations of a C2′-epimerized sterol-bound AmB structure showed a disruption of hydrogen-bonding interactions between the C2′-OH and 3β-OH (Extended Data Fig. 4d).
We thus synthesized C2′epiAmB through an 11-step route (Extended Data Fig. 6a and Supplementary Fig. 1). Consistent with our hypothesis, ITC heat exchange showed that, in contrast to AmB, which binds both Erg and Chol, C2′epiAmB shows no binding with Chol with retained, albeit diminished, capacity to bind Erg (Fig. 2c). These results are consistent with UV–Vis-based sterol binding assays (Fig. 2d,e). This lack of Chol binding translated to substantially decreased toxicity of C2′epiAmB relative to AmB against human primary renal cells (Fig. 2f), as well as human red blood cells, K562 cells, HepG2 liver cells, H9c2 myocardial cells and SH-SY5Y neural cells (Extended Data Fig. 7b-g). Specifically, in contrast to AmB, C2′epiAmB showed no toxicity against any of these cells even up to the highest concentration tested (50 μM, 500 μM for red blood cells). The renal-sparing nature of C2′epiAmB was also evaluated in mice. In contrast to AmB, which caused substantial increases in renal injury biomarkers32,33 after dosing at 4 mg kg−1 and moderate to severe histopathological changes consistent with renal damage following dosing as low as 2 mg kg−1, C2′epiAmB causes no elevation of renal biomarkers and little to no change in renal histopathology even at doses of ≥45 mg kg−1 (Fig. 2g,h).
Encouraged by these results, we next questioned whether the retained, albeit diminished, capacity for C2′epiAmB to bind Erg would be sufficient to retain potent antifungal activity against human pathogens. Both in vitro and in vivo studies revealed that C2′epiAmB is less potent than AmB (Fig. 2i and Extended Data Fig. 6b). Notably, however, this decrease in potency varied substantially between the different pathogens. In some strains the decrease was small (Candida krusei, minimum inhibitory concentrations (MICs): 0.5 μM for AmB and 1 μM for C2′epiAmB), whereas in others it was large (Aspergillus fumigatus 91, MICs: 1 μM for AmB and >32 μM for C2′epiAmB). This suggested that factor(s) other than the strength of Erg binding may also be driving this loss of potency. Mechanistic and biophysical studies confirmed that C2′epiAmB primarily kills fungi through the same sterol sponge mechanism and retains AmB-like in cellulo stability (Extended Data Fig. 6c-j).
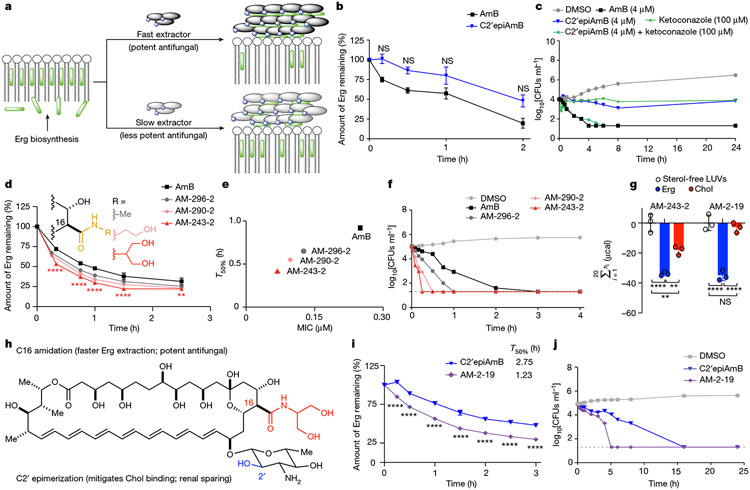
Given the retained sterol extraction mechanism and pathogen-dependent variation in potency, we reasoned that C2′epiAmB may extract Erg more slowly than AmB thus enabling Erg biosynthesis to variably compete with Erg extraction in different pathogen strains (Fig. 3a). Evaluation of Erg extraction rates and killing kinetics confirmed that C2′epiAmB extracts Erg and kills fungi more slowly than AmB (Fig. 3b,c). Co-administration of C2′epiAmB and an Erg biosynthesis inhibitor, ketoconazole, restored AmB-like potency and killing kinetics (Fig. 3c). Such co-administration also restored AmB-like potency against Aspergillus strains that were highly resistant to C2′epiAmB (A. fumigatus 91 and A. fumigatus 1163; Extended Data Fig. 6k-n). These data are all consistent with the rate competition model (Fig. 3a).
Fig. 3 ∣. Kinetics of Erg extraction enabling tuning of antifungal efficacy.
a, Competitive kinetics model of sterol encapsulation for AmB analogues. b, C2′epiAmB extracts Erg more slowly than AmB from C. albicans SN250 at 5 μM (n = 4 for 0.167 and 0.5 h and n = 3 for 1.0 and 2.0 h time points). Pairwise comparison with AmB at each time point using two-way ANOVA with Tukey’s multiple comparison test. c, Cotreatment of C. albicans SN250 with the Erg biosynthesis inhibitor ketoconazole promotes faster killing and restores potency for C2′epiAmB. CFUs, colony-forming units. d, At 5 μM, hyper potent AmB amides extract Erg more quickly than the parent AmB (n = 3 per time point) from C. albicans SN250. Pairwise comparison with AmB at each time point using two-way ANOVA with Tukey’s multiple comparison test; **P = 0.0064, ****P < 0.0001. e, Kinetics versus efficacy for C. albicans SN250, showing T50% (time required for 50% Erg extraction) versus observed MIC. f, Rate of cell killing at 4 μM (n = 2 per sample per time point) for C. albicans SN250. g, ITC data showing retained Chol binding for AM-243-2 and selectivity for Erg over Chol for AM-2-19 (n = 3). Pairwise comparison using two-way ANOVA with Tukey’s multiple comparison test; **P = 0.0012, **P = 0.0038, ****P < 0.0001. h, The hybrid design strategy combines the renal-sparing nature of C2′ epimerization and potency-promoting C41 amidation in AM-2-19. i, AM-2-19 extracts Erg more quickly than C2′epiAmB (5 μM; n = 3 per time point) from C. albicans SN250. Pairwise comparison with C2′epiAmB at each time point using two-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001. j, AM-2-19 kills C. albicans SN250 more quickly than C2′epiAmB (4 μM). Results are means ± s.d. For c,f,j a nominal value of 20 CFUs ml−1 was assigned on the basis of the dilution factor for plates with no fungal growth.
Guided by these insights, rather than targeting increased sterol binding affinity through restabilizing the AmB–sterol complex, which we reasoned might lead to recovery of Chol binding and renal toxicity, we sought to selectively increase Erg extraction kinetics. In this vein, we were intrigued by observations that converting the carboxylic acid at the C16 position of AmB into a non-ionizable functional group can in some cases increase antifungal potency34-36, and we questioned whether such effects are caused by an increased rate of Erg extraction in the corresponding non-amphoteric derivatives. Accordingly, we found that a series of more potent AmB-C16 amides extract Erg more quickly than AmB (Fig. 3d; T50% values: 0.92 h for AmB, 0.65 h for AM-296-2, 0.55 h for AM-290-2 and 0.42 h for AM-243-2). This trend in Erg extraction rates matched the trends in increasingly lower MICs (Fig. 3e and Extended Data Fig. 7a; Candida albicans SN250 MICs: 0.25 μM for AmB, 0.125 μM for AM-296-2, 0.096 μM for AM-290-2 and 0.067 μM for AM-243-2) and in faster killing kinetics (Fig. 3f). These increases in potency were not accompanied by increases in Erg binding by ITC (AM-243-2 ΔΔHErg = −33.9 ± 1.2 μcal, ΔΔHChol = −18.1 ± 2.9 μcal; AmB ΔΔHErg = −42.4 ± 11.2 μcal, ΔΔHChol = −17.6 ± 3.3 μcal; ΔΔHsterol = ΔHsterol – ΔHsterol-free). Thus, increasing Erg extraction kinetics without increasing Erg binding affinity can drive increased fungicidal activity.
AM-243-2 is toxic to human cells, consistent with retained Chol binding (Fig. 3g and Extended Data Fig. 7b-f). In contrast to C2′epiAmB, previously reported C16-modified AmB urea derivatives with an improved therapeutic index turned out to also retain the capacity for Chol binding through the sensitive UV–Vis assay reported herein and had unacceptable toxicity in more advanced preclinical studies (Extended Data Fig. 6o,p). On the basis of the structural insights described above, we reasoned that controlled destabilization of the clathrate–sterol complex through C2′ epimerization would preclude the binding of Chol guests irrespective of further modifications at C16. We therefore reasoned that combining this Chol binding and thus toxicity-eliminating C2′ epimerization with the Erg extraction rate and thus potency-increasing C16 amidation would selectively increase Erg extraction kinetics, providing a new class of renal-sparing and potent polyene antifungals.
We paired the C2′ epimerization with the most potency-promoting serinol side chain to form the hybrid derivative AM-2-19 (Fig. 3h). AM-2-19 binds to Erg but not to Chol or other sterols lacking a diene motif in the B-ring (Fig. 3g and Extended Data Fig. 7j,k,n-p), is non-toxic to primary human renal and other mammalian cells (Extended Data Fig. 7b-g) and is renal sparing in mice (Extended Data Fig. 7i). AM-2-19 also extracts Erg more quickly and kills fungi more quickly than C2′epiAmB (Fig. 3i,j; T50% values: 2.75 h for C2′epiAmB and 1.23 h for AM-2-19) and recovers excellent potency against a panel of pathogenic fungi, including Aspergillus strains that had proved to be highly resistant to both C2′epiAmB and C2′deOAmB (Fig. 2i and Extended Data Fig. 7l,n). It is notable that both of these chemical modifications are positioned on the portion of AmB that is conserved in all of the glycosylated polyene macrolide natural products that operate through the same sterol sponge mechanism5.
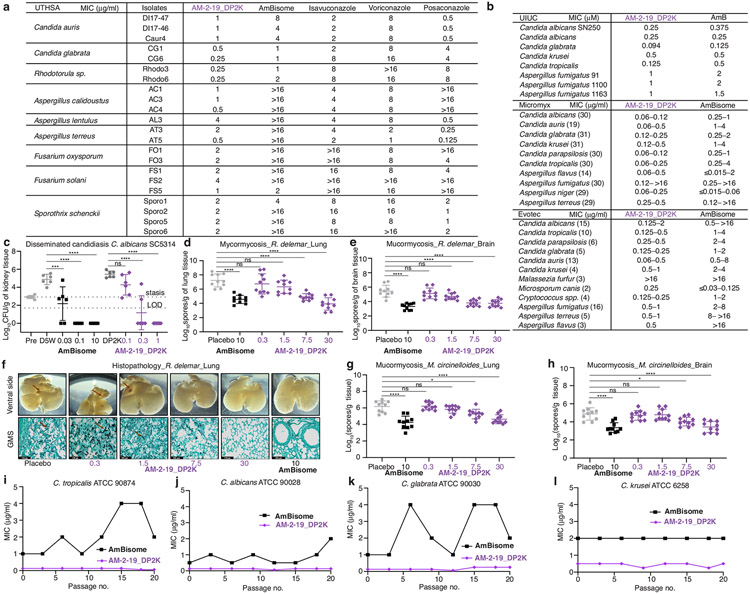
Encouraged by the restored efficacy and retained tolerability, we tested AM-2-19 in a series of translation-oriented preclinical studies. AM-2-19 showed no evidence of human ether-a-go-go related gene (hERG) potassium current inhibition, genotoxicity or drug–drug interactions (Extended Data Fig. 7h and Supplementary Information). To maintain stable and plasma-compatible aqueous solutions, a simple micellar formulation of AM-2-19 as a 1:3 mixture with commercially available block co-polymer distearoyl-rac-glycerol-polyethylene glycol 2000 (DP2K) was developed (AM-2-19_DP2K, Methods). In vivo drug safety evaluation revealed little to no elevation of clinical biomarkers or histopathological changes in the kidney up to 45 mg kg−1 single dose of AM-2-19_DP2K in healthy female CD-1 mice (Fig. 4a and Extended Data Fig. 7i).
Fig. 4 ∣. In vivo tolerability and efficacy of AM-2-19_DP2K.
a, Histopathological analysis of kidneys collected 24 h post single IV dosing from mice treated with AmBisome (n = 3 per group) or AM-2-19_DP2K (n = 4 per group) (A, mixed cell infiltrate, interstitial; B, tubular basophilia, cortex; C, tubular cellular casts, cortex; D, tubular cellular casts, medulla; E, tubular degeneration and necrosis, cortex; F, tubular degeneration and necrosis, medulla; G, tubular dilatation, cortex; H, tubular protein casts, cortex; I, tubular protein casts, medulla; J, vascular congestion, medulla). b, Representative examples of in vitro efficacy of AM-2-19_DP2K and the comparator AmBisome tested at UT Health San Antonio (number of isolates in parenthesis; 100% inhibition reported). c, In vivo efficacy of AM-2-19_DP2K and the comparator AmBisome in a neutropenic disseminated candidiasis mouse model infected with C. glabrata ATCC2001 after 5 days of daily IV dosing (n = 6 per group). LOD, limit of detection. One-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0008, ****P < 0.0001, **P = 0.0036. d, Efficacy of AM-2-19_DP2K and the comparator AmBisome in a neutropenic disseminated aspergillosis mouse model infected with A. fumigatus 1163 after 4 days of daily IV dosing (n = 6 per group). One-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001. e, Neutropenic invasive pulmonary aspergillosis mouse model infected with A. fumigatus ATCC 204305 after 4 days of daily IV dosing (n = 8 per group). One-way ANOVA with Tukey’s multiple comparison test; *P = 0.0117, **P = 0.0032, ****P < 0.0001. f, Neutropenic disseminated aspergillosis mouse model infected with A. terreus 49 after 10 days of daily IV dosing (n = 10 per group). One-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0008, ****P < 0.0001. g,h, Survival curves of a neutropenic invasive pulmonary mucormycosis mouse model infected with R. delemar (n = 10 per group; g) and Mucor circinelloides (n = 10 per group; h) after 4 days of daily IV dose. Pairwise comparisons with the placebo using a log-rank (Mantel–Cox) test. Dosing units are milligrams per kilogram. Results are means ± s.d. Fungal burdens below the limit of detection were given a nominal value of 1 CFU g−1. Concentrations (a–h) listed reflect the mass of active ingredients.
We also tested the antifungal activity of AM-2-19_DP2K head-to-head with AmBisome and other clinical antifungal comparators against >500 different pathogenic fungal strains in 4 different research laboratories (Fig. 4b and Extended Data Fig. 8b). The potency and pathogen scope of AM-2-19_DP2K generally match or surpass those of AmBisome. AM-2-19_DP2K also showed potent antifungal activity against several isolates that were relatively resistant to AmBisome and some or all of the leading azoles (Extended Data Fig. 8a). Spontaneous mutation evaluation revealed that AM-2-19_DP2K retains its potency after multiple passages equal to or better than AmBisome (Extended Data Fig. 8i-l).
The in vivo efficacy of AM-2-19_DP2K was evaluated in candidiasis, aspergillosis and mucormycosis mouse models. In neutropenic mice infected with Candida glabrata ATCC2001 and treated daily with AM-2-19_DP2K for 5 days (intravenous (IV)) a dose-dependent decrease of renal fungal burden was recorded with complete eradication at ≥10 mg kg−1 dose (Fig. 4c). In immunocompetent mice infected with C. albicans SC5314, a similar dose-dependent decrease of fungal burden (every 24 hours(q24h), 7 days) and eradication at ≥0.3 mg kg−1 dose was recorded (Extended Data Fig. 8c).
The antifungal potency against Aspergillus was evaluated in both disseminated (Fig. 4d,f) and pulmonary (Fig. 4e) infection mouse models, in which we observed a dose-dependent decrease in fungal burden and, in some cases, eradication of fungal burden at higher doses, including against C2′epiAmB-resistant A. fumigatus 1163 (Fig. 4d). We also evaluated the antifungal potency of AM-2-19_DP2K against Aspergillus terreus 49, which is highly resistant to AmBisome but is sensitive to AM-2-19_DP2K in vitro (MIC values: 8 μg ml−1 for AmBisome and 0.5 μg ml−1 for AM-2-19_DP2K; Extended Data Fig. 8b). The fungal burden in AmBisome-treated mice after daily dosing for 10 days remained above the level of stasis, whereas AM-2-19_DP2K-treated animals demonstrated a dose-dependent decrease of kidney fungal burden (Fig. 4f). Thus, the exceptional potency of AM-2-19_DP2K in vitro against drug-resistant strains translates to superior in vivo efficacy (Fig. 4b and Extended Data Fig. 8a).
In the Rhizopus delemar-induced mouse mucormycosis model, AM-2-19_DP2K-treated animals showed dose-dependent improved survival (Fig. 4g; minimum inhibitory concentration for 90% inhibition (MIC90) values: 0.0315 μg ml−1 for AmBisome and 0.250 μg ml−1 for AM-2-19_DP2K) and a dose-dependent decrease in fungal burden in lung and brain tissues (Extended Data Fig. 7d,e). Histopathological examination of lungs collected from mice infected with R. delemar and treated with either AM-2-19_DP2K or AmBisome showed considerable improvement in lung histology with fewer fungal abscesses and improved architecture that was directly proportional to the dose used in treatment when compared to placebo-treated mice (Extended Data Fig. 8f). AM-2-19_DP2K also demonstrated dose-dependent efficacy that was comparable to that of AmBisome in prolonging survival (Fig. 4h; MIC90 values: 0.0625 μg ml−1 for AmBisome and 0.125 μg ml−1 for AM-2-19_DP2K) and decreasing tissue fungal burden (Extended Data Fig. 8g,h) of neutropenic mice intratracheally infected with Mucor circineloides, another widely isolated Mucorales from mucormycosis37.
Although no pressure for Erg selectivity is present in nature, and it remained unclear whether it was possible to achieve, we have provided a structural rationale for imparting selectivity for Erg over Chol and demonstrated that the selectivity predicted by atomistic-resolution structural models translates to renal-sparing properties in primary human renal cells and in mice. Thus, advanced understanding of the mechanism by which polyene macrolide natural products kill fungal and human cells enabled the selective tuning of sterol extraction kinetics to yield a new polyene antifungal that is both renal sparing and highly potent in clinically relevant animal models of invasive fungal infections. These insights more broadly suggest a conceptual framework for rationally optimizing other members of the large family of glycosylated polyene macrolide natural products and potentially other resistance-evasive antimicrobials24 including those that have recently been shown to operate through supramolecular structures that target specific lipids9.
Methods
General information
Unless mentioned oxtherwise, commercial reagents were stored and used as per the manufacturer’s recommendation. The sample size of the experiments was selected on the basis of literature precedence and preliminary experiments to adequately show the difference in outcome between different groups. No statistical methods were used to predetermine sample size. For in vivo experiments, animals were randomly assigned to different groups. The species and the sex of the animals are specified in the protocol. Histopathological analysis of tissues was carried out following the blind evaluation technique. All experiments involving mammalian cell lines were carried out following a BSL2 safety protocol and the set up was approved by the Division of Research Safety, University of Illinois at Urbana-Champaign. Data and statistics were analysed using Microsoft Excel for Mac (v.16.76 (23081101)) and GraphPad Prism 9 (v.9.4.1) on the Mac operating system. Chemical structures were drawn using Chemdraw 21.0.0. NMR data were analysed using MestReNova 14.2.3 and NMRFAM-SPARKY v.1.470. SSNMR data were processed using NMRPipe System v.10.9, VnmrJ (v.2.1B), SpinSight (v.4.3.2) and MATLAB v.R2017b. Xplor-NIH (3.0) was used for simulated annealing calculations. Visual Molecular Dynamics v.1.9.4 was used for molecular representations. NAMD2 (v.2.14) was used molecular dynamics simulations. ImageJ v.1.53a was used for histopathology image processing. Final figures and images were processed using Adobe Illustrator 26.1.
Isotopic expression and purification of AmB and Erg
Isotopically 13C-enriched AmB was expressed from cultures of Streptomyces nodosus (ATCC 14899) and isotopically 13C-enriched Erg was expressed from cultures of Saccharomyces cerevisiae as previously described3,4,38,39. Both compounds were purified as previously described3,4. Natural-abundance Erg and Chol were recrystallized in acetone before use.
Preparation of AmB–sterol complex samples for SSNMR
To obtain well-resolved SSNMR data suitable for structure determination, it was necessary to develop a new sample preparation method. First, a 15 mM solution of reagent-grade AmB was prepared in dimethylsulfoxide (DMSO), and a 70 mM solution of Erg was prepared in CDCl3. The concentrations of the stock solutions were determined by UV–Vis absorbance (406 nm, 164 mM−1 cm−1 extinction coefficient for AmB (ref. 40); 282 nm, 10.4 mM−1 cm−1 extinction coefficient for Erg). The 70 mM Erg stock was diluted to 1 mM in DMSO. A 900 μl volume of the 1 mM Erg stock was added to a small volume of AmB stock to obtain a 1:3 molar ratio of AmB to sterol. The DMSO mixture was added to 30 ml of rapidly stirring 1× PBS and stirred at room temperature for 1 min. Next, the sample was pelleted by ultracentrifugation at 40,000 r.p.m. for 1 h. After removing the supernatant, pellets were transferred and centrifuged in an Eppendorf table-top centrifuge at 10,000 r.p.m. for 45 min. Samples were centrifuged two additional times, one ultracentrifugation at 50,000 r.p.m. for 1 h and a final ultracentrifugation at 80,000 r.p.m. for 1 h. After the final centrifugation, samples were transferred to 3.2-mm or 1.6-mm magic-angle spinning rotors using a custom-designed rotor packing device41.
Paramagnetic relaxation enhancement sample preparation
Liposomal vesicles with and without 5% 16-DOXYL-labelled POPC were prepared as previously described3.
13C 1D and 13C–13C 2D SSNMR spectroscopy
Magic-angle spinning (MAS) SSNMR experiments were conducted at 14.1 T (600 MHz), 17.6 T (750 MHz) and 21.1 T (900 MHz) on InfinityPlus (600 MHz), Varian Instruments VNMRS (750 MHz), and Bruker Avance III HD (900 MHz) spectrometers using 3.2-mm Balun (Varian) and 1.6-mm FastMAS probes (Varian and Phoenix NMR). Spinning was controlled with a Varian or Phoenix MAS controller to 23,809 Hz (750 MHz 1H frequency), 12,500 Hz (600 MHz 1H frequency) and 16,900 Hz (900 MHz 1H frequency). Typical pulse widths were about 1.7–2.5 μs for 1H, and about 1.5–3 μs for 13C. High-power decoupling was used during evolution and acquisition, and sample temperature was maintained at 0 ± 5 °C. Spectra were processed with 20 Hz and 75–120 Hz line broadening for 1D and 2D experiments, respectively. Chemical shifts were externally referenced using adamantane with the downfield peak set to 40.48 ppm (ref. 42). Chemical shifts for each sample were obtained using 50 or 100 ms DARR or combined -driven (CORD) mixing as well as 13C–13C supercycled POST-C5 (SPC-5) with a short mixing time to obtain one- and two-bond correlations13,16. Distance restraints were obtained from 500-ms CORD and 12.6-ms PAR spectra12,15. Spectra were processed with NMRPipe with back linear prediction and polynomial baseline correction applied to the direct dimension. Lorentzian-to-Gaussian apodization, phase-shifted sine bells and zero filling were applied to both dimensions before Fourier transformation and phase correction. The spectra were analysed using NMRFAM-Sparky43.
REDOR experiments
13C{19F} REDOR experiments were conducted at 17.6 T (750 MHz 1H frequency) on an Agilent Technologies VNMRS spectrometer with a 1.6-mm HFXY FastMAS probe. Samples were packed into 1.6-mm FastMAS rotors using low-19F-background spacers and caps. The spinning was controlled at 16,667 ± 10 Hz by a Varian MAS controller. Broadband 1D 13C{19F} REDOR experiments44 were carried out as described previously with a hard refocusing pulse applied to the 13C channel and dipolar recoupling, rotor-synchronized 180° pulses applied to the 19F channel14. Typical 90° pulse widths after optimization were about 3 μs for 1H, about 2 μs for 13C and about 2.5–3 μs for 19F. The cross-polarization (CP) contact time used was 1.2 ms. Two-pulse phase modulation (TPPM) decoupling of 66 kHz was applied during the REDOR period and 60 kHz small phase incremental alternation (SPINAL) decoupling was used during acquisition. Typical refocusing 13C 180° pulse widths were about 5–6 μs. 19F 180° pulse widths were about 6 μs. For frequency-selective (FS) experiments, a Gaussian refocusing 13C pulse was applied with pulse widths of 500–1,200 μs. REDOR dephasing times were varied from 0.14 to 15.6 ms. Spectra obtained from the frequency-selective experiments45 were processed in VNMRJ using 0.5–1.0-ppm line broadening. The intensities of the S and S0 peaks were measured in VNMRJ and the ΔS/S0 values were plotted and fitted to Bessel functions to obtain the dipolar couplings46. Fitting was carried out using MATLAB v.R2017b (ref. 47).
1H–13C spin-diffusion experiments
1H–13C spin-diffusion correlation experiments were carried out as previously described3,48.
Paramagnetic relaxation enhancement experiments
T1 values were measured using a standard T1 inversion-recovery pulse sequence with a 1.5-s pulse delay and a CP contact time of 1.8 ms. Experiments were conducted on a 600 MHz InfinityPlus spectrometer with temperatures maintained at 20 ± 5 °C. Data were processed and fitted with Varian SpinSight software v.4.3.2, and longitudinal 13C paramagnetic relaxation enhancement (PRE) values were calculated as previously described3.
Xplor-NIH calculations
AmB parameters were taken from ref. 49, and the starting geometry was obtained from comparing chemical shifts to geometries calculated by density functional theory as previously described4. CHARMM36 parameter files were used for the Erg molecules, and starting structures were obtained from Cambridge Crystallographic Data Centre no. 146357. Xplor-NIH v.3.0 was used for structure calculations.
The structure calculation used portions of the probabilistic assignment algorithm for automated structure determination in addition to manually assigned cross-peaks to generate restraint tables4,50. Totals of 125 AmB monomers and 250 Erg molecules were arranged into a 5 × 5 × 5 lattice of subunits with each subunit containing 1 AmB molecule and 2 Erg molecules, corresponding to the 2 Erg ring states identified in the SSNMR spectra (Extended Data Fig. 2). Each subunit corresponded to a unique segment ID in the Xplor-NIH structure calculations with AmB assigned to resid 1, Erg state A assigned to resid 2 and Erg state B assigned to resid 3. The 1 AmB, 2 Erg subunit was used as the primary subunit from which the coordinates of the remaining 125 subunits were generated by translations along the three Cartesian axes within Xplor-NIH’s strict symmetry implementation11. Weighted intermolecular AmB–AmB and AmB–Erg distance restraints from the experimental data were used to maintain lattice packing consistent with the staggered AmB–AmB interactions described in Extended Data Fig. 5 and the interactions between the Erg rings and the conserved GPM motif described in Extended Data Fig. 3. Ambiguous distance restraints from 13C–13C 2D 12.6-ms PAR experiments carried out on a 13C[AmB]–13C[Erg] complex, ambiguous restraints from 13C–13C 2D 500-ms CORD experiments carried out on a 13C[AmB]–C6F[Erg] complex and ambiguous and unambiguous 13C–19F restraints from FS-REDOR experiments carried out on a 13C[AmB]–C6F[Erg] complex were included in the calculation (Supplementary Table 8).
The annealing calculation of 1,500 structures was carried out as described previously4. The calculation was run on HTCondor through the Center for High Throughput Computing at the University of Wisconsin Madison.
Molecular dynamics simulations
The system was prepared using the lowest-energy AmB–Erg lattice (Fig. 1a) from the calculation reported in Extended Data Table 1. First, the system was solvated with water, and 150 mM NaCl was added. The atoms from the sponge were constrained using the fixedAtoms flag in the NAMD2 configuration to maintain the experimentally resolved sponge structure and allowing water sampling about the sponge model during a 2,000-step minimization and a 1-ns production run. The same protocol was carried out on a directly epimerized AmB sponge, preserving all sponge characteristics of the initial model, with the epimerized groups fixed only during the production run. The simulation was carried out using the molecular dynamics software package NAMD2 (ref. 51). The simulation parameters for each AmB and epimerized AmB were obtained from the CHARMM General Force field52. In addition, the solvent used a transferable intermolecular water potential with 3 points (TIP3P) model for water53. The simulation was carried out using an isothermal–isobaric ensemble at 310 K with a pressure of 1.0 atm. The Nosé–Hoover Langevin piston method54 was used to maintain a constant pressure, with temperature maintained constant through Langevin dynamics with a damping coefficient of 0.5 ps−1. A 12-Å cutoff was used for nonbonded interactions with a smoothing function implemented farther than 10 Å. The integration time step of 2 fs was used, and the bond distances of the hydrogen atoms were constrained using the SHAKE algorithm55. For long-range electrostatic calculations, the particle mesh Ewald method56 was used, with a grid density of less than 1 Å−3. Visualization and analyses of the simulations were carried out using Visual Molecular Dynamics57.
Synthesis of amide derivatives
Freshly distilled triethylamine was added dropwise to a solution of AmB (or C2′epiAmB) (10 mg; 0.01 mmol) and amine (or salt) (3 equivalents) in DMF (500 μl until pH 9 was reached (by pH paper). The reaction mixture was stirred for 15 min at room temperature. Solid PyBOP (1.5 equivalents; 8.4 mg) was added under a nitrogen atmosphere, and the sealed vial was stirred for 3 h and the progress was monitored by analytical HPLC.
Once completed (confirmed by LC–MS), the product was precipitated and washed with anhydrous diethyl ether (10 ml). The suspension was centrifuged at 3,000g for 5 min. The solvent was decanted out and the pellet was dissolved in DMSO and filtered through a 0.2-mm syringe filter for purification on a C18 Prep HPLC system. The pure product was dried on a lyophilizer as yellowish powder and stored at −20 °C under a nitrogen atmosphere.
Preparation of amide acetate salt
A 50 mg quantity of HPLC- purified amide was added to 2.5 ml of milliQ water followed by 200 μl of 0.5 (M) acetic acid. After addition of acid, the solution was vortexed. Following sonication for 1–2 min, the solution was stirred at 800 r.p.m. for 15 min at 23 °C (room temperature) and the clear solution was lyophilized for 24–36 h to form a fluffy yellow solid salt.
Preparation of AM-2-19_DP2K
In a clean 7-ml glass vial, 20 mg (7.5 mmol) of DP2K solid powder was added to 1 ml of D5W, and then 2.5 mg (2.5 mmol) of AM-2-19 acetate salt (AM-2-19-OAc) was added and the mixture was vortexed for 30 s and sonicated as necessary to dissolve all solid residues. The mixture was stirred for 30 min at 50 °C, allowed to cool to 23 °C (room temperature) and then filtered through a 0.2-mm syringe disc filter before use.
Sterol UV–Vis binding assay
Erg solution.
A stock solution of recrystallized Erg from chloroform (about 70 mM) was prepared and the concentration was determined by UV (ε = 10,400 M−1 cm−1 in ethanol). This stock solution was diluted to 1 mM and 0.1 mM using DMSO.
Chol solution.
A stock solution of recrystallized Chol was prepared (100–200 mM) in chloroform. This stock solution was diluted to 1 mM and 0.1 mM with DMSO.
Compound solution.
AmB or its derivatives (about 5 mg) were dissolved in DMSO (0.2–0.5 ml) to prepare the stock solution (≥10 mM). The final concentration was measured by UV–Vis (for AmB and AmB amides, ε = 164,000 M−1 cm−1 in methanol; for AM-2-19, ε = 155,000 M−1 cm−1 in methanol). The stock was diluted to 1 mM in DMSO.
Sterol complexes were prepared by mixing the stock solutions as shown in Supplementary Table 9.
Cell viability assay
Primary human renal proximal tubule epithelial cell.
Primary human renal proximal tubule epithelial cells were purchased from ATCC (PCS-400-010) and cultured immediately following arrival. No additional authentications or tests were performed on any cell lines. Complete growth medium was prepared as per ATCC recommendations using renal epithelial basal medium (ATCC, PCS-400-030), renal epithelial growth kit (ATCC, PCS-400-040) and Pen-Strep.
Human hepatocellular epithelial cells (HepG2).
Human hepatocellular epithelial cells (HepG2) were purchased from ATCC (HB-8065) and cultured immediately following arrival. Complete minimum essential medium (MEM) was prepared as per ATCC recommendations by combining MEM (1 l), fetal bovine serum (FBS; 113.64 ml; Gemini), Pen-Strep (11.36 ml) and non-essential amino acids (11.63 ml).
Human neuroblastoma cells (SH-SY5Y).
Human neuroblastoma epithelial cells (SH-SY5Y) were purchased from ATCC (CRL-2266) and cultured immediately following arrival. Complete MEM was prepared as per ATCC recommendations by combining MEM (1 l), FBS (113.64 ml; Gemini), Pen-Strep (11.36 ml) and non-essential amino acids (11.63 ml).
Human haematopoietic cells (K562).
Human haematopoietic cells (K562) were purchased from ATCC (CRL-3344) and cultured immediately following arrival. Complete Iscove’s modified Dulbecco’s medium (IMDM) was prepared as per ATCC recommendations by combining IMDM (1 l), FBS (113.64 ml; Gibco) and Pen-Strep (11.36 ml).
Rat myocardial cells (H9c2).
Rat myocardial cells (H9c2) were purchased from ATCC (CRL-1446) and cultured immediately following arrival. Complete Dulbecco’s modified Eagle’s medium (DMEM) was prepared as per ATCC recommendations by combining DMEM (1 l), FBS (113.64 ml; Gemini), Pen-Strep (11.36 ml) and non-essential amino acids (11.63 ml).
Sterile complete growth medium was stored at 4 °C and used within 30 days of preparation. Cells were seeded in T75 flasks and incubated at 37 °C under a 5% CO2 atmosphere. Cell viability was quantified using fluorescence-based alamarBlue (Thermo Fisher, DAL1025).
Compound stock solutions were prepared in DMSO as 100× the target concentration. The experiment was set up on a 96-well plate, and the final concentrations ranged between 0.5 and 50 μM. DMSO and puromycin (10 mg ml−1) were used as controls.
After trypsinization (except for K562), the inoculum density was adjusted to 5 × 105 cells ml−1 (for K562, the density was 12.5 × 105 cells ml−1). In a tissue-culture-treated 96-well plate, a 20-μl cell suspension (104 cells per well) was seeded and incubated at 37 °C for 3 h before the addition of the compound. In each well, the total volume was adjusted to 100 μl using the growth medium and DMSO (1%). Plates were incubated at 37 °C under a 5% CO2 atmosphere for 24 h. To assess cell viability, 10 μl alamarBlue was added to each well, mixed gently and incubated for 3 h at 37 °C, and fluorescence intensity was recorded (excitation 537 nm; emission 585 nm). Experiments were carried out in triplicate and the reported results represent an average of three experiments.
Isothermal calorimetry studies
Potassium efflux assays
Efflux studies were carried out as previously reported5.
MIC assay
The protocol for MIC determination on Candida strains was adapted from that of the Clinical and Laboratory Standards Institute (CLSI)58. The protocol for MIC determination on Aspergillus contained changes based on a CLSI protocol59.
Erg extraction experiment
This assay was adapted on the basis of an earlier report36 with slight modifications to accommodate C. albicans SN250, which was cultured and inoculated in RPMI-1640 medium.
Spontaneous mutation study
General information.
In this study Candida spp. were exposed to AM-2-19_DP2K, and AmBisome (liposomal AmB) as the comparator, using gradient agar plates. The experimental design was based on that of ref. 60, which monitored resistance development from passaging on gradient agar plates. Every third passage, yeasts were collected and subjected to broth microdilution to assess whether resistance was increasing for the passaged isolates following procedures from the CLSI58,61.
Agar serial passage.
Mueller Hinton agar (MHA) drug gradient plates were created by pouring two overlapping layers of medium. The first layer containing 15 ml of drug-free MHA was poured into 90 × 90 square Petri dishes while on an incline. Once solidified, the plate was placed flat, and a second layer was poured containing AM-2-19_DP2K or AmBisome at a concentration capable of inhibiting growth for each strain but allowing for some growth past the edge of the plate containing no drug into the start of the drug gradient. Following each passage, the leading edge of growth was resuspended into RPMI to create a 0.5 McFarland standard (1–5 × 105 CFUs ml−1) and a 100-μl aliquot was spread onto a fresh passage plate using sterile L-spreaders. The inoculum from each passage was cryopreserved at −80 °C. Drug concentrations were adjusted on the basis of how well the strains grew on the previous passage plate. Twenty consecutive serial passages were completed for each drug–strain combination. At every third passage, the inoculum suspension was plated for identification by matrix-assisted laser desorption/ionization and diluted into RPMI for broth microdilution susceptibility testing to evaluate the MIC for the test agents, AmB, caspofungin and fluconazole in accordance with CLSI guidelines. Fresh suspensions of the parent isolates and relative ATCC QC strains were included for each broth microdilution MIC.
Broth microdilution MIC procedure.
MIC values were determined using a broth microdilution procedure described by CLSI60. Automated liquid handlers (Multidrop 384, Labsystems; Biomek 2000 and Biomek FX, Beckman Coulter) were used to conduct serial dilutions and liquid transfers. All wells in columns 2–12 of a standard 96-well microdilution plate (Costar 3795) were filled with 150 μl of the appropriate diluent. Then, 300 μl volumes of the test and comparator compounds were added to the wells of column 1 of the plates at 40× the highest final concentration to be tested. Serial twofold dilutions were made across the rows to column 11 using the Biomek 2000 or by hand. The wells of column 12 contained no drug and served as the growth control wells. This plate served as the ‘mother’ plate from which MIC assay plates or ‘daughter’ plates were made. The daughter plates were loaded with 185 μl per well of medium using the Multidrop 384. The daughter plates were then created using the Biomek FX, which transferred 5 μl of drug solution from each well of a mother plate to each corresponding well of the daughter plate in a single step. A standardized inoculum of each test organism was prepared per CLSI methods58,61. The plates were then inoculated with 10 μl of the diluted inoculum using the Biomek 2000 from low to high drug concentration, resulting in a final concentration of approximately 0.5 to 2.5 × 103 CFUs ml−1. An uninoculated plate was incubated for the purpose of assessing solubility of the drug in the test medium. The plates were stacked 3 to 4 high, covered with a sterile lid on the top plate, and incubated aerobically at 35 °C for 24 h. An uninoculated solubility control was observed for evidence of drug precipitation and to assess medium sterility. The MIC was read and recorded as the lowest concentration of drug that inhibited visible growth of the organism except for caspofungin and fluconazole, which were read at 50% inhibition.
In vivo efficacy
Neutropenic mouse model of disseminated candidiasis.
Healthy female CD-1 mice (21–28 g; 5–7 weeks old) were treated with the immunosuppressant cyclophosphamide on day −3 (200 mg kg−1) and day +1 (150 mg kg−1). Animals were infected on day 0 by IV injection of 200 μl C. glabrata ATCC2001 inoculum (about 1 × 108 CFUs ml−1) into the lateral tail vein. Pretreatment groups were euthanized 24 h after infection to assess the initial fungal burden, and the rest of the groups received drug treatment 24 h post infection. Placebo control groups were treated daily with 5 ml kg−1 D5W (for AmBisome) and DP2K in D5W (for AM-2-19_DP2K) for 5 days. AmBisome was used as the comparator, and the animals were administered three different daily doses (q24h) of 0.1, 1 and 10 mg kg−1 (n = 6 per group) for 5 days. Four different dosing groups of AM-2-19_DP2K were administered 1, 3, 10 and 30 mg kg−1 (n = 6 per group) of daily IV dose for 5 days. Animals were closely monitored for any clinical signs or symptoms. At the clinical endpoint (after day 5), groups were euthanized, blood was collected through terminal intra-cardiac puncture for plasma separation, and kidneys were collected for quantitative analysis of fungal burden by quantitative culture onto SAB/Dextrose agar containing chloramphenicol at 37 °C 24–48 h.
Mice were allowed to acclimatize for a minimum of 7 days before any procedures. Mice were housed in sterile individual ventilated cages exposing animals at all times to HEPA-filtered air. Mice had free access to food and water and were housed on aspen chip bedding. The room temperature was 22 °C ± 1 °C, with a relative humidity of 50–60% and a maximum background noise of 56 dB. Mice were exposed to 12 h light/dark cycles with dawn and dusk phases. Studies were approved by Evotec onsite Alderley Park UK, Animal Welfare Ethical Review Body (AWERB), with subsequent approval by UK Home Office regulators.
Neutropenic mouse model of disseminated aspergillosis.
Healthy male CD-1 mice (about 25–30 g; 5–7 weeks old) were treated with the immunosuppressant cyclophosphamide on day −3 (200 mg g−1). Animals were infected on day 0 by IV injecting 200 μl of A. fumigatus 1163 inoculum (about 6.0 × 105 CFUs ml−1) into the lateral tail vein. Pretreatment groups were euthanized 5 h after infection to assess the initial fungal burden, and the rest of the groups received treatments. Control groups were treated daily with 5 ml kg−1 D5W (for AmBisome) and DP2K in D5W (AM-2-19) for 4 days. AmBisome was used as the comparator and three different daily IV doses (q24h) of 0.1, 1 and 10 mg kg−1 (n = 6 per group) were tested for 4 days. Four different doses of AM-2-19_DP2K, 0.1, 1, 10 and 30 mg kg−1 (n = 6 per group), were administered as a daily IV dose for 4 days. Animals were closely monitored for any clinical signs. At the clinical endpoint (101 h post infection), groups were euthanized, blood was collected through terminal intra-cardiac puncture for plasma separation and kidneys were collected for quantitative analysis of fungal burden by quantitative culture onto SAB/Dex agar containing chloramphenicol at 37 °C 24–48 h.
Mice were allowed to acclimatize for minimum of 7 days before any procedures. Mice were housed in sterile individual ventilated cages exposing animals at all times to HEPA-filtered air. Mice had free access to food and water and were housed on aspen chip bedding. The room temperature was 22 °C ± 1 °C, with a relative humidity of 50–60% and a maximum background noise of 56 dB. Mice were exposed to 12 h light/dark cycles with dawn and dusk phases. Studies were approved by Evotec onsite Alderley Park UK, AWERB, with subsequent approval by UK Home Office regulators.
Neutropenic mouse model of invasive pulmonary aspergillosis.
Healthy male CD-1 mice (about 25–30 g; 5–7 weeks old) were treated with the immunosuppressant cyclophosphamide (150 mg kg−1) on day −4 and day −1 and 175 mg kg−1 of cortisone acetate on day −1. Ceftriaxone (50 mg kg−1, q24h) prophylaxis was maintained from day −4 to day +4. Animals were infected on day 0 intranasally into both nares with 40 μl of A. fumigatus ATCC204305 inoculum (about 6.5 × 105 CFUs ml−1). Pretreatment groups were euthanized 5 h after infection to assess the initial fungal burden, and the rest of the groups received treatment. Placebo control groups were treated daily with 5 ml kg−1 of D5W (for AmBisome) and DP2K in D5W (for AM-2-19) for 4 days. AmBisome was used as the comparator and three different daily IV doses (q24h) of 0.1, 1 and 10 mg kg−1 (n = 8 per group) were administered for 4 days. Five different doses of AM-2-19_DP2K, 0.3, 1, 3, 10 and 30 mg kg−1 (n = 8 per group), were administered as a single daily IV dose for 4 days. Animals were closely monitored for any clinical signs or symptoms. At the clinical endpoint (101 h post infection), groups were euthanized, blood was collected through terminal intra-cardiac puncture for plasma separation and lungs were removed for quantitative PCR analysis.
Mice were allowed to acclimatize for minimum of 7 days before any procedures. Mice were housed in sterile individual ventilated cages exposing animals at all times to HEPA-filtered air. Mice had free access to food and water and were housed on aspen chip bedding. The room temperature was 22 °C ± 1 °C, with a relative humidity of 50–60% and a maximum background noise of 56 dB. Mice were exposed to 12 h light/dark cycles with dawn and dusk phases. Studies were approved by Evotec onsite Alderley Park UK, AWERB, with subsequent approval by UK Home Office regulators.
Neutropenic mouse model of disseminated aspergillosis.
Healthy male CD-1 mice (about 25–30 g; 5–7 weeks old) were treated with the immunosuppressant cyclophosphamide (200 mg kg−1) on day −3. Animals were infected on day 0 by IV injection of 200 μl A. terreus 49 inoculum (about 9.3 × 105 CFUs ml−1) into the lateral tail vein. Pretreatment groups were euthanized 24 h after infection to assess the initial fungal burden, and the remaining groups received treatments. Control groups were treated daily with 5 ml kg−1 D5W (for AmBisome) and DP2K in D5W (for AM-2-19) for 10 days. AmBisome was used as the comparator and three single daily IV doses (q24h) of 0.1, 1 and 10 mg kg−1 (n = 10 per group) were administered for 10 days. Five different doses of AM-2-19_DP2K, 0.3, 1, 3, 10 and 30 mg kg−1 (n = 10 per group), were administered as a single daily IV dose for 10 days. Animals were closely monitored for any clinical signs or symptoms. At the clinical endpoint (240 h post infection), groups were euthanized, blood was collected through terminal intra-cardiac puncture for plasma separation, and kidneys were collected for quantitative CFU analysis of fungal burden by quantitative culture onto SAB/Dextrose agar containing chloramphenicol at 37 °C 24–48 h.
Mice were allowed to acclimatize for minimum of 7 days before any procedures. Mice were housed in sterile individual ventilated cages exposing animals at all times to HEPA-filtered air. Mice had free access to food and water and were housed on aspen chip bedding. The room temperature was 22 °C ± 1 °C, with a relative humidity of 50–60% and a maximum background noise of 56 dB. Mice were exposed to 12 h light/dark cycles with dawn and dusk phases. Studies were approved by Evotec onsite Alderley Park UK, AWERB, with subsequent approval by UK Home Office regulators.
Neutropenic mouse model for invasive pulmonary mucormycosis.
Healthy male ICR mice (about 23–25 g; about 6–8 weeks) were immunosuppressed with intraperitoneal injection of cyclophosphamide (200 mg kg−1) and subcutaneous administration of cortisone acetate (500 mg kg−1) on day −2 and day +3, relative to intratracheal infection with Rhizopus delemar 99–880 (2.5 × 105 spores per 25 μl) or M. circinelloides f. jenssenii DI15-131 (2.5 × 106 spores per 25 μl) as previously described62, both obtained from the Fungus Testing Laboratory at the University of Texas Health Sciences Center at San Antonio. To prevent bacterial superinfection, animals were given 50 μg ml−1 enrofloxacin (Baytril; Bayer) mixed with drinking water from day −3 to day 0 and then switched to daily subcutaneous injection of ceftazidime (5 μg per dose) on day 0 until day +9. Treatment involved IV administration of a single daily dose of 10 mg kg−1 AmBisome or AM-2-19_DP2K at doses of 0.3, 1.5, 7.5 and 30 mg kg−1 beginning at 16 h after challenge continuing for 4 days. Placebo control mice (infected and untreated) received DP2K in D5W (for AM-2-19) for the duration of treatment. The primary efficacy endpoint was a Kaplan–Meier survival time up to 21 days post challenge. Ten mice per group were used, and the experiment was repeated (for a total of 20 mice per group) to allow for a 90% power to detect a 50% difference in survival time (assessed by log-rank test).
As a secondary endpoint and in an independent experiment, lungs and brains (primary and secondary target organs, respectively)62,63 were collected from euthanized mice (n = 10 per group) after 6 h from the last dose on day +4 and processed for fungal burden by conidial equivalent using quantitative PCR64. Histopathological examination was conducted on the same organs processed for tissue fungal burden and stained with haematoxylin and eosin and Grocott’s methenamine silver.
Studies were approved by the Institutional Animal Care and Use Committee of the Lundquist Institute at Harbor-UCLA Medical Center. The Biological Resources Center office is responsible for managing and administering a centralized program of laboratory animal care that is in compliance with the federal Animal Welfare Act, the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals, the Public Health Service Policy on Human Care and Use of Laboratory Animals and the US Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training. Relative humidity acceptable for all species was set between 30 and 70%. Room temperature for rodents was kept between 20 and 26 °C. Mice had free access to food and water. Regular 12-h diurnal light/dark cycles were provided by time-controlled lighting systems in most facilities.
Statistics
Experiments were carried out in biological duplicate or triplicate, and the error represents s.d. One-way and two-way ANOVAs were carried out with Tukey’s multiple comparison test: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. For the mucormycosis animal studies, the nonparametric log-rank test was used to determine differences in survival times. A P value < 0.05 was considered significant.
Animal research ethics
The in vivo toxicological studies in mice were approved by the Institutional Animal Care and Use Committee at the University of Illinois at Urbana-Champaign. Survival studies in the mucormycosis mouse model were approved by the Institutional Animal Care and Use Committee of the Lundquist Institute at Harbor-UCLA Medical Center. The efficacy study in the candidiasis mouse model was approved by the Animal Research Committee of the William S. Middleton Memorial Veterans Affairs Hospital. The animals used were maintained in accordance with the criteria of the American Association for Accreditation of Laboratory Animal Care. In vivo efficacy studies carried out in collaboration with Evotec were approved by Evotec onsite Alderley Park UK, AWERB, with subsequent approval by UK Home Office regulators.
Extended Data
Extended Data Fig. 1 ∣. Cholesterol binding primarily drives renal toxicity of AmB, not ion channel formation.
a, Synthesis of C35MeOAmB, a non-ion-channel forming antifungal probe starting from AmB. b, C35MeOAmB binds cholesterol during UV-Vis binding assay. c, C35MeOAmB does not permeabilize human red blood cells. Pre-complexation with cholesterol mitigates renal toxicity of both d, AmB and e, C35MeOAmB against human primary renal cells. Cholesterol (53 mg/g) in β-Me cyclodextrin (MβCD) obtained from Sigma Aldrich (C495; lot no SLCJ3255). Results are means ± SD (n = 3 biological replicates/time point). In d, all pairwise comparisons with corresponding (1:0) at each concentration were performed using two-way ANOVA with Tukey’s multiple comparison test; *P = 0.028, **P = 0.0086, **P = 0.0019, ***P = 0.0004, ***P = 0.0003, ****P < 0.0001. In e, all pairwise comparisons with corresponding (1:0) at each concentration were performed using two-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0004, ****P < 0.0001.
Extended Data Fig. 2 ∣. AmB in one state binds to multiple sterols in AmB-sterol complexes.
a, UV-vis spectra of AmB (black) and AmB upon titration with increasing molar ratios of Erg (0.5, yellow, 1, green, 2, cyan, and 3, blue). The red shift and “finger”-like pattern at higher sterol ratios is consistent with the AmB polyenes separating to accommodate sterols. b, AmB:sterol stoichiometric ratios (average of three replicates) after three rounds of complex washing as described in Methods and a picture of AmB-sterol complexes undergoing sterol washing in chloroform. c, 13C-13C 2D 50 ms DARR spectrum of 13C-AmB:Chol showing AmB in one state. d, 13C-13C 2D SPC5 spectrum with one bond correlations in green and two-bond correlations in blue for a 13C-AmB:13C-Erg sample with Erg crosspeaks highlighted. The largest chemical shift difference observed for each Erg carbon is summarized graphically (SI Table 7). UV-Vis sterol binding interaction between AmB and sterols from ergosterol biosynthetic pathway e, lanosterol, f, zymosterol, and g, episterol.
Extended Data Fig. 3 ∣. Erg interacts with AmB through contacts between the sterol rings and conserved GPM motif.
a, A diagram summarizing AmB-Erg interactions shown in c, d, and e, highlighting 13C-13C and 19F-13C interactions between the Erg A and B rings and the conserved C11-C20 motif and mycosamine. b,13C detected 1H-1H polarization transfers from water to the 13C-AmB C13/C1’ signal for homogenized 13C-AmB (black line) and 13C-AmB:Erg complexes (blue lines). Each data point comes from one peak (n = 1). Error bars represent uncertainties from the signal-to-noise ratio of each spectrum. c, Dephasing curves (red stars for experimental data and black lines for simulated curves) from 13C{19F} frequency selective REDOR experiments performed on a 13C-AmB:C6F-Erg sample and corresponding distances calculated from the dipolar couplings. Data presented as mean +/− SD of n = 11, 5, and 10 technical replicates, respectively. Error bars indicate uncertainty from the spectrum signal-to-noise ratio. d, AmB chemical shift perturbations relative to Erg bound 13C-AmB for the apo AmB states, previously reported in Nat. Chem. Bio. 28, 972 (2021), and 13C-AmB bound to 13C-Erg, C6F-Erg, and Chol (SI Table 6). e, Overlay of 13C-13C 2D PAR spectra collected at 8.4 ms (blue), 12.6 ms (magenta), and 16.8 ms (cyan) mixing times obtained from a 13C-AmB:13C-Erg sample highlighting interactions between the two primary Erg ring states and AmB.
Extended Data Fig. 4 ∣. SSNMR AmB-Erg structure and water MD simulations.
a, An overlay of the AmB structures from the 10 lowest energy lattices with the average and standard deviation of the dihedral angle at the mycosamine attachment b, Overlay of AmB and one Erg from the minimal subunit for the 6 structures, taken from the 10 lowest energy lattices of the calculation, in which the 3β-hydroxyl oxygen is within 4 Å of the C2’ hydroxyl oxygen. Water interactions from MD simulations for Erg-bound AmB, c, and Erg-bound C2’epiAmB, d, sponges. left, overlays of observed water molecules within the sponges highlighting regions of high water propensity. Right, single models taken from the overlays on the left showing representative positions of persistent waters and distances (in Å) consistent with hydrogen bonding interactions. See Extended Data Table 1 for SSNMR structure statistics.
Extended Data Fig. 5 ∣. AmB 13C-13C Correlations in the AmB-Erg Complex.
a, A diagram summarizing AmB-AmB 13C-13C correlations observed in 500 ms CORD and 12.6 ms PAR spectra between AmB carbons with intramolecular distances >8.5 Å. Blue ovals represent sterol molecules. b, 13C-13C 2D 500 ms CORD and 12.6 ms PAR spectra obtained from 13C-AmB:C6F-Erg and 13C-AmB:13C-Erg samples, respectively, highlighting intermolecular AmB-AmB interactions.
Extended Data Fig. 6 ∣. Mechanistic probing of C2’epiAmB’s decreased potency.
a, Synthesis of C2’epiAmB starting from AmB. b, Evaluations of C2’epiAmB efficacy in disseminated candidiasis mice model infected with C. albicans SN250, 24 h post single IV dose (n = 3 mice/group). Both compounds were administered as 1:2 deoxycholate in D5W to improve aqueous solubility. All dose units are mg/kg. Pairwise comparisons with 24 h placebo group were performed using (one-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0003; ****P < 0.0001. c, Structure and numbering system of skip labeled 13C-erg. Sterol sequestration mechanism of antifungal action is conserved in C2’epiAmB as probed in d, PRE experiments, showing a decrease in the PRE effects of resolved Erg resonances in the presence of C2’epiAmB, indicating a decrease in sterol proximity to doxyl-labeled lipids (samples 40:1 POPC/ 13C-Erg ± 5 mol% 16-DOXYL-PC; Data presented as mean +/− SD of 3 measurements. Error bars indicate uncertainty from the spectrum signal-to-noise ratio.) and decreased efficacy upon pre-complexation with ergosterol for both e, AmB (n = 3 biological replicates/conc.) and f, C2’epiAmB (n = 3 biological replicates/conc.). Both molecules have similar stability profiles in g, C. albicans SN250 (n = 4 biological replicates/conc.) and h, A. fumigatus 1163 cultures (for 8 μM AmB n = 3, for all other n = 4 biological replicates). Both molecules (3 μM) have similar ion channel forming ability and have similar time-delay between compound addition and efflux in i, wild-type C. albicans SN250 and j, instant ion-channel formation in protoplast. Improvement of C2’epiAmB efficacy upon cotreatment with erg biosynthetic inhibitors Ketoconazole and Posaconazole against k, l, A. fumigatus 1163 and m, n, A. fumigatus 91 reveals that the extraction rate-driven kinetic model of efficacy is conserved against moulds. o, Unlike C2’epiAmB, AmBMU, though better tolerated than AmB, retains cholesterol binding and p, causes kidney damage in mice (n = 4 mice/group). Pairwise statistical analyses were performed using two-way ANOVA with Tukey’s multiple comparison test; *P = 0.0422; ***P = 0.0008, ***P = 0.0005, ***P = 0.001, ****P < 0.0001. Results are means ± SD.
Extended Data Fig. 7 ∣. Combining C2’epimerization and C16 amidation results in a renal sparing potent antifungal.
a. in vitro efficacy of potent AmB amides. Head-to head comparison of in vitro toxicity of AmB, C2’epiAmB, AM-243-2 and AM-2-19 against b, H9C2 cells (rat cardiomyocyte; n = 3 biological replicates/conc.) c, Hep-G2 cells (human liver cell; n = 3 biological replicates/conc.) d, K562 (human red blood cells progenitors; n = 3 biological replicates/conc.) e, SHSY-5Y (human neural blastoma; n = 3 biological replicates/conc.) and f, RPTEC (primary renal proximal tubule epithelial cells; n = 3 biological replicates/conc.). C2’epiAmB and AM-2-19 both g, do not lyse human red blood cells (n = 3 biological replicate/conc.), and h, retain an AmB-like drug-drug interaction profile and i, do not elevate kidney injury biomarkers after 24 h of single IV 45 mg/kg dose (n = 4 mice/group). Pairwise statistical analyses were performed using two-way ANOVA with Tukey’s multiple comparison test; **P = 0.0022, ****P < 0.0001. AM-2-19 binds j, ergosterol but not k, cholesterol in the UV-Vis binding assay and is l, highly efficacious against pathogens that were resistant to C2’epiAmB (100% inhibition reported). m, Comparison of solution stability between AM-2-19 and C2’deOAmB in PBS buffer at pH 6. Pairwise two-way ANOVA with Tukey’s multiple comparison tests was performed at each time point; ***P = 0.0005, ****P < 0.0001. AM-2-19 does not bind n, lanosterol, o, zymosterol, and shows little to no binding with p, episterol. q, Head-to-head in vitro efficacy comparison between AM-2-19, C2’deOAmB, Natamycin and Nystatin. Data in j and k are representative of at least 3 independent experiments.
Extended Data Fig. 8 ∣. AM-2-19_DP2K is highly efficacious and resistance evasive.
a, Representative examples of several drug-resistant strains that are susceptible to AM-2-19_DP2K treatment, unlike its comparators. The study was conducted at UT Health, San Antonio, and 100% inhibition was reported. b, AM-2-19_DP2K is highly efficacious against hundreds of clinical fungal isolates tested at different locations (number of isolates in parenthesis; 100% inhibition reported). For AM-2-19_DP2K and comparators, dosing concentrations (μM and μg/ml) are reported based on the active pharmaceutical ingredient (API). The MIC values of DSG-PEG 2000 (DP2K), tested at UIUC, were >16 μM against all isolates. AmB or AmBisome® is used as a comparator because, unlike AM-2-19, AmB does not form a homogenous solution with DSG-PEG 2000. c, in vivo efficacy of AM-2-19_DP2K and comparator AmBisome® was evaluated in male CD-1 non-neutropenic disseminated candidiasis model infected with C. albicans SC5314 after 7 days of daily IV dosing (n = 6 mice/group). Pairwise statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0008, ****p < 0.0001. Dose dependent mitigation of fungal burden on day 4 in neutropenic mucormycosis model, infected with R. delemar (first dose 48 h post infection; q24h; n = 10 mice/group) was measured by qPCR for d, lung (one-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001) and e, brain tissues (one-way ANOVA with Tukey’s multiple comparison test; ****P < 0.0001). f, Gross pathology, and histological examination of lung tissues harvested from mice infected with R. delemar, and treated with placebo, AM-2-19_DP2K or AmBisome®. Notice the diffused filamentation in the lung from placebo or 0.3 mg/kg of AM-2-19_DP2K treated mice. Fewer/no hyphae are evident in lungs of mice treated with AM-2-19_DP2K 1.5, 7.5 and 30 mg/kg or AmBisome®. Arrows in upper panel refer to fungal abscesses. Scale bar 100 μm for histopathological analysis (bottom). Tissue fungal burden of g, lungs (one-way ANOVA with Tukey’s multiple comparison test; *P = 0.0147, ****P < 0.0001) and h, brain (one-way ANOVA with Tukey’s multiple comparison test; *p = 0.0243, ****P < 0.0001) of mice (n = 10 mice/group) infected with M. circinelloides, treated with either drug and euthanized on Day +4 post infection. Spontaneous mutation frequency study indicates AmBisome-like resistance-refractory property of AM-2-19_DP2K against i, C. tropicalis ATCC 90874, j, C. albicans ATCC90028 k, C. glabrata ATCC 90030 and l, C. krusei ATCC 6258 after 20 passage and MIC recorded after every three passages. All dosing units are mg/kg. In mice, fungal burdens below the limit of detection (LOD) were given a nominal value CFU/g = 1. Results are means ± SD. In mice, fungal burdens below the limit of detection (LOD) were given a nominal value of CFU/g = 1.
Extended Data Table 1 ∣.
SSNMR structure calculation statistics
| AmB-Erg lattice | |
|---|---|
| NMR distance and dihedral restraints per lattice | |
| Distance restraints | |
| Total | 512 |
| 13C-13C restraints | 501 |
| Up to 6 Å | 94 |
| Up to 7.5 Å | 407 |
| FS-REDOR 13C-19F | 11 |
| Total dihedral angle restraints | 81 |
| Structure statistics from 33 lowest energy structures | |
| Violations (mean and (s.d.)) | 19.5 (2.2) |
| Distance restraints ≥ 0.5 Å | 3.3 (2.2) |
| Dihedral angle restraints ≥ 5° | 0.0 (0.0) |
| Bond angle restraints | 16.2 (0.6) |
| Improper restraints | 0.0 (0.0) |
| Deviations from idealized geometry | |
| Max. bond angle violation (°) | 9.9 |
| Average r.m.s.d of 10 lowest energy lattices (Å) | 0.54a |
Overlaid over segid 222 resid 1 (the center AmB of the 5x5x5 lattice)
SSNMR restraints and refinement statistics for the structure calculation presented in Figs. 1a, 2a, and Extended Data Fig. 4a,b.
Supplementary Material
Acknowledgements
We thank L. Zhu for assistance with the NMR experiment set up; J. G. Weers, A. Blake, S. Ekaputri and T. Tyrikos-Ergas for helpful discussions; Y. Gu for help with the mucormycosis animal studies; the SCS NMR Lab at University of Illinois Urbana-Champaign for technical support; and the Beckman Institute at University of Illinois Urbana-Champaign for the fellowship of A.M. This work was supported by US NIH grants 5R01-AI135812-04 and R35-GM118185 to M.D.B., R01-GM112845 and R01-GM123455 to C.M.R. and R01-AI063503 to A.S.I.. Studies carried out at the National Magnetic Resonance Facility at Madison were supported by NIH grant P4-GM136463. The Bruker 500-MHz NMR spectrometer was obtained with the financial support of the Roy J. Carver Charitable Trust, Muscatine, Iowa, USA. The work was also supported by Sfunga Therapeutics.
Footnotes
Competing interests A.M., J.Z., S.Y., C.M.R. and M.D.B. are inventors on patents PCT/US20/45566, PCT/US20/45399, PCT/US 2021/45205, PCT/US2020/45387 and/or UIUC2022-022-01, submitted by University of Illinois Urbana-Champaign. M.D.B. and K.A.M. disclose consulting income and equity in Sfunga Therapeutics.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-023-06710-4.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-023-06710-4.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data that support the findings of this study are available within the paper and its Supplementary Information. SSNMR source data are provided in BMRbig entry IDs bmrbig84 and BMRB 21102. Molecular dynamics simulation trajectories are provided for Extended Data Fig. 4c,d at https://usegalaxy.org/u/taras.pogorelov/h/amb-simulations-2023. Further requests can be directed to the corresponding authors. Source data are provided with this paper.
References
- 1.Katzung BG, Masters SB & Trevor AJ Basic & Clinical Pharmacology 12th edn (McGraw Hill, 2012). [Google Scholar]
- 2.Gray KC et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl Acad. Sci. USA 109, 2234–2239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson TM et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol 10, 400–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewandowska A. et al. Fungicidal amphotericin B sponges are assemblies of staggered asymmetric homodimers encasing large void volumes. Nat. Struct. Mol. Biol 28, 972–981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X. et al. Sterol sponge mechanism is conserved for glycosylated polyene macrolides. ACS Cent. Sci 7, 781–791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revie NM & Cowen LE Glycosylated polyene macrolides kill fungi via a conserved sterol sponge mechanism of action. ACS Cent. Sci 7, 706–708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Fungal Priority Pathogens list to Guide Research, Development and Public Health Action (World Health Organization, 2022). [Google Scholar]
- 8.MacAlpine J. et al. A small molecule produced by Lactobacillus species blocks Candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat. Commun 12, 6151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla R. et al. Teixobactin kills bacteria by a two-pronged attack on the cell envelope. Nature 608, 390–396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilcock BC, Endo MM, Uno BE & Burke MD C2′-OH of amphotericin B plays an important role in binding the primary sterol of human cells but not yeast cells. J. Am. Chem. Soc 135, 8488–8491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwieters CD, Bermejo GA & Clore GM Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 27, 26–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Paëpe G, Lewandowski JR, Loquet A, Böckmann A & Griffin RG Proton assisted recoupling and protein structure determination. J. Chem. Phys 129, 245101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takegoshi K, Nakamura S & Terao T 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett 344, 631–637 (2001). [Google Scholar]
- 14.Shcherbakov AA & Hong M Rapid measurement of long-range distances in proteins by multidimensional 13C–19F REDOR NMR under fast magic-angle spinning. J. Biomol. NMR 71, 31–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou G, Yan S, Trébosc J, Amoureux J-P & Polenova T Broadband homonuclear correlation spectroscopy driven by combined R2nv sequences under fast magic angle spinning for NMR structural analysis of organic and biological solids. J. Magn. Reson 232, 18–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohwy M, Rienstra CM, Jaroniec CP & Griffin RG Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double quantum spectroscopy. J. Chem. Phys 110, 7983–7992 (1999). [Google Scholar]
- 17.Kasai Y. et al. Synthesis of 6-F-ergosterol and its influence on membrane-permeabilization of amphotericin B and amphidinol 3. Org. Biomol. Chem 9, 1437–1442 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Pinalli R. et al. The origin of selectivity in the complexation of N-methyl amino acids by tetraphosphonate cavitands. J. Am. Chem. Soc 138, 8569–8580 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Moudrakovski IL, Udachin KA, Alavi S, Ratcliffe CI & Ripmeester JA Facilitating guest transport in clathrate hydrates by tuning guest-host interactions. J. Chem. Phys 142, 074705 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Biler M, Crean RM, Schweiger AK, Kourist R & Kamerlin SCL Ground-state destabilization by active-site hydrophobicity controls the selectivity of a cofactor-free decarboxylase. J. Am. Chem. Soc 142, 20216–20231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruben EA et al. Ground state destabilization from a positioned general base in the ketosteroid isomerase active site. Biochemistry 52, 1074–1081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaral M. et al. Protein conformational flexibility modulates kinetics and thermodynamics of drug binding. Nat. Commun 8, 2276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiller JB et al. Structure determination of high-energy states in a dynamic protein ensemble. Nature 603, 528–535 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q. et al. Synthetic group A streptogramin antibiotics that overcome Vat resistance. Nature 586, 145–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan KP, Singh K, Hazra A & Madhusudhan MS Peptide bond planarity constrains hydrogen bond geometry and influences secondary structure conformations. Curr. Res. Struct. Biol 3, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black K, Liu P, Xu L, Doubleday C & Houk KN Dynamics, transition states, and timing of bond formation in Diels–Alder reactions. Proc. Natl Acad. Sci. USA 109, 12860–12865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nendel M, Tolbert LM, Herring LE, Islam MN & Houk KN Strained allenes as dienophiles in the Diels–Alder reaction: an experimental and computational study. J. Org. Chem 64, 976–983 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Kawase T & Kurata H Ball-, bowl-, and belt-shaped conjugated systems and their complexing abilities: exploration of the concave–convex π–π interaction. Chem. Rev 106, 5250–5273 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Koren AB, Curtis MD, Francis AH & Kampf JW Intermolecular interactions in π-stacked conjugated molecules. Synthesis, structure, and spectral characterization of alkyl bithiazole oligomers. J. Am. Chem. Soc 125, 5040–5050 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Jarzembska KN et al. Controlled crystallization, structure, and molecular properties of Iodoacetylamphotericin B. Crystal Growth Design 12, 2336–2345 (2012). [Google Scholar]
- 31.Schmidt RR in Comprehensive Organic Synthesis (eds Trost BM & Fleming I) vol. 6, ch. 1 (Elsevier, 1991). [Google Scholar]
- 32.Han WK, Bailly V, Abichandani R, Thadhani R & Bonventre JV Kidney injury molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 62, 237–244 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Griffin BR, Faubel S & Edelstein CL Biomarkers of drug-induced kidney toxicity. Ther. Drug Monit 41, 213–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H. et al. Design, synthesis, and structure–activity relationship studies of bisamide derivatives of amphotericin B with potent efficacy and low toxicity. J. Med. Chem 65, 8897–8913 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Volmer AA, Szpilman AM & Carreira EM Synthesis and biological evaluation of amphotericin B derivatives. Nat. Prod. Rep 27, 1329–1349 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Davis SA et al. Nontoxic antimicrobials that evade drug resistance. Nat. Chem. Biol 11, 481–487(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roden MM et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis 41, 634–653 (2005). [DOI] [PubMed] [Google Scholar]
References
- 38.Seo S. et al. Biosynthesis of sitosterol, cycloartenol, and 24-methylenecycloartanol in tissue cultures of higher plants and of ergosterol in yeast from [1,2-13C2]- and [2-13C2H3]-acetate and [5-836 13C2H2]MVA. J. Chem. Soc. Perkin 1 10.1039/P19880002407 (1988). [DOI] [Google Scholar]
- 39.Umegawa Y. et al. Head-to-tail interaction between amphotericin B and ergosterol occurs in hydrated phospholipid membrane. Biochemistry 51, 83–89 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Palacios DS, Anderson TM & Burke MD A post-PKS oxidation of the amphotericin B skeleton predicted to be critical for channel formation is not required for potent antifungal activity. J. Am. Chem. Soc 129, 13804–13805 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hisao GS et al. An efficient method and device for transfer of semisolid materials into solid-state NMR spectroscopy rotors. J. Magn. Reson 265, 172–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morcombe CR & Zilm KW Chemical shift referencing in MAS solid state NMR. J. Magn. Reson 162, 479–486 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Lee W, Tonelli M & Markley JL NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cegelski L. REDOR NMR for drug discovery. Bioorg. Med. Chem. Lett 23, 5767–5775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaroniec CP, Tounge BA, Herzfeld J & Griffin RG Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate 13C–15N distance measurements in uniformly 13C,15N-labeled peptides. J. Am. Chem. Soc 123, 3507–3519 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Bak M, Rasmussen JT & Nielsen NC SIMPSON: a general simulation program for solid-state NMR spectroscopy. J. Magn. Reson 147, 296–330 (2000). [DOI] [PubMed] [Google Scholar]
- 47.MATLAB v.R2017b (MathWorks, 2017). [Google Scholar]
- 48.Huster D, Yao X & Hong M Membrane protein topology probed by 1H spin diffusion from lipids using solid-state NMR spectroscopy. J. Am. Chem. Soc 124, 874–883 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Neumann A, Wieczor M, Zielinska J, Baginski M & Czub J Membrane sterols modulate the binding mode of amphotericin B without affecting its affinity for a lipid bilayer. Langmuir 32, 3452–3461 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Kuszewski JJ, Thottungal RA, Clore GM & Schwieters CD Automated error-tolerant macromolecular structure determination from multidimensional nuclear Overhauser enhancement spectra and chemical shift assignments: improved robustness and performance of the PASD algorithm. J. Biomol. NMR 41, 221–239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips JC et al. Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanommeslaeghe K & MacKerell AD Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model 52, 3144–3154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935 (1983). [Google Scholar]
- 54.Martyna GJ, Tobias DJ & Klein ML Constant pressure molecular dynamics algorithms. J. Chem. Phys 101, 4177–4189 (1994). [Google Scholar]
- 55.Kräutler V, van Gunsteren WF & Hünenberger PH A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem 22, 501–508 (2001). [Google Scholar]
- 56.Darden T, York D & Pedersen L Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089–10092 (1993). [Google Scholar]
- 57.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts 4th edn, CLSI document M27 (Clinical and Laboratory Standards Institute, 2017). [Google Scholar]
- 59.Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi 3rd edn, CLSI document M38 (Clinical and Laboratory Standards Institute, 2017). [Google Scholar]
- 60.Locke JB, Almaguer AL, Zuill DE & Bartizal K Characterization of in vitro resistance development to the novel echinocandin CD101 in 1Candida species. Antimicrob. Agents Chemother 60, 6100–6107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Performance Standards for Antifungal Susceptibility Testing of Yeasts 1st edn, CLSI Supplement M60 (Clinical and Laboratory Standards Institute, 2017). [Google Scholar]
- 62.Luo G. et al. Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob. Agents Chemother 57, 3340–3347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo G. et al. Isavuconazole therapy protects immunosuppressed mice from mucormycosis. Antimicrob. Agents Chemother 58, 2450–2453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ibrahim AS et al. Caspofungin inhibits Rhizopus oryzae 1,3-β-d-glucan synthase, lowers burden in brain measured by quantitative PCR, and improves survival at a low but not a high dose during murine disseminated zygomycosis. Antimicrob. Agents Chemother 49, 721–727 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available within the paper and its Supplementary Information. SSNMR source data are provided in BMRbig entry IDs bmrbig84 and BMRB 21102. Molecular dynamics simulation trajectories are provided for Extended Data Fig. 4c,d at https://usegalaxy.org/u/taras.pogorelov/h/amb-simulations-2023. Further requests can be directed to the corresponding authors. Source data are provided with this paper.