Abstract
Arbuscular mycorrhiza (AM) is a widespread symbiotic association between plants and fungal microsymbionts that supports plant development under nutrient-limiting and various stress conditions. In this study, we focused on the overlapping genetic program activated by two commonly studied microsymbionts in addition to identifying AM-related genes. We thus applied 16,086 probe microarrays to profile the transcriptome of the model legume Medicago truncatula during interactions with Glomus mosseae and Glomus intraradices and specified a total of 201 plant genes as significantly coinduced at least 2-fold, with more than 160 being reported as AM induced for the first time. Several hundred genes were additionally up-regulated during a sole interaction, indicating that the plant genetic program activated in AM to some extent depends on the colonizing microsymbiont. Genes induced during both interactions specified AM-related nitrate, ion, and sugar transporters, enzymes involved in secondary metabolism, proteases, and Kunitz-type protease inhibitors. Furthermore, coinduced genes encoded receptor kinases and other components of signal transduction pathways as well as AM-induced transcriptional regulators, thus reflecting changes in signaling. By the use of reporter gene expression, we demonstrated that one member of the AM-induced gene family encoding blue copper binding proteins (MtBcp1) was both specifically and strongly up-regulated in arbuscule-containing regions of mycorrhizal roots. A comparison of the AM expression profiles to those of nitrogen-fixing root nodules suggested only a limited overlap between the genetic programs orchestrating root endosymbioses.
Legume plants establish two different endosymbioses with soil microorganisms: the nitrogen-fixing root nodule symbiosis and the arbuscular mycorrhiza (AM). Nodulation is almost exclusively restricted to legumes and requires the organogenesis of a root nodule that houses the rhizobial prokaryotes capable of symbiotic nitrogen fixation (Schultze and Kondorosi, 1998). In contrast, more than 80% of higher plants enter an AM symbiosis with fungi of the phylum Glomeromycota (Schüssler et al., 2001), including Glomus mosseae and Glomus intraradices as prominent representatives. The AM interaction is characterized by the transfer of minerals, in particular phosphorus, from the soil to the plant in exchange for photosynthates allocated to the fungus.
To initiate the symbiotic interaction, fungal hyphae from an extraradical mycelium penetrate the root epidermis through an appressorium and subsequently proliferate in the inner cortex (Harrison, 1997; Strack et al., 2003). These intraradical hyphae terminate in highly branched, intracellular structures designated arbuscules (Bonfante and Perotto, 1995), which are surrounded by the periarbuscular membrane. With respect to phosphorus and mineral acquisition, arbuscules are regarded as the major site of nutrient uptake, and mycorrhiza-specific phosphate transporters are located at the periarbuscular interface (Harrison et al., 2002). Apart from this novel symbiotic compartment, intraradical hyphae are assumed to be important for the allocation of carbohydrates to the fungus (Shachar-Hill et al., 1995; Bago et al., 2000). Although, in contrast to nodulation, a de novo plant organ is not formed in AM, colonization of roots by endosymbiotic fungi creates an additional carbon sink that alters the physiology of and the metabolite allocation to this symbiotic root system (Wright et al., 1998).
Both rhizobial and fungal microsymbionts colonize plant cells during nodule and AM symbioses, but they remain separated by perisymbiotic membranes controlling nutrient exchange (Provorov et al., 2002). Due to analogies in the infection process, an overlap in the activation of gene expression (vanRhijn et al., 1997; Journet et al., 2001; Brechenmacher et al., 2004; Sanchez et al., 2004) and in the recruitment of signal transduction cascades leading to nodulation and mycorrhization (Cullimore and Dénarié, 2003) is not surprising.
A key goal in legume research has been the identification of genes expressed during the development and function of root endosymbioses, an approach that profited from research in the two model legumes Medicago truncatula Gaertn and Lotus japonicus (Weidner et al., 2003). In contrast to studies dedicated to nodulation, targeted approaches addressing AM have so far revealed a markedly smaller number of genes activated in arbuscules (Franken and Requena, 2001), e.g. the phosphate transporter MtPt4 (Harrison et al., 2002), the plasma-membrane H+-ATPase Mtha1 (Krajinski et al., 2002), the germin-like protein MtGlp1 (Doll et al., 2003), the glutathione S-transferase MtGst1 (Wulf et al., 2003), and the Ser carboxypeptidase MtScp1 (Liu et al., 2003a). In the era of genomics, a more comprehensive view of gene induction during AM should be possible, and experiments making use of suppression subtractive hybridization (SSH) cDNA libraries, cDNA-array hybridizations, and real-time reverse transcription (RT)-PCR experiments identified several dozens of mycorrhiza-related M. truncatula genes (Liu et al., 2003a; Wulf et al., 2003; Brechenmacher et al., 2004; Küster et al., 2004; Manthey et al., 2004; Weidmann et al., 2004). On the fungal side, SSH approaches in particular facilitated an identification of Glomus genes expressed during appressorium formation (Requena et al., 2003; Brechenmacher et al., 2004; Breuninger and Requena, 2004), complementing knowledge on AM gene expression during early stages of the symbiosis. Regardless of the progress made, and largely due to the obligate biotrophy of Glomus spp. fungi (Franken and Requena, 2001) as well as the presence of different stages of AM formation in mycorrhizal roots (Gianinazzi-Pearson and Brechenmacher, 2004), knowledge on genes activated during AM is still limited in relation to more than a thousand genes identified as up-regulated during different steps of root nodule initiation and function using different cDNA-based macro- and microarrays (Colebatch et al., 2002, 2004; Fedorova et al., 2002; El Yahyaoui et al., 2004; Kouchi et al., 2004; Lee et al., 2004).
To reduce cross-hybridization, cDNA-based arrays are increasingly replaced by 50 to 70-mer oligonucleotide microarrays or in situ synthesized gene chips (Meyers et al., 2004). These arrays are limited in scope only by the number of sequences available for a given organism. Due to their comprehensive character, such microarrays allow us to take up the challenge of specifying the network of genes orchestrating AM formation in legumes to elucidate developmental and physiological processes relevant for this important endosymbiosis.
Expression profiling in mycorrhizal roots so far mainly focused on interactions with a particular AM fungus, leading to the definition of marker genes that subsequently had to be verified in other associations. Since there is evidence that AM fungi are characterized by different degrees of colonization as well as altered carbon allocation and symbiotic efficiency (Klironomos and Hart, 2002; Lerat et al., 2003; Smith et al., 2003b; Munkvold et al., 2004), together leading to an induction of different sets of host genes (Burleigh et al., 2002), the common genetic program for AM interactions still needs to be defined. To identify genuine mycorrhiza-related plant genes as opposed to genes activated by a particular AM fungus, we took advantage of 16,086 probe oligo microarrays, to date the most comprehensive representation of M. truncatula genes. We specified 201 M. truncatula genes as significantly induced during mycorrhization with the 2 commonly studied AM fungi G. mosseae and G. intraradices. Our global view of the M. truncatula AM transcriptome provides insights into the genetic program orchestrating this root endosymbiosis and facilitates future studies targeted at identifying functions for the encoded gene products during AM.
RESULTS AND DISCUSSION
AM Formation Significantly Alters the Transcriptome of M. truncatula Roots
To select mycorrhizal roots infected at a comparable level with the two commonly studied arbuscular mycorrhizal fungi G. mosseae and G. intraradices, random samples from root systems were stained for fungal structures 28 d post inoculation, and only those roots with similar degrees of mycorrhization were used for RNA isolation. Subsequently, the expression of marker genes for colonization intensity (MtPt4; Harrison et al., 2002) and nodulation (ENOD18; Hohnjec et al., 2003) was checked by real-time RT-PCR. Only those RNA pools with strongest MtPt4 induction and no detectable ENOD18 expression (data not shown) were selected to synthesize targets for hybridization experiments. As controls, nonmycorrhizal roots grown under phosphate limitation were used and here, the absence of MtPt4 and ENOD18 expression was confirmed by RT-PCR (data not shown).
Based on the analysis of 2 biological replicates, we identified several hundred M. truncatula genes as at least 2-fold differentially expressed in either interaction with a statistical significance of P < 0.05 (Table I), and these genes are included in Supplemental Table I. When comparing these gene expression profiles, we specified 203 genes as coinduced at least 2-fold in G. mosseae- as well as in G. intraradices-colonized M. truncatula roots (Supplemental Table II) and 176 genes as corepressed (Supplemental Table I). For 31 coinduced genes exclusively represented by expressed sequence tags (ESTs) from AM roots, we verified their origin using different approaches (Supplemental Table III). It turned out that two tentative consensus sequences (TCs) were derived from fungal ESTs, and these TCs were not considered further. A similarly low rate of fungal sequences among mycorrhiza-specific TCs was reported by Liu et al. (2003a) for the M. truncatula-Glomus versiforme interaction. Our strategy to focus only on those 201 plant genes significantly coregulated at least 2-fold in the 2 interactions studied is supported by the fact that within the list of coinduced genes, we identified well-studied AM-related marker genes. These include the phosphate transporter MtPt4 (Harrison et al., 2002), the germin-like protein MtGlp1 (Doll et al., 2003), the glutathione S-transferase MtGst1 (Wulf et al., 2003), the Ser carboxypeptidase MtScp1 (Liu et al., 2003a), the hexose transporter MtSt1 (Harrison, 1996), the 1-deoxy-d-xylulose 5-phosphate synthase MtDXS2 (Walter et al., 2002), and a multifunctional Nodulin 26-like aquaporin (Brechenmacher et al., 2004).
Table I.
Overview of the results obtained from AM transcriptome profiling
To study gene expression in AM, whole root systems of M. truncatula colonized with either G. mosseae or G. intraradices were harvested 4 weeks postinoculation. The table lists the number of probes with a log2 activation or repression ratio M larger than 1 or smaller than −1 (2-fold induction or repression), respectively, in relation to noninoculated reference samples grown under conditions of phosphate limitation. For each interaction, two independent biological replicates were studied. The values are based on probes in which at least 5 to 8 replicate spots remained after flagging for empty and poor spots and in which associated P-values were P < 0.05.
| Samples
|
Induced Probes
|
Repressed Probes
|
||
|---|---|---|---|---|
| M > 1 | Maximum M | M < −1 | Maximum M | |
| G. mosseae-colonized roots | 654 | 5.85 | 433 | −7.76 |
| G. intraradices-colonized roots | 757 | 7.21 | 621 | −6.86 |
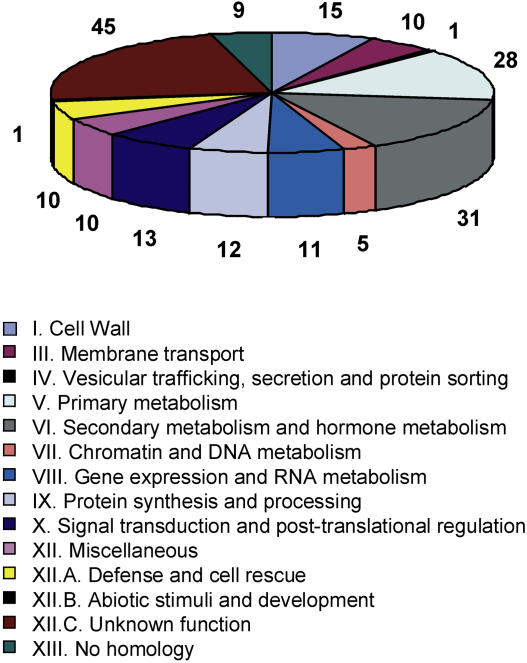
Based on comparisons to the current releases of the PIR and TrEMBL databases as well as Interpro searches, we reannotated the genes that were differentially expressed in both AM interactions. Subsequently, the proteins encoded by AM-induced M. truncatula genes were grouped into functional categories according to Journet et al. (2002). This classification (Fig. 1) illustrates that the genes identified in this study as transcriptionally activated in AM roots specify proteins relevant for different cellular, metabolic, and regulatory processes of AM formation in M. truncatula, and these are discussed in the subsequent paragraphs.
Figure 1.
Classification of 201 plant genes found to be at least 2-fold induced in M. truncatula roots colonized by the arbuscular mycorrhizal fungi G. mosseae and G. intraradices. Proteins encoded by AM-induced genes were grouped into the functional categories as defined by Journet et al. (2002). The number of genes allocated to each functional category is indicated, and the functional categories are defined.
AM-Induced Genes Associated with Cell Wall Degradation and Modification
The colonization of a root by AM fungi is accompanied by the reorganization of cell walls and extracellular matrices during (1) the penetration of the epidermis subsequent to appressoria formation, (2) the inter- and intracellular growth of fungal hyphae in the root cortex, and (3) the differentiation of membrane and cell wall structures surrounding arbuscules. It thus makes sense that 15 coinduced genes (Table II) encode a range of enzymes implicated in cell wall degradation and modification, catalytic functions that were reported to be relevant during interactions of plants with AM fungi (Peretto et al., 1995; Liu et al., 2003a).
Table II.
AM-induced M. truncatula genes related to cell wall degradation and modification
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Oligo ID, Identifier of M. truncatula 70-mer oligonucleotides. TIGR ID, Identifier in the TIGR M. truncatula Gene Index. Annotation, Updated annotations according to blast 2x hits in PIR and TrEMBL databases as well as Interpro searches. FC, Functional categories as defined by Journet et al. (2002) and in Figure 1. GM and GI, log2 expression ratios in G. mosseae- and G. intraradices-colonized roots. E-Northern, Expression profiles from the TIGR M. truncatula Gene Index. Myc-specific, TIGR TC is exclusively composed of ESTs from AM roots; Myc-induced, TIGR TC is predominantly composed of ESTs from AM roots; Sym-induced, TIGR TC is predominantly composed of ESTs from root nodules, nodulated roots and AM roots; Myc-expressed, TIGR TC is composed of at least one EST from AM roots. Literature, Found to be AM induced in other expression profiling studies; M, Manthey et al. (2004).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT008649 | TC87796 | Pro-rich protein | I | 2.22 | 1.18 | ||
| MT007463 | TC77106 | Osmotin/thaumatin-like protein | I | 2.01 | 1.18 | Myc-expressed | |
| MT003502 | TC88229 | β-1,3-Glucanase | I | 1.97 | 1.41 | ||
| MT001341 | TC78420 | Pectinesterase | I | 1.56 | 1.41 | Myc-expressed | |
| MT003194 | TC81637 | Endo-1,4-β-glucanase | I | 1.51 | 1.83 | ||
| MT000669 | TC77589 | Arabinogalactan | I | 1.42 | 1.08 | M | Myc-expressed |
| MT002404 | TC88957 | Polygalacturonase | I | 1.34 | 2.17 | ||
| MT000216 | TC76827 | Pro-rich protein | I | 1.29 | 1.54 | Myc-expressed | |
| MT006917 | TC82059 | Pectinesterase | I | 1.22 | 2.04 | ||
| MT007159 | TC85575 | Arabinogalactan | I | 1.22 | 1.14 | ||
| MT000950 | TC86689 | Endo-1,3–1,4-β-d-glucanase | I | 1.16 | 1.58 | Sym-induced | |
| MT007032 | TC85309 | Extensin | I | 1.11 | 1.64 | Sym-induced | |
| MT008602 | TC87560 | α-d-Xylosidase | I | 1.11 | 1.47 | ||
| MT014699 | TC89301 | Reversibly glycosylated polypeptide (RGP1) | I | 1.04 | 1.20 | Myc-expressed | |
| MT002974 | TC80800 | Pectate lyase | I | 1.02 | 1.58 |
Concerning cell wall degrading enzymes, 3 different (endo)-glucanases with different preferences for sugar bonds (TC88229, TC81637, and TC86689) as well as 4 different pectolytic or polygalacturonate-degrading enzymes (TC78420, TC88957, TC82059, and TC80800) were coinduced. In addition to an α-d-xylosidase (TC87560) involved in the degradation of complex carbohydrates, these enzymatic functions could modify the extracellular matrix during inter- or intracellular fungal spread as well as during the formation of the periarbuscular matrix, as proposed for the MtCel1 gene in G. versiforme-colonized roots (Liu et al., 2003a). The identification of a gene encoding a reversibly glycosylated polypeptide (TC89301) suggests that biosynthesis of xyloglucan and other hemicelluloses is relevant during colonization of roots with AM fungi.
Apart from carbohydrate modification, the identification of 3 genes encoding different Pro-rich proteins and extensins (TC87796, TC76827, and TC85309) as well as 2 genes specifying arabinogalactans (TC77589 and TC88575) point to cell wall alterations by the incorporation of structural or glycosylated proteins, similar to the observation of van Buuren et al. (1999), who localized arabinogalactan gene expression in arbuscule-containing cells of M. truncatula roots. Cell wall modification is also evident from the identification of a gene encoding an osmotin/thaumatin-like protein (TC77106). These proteins are known to be resistant to proteases as well as to denaturation, and the corresponding genes are described to be induced in response to osmotic stress or in response to fungi (Ng, 2004).
AM-Induced Genes Associated with Protein Degradation and Plant Defense
Cellular processes related to protein synthesis and processing as well as defense and cell rescue (Table III) were reported to be activated during AM (Dumas-Gaudot et al., 1994; Gianinazzi-Pearson et al., 1996; Salzer et al., 2000).
Table III.
AM-induced M. truncatula genes related to protein degradation and plant defense
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, Found to be AM induced in other expression profiling studies; K, Küster et al. (2004); W, Wulf et al. (2003); L, Liu et al. (2003a).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT009186 | TC85938 | Ser carboxypeptidase | IX | 4.36 | 2.80 | L | Myc-specific |
| MT009185 | TC85937 | Ser carboxypeptidase MtScp1 | IX | 3.99 | 4.46 | L | Myc-specific |
| MT002165 | TC79071 | Ser carboxypeptidase | IX | 2.39 | 2.65 | ||
| MT012331 | TC91847 | Translation initiation factor | IX | 2.21 | 2.35 | ||
| MT015804 | TC89543 | Subtilisin-like proteinase | IX | 1.76 | 1.11 | ||
| MT007217 | AW329656 | Ribosomal protein L5 | IX | 1.75 | 1.18 | ||
| MT011611 | TC77675 | Signal peptidase | IX | 1.67 | 2.29 | W, K | Myc-specific |
| MT015663 | TC88029 | Signal peptidase | IX | 1.55 | 1.21 | Sym-induced | |
| MT015467 | TC86655 | 40S ribosomal protein S12 | IX | 1.51 | 1.82 | Myc-expressed | |
| MT002953 | TC80768 | Prolyl oligopeptidase | IX | 1.44 | 1.15 | ||
| MT007237 | TC76882 | 40S ribosomal protein S21 | IX | 1.33 | 1.14 | Myc-expressed | |
| MT014816 | TC90718 | Cys protease | IX | 1.21 | 2.08 | Sym-induced | |
| MT014645 | TC77480 | Cys-rich antifungal protein 1 | XII.A | 4.61 | 5.34 | K | Myc-specific |
| MT006798 | TC78015 | Miraculin-trypsin inhibitor Kunitz | XII.A | 4.32 | 4.12 | W | Myc-specific |
| MT013028 | TC83316 | Protease inhibitor | XII.A | 3.62 | 2.97 | Myc-specific | |
| MT015420 | TC86086 | Kunitz-type proteinase inhibitor | XII.A | 3.53 | 4.39 | K | Sym-induced |
| MT015000 | TC84602 | Kunitz-type proteinase inhibitor | XII.A | 2.62 | 4.34 | K | Myc-specific |
| MT008080 | TC85804 | Polygalacturonase-inhibiting protein | XII.A | 2.18 | 2.16 | Sym-induced | |
| MT007836 | TC86633 | LRR protein precursor | XII.A | 1.73 | 1.83 | ||
| MT007790 | TC86418 | SAM:salicylic acid carboxylmethyltransferase | XII.A | 1.29 | 2.05 | Myc-expressed | |
| MT015651 | TC79023 | Resistance protein | XII.A | 1.12 | 1.14 | ||
| MT002474 | TC78600 | Xyloglucan-specific fungal endoglucanase inhibitor | XII.A | 1.07 | 2.07 |
Concerning protein synthesis, we found evidence for the AM-induced expression of different ribosomal proteins, a finding consistent with an observation of Journet et al. (2001) based on digital expression profiling and, most interestingly, the strong coinduction of a translation initiation factor (TC91847). With respect to protein processing, 3 Ser carboxypeptidases (TC85938, TC85937, and TC79071), a subtilisin proteinase (TC89543), a Cys protease (TC90718), and an oligopeptidase (TC80768) were coinduced. Among these, the arbuscule-induced Ser carboxypeptidase MtScp1 (TC85937; Liu et al., 2003a) was activated most strongly, and 2 signal peptidases (TC77675 and TC88029), one of them also identified by Wulf et al. (2003), could be involved in AM-specific protein translocation to subcellular compartments. In view of the strong induction of proteases, it is tempting to speculate that protein processing is important for arbuscular formation or a reestablishment of cellular structures after arbuscule degradation, processes that occur in parallel during AM.
With respect to defense and cell rescue, we detected 4 different (Kunitz-type) protease inhibitors (TC78105, TC83316, TC86086, and TC84602), a Cys-rich antifungal protein (TC77480), and a xyloglucan-specific fungal endoglucanase inhibitor (TC78600). These proteins could fine-tune protease activity during arbuscule degradation or modulate plant defense responses elicited by the intraradical presence of fungal hyphae. Defense-related gene expression is well documented in AM roots (Salzer et al., 2000) and was also found in other transcriptome profiling studies (Liu et al., 2003a; Brechenmacher et al., 2004).
AM-Induced Genes Encoding Nutrient Transporters
In arbuscular mycorrhizae, the extraradical fungal mycelium acts as an extension of the roots and reaches beyond the root depletion zone, enabling a thorough exploration of the soil for limiting nutrients such as phosphorus and minerals (Smith and Read, 1997; Pfeffer et al., 1999). The extensive periarbuscular membrane interface containing plant and fungal H+-ATPases (Gianinazzi-Pearson et al., 2000; Krajinski et al., 2002; Requena et al., 2003) supports the suggestion that nutrient transfer in particular takes place at this interface, although some studies have shown that nutrient transfer may additionally occur at the intracellular hyphae (Gianinazzi-Pearson, 1996). Nevertheless, the molecular mechanisms underlying the translocation of nutrients in AM are presently not well understood.
In our global profiling approach, we anticipated to obtain information on transport processes relevant for nutrient allocation between fungus and host. We identified 10 genes encoding putative membrane transport proteins (Table IV) as coinduced at least 2-fold in both mycorrhizal associations, and among these, 3 that were previously described to be mycorrhiza induced. Acting as a marker gene, the arbuscule-specific phosphate transporter MtPt4 was strongly induced in both endosymbioses. MtPt4 transcripts and proteins were shown to be most prominent in mature arbuscules, and MtPt4 expression was reported to be positively correlated with the extent of G. versiforme colonization (Harrison et al., 2002). Another mycorrhiza-induced gene (TC86110) encodes a previously identified membrane-intrinsic multifunctional aquaporin (Küster et al., 2004; Manthey et al., 2004; Sanchez et al., 2004). One of the two coinduced sugar transporters (TC77798 and TC87421) represents the Mtst1 gene that is known to be highly expressed in arbuscule-containing cells and in cortical cells surrounding infected areas (Harrison, 1996).
Table IV.
AM-induced M. truncatula genes related to membrane transport
Gene activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, found to be AM induced in other expression profiling studies: M, Manthey et al. (2004); K, Küster et al. (2004); H96, Harrison (1996); S, Sanchez et al. (2004); H02, Harrison et al. (2002); W, Wulf et al. (2003).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT009707 | TC85743 | Inorganic phosphate transporter MtPt4 | III | 4.98 | 5.06 | H02, W, K | Myc-specific |
| MT002501 | TC78158 | High-affinity nitrate transporter | III | 4.76 | 1.03 | ||
| MT009589 | TC78157 | High-affinity nitrate transporter | III | 2.51 | 1.14 | Myc-expressed | |
| MT007526 | TC86110 | Multifunctional Nodulin 26-like aquaporin | III | 2.06 | 1.97 | K, M, S | Myc-expressed |
| MT008596 | TC87421 | Hexose transporter MtSt1 | III | 1.42 | 2.05 | H96 | Myc-expressed |
| MT002866 | TC88701 | Manganese transporter MtZIP7 | III | 1.32 | 1.76 | ||
| MT003256 | TC80954 | High-affinity nitrate transporter | III | 1.21 | 1.91 | ||
| MT001072 | TC77763 | Proton pump interactor | III | 1.14 | 3.07 | Myc-expressed | |
| MT007819 | TC77798 | Hexose transporter | III | 1.12 | 1.22 | Myc-expressed | |
| MT006556 | TC84545 | Nitrate transporter | III | 1.01 | 1.18 |
Four coinduced genes encoded the first AM-related nitrate transporters (TC78157, TC78158, TC80954, and TC84545) identified in M. truncatula. These findings add to a report on an AM-induced nitrate transporter in tomato (Hildebrandt et al., 2002) and suggest mechanisms not only supporting the uptake of ammonium but also the acquisition of nitrate during AM. So far, only an M. truncatula nitrate transporter gene (TC79437) down-regulated in roots colonized by G. mosseae was identified (Burleigh, 2001) and in our experiments, this repression was confirmed not only in G. mosseae- but also in G. intraradices-colonized roots. Concerning ion transporters of the MtZIP family (Burleigh et al., 2003), we describe the first mycorrhiza-induced manganese transporter (MtZIP7, TC88701) whose transport properties were recently characterized in yeast expression systems (López-Millán et al., 2004).
It is assumed that a modulation of transporter gene expression may be related to changes in the internal micro- or macronutrient concentrations (Liu et al., 1998). Thus, the coordinated suppression and induction of genes seems to be a common feature reflecting a switch in nutrient supply from direct root uptake to symbiotic uptake. In mycorrhizal roots, the symbiotic pathway accounts for most if not all of the total inorganic phosphate (Pi) acquisition (Pearson and Jakobsen, 1993) and recent studies support a hypothesis that in the M. truncatula-G. intraradices symbiosis, the entire Pi was delivered via the mycorrhizal rather than the direct pathway (Smith et al., 2003a). In accordance with this, the 2 high-affinity phosphate transporters, MtPT1 and MtPT2, representing the direct pathway of Pi acquisition and shown to be down-regulated during both G. versiforme and G. intraradices AM (Versaw et al., 2002), were both repressed in G. intraradices AM. On the other hand, these genes were still slightly induced or not regulated during G. mosseae AM, suggesting possible differences in the regulation of phosphate transporters in different AM interactions. Apart from these genes, our study revealed an additional strongly cosuppressed Pi transporter gene (TC84790), which could be a representative of the direct uptake system in addition to MtPT1 and MtPT2.
Even on the basis of approximately 16,000 M. truncatula probes, MtPt4 remains the only phosphate transporter (PT) gene strongly induced in both AM analyzed, while several other members of the PT gene family were not. This exclusive focus on one single symbiotic PT gene agrees with Paszkowski et al. (2002), who identified an entire set of 13 high-affinity Pi transporter genes in Oryza sativa, in which only one (OsPT11) responded to mycorrhizal colonization. At present, this is also the case for Lycopersicon esculentum (LePT1) and Solanum tuberosum (StPT3; Liu et al., 1998; Rosewarne et al., 1999; Rausch et al., 2001). Thus, the evolution of a single AM-induced phosphate transporter gene might reflect a common strategy.
In contrast, this principle seems not to be favored for the regulation of nitrate transporter genes. Here, stringent conditions uncovered at least 4 genes as being coinduced in AM, while other nitrate transporter genes were down-regulated to different extents (TC88300 and TC82201). A similar situation of up- and down-regulation is evident for the hexose transporter gene family. While MtSt1 (Harrison, 1996) and the hexose transporter specified by TC77798 are coinduced, we identified counterparts in TC77077 and TC83509, both down-regulated by AM fungi. Surprisingly, the hexose transporter gene represented by TC77077 was previously described to be up-regulated during AM formation between M. truncatula and G. versiforme using macroarray hybridizations (Liu et al., 2003a). This raises the possibility that different AM fungi might activate different members of the hexose transporter gene family during symbiotic uptake mechanisms. Alternatively, different sugar transporters might be recruited during specific stages of the symbiosis. Whereas it seems likely that AM-induced mineral transporters function as nutrient importers to supply minerals to the plant, the situation is more complex with respect to up-regulated sugar transporters. Here, either an export function to supply hexoses to the symbiotic fungus or an import function to fine-tune the amount of carbohydrates allocated to the mycorrhizal fungus has to be considered.
AM-Induced Genes Associated with Primary Metabolism
The establishment of an arbuscular mycorrhiza significantly alters the metabolism of a plant root. In accordance with this, 28 genes up-regulated in both AM interactions encoded gene products related to primary metabolism (Table V). In plants, Suc serves as the major transport molecule for source-to-sink carbon allocation (Hawker, 1985). Since Suc is a stable disaccharide, it has to be cleaved into hexoses prior to further metabolic degradation by either Suc synthases or different invertases (Copeland, 1990), both regarded as key enzymes responsible for carbon partitioning. In roots colonized by AM fungi, there is evidence for an up-regulation of Suc synthase, soluble acid invertase, and alkaline invertase in regions surrounding arbuscules and even within arbuscule-containing cells (Blee and Anderson, 2002; Hohnjec et al., 2003; Ravnskov et al., 2003). Cleavage of Suc by either enzyme yields hexoses that after phosphorylation enter glycolysis and the tricarboxylic acid cycle. Opposite to the induction of genes corresponding to this pathway in root nodules (El Yahyaoui et al., 2004), the expression levels of most genes related to glycolysis and the tricarboxylic acid cycle are not significantly altered in AM on the basis of pooled tissue samples.
Table V.
AM-induced M. truncatula genes related to primary metabolism
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, Found to be AM induced in other expression profiling studies; K, Küster et al. (2004).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT015669 | TC88539 | MtBcp1 | V | 4.28 | 4.89 | K | Myc-specific |
| MT000134 | TC76657 | PSII oxygen-evolving complex protein 2 | V | 3.95 | 3.62 | Myc-expressed | |
| MT003225 | TC87415 | Bcp | V | 2.94 | 2.35 | Myc-specific | |
| MT004678 | TC82671 | Cys synthase | V | 2.81 | 4.54 | ||
| MT010383 | TC90212 | Leucoanthocyanidin dioxygenase | V | 2.36 | 1.28 | Sym-induced | |
| MT004097 | TC87185 | Allyl alcohol dehydrogenase | V | 2.36 | 2.12 | Myc-induced | |
| MT015668 | TC88539 | MtBcp1 | V | 1.94 | 2.68 | K | Myc-specific |
| MT015793 | TC81478 | UDP-glucoronosyl/UDP-glucosyl transferase | V | 1.91 | 1.62 | ||
| MT008396 | TC78334 | l-Ascorbate oxidase | V | 1.72 | 1.67 | ||
| MT000147 | TC76664 | Patatin | V | 1.70 | 1.27 | ||
| MT002011 | TC88442 | β-Hydroxyacyl-ACP dehydratase | V | 1.60 | 1.86 | ||
| MT004744 | TC90058 | Putative dehydrogenase | V | 1.42 | 1.31 | Myc-expressed | |
| MT000961 | TC86557 | Branched-chain amino acid aminotransferase | V | 1.40 | 2.56 | Myc-expressed | |
| MT008259 | TC87200 | Acetylornithin aminotransferase | V | 1.38 | 1.40 | ||
| MT000454 | TC86072 | Cys synthase | V | 1.37 | 1.71 | ||
| MT007664 | TC86426 | l-Ascorbate oxidase | V | 1.37 | 1.48 | Myc-expressed | |
| MT007807 | TC86231 | Protein disulfide isomerase | V | 1.34 | 1.34 | ||
| MT004004 | TC77871 | Malonyl-CoA: acyl carrier protein transacylase | V | 1.33 | 1.71 | Myc-expressed | |
| MT002830 | TC80501 | Chlorophyll b synthase | V | 1.28 | 2.43 | ||
| MT004707 | TC81108 | Glucosyltransferase | V | 1.22 | 1.72 | ||
| MT015293 | TC85778 | NFU1 iron-sulfur cluster assembly factor | V | 1.21 | 1.66 | ||
| MT010141 | TC80652 | α-Fucosidase | V | 1.20 | 1.66 | ||
| MT015305 | TC85814 | Ω-6 desaturase | V | 1.20 | 1.31 | Myc-expressed | |
| MT000873 | TC77795 | Pyruvate dehydrogenase E1 α-subunit | V | 1.19 | 1.59 | Myc-expressed | |
| MT000207 | TC76829 | Fructokinase | V | 1.14 | 1.49 | Sym-induced | |
| MT007901 | TC77591 | Putative phosphatase | V | 1.10 | 1.16 | Sym-induced | |
| MT000450 | TC77284 | Bcp | V | 1.05 | 1.51 | Sym-induced | |
| MT007472 | TC86035 | Triacylglycerol lipase | V | 1.02 | 1.15 | Sym-induced |
On the other hand, 4 genes encoding enzymes related to fatty acid metabolism were induced: malonyl-CoA:Acyl carrier protein transacylase (TC77871), β-hydroxyacyl-ACP dehydratase (TC88442), Ω-6 desaturase (TC85814), and triacylglycerol lipase (TC86035). These enzymes might be implicated in the biosynthesis and modification as well as the metabolic degradation of lipids from the membranes surrounding arbuscules. Since arbuscules are transient structures with a life span of only a few days, both establishment and degradation of periarbuscular membranes occur in parallel in mycorrhizal roots and require both an active fatty acid biosynthesis and the degradation of fatty acids.
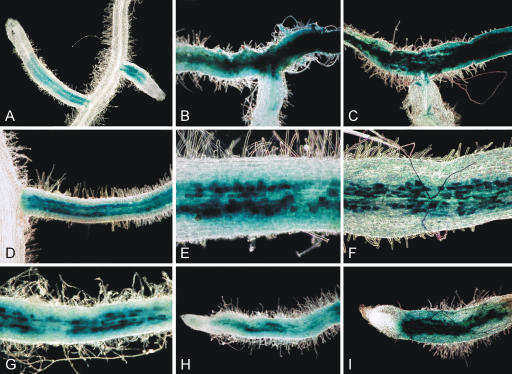
Finally, 3 different members (TC88539, TC87415, and TC77284) of a gene family encoding different blue copper proteins (Bcp) were coinduced, 2 of which were already identified on the basis of cDNA microarrays (Küster et al., 2004). Bcps contain a single copper atom and are implicated in electron transfer reactions. The most characterized members of this class of proteins are chloroplastic plastocyanins that exchange electrons with cytochromes. So far, the role of Bcps in AM roots is elusive, but it is tempting to speculate that they are involved in electron transfer processes taking place in the network of plastids surrounding arbuscules in mycorrhizal roots. Here, plastids are known to be essential compartments for fatty acid biosynthesis, nitrogen assimilation, and starch deposition (Fester et al., 2001). The cell-specific movement of plastids to a close vicinity of arbuscules is associated with extensive cytoskeleton reorganizations, supported by an arbuscule-related expression of the M. truncatula β-tubulin gene MtTubb1 (Manthey et al., 2004). These correlations prompted us to inspect the genomic organization of the Bcp gene family and we found that 7 genes encoding Bcps are located in tandem in a region of the M. truncatula genome represented by bacterial artificial chromosome (BAC) mth2-15c20 (GenBank accession no. AC126009). For one of these genes, designated MtBcp1 (TC88539), 1,180 bp of promoter sequence were PCR-amplified and used to evaluate the cellular localization of gusAint expression in transgenic roots mycorrhized with G. intraradices. As shown in Figure 2, β-glucuronidase (GUS) staining is most prominent in regions where arbuscule formation takes place and correlates with the degree of mycorrhization. Intensely mycorrhized areas characterized by a high density of arbuscule-containing cells exhibited strongest reporter gene expression in the whole root tissue including outer cortical cells. Counterstaining with ink demonstrated that, depending on the mycorrhization status, pMtBcp1-driven gusAint expression is strongest in the arbuscule-containing cells but is, although to a lesser extent, additionally present in adjacent cortical cells. Thus, the observed promoter activity indicates a correlation of MtBcp1 expression with root colonization and supports the assumption that Bcps serve as mediators of electron transfer processes in arbuscule-containing cells and their close vicinity.
Figure 2.
Histochemical localization of GUS activity in transgenic roots of M. truncatula colonized with G. intraradices and expressing the −1,181/-2 pMtBcp1-gusAint fusion. Prior to dark-field microscopy, roots shown in A, B, D, E, G, and H were stained for GUS activity exclusively, while those in C, F, and I were additionally stained with ink to visualize fungal structures. A, Distinct GUS activity in cortical cells surrounding the vasculature of young, lateral roots. B, Root exhibiting strongest GUS staining on the right, extending to staining of lesser extent on the left. Here, intensive coloration is confined to inner cortical cells. C, Double-staining of the same mycorrhizal root fragment as shown in B. Fungal structures (arbuscules and hyphae) are most prominent in the intensely infected area on the right and decrease toward the left. Parts of the root that do not exhibit GUS coloration are free of fungal infection. D, E, G, and H, Strong local GUS expression represented by single cells of the innermost cortical layers and surrounded by cells that are stained to a lower extent and do not exhibit distinct dark spots. F, Double-stained root fragment. The ink staining reveals arbuscule-containing cells that overlay the single blue spots of intensively GUS coloration. I, Lateral root tip with infected parts corresponding to pMtBcp1-activated cells, surrounded by outer cortical cells exhibiting GUS staining without harboring fungal structures.
AM-Induced Genes Associated with Secondary Metabolism and Hormone Action
Secondary metabolism and phytohormone biosynthesis are processes with major relevance for AM roots (Fester et al., 1999; Strack et al., 2003). In our study, apart from the AM-induced glutathione S-transferase gene MtGst1 (Bestel-Corre et al., 2002; Wulf et al., 2003) that might be involved in detoxification processes, 30 AM-activated genes (Table VI) encoded enzymes involved in the synthesis of secondary metabolites such as terpenes (TC76892 and TC80795), flavonoids (TC81553, TC85753, TC87789, and TC88553), ascorbate (TC78334), and amygdalin (TC76723), compounds known to be present in AM (Liu et al., 2003b; Strack et al., 2003).
Table VI.
AM-induced M. truncatula genes related to secondary metabolism and hormone action
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, Found to be AM induced in other expression profiling studies; K, Küster et al. (2004); S, Sanchez et al. (2004); W, Wulf et al. (2003).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT009013 | TC85868 | Glutathione S-transferase MtGst1 | VI | 5.02 | 5.86 | W, S, K | Myc-specific |
| MT004625 | TC81595 | Ent-kaurene synthase A | VI | 3.25 | 2.05 | Myc-expressed | |
| MT006682 | TC84637 | Cytochrome P450 | VI | 3.12 | 1.12 | ||
| MT000228 | TC85641 | Cytochrome P450 | VI | 2.45 | 2.61 | Myc-expressed | |
| MT000634 | TC77465 | Auxin-regulated protein GH3 | VI | 2.44 | 3.13 | ||
| MT002829 | TC89260 | Cytochrome P450 | VI | 2.43 | 3.00 | K | Myc-specific |
| MT003018 | TC88553 | Flavanone 3-hydroxylase | VI | 2.43 | 1.64 | ||
| MT015424 | TC76892 | Terpene synthase | VI | 2.38 | 2.45 | Sym-induced | |
| MT007595 | TC77355 | SRG1-like oxidoreductase (ethylene-forming enzyme) | VI | 2.23 | 2.68 | ||
| MT000541 | TC86263 | Δ-Aminolevulinic acid dehydratase | VI | 2.16 | 2.01 | Myc-expressed | |
| MT007257 | TC76942 | Narbonin | VI | 1.84 | 1.45 | Sym-induced | |
| MT000424 | TC77154 | Narbonin | VI | 1.81 | 1.30 | Sym-induced | |
| MT003200 | TC80866 | Narbonin | VI | 1.69 | 1.37 | ||
| MT008518 | TC78110 | SRG1-like oxidoreductase (ethylene-forming enzyme) | VI | 1.50 | 1.12 | Sym-induced | |
| MT006856 | TC78989 | Cytochrome P450 | VI | 1.49 | 1.38 | Myc-expressed | |
| MT003010 | TC89135 | Cytochrome P450 | VI | 1.42 | 1.52 | Myc-expressed | |
| MT003219 | TC80795 | 5-α-Taxadienol-10-β-hydroxylase | VI | 1.35 | 1.05 | Sym-induced | |
| MT002731 | TC78764 | 1-Aminocyclopropane-1-carboxylic acid oxidase | VI | 1.33 | 1.79 | Sym-induced | |
| MT015692 | TC88126 | Cytochrome P450 | VI | 1.27 | 1.73 | ||
| MT000598 | TC85270 | Benzoyl-CoA:benzyl alcohol benzoyl transferase | VI | 1.25 | 1.38 | Myc-expressed | |
| MT015525 | TC78048 | GA-regulated protein GASA4 | VI | 1.24 | 1.54 | ||
| MT003683 | TC81553 | Flavonoid 3′-hydroxylase | VI | 1.23 | 1.04 | ||
| MT015597 | TC87789 | UDP-glycose:flavonoid glycosyltransferase | VI | 1.21 | 1.43 | ||
| MT005972 | TC78620 | Cytochrome P450 | VI | 1.19 | 1.38 | Myc-expressed | |
| MT003693 | TC89459 | Oxidase-like protein | VI | 1.18 | 1.29 | ||
| MT007295 | TC85753 | Flavon momnoamine oxidase | VI | 1.11 | 1.02 | Sym-induced | |
| MT013911 | TC90420 | Chalcone O-methyltransferase | VI | 1.11 | 1.43 | Sym-induced | |
| MT002771 | TC78589 | DXS2 | VI | 1.08 | 1.27 | Myc-expressed | |
| MT000153 | TC76723 | Amygdalin hydrolase isoform AH I | VI | 1.05 | 2.51 | Sym-induced | |
| MT000313 | TC77051 | Mevalonate disphosphate decarboxylase | VI | 1.03 | 1.72 | Sym-induced | |
| MT002711 | TC78989 | Cytochrome P450 | VI | 1.03 | 1.44 | Myc-expressed |
Important members of the terpene family are carotenoid tetraterpenes, and their biosynthesis was reported to be partially controlled on the transcriptional level in mycorrhizal roots (Fester et al., 2002). It is well documented that cleavage products deriving from carotenoids are known to form the yellow pigment that is characteristic of AM roots in many plants (Strack et al., 2003). Two genes related to the biosynthesis of terpenes were identified here: TC77051 encoding mevalonate disphosphate decarboxylase, an important component of the mevalonate pathway of carotenoid production, and TC78589 encoding 1-deoxy-d-xylulose 5-phosphate synthase 2 (DXS2; Walter et al., 2002), a key enzyme of the mevalonate-independent pathway of carotenoid biosynthesis (Strack et al., 2003). Although either pathway results in the formation of isopentenyl diphosphate, the key intermediate of terpene biosynthesis, only the DXS2-related pathway was previously reported to be AM induced and related to mycorrhadicin production (Walter et al., 2002).
Strikingly, and in line with the induction of a range of genes associated with secondary metabolism, 8 different cytochrome P450 genes were identified as AM induced. P450-type cytochromes comprise a range of different families, and are, for example, involved in the oxidation of different isoflavonoids, phenylpropanoid metabolites characteristic of legumes (Dixon and Sumner, 2003). Recently, Liu et al. (2003b) reported on P450 cytochromes that function as isoflavone 2′- and 3′-hydroxylases, and similar to our observations, one of them showed elevated expression during AM. Interestingly, the action of hydroxylated isoflavonoids, e.g. the phytoalexin medicarpin, is related to pathogen- as well as insect-induced responses, respectively (Dixon, 1999), and the synthesis of the phytoalexin medicarpin was shown to be transiently induced also during AM (Harrison and Dixon, 1993).
Concerning phytohormone biosynthesis and action, different coinduced genes can be related to GA3, auxin, and ethylene. In case of GA3, genes were up-regulated that specify the GA biosynthesis enzyme ent-kaurene synthase A (TC81595) and the GA-regulated protein GASA 4 (TC78048), indicating the synthesis of GA3 in AM roots. This is opposite to the situation in root nodules, where a down-regulation of these genes was reported (El Yahyaoui et al., 2004). With respect to auxin hormones, the auxin-regulated protein GH3 (TC77465) was induced. Finally, AM-induced genes involved in ethylene biosynthesis were identified: 2 ethylene-forming enzymes (TC77355 and TC78110) and an aminocyclopropane carboxylic acid oxidase (TC78764). Whereas auxin biosynthesis was reported to occur in AM roots (Kaldorf and Ludwig-Müller, 2000), data on the effect of ethylene and GA3 on AM formation are conflicting and rather limited (El Ghachtouli et al., 1996; Geil and Guinel, 2002). Nevertheless, the expression data reported here support the involvement of these phytohormones at least during particular stages of AM, as shown for cytokinins, abscisic acid, or jasmonic acid (Strack et al., 2003).
AM-Induced Genes Encoding Components of Signal Transduction Pathways
With dmi1, dmi2, and dmi3, 3 plant genes relevant for the common early stages of signal transduction during nodulation and AM formation were identified by positional cloning (Cullimore and Dénarié, 2003). Apart from the well-studied dmi genes, several mutations with specific defects in AM development were identified (Marsh and Schultze, 2001), but in most cases, the genes responsible for the mycorrhiza-deficient phenotypes are not known. Thus, there is a need to increase the knowledge on genetic determinants of plant signal perception prior to the action of dmi genes as well as on genes involved in signaling during later stages of AM development.
In this study, we identified 13 such genes as induced in both G. mosseae- and G. intraradices-colonized roots, and these can be related to different aspects of plant signal perception and signal transduction (Table VII). First, 2 genes (TC78350 and TC87043) encoding different lectins were strongly up-regulated in AM, and this expression pattern is supported by data from cDNA-based microarrays (Küster et al., 2004), by the identification of corresponding ESTs in a G. intraradices SSH library (Wulf et al., 2003), and by digital expression profiling (Supplemental Table II). Legume lectins were described as determinants of specificity during nodule symbioses by gain-of-function studies (van Rhijn et al., 1998), and their strong activation in AM roots points to a role during signal perception also in AM.
Table VII.
AM-induced M. truncatula genes related to signal transduction
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, Found to be AM induced in other expression profiling studies; K, Küster et al. (2004).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT003520 | TC78350 | Man/Glc-binding lectin | X | 4.35 | 5.20 | Myc-induced | |
| MT013816 | TC87043 | Man/Glc-binding lectin | X | 3.87 | 2.92 | K | Myc-specific |
| MT006551 | TC82283 | Putative phytosulfokine LRR-type receptor kinase | X | 2.55 | 4.02 | ||
| MT004715 | TC80630 | Ser/Thr protein kinase | X | 2.05 | 1.71 | Myc-specific | |
| MT014872 | TC89285 | NBS/LRR resistance protein | X | 1.99 | 3.68 | Myc-specific | |
| MT005914 | TC93498 | 33-kD secretory protein | X | 1.78 | 1.37 | Myc-expressed | |
| MT014665 | TC86597 | Ser/Thr receptor kinase | X | 1.77 | 2.52 | Sym-induced | |
| MT003662 | TC80490 | Ser/Thr protein kinase | X | 1.28 | 1.04 | Myc-expressed | |
| MT005857 | TC92439 | Phosphoinositol-specific phospholipase C | X | 1.24 | 4.89 | ||
| MT002595 | TC80104 | LRR receptor-like protein kinase | X | 1.15 | 3.03 | Sym-induced | |
| MT006436 | TC93373 | ERG GTPase | X | 1.14 | 1.05 | Myc-induced | |
| MT007943 | TC86792 | Putative two-component response regulator | X | 1.08 | 2.33 | Myc-expressed | |
| MT003494 | TC88105 | Ser/Thr protein kinase | X | 1.02 | 1.59 |
Concerning AM-related receptors, genes encoding a Ser/Thr receptor kinase (TC86597) and a Leu-rich repeat (LRR) receptor-like protein kinase (TC80104) were identified. LRR-type receptor-like kinases are characterized by extracellular LRR domains mediating protein-protein interactions. These receptors act by binding extracellular ligands and transducing this signal to intracellular protein kinase domains (Chen, 2001). In addition to classical receptor-like kinases, a gene (TC93498) encoding a homolog of the Arabidopsis (Arabidopsis thaliana) 33-kD Cys-rich secretory proteins deserves attention. These proteins resemble extracellular domains of receptor-like protein kinases, but in contrast to these, they do not contain a transmembrane and an intracellular protein kinase domain. Thus, it was proposed that they may interact with membrane-bound receptor-like protein kinases during signal perception (Chen, 2001). The identification of a putative phytosulfokine LRR-type receptor kinase (TC82283) seems also to be noteworthy, since phytosulfokine constitutes a sulfated intercellular peptide signal during cellular differentiation and proliferation in plants (Matsubayashi et al., 2002).
With respect to signal transduction cascades initiated after signal perception, we identified an AM-induced gene (TC92439) encoding a phosphoinositol-specific phospholipase C, an enzyme that generates the second messengers inositol triphosphate and diacylglycerol through hydrolysis of membrane-bound PIP2. The second messengers initiate further signal transduction events, e.g. the release of Ca2+ from intracellular stores, a process relevant during initial stages of Nod-factor perception during nodulation. Since plant phosphoinositol-specific phospholipase Cs mediate different stress and pathogen responses (Repp et al., 2004), an involvement in signaling during AM is also possible. Downstream components of signal transduction cascades are protein kinases that, for example, facilitate signal amplification, and here we identified 3 different AM-induced genes (TC80630, TC80490, and TC88105) coding for Ser/Thr protein kinases with possible relevance for this process.
During initial stages of AM formation, Myc-factors are postulated to be perceived by the plant (Cullimore and Dénarié, 2003), but the molecular processes mediating this perception are unknown. Subsequent to the very early stages of signaling, diffusible low-Mr compounds activate expression of the early nodulin gene ENOD11 (Kosuta et al., 2003), and the AM-induced genes reported here are candidates for legume genes required for the recognition and transduction of such signals.
AM-Induced Genes Encoding Transcriptional Regulators
As detailed above, the colonization of plant roots by AM fungi results in an extensive reorganization of cellular structures and in specific alterations of metabolism. These changes prerequisite differential gene expression, a process primarily mediated by transcriptional regulators. In accordance with this, 11 coinduced genes encoding transcription factors were identified (Table VIII). Highest transcript accumulation was detected for a Myb-family transcription factor (TC78253) already identified as induced by different AM fungi using macroarrays (Liu et al., 2003a; Küster et al., 2004; Sanchez et al., 2004). Similarly, a TINY AP2 domain transcription factor gene (TC78355) induced during M. truncatula-G. versiforme associations (Liu et al., 2003a) exhibited elevated expression in G. mosseae and G. intraradices AM.
Table VIII.
AM-induced M. truncatula genes encoding transcriptional regulators
Genes activated more than 2-fold in both AM interactions are sorted according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined for Table II. Literature, Found to be AM induced in other expression profiling studies; K, Küster et al. (2004); S, Sanchez et al. (2004); L, Liu et al. (2003a).
| Oligo ID | TIGR ID | Annotation | FC | GM | GI | Literature | E-Northern |
|---|---|---|---|---|---|---|---|
| MT001930 | TC78235 | Myb-like transcription factor | VIII | 5.15 | 3.09 | L, S, K | Myc-specific |
| MT007392 | TC77052 | Myb-family transcription factor | VIII | 2.15 | 1.26 | Myc-expressed | |
| MT011784 | TC91273 | Homeodomain-zip protein | VIII | 1.96 | 1.27 | Myc-expressed | |
| MT006389 | TC92797 | Zinc-finger, CCHC type | VIII | 1.86 | 1.47 | ||
| MT005076 | TC92211 | AT-rich interaction domain protein | VIII | 1.82 | 1.72 | Sym-induced | |
| MT005698 | TC92089 | YABBY protein transcription factor | VIII | 1.74 | 3.36 | Myc-specific | |
| MT001444 | TC78204 | Zinc-finger RNA binding protein | VIII | 1.70 | 1.16 | Myc-expressed | |
| MT002458 | TC79248 | Zinc-finger RNA binding protein | VIII | 1.22 | 1.14 | Myc-expressed | |
| MT005542 | TC92282 | Zinc-finger, C2H2 type | VIII | 1.20 | 1.94 | Myc-specific | |
| MT001680 | TC78355 | TINY-like protein | VIII | 1.16 | 2.91 | L | Sym-induced |
| MT003698 | TC81463 | bZIP transcription factor | VIII | 1.04 | 1.50 | Sym-induced |
Among the other regulatory genes, we identified another putative Myb family transcription factor (TC77052). So far, only the Myb gene Mt-phan was described in M. truncatula, and this gene was expressed in lateral root initials, in nematode-induced giant cells, and in root nodules (Koltai et al., 2001). Apart from genes encoding Myb transcription factors, a homeobox-Leu zipper (TC91273) and a basic-Leu zipper (bZIP) transcription factor (TC81463) were identified. Both families of proteins are involved in pathogen defense (Zhou et al., 2000) and hormone action (Fukazawa et al., 2000). Interestingly, members of the TGA class of bZIP factors were discussed to regulate auxin-induced glutathione S-transferases (Johnson et al., 2001), and with MtGst1, an AM-induced glutathione S-transferase (TC85868) was recently characterized by Wulf et al. (2003).
TC92089 corresponds to a strongly induced gene encoding a YABBY transcription factor. These regulators form a small protein family known to be responsible for the specification of abaxial cell fate in Arabidopsis lateral organs as well as axis formation (Bowman et al., 2002). Additional genes (e.g. TC92282) encoded DNA- or RNA-binding proteins containing zinc fingers, suggesting a role for these proteins in the regulation of gene expression in AM.
From 5 genes that were activated more than 2-fold in the opposite direction in the 2 AM interactions studied (Supplemental Table I), 3 genes specified a Myb transcription factor (TC86301), a RING zinc finger protein (TC89100), and an AP2-domain DNA-binding protein (TC88292). The identification of differentially regulated genes encoding putative transcription factors supports the observation that in addition to common genetic mechanisms, specific sets of host genes are induced by different AM fungi.
In Arabidopsis, different families of transcription factors, each containing distinct DNA binding domains, were implicated in plant stress responses since their expression is modulated under particular stress conditions (Shinozaki and Yamaguchi-Shinozaki, 2000). This supports the notion that in addition to mediating early signal transduction during fungal colonization, the AM-induced transcription factors reported here could regulate cellular responses evoked by the intraradical presence of these two microsymbionts, processes certainly placed downstream of the initial recognition of mycorrhizal symbionts. With respect to this, there is increasing evidence for the existence of regulatory mechanisms that govern arbuscule-specific gene expression (Harrison et al., 2002; Doll et al., 2003; Liu et al., 2003a; Hohnjec et al., 2003; Wulf et al., 2003; Vieweg et al., 2004), and together with the components of signal transduction mentioned above, the AM-induced transcriptional regulators could e.g. mediate gene expression in arbuscule-containing cells.
AM-Induced Genes Encoding Proteins of Unknown Function or No Homology
A total of 54 genes up-regulated in AM roots either encode gene products matching hypothetical gene products, proteins only characterized by Interpro domains, or proteins with no homology (Supplemental Table II). Among these are four putative F-box containing proteins and a putative ubiquitin C-terminal hydrolase, possibly related to protein degradation and processing via the ubiquitin ligase complex. Protein degradation apart from general protein turnover processes is known to play important regulatory roles (Hellmann and Estelle, 2002) by controlling protein degradation during the differentiation of symbiotic structures. Heavy metal detoxification is another important feature of mycorrhizal roots, also with respect to phytoremediation (Vassilev et al., 2004). A coinduced gene (TC78576) encoding a putative heavy metal transport/detoxification protein is thus of interest, and the encoded protein could be involved in the uptake of heavy metals from the soil.
Digital Expression Profiling Generally Validates AM-Related Gene Expression
Digital expression profiling approaches have become increasingly popular not only for identification of differentially expressed genes, but also for the validation of high-throughput expression profiling data (Alba et al., 2004). In the case of M. truncatula, almost 190,000 ESTs derived from more than 40 different conditions are deposited in The Institute for Genomic Research (TIGR) M. truncatula Gene Index, and among these, 21,049 ESTs are derived from arbuscular mycorrhiza roots (http://www.tigr.org/tdb/tgi/mtgi). As shown in Supplemental Table II, apart from typical mycorrhiza-specific marker genes (e.g. MtPt4, Harrison et al., 2002; MtDxs2, Walter et al., 2002; MtScp1, Liu et al., 2003a; MtGst1, Wulf et al., 2003; MtGlp1, Doll et al., 2003), most genes reported here to be up-regulated in AM roots are either mycorrhiza specific, mycorrhiza induced, or are at least expressed in cDNA libraries constructed from mycorrhizal roots. In addition, digital expression profiling indicated that several genes are up-regulated in endosymbioses including AM. Due to the different biological material and the different growth conditions of plants used for cDNA library construction and considering that with G. versiforme a different fungus was used to obtain 7,351 of the 21,049 AM ESTs (Liu et al., 2003a), these in silico data generally support our expression profiles.
Real-Time RT-PCR Experiments Support the AM-Induced Expression of Selected Genes
Using 4 additional biological samples of G. mosseae and G. intraradices colonized roots in comparison to nonmycorrhized control roots, we performed real-time RT-PCR experiments to verify the expression of 20 coinduced genes corresponding to a range of functional categories and expression ratios (Table IX). Based on these samples, we confirmed the AM-induced expression for 19 of 20 genes tested. Whereas 15 genes were coinduced more than 2-fold in either interaction, 4 genes were identified as coinduced more than 1.7-fold in G. mosseae and more than 2-fold in G. intraradices colonized roots (Table IX). In some cases, expression ratios based on real-time RT-PCR were significantly higher than those ratios obtained from microarray hybridizations, which is a common phenomenon for specifically expressed genes. In other cases, expression ratios obtained by real-time RT-PCR were comparably low. Similar to results reported by Manthey et al. (2004), this was evident in particular for those genes only weakly induced on the basis of microarray hybridizations. A low induction ratio had to be expected for several genes since in contrast, for example, to nodulation where many nodule-induced genes do not exhibit a basal transcription in roots, a significant expression level can be detected in nonmycorrhized tissues for many AM-related genes. In summary, our real-time RT-PCR data, obtained from samples, support our microarray-based expression profiles. The differences observed are likely to result from the fact that the extent and efficiency of mycorrhizal colonization is difficult to control and that pooled tissue samples have been studied, making it inevitable to average over different cell types.
Table IX.
Verification of AM-induced M. truncatula genes by real-time RT-PCR
The expression of 20 AM-induced genes belonging to a range of functional categories was verified by real-time RT-PCR. Expression ratios are given as log2 values to allow a comparison to the expression ratios derived from microarray hybridizations. All expression ratios except one are significant with P < 0.05. Abbreviations are as defined in Table II.
| Oligo ID
|
TIGR ID
|
Annotation
|
FC
|
Microarray
|
Real-Time RT-PCR
|
||
|---|---|---|---|---|---|---|---|
| GM | GI | GM | GI | ||||
| MT003502 | TC88229 | β-1,3-Glucanase | I | 1.97 | 1.41 | 1.30 | 1.32 |
| MT001341 | TC78420 | Pectinesterase | I | 1.56 | 1.41 | 1.93 | 1.41 |
| MT002501 | TC78158 | High-affinity nitrate transporter | III | 4.76 | 1.03 | 0.93 | 1.12 |
| MT009589 | TC78157 | High-affinity nitrate transporter | III | 2.51 | 1.14 | 1.35 | 1.30 |
| MT002866 | TC88701 | Manganese transporter MtZIP7 | III | 1.32 | 1.76 | 2.74 | 2.46 |
| MT006556 | TC84545 | Nitrate transporter | III | 1.01 | 1.18 | 0.74 | 1.37 |
| MT015669 | TC88539 | MtBcp1 | V | 4.28 | 4.89 | 12.03 | 9.95 |
| MT003225 | TC87415 | Bcp | V | 2.94 | 2.35 | 2.36 | 1.53 |
| MT004004 | TC77871 | Malonyl-CoA:acyl carrier protein transacylase | V | 1.33 | 1.71 | 1.40 | 1.00 |
| MT000207 | TC76829 | Fructokinase | V | 1.14 | 1.49 | 0.20a | 0.85 |
| MT004625 | TC81595 | Ent-kaurene synthase A | VI | 3.25 | 2.05 | 0.87 | 1.90 |
| MT000634 | TC77465 | Auxin-regulated protein GH3 | VI | 2.44 | 3.13 | 4.36 | 3.33 |
| MT002731 | TC78764 | 1-Aminocyclopropane-1-carboxylic acid oxidase | VI | 1.33 | 1.79 | 0.78 | 1.51 |
| MT002771 | TC78589 | DXS2 | VI | 1.08 | 1.27 | 1.59 | 1.90 |
| MT001930 | TC78235 | Myb-like transcription factor | VIII | 5.15 | 3.09 | 13.75 | 11.18 |
| MT005542 | TC92282 | Zinc-finger, C2H2 type | VIII | 1.20 | 1.94 | 3.76 | 9.95 |
| MT014816 | TC90718 | Cys protease | IX | 1.21 | 2.08 | 10.20 | 7.48 |
| MT003520 | TC78350 | Man/Glc-binding lectin | X | 4.35 | 5.20 | 4.42 | 3.93 |
| MT004715 | TC80630 | Ser/Thr protein kinase | X | 2.05 | 1.71 | 1.52 | 1.03 |
| MT013028 | TC83316 | Protease inhibitor | XII.A | 3.62 | 2.97 | 6.79 | 5.39 |
Expression ratio not significant.
M. truncatula Genes Down-Regulated in AM Are Largely Related to Stress Responses
In total, 176 genes were identified as down-regulated more than 2-fold in response to both G. intraradices and G. mosseae colonization (Supplemental Table I), and the 50 most strongly down-regulated genes are listed in Table X. Since nonmycorrhizal roots used as controls were grown under conditions of phosphate limitation (20 μm), it is not surprising that several of the AM down-regulated genes coded for proteins involved in stress responses.
Table X.
M. truncatula genes repressed in AM roots
The 50 genes most strongly repressed in both AM interactions are listed according to the functional categories as defined by Journet et al. (2002) and are sorted within these classes according to the induction level in G. mosseae-colonized roots. Abbreviations are as defined in Table II.
| Oligo ID | TIGR ID | Annotation | FC | GM | GI |
|---|---|---|---|---|---|
| MT007074 | TC76646 | Caffeic acid O-methyltransferase | I | −4.73 | −2.96 |
| MT005666 | TC83381 | Caffeic acid O-methyltransferase | I | −3.80 | −2.75 |
| MT000246 | TC85734 | Pro-rich protein | I | −3.15 | −1.11 |
| MT000967 | TC77984 | Expansin-related protein | I | −2.58 | −1.22 |
| MT003596 | TC88633 | Isoprenylated heavy metal transport/detoxification protein | III | −3.66 | −2.73 |
| MT003439 | TC90076 | RAB1 GTP-binding protein | IV | −2.92 | −1.71 |
| MT015170 | TC76593 | Δ-1-Pyrroline-5-carboxylate synthetase | V | −4.58 | −3.17 |
| MT014053 | TC76589 | Δ-1-Pyrroline-5-carboxylate synthetase | V | −4.52 | −3.48 |
| MT015169 | TC76589 | Δ-1-Pyrroline-5-carboxylate synthetase | V | −4.37 | −4.58 |
| MT000507 | TC77268 | Cinnamoyl-CoA reductase | V | −3.54 | −2.30 |
| MT000869 | TC77949 | Putative copper-binding protein | V | −2.96 | −3.78 |
| MT005126 | TC78918 | Suc synthase MtSucS2 | V | −2.70 | −2.38 |
| MT011868 | TC82744 | Suc synthase MtSucS4 | V | −2.68 | −2.35 |
| MT007444 | TC86064 | Glutamate decarboxylase | V | −2.59 | −2.07 |
| MT014842 | TC90894 | Anthocyanin 5-aromatic acyltransferase-like protein | V | −2.52 | −2.56 |
| MT007047 | TC76593 | Δ-1-Pyrroline-5-carboxylate synthase | V | −2.49 | −2.50 |
| MT012479 | TC83483 | Malate dehydrogenase | V | −2.38 | −3.89 |
| MT004582 | TC82770 | Chalcone reductase | VI | −5.46 | −3.65 |
| MT003716 | TC80105 | Ferritin | VI | −4.05 | −2.93 |
| MT008389 | TC78616 | Probable stress and abscisic acid induced protein | VI | −3.31 | −2.31 |
| MT009487 | TC88609 | NAC domain protein NAC2 | VIII | −2.98 | −2.52 |
| MT008226 | TC78257 | Homeobox-Leu zipper protein | VIII | −2.97 | −1.78 |
| MT000799 | TC86417 | NAM-like regulatory protein | VIII | −2.59 | −2.28 |
| MT014768 | TC90326 | bZIP transcription factor | VIII | −2.45 | −2.52 |
| MT004368 | TC91563 | γ-Glutamyltranspeptidase | IX | −3.89 | −3.41 |
| MT000029 | TC76371 | Protein kinase C, phorbol ester/diacylglycerol binding | X | −4.13 | −1.66 |
| MT012651 | TC83559 | Peptidoglycan-binding LysM receptor kinase | X | −3.60 | −3.13 |
| MT007340 | TC85793 | Annexin | X | −3.52 | −3.27 |
| MT014127 | TC85429 | Nonspecific lipid-transfer protein | XII | −4.07 | −3.07 |
| MT015187 | TC85327 | Proteinase inhibitor | XII.A | −2.82 | −2.88 |
| MT015189 | TC85327 | Proteinase inhibitor | XII.A | −2.54 | −3.88 |
| MT014313 | TC76866 | Late embryogenesis abundant protein | XII.B | −7.76 | −3.61 |
| MT014312 | TC76867 | Late embryogenesis abundant protein | XII.B | −6.77 | −4.20 |
| MT005274 | TC84691 | Late embryogenesis abundant protein | XII.B | −6.43 | −3.56 |
| MT014784 | TC81698 | Desiccation-responsive low-temperature-induced protein | XII.B | −5.53 | −3.50 |
| MT007558 | TC85950 | Late embryogenesis abundant protein | XII.B | −5.47 | −3.12 |
| MT000542 | TC77327 | Dehydrin-like protein | XII.B | −5.38 | −3.08 |
| MT007538 | TC85220 | Late embryogenesis abundant protein | XII.B | −5.03 | −3.17 |
| MT008432 | TC78666 | Alkaline α-galactosidase II | XII.B | −3.70 | −2.51 |
| MT015129 | TC76537 | Cold acclimation responsive protein | XII.B | −3.41 | −2.42 |
| MT007556 | TC85949 | Seed maturation protein LEA 4 | XII.B | −3.28 | −3.89 |
| MT007269 | TC76315 | Cold acclimation response protein | XII.B | −2.65 | −3.26 |
| MT014258 | TC76698 | Dehydrin-like protein | XII.B | −2.62 | −2.32 |
| MT000966 | TC86847 | Thaumatin-like protein | XII.B | −2.46 | −1.29 |
| MT004870 | TC89522 | Hypothetical protein | XII.C | −6.26 | −4.15 |
| MT007837 | TC77899 | Desiccation-responsive protein | XII.C | −3.26 | −3.15 |
| MT001853 | TC88006 | Hypothetical protein | XII.C | −2.77 | −2.79 |
| MT012402 | TC92349 | Hypothetical protein | XII.C | −2.63 | −3.24 |
| MT002839 | TC80357 | Hypothetical protein | XII.C | −2.47 | −2.07 |
| MT005919 | TC86466 | No significant homology | XIII | −2.64 | −4.98 |
It was interesting that several gene families were identified as activated in phosphate-starved roots, e.g. encoding two caffeic acid O-methyltransferases involved in the biosynthesis of lignin cell wall precursors (Zubieta et al., 2002), 4 Δ-1-pyrroline-5-carboxylate synthetases (the first enzymes of the Pro biosynthetic pathway) known to be induced under salt stress (Ginzberg et al., 1998), and 5 different late embryogenesis abundant proteins relevant for desiccation tolerance (Wise and Tunnacliffe, 2004). Further examples for genes related to stress responses are a desiccation-responsive protein (TC77899), a dehydrin-like protein (TC77327), and a desiccation-responsive low temperature-induced protein (TC81698). The observation that the expression level of different stress-induced genes is obviously much lower in AM-colonized roots adds to reports that efficient mycorrhization increases the capability of plants to alleviate abiotic stress conditions (Smith and Read, 1997). With respect to metabolism, the identification of 2 phosphate starvation-induced genes encoding the Suc synthases MtSucS2 and MtSucS4 is in accordance with real-time RT-PCR experiments showing elevated expression of these genes only in phosphate-starved roots (Hohnjec et al., 2003).
In addition to transcription factors, the identification of a down-regulated peptidoglycan-binding LysM receptor kinase (TC83559) is intriguing, since LysM receptor kinases mediate Nod-factor perception during nodulation. Although there are common steps in early signaling during nodulation and mycorrhization, the initial signal perception of the host is specific for either symbiosis and requires a Nod-factor or a hypothetical Myc-factor (Cullimore and Dénarié, 2003). The strong down-regulation of this LysM-type receptor kinase in AM argues against a role during mycorrhization, but might indicate a function related to nodule initiation.
AM-Induced Gene Expression Is Not Mediated by Phosphate
To assess if the AM-induced genes identified in this study are also activated by exogenously supplied phosphate, we studied gene expression in M. truncatula roots in response to high phosphate concentrations (2 mm). Although phosphate acquisition is one of the major benefits of the plant in mycorrhiza symbioses, these experiments revealed that from 201 genes found to be coinduced in 2 AM interactions, only 8 genes were up-regulated more than 2-fold in roots grown under conditions of high phosphate supply (Supplemental Table II). Among these were 1 of the 2 high-affinity nitrate transporter (TC78158) and 1 of the 4 Kunitz-type protease genes (TC78015), indicating a differential response of members of gene families to external phosphate concentrations. The low overlap between AM- and phosphate-induced gene expression shows that the transcriptional changes observed are largely due to the colonization of roots by AM fungi and cannot be regarded as a mere consequence of a mycorrhiza-improved phosphorus nutrition, a finding in accordance with the observations by Liu et al. (2003a) for the M. truncatula-G. versiforme symbiosis.
AM- and Nodulation-Induced Gene Expression Overlap Only to a Limited Extent
Since there is evidence for common gene expression during AM and nodulation in the early stages of these interactions, we compared the AM-induced transcription profiles with those derived from nitrogen-fixing root nodules. Although large-scale transcriptomics studies during M. truncatula nodulation were published (El Yahyaoui et al., 2004; Manthey et al., 2004), expression data derived from different probe sets are difficult to relate, since oligonucleotide probes can match multiple EST clusters, depending on the clustering strategies applied.
To assess the overlap between nodulation- and mycorrhization-related gene expression, we performed reference hybridizations using mature, nitrogen-fixing nodules and uninfected roots of comparable age. A relation of the expression profiles revealed that from 201 plant genes significantly induced in AM roots, 27 were also up-regulated at least 2-fold in mature root nodules (Supplemental Table II). Among these were 3 nodulin genes encoding a multifunctional Nodulin 26-like aquaporin known to be up-regulated in both root nodules and AM (TC86110; Brechenmacher et al., 2004), an MtN21-like putative membrane protein (TC78291; Gamas et al., 1996), and a symbiosome membrane nodulin (TC90920). From the up-regulation of the symbiosome membrane nodulin gene originally identified in soybean in response to Bradyrhizobium japonicum inoculation (Winzer et al., 1999), it can be inferred that the peribacteroid and periarbuscular membranes share common structural properties.
An induction of TC88957 encoding a polygalacturonase and TC86689 encoding an endo-1,3 to 1,4-β-d-glucanase indicates the recruitment of similar cell wall modifying enzymes in root nodules and AM, whereas an activation of the auxin-regulated protein GH3 (TC77465) is consistent with the synthesis of auxin in root nodules. Interestingly, different TCs encoding enzymes involved in protein processing were identified: TC85937 representing the Ser carboxypeptidase MtScp1, TC90718 specifying a Cys protease, and TC88029 that codes for a signal peptidase. The identification of a symbiosis-induced signal peptidase is intriguing, since this either indicates an involvement of similar genes in the translocation of proteins across perisymbiotic membranes or extends the hypothesis of Mergaert et al. (2003) that cleaved signal peptides of conserved structure might act as signals in root nodules. Finally, one gene (TC79248) encoding a zinc-finger RNA binding protein was identified that represents a candidate for a symbiotic regulator of gene expression.
The comparably low overlap in gene induction during mature stages of two different root endosymbioses is reminiscent of the observations reported by Manthey et al. (2004) and El Yahyaoui et al. (2004) on the basis of less comprehensive M. truncatula cDNA microarrays. In conclusion, only a limited overlap in transcriptional activation exists between mature stages of the two root endosymbioses arbuscular mycorrhiza and root nodule.
CONCLUSIONS AND PERSPECTIVES
Following on different cDNA-based arrays (Liu et al., 2003a; VandenBosch and Stacey, 2003; Küster et al., 2004), 70-mer oligonucleotide microarrays designated Mt16kOLI1 now constitute the most comprehensive expression profiling tool available for this model plant. Using these microarrays, we here present a global overview on common gene expression in AM by studying the interaction of M. truncatula with two different mycorrhizal fungi. By focusing on those genes that were reproducibly induced in the two interactions studied, gene expression related to the inoculum or the fungal species should have been minimized, allowing us to specify genuine mycorrhiza-related genes. On the other hand, AM formation is an asynchronous process suffering from dilution effects and a mixture of different developmental stages. Although it is likely that more AM-related genes can be identified by applying cell-specific expression profiling techniques, our collection of AM-related genes already forms a comprehensive resource for functional characterizations in the T-DNA or TILLING mutant collections currently established for different legume species.
MATERIALS AND METHODS
Plant Production
Medicago truncatula Gaertn cv Jemalong genotype A17 seeds were surface-sterilized and scarified as described previously (Hohnjec et al., 2003). Subsequently, they were spread on 0.8% (w/v) water agar (Sigma agar-agar, Sigma-Aldrich, Steinheim, Germany) in petri dishes wrapped with parafilm and aluminum foil. Seeds were vernalized at 4°C for 4 d, transferred to RT for 2 d, and finally incubated for one more day at RT in the light. The seedlings were grown in 27-cm3 pots in clay granules (Seramis, Masterfoods, Verden, Germany) at 22°C with 60% relative humidity and an 18-h/6-h-light/-dark alteration until the plantlets developed at least one trifoliate. Afterward, they were transferred to 600-cm3 pots half-filled with clay granules that were covered with approximately 1 cm of sand. An inoculum containing Glomus mosseae (Granular AMF Inoculum G. mosseae BEG 12, Biorize R&D, Dijon, France) or Glomus intraradices (Premier Tech Biotechnologies, Rivière-de-Loup, Québec, Canada), respectively, was directly applied to the root system before pots were filled to a maximum volume with clay granules. Control plants were treated comparably except for inoculum application. Plants were watered twice a week with one-half-strength Hoagland solution containing 20 μm phosphate (Arnon and Hoagland, 1940). At 28 d post inoculation, mycorrhizal roots as well as nonmycorrhizal control roots were harvested and immediately frozen in liquid nitrogen. At this time, randomly selected mycorrhizal roots were stained for colonization measurements using the gridline intersection method according to McGonigle et al. (1990). The percentage of root length colonization (AMF hyphae, spores, vesicles, or arbuscules) ranged between 50% and 80%, whereas the relative arbuscule frequency within colonized fragments varied between 55% and 75%, respectively.
To obtain phosphate supplied roots, plants were grown under the conditions described above and fertilized twice a week using one-half-strength Hoagland solution (Arnon and Hoagland, 1940) supplemented with 2 mm phosphate. Twenty-eight days after being transferred into 600-m3 pots, roots grown under 2 mm phosphate, and corresponding control roots grown under conditions of phosphate limitation (one-half-strength Hoagland solution, 20 μm phosphate) were harvested and immediately frozen in liquid nitrogen.
To collect nitrogen-fixing root nodules, vernalized seedlings were grown aeroponically using a nitrogen-rich medium (Journet et al., 2001) with 40 plants/250 L caisson. After 2 weeks, this medium was exchanged for a medium lacking combined nitrogen. Three days later, plants were inoculated with a Sinorhizobium meliloti RCR2011 pXLGD4 (Journet et al., 2002) culture diluted to a final concentration of approximately 5 × 105 bacteria/milliliter medium. After 20 d, nitrogen-fixing root nodules were collected. As a control, noninoculated roots grown to the same age in nitrogen-rich medium (Gallusci et al., 1991) were harvested from approximately 4 cm below the crown. All collected tissues were immediately frozen in liquid nitrogen.
Isolation of Total RNA, Real-Time RT-PCR Experiments, and Genomic PCR-Amplifications
Total RNA was prepared using the RNeasy Plant Mini kit and DNase I on-column digestion according to the manufacturer's instructions (Qiagen, Hilden, Germany) from different pools of six to eight roots (or different root nodule pools) to provide biological replicates for expression profiling experiments. The resulting RNA preparations were concentrated to 1.25 μg/microliter using Microcon-30 columns (Millipore, Schwalbach, Germany) and stored at −80°C until use. The integrity of total RNA was checked on agarose gels and its quantity as well as purity was determined spectrophotometrically.
Real-time RT-PCR experiments were performed according to a protocol reported by Hohnjec et al. (2003) using gene-specific primers (Supplemental Table IV). The results for Glomus-inoculated versus nonmycorrhizal roots were averaged over four biological replicates and the expression ratios were tested for statistical significance using a Student's t test.
To prove the plant origin of AM-induced genes, gene-specific primers (Supplemental Table IV) were used to PCR-amplify the corresponding genomic regions from total M. truncatula DNA (isolated from leaves) using Taq DNA polymerase (Qiagen) as recommended by the manufacturer.
Scope, Layout, and Printing of Mt16kOLI1 Microarrays
Mt16kOLI1 microarrays contain 16,086 70-mer oligonucleotide probes (Qiagen) representing all TCs of the TIGR M. truncatula Gene Index 5 (http://www.tigr.org/tdb/mtgi) as well as different GAPDH controls (Küster et al., 2004). An assessment of background levels and unspecific hybridizations was performed on the basis of 226 probes containing spotting buffer and 12 70-mer probes that serve as negative controls. The layout of Mt16kOLI1 microarrays comprises 48 grids arranged in 12 metarows and 4 metacolumns. Each grid contains 702 spots in 27 rows and 26 columns with 24 columns carrying 27 probes and 2 columns carrying 20 probes. For each M. truncatula probe, duplicate spots are present in the same grids throughout Mt16kOLI1 microarrays.
Oligonucleotides were dissolved in 1.5 m betaine, 3× SSC to a concentration of 40 μm and were printed onto QMT epoxy slides (Quantifoil, Jena, Germany) using a MicroGrid II 600 spotter (BioRobotics, Cambridge, UK) with 48 SMP3 stealth pins (TeleChem International, Sunnyvale, CA). We estimate that each spot contains approximately 300 fmol of oligonucleotides. DNA was cross-linked to the slide by incubation for 105 min at 80°C. Slides were kept in sealed plastic bags containing desiccation packs at 18°C to 22°C for up to 6 months with no loss of quality. For validation of microarray printing and efficient coupling to the slide, one microarray from each series was hybridized with Alexa 555-labeled random nonamers (Molecular Probes, Leiden, The Netherlands) as described (Küster et al., 2004). Mt16kOLI1 array definition files were deposited in ArrayExpress under accession A-MEXP-85.
Cy-Labeling of Hybridization Targets, Hybridization, and Image Acquisition
Twenty micrograms of total RNA was used to synthesize Cy3- or Cy5-labeled cDNA targets according to Küster et al. (2004) using a mixture of 2.5 μg of double-anchored oligo(dT)15VN primers and 5 μg of random hexamers. Labeled cDNA was purified using CyScribe GFX columns (Amersham Biosciences, Freiburg, Germany), and labeling efficiency was checked as described in Küster et al. (2004). Immediately before use, microarrays were rinsed once for 5 min in 0.1% (v/v) Triton X-100, twice for 2 min each in 250 mL MilliQ water containing 29 μL 32% (v/v) HCl, once for 10 min in 0.1 m KCl, and once for 1 min in MilliQ water (at 20°C each). Slides were blocked for 15 min at 50°C in 200 mL 1× QMT (Quantifoil) solution containing 46 μL 32% (v/v) HCl. Subsequently, slides were rinsed in MilliQ water (20°C, 1 min) and dried by centrifugation (185g, 5 min, 20°C). Hybridization was performed in an ASP hybridization station (Amersham Biosciences) in a sample volume of 250 μL DIG EasyHyb solution (Roche, Mannheim, Germany) supplemented with 15 μg sonicated salmon sperm DNA (Amersham Biosciences). Immediately before injection, samples were denatured for 5 min at 65°C. After 16 h of hybridization at 42°C, microarrays were washed once in 2× SSC, 0.2% (w/v) SDS (1 min, 42°C), twice in 0.2× SSC, 0.1% (w/v) SDS (1 min, 20°C), twice in 0.2× SSC (1 min, 20°C), and once in 0.1× SSC (1 min, 18°C). Slides were dried by centrifugation (185g, 5 min, 20 C) and scanned with a resolution of 10 μm using the ScanArray 4000 (PerkinElmer, Boston). Original expression profiling data were deposited in the ArrayExpress database under accession numbers E-MEXP-218, E-MEXP-237, and E-MEXP-238.
Analysis of Image Data from Microarray Hybridizations
Image processing was performed using the ImaGene 5.5 software (BioDiscovery, Los Angeles). The mean intensities of signal pixels and the mean intensities of local background pixels were calculated for each spot in both channels, and spots were flagged “empty” in the case of R ≤ 0.7 for both channels. The R ≤ 0.7 threshold resulted in the removal of signals corresponding to negative controls and empty wells. In addition, manual flags were set for spots that were within hybridization artefacts. Data files were imported into the EMMA1.1 array analysis software (Dondrup et al., 2003), and flagged spots were discarded during import. Subsequent to Lowess normalization with a floor value of 20, regulated genes were identified using a t-statistics. Genes were regarded as significantly differentially expressed if P ≤ 0.05 and M ≥ 1 or if P ≤ 0.05 and M ≤ −1 with M specifying the expression ratio.
Construction and Histological Analysis of Transgenic Hairy Roots
The promoter of the MtBcp1 gene (TC88539) was PCR-amplified with Pwo DNA polymerase (Roche) from BAC mth2-15c20 (GenBank accession no. AC126009) using gene-specific primers containing appropriate restriction sites. The amplified fragment covered the −1,181/−2 region (position 29,680 to 28,501 of BAC mth2-15c20) relative to the start codon and was cloned as SphI/EcoRI fragment in front of the gusAint gene of pGUSINT (Hohnjec et al., 2003). Subsequently, the promoter-gusAint fusion was excised using SpeI, filled in with Klenow polymerase, and cloned into the SmaI site of the binary plasmid pRedRoot (Limpens et al., 2004). The resulting plasmid was electroporated into Agrobacterium rhizogenes ARqua1, and this strain was used to generate hairy roots on M. truncatula cv Jemalong A17 according to Vieweg et al. (2004). Four weeks after mycorrhization with G. intraradices, GUS assays were performed as described by Hohnjec et al. (2003) and subsequent stainings for fungal structures as described by Vierheilig et al. (1998), except that transgenic roots were preselected using dsRed fluorescence (Leica MZ FL III, Wetzlar, Germany). Examination of tissues was performed by light microscopy and documented with a digital camera (Olympus C-4040Z, Hamburg, Germany).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AC126009.
Supplementary Material
Acknowledgments
We are grateful to Tim Kahlke and Michael Dondrup (Bielefeld University) for integrating Mt16kOLI1 layouts and features into the ArrayLIMS and the EMMA 1.1 software. We thank Matthew McIntosh (Bielefeld University) for support during manuscript polishing.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1084 MolMyk: Molecular Basics of Mycorrhizal Symbioses projects Ku–1478/1–2 and Pu 28/25–3); and grant BIZ 7. N. Hohnjec and H. Küster acknowledge financial support of the International Graduate School in Bioinformatics and Genome Research (Bielefeld University).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056572.
References
- Alba R, Zhangjun F, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D'Ascenzo M, Gordon JS, Rose JK, et al (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50: 463–483 [Google Scholar]
- Bago B, Pfeffer PE, Shachar-Hill Y (2000) Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol 124: 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestel-Corre G, Dumas-Gaudot E, Poinsot V, Dieu M, Dierick JF, van Tuinen D, Remacle J, Gianinazzi-Pearson V, Gianinazzi S (2002) Proteome analysis and identification of symbiosis-related proteins from Medicago truncatula Gaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis 23: 122–137 [DOI] [PubMed] [Google Scholar]
- Blee KA, Anderson AJ (2002) Transcripts for genes encoding soluble acid invertase and sucrose synthase accumulate in root tip and cortical cells containing mycorrhizal arbuscules. Plant Mol Biol 50: 197–211 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Perotto S (1995) Strategies of arbuscular mycorrhizal fungi when infecting host plants. New Phytol 130: 3–21 [Google Scholar]
- Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Weidmann S, van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V (2004) Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14: 253–262 [DOI] [PubMed] [Google Scholar]
- Breuninger M, Requena N (2004) Recognition events in AM symbiosis: analysis of fungal gene expression at the early appressorium stage. Fungal Genet Biol 41: 794–804 [DOI] [PubMed] [Google Scholar]
- Burleigh SH (2001) Relative quantitative RT-PCR to study the expression of plant nutrient transporters in arbuscular mycorrhizas. Plant Sci 160: 899–904 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Cavagnaro T, Jakobsen I (2002) Functional diversity of arbuscular mycorrhizas extends to the expression of plant genes involved in P nutrition. J Exp Bot 53: 1593–1601 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Kristensen BK, Ellegaard Bechmann I (2003) A plasma membrane zinc transporter from Medicago truncatula is up-regulated in roots by Zn fertilization, yet down-regulated by arbuscular mycorrhizal colonization. Plant Mol Biol 52: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Chen Z (2001) A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol 126: 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15: 411–420 [DOI] [PubMed] [Google Scholar]
- Copeland L (1990) Enzymes of sucrose metabolism. Methods Plant Biochem 3: 73–85 [Google Scholar]
- Cullimore J, Dénarié J (2003) How legumes select their sweet talking symbionts. Science 302: 575–578 [DOI] [PubMed] [Google Scholar]
- Dixon RA (1999) Isoflavonoids: biochemistry, molecular biology and biological functions. In U Sankawa, ed, Comprehensive Natural Products Chemistry, Vol 1. Elsevier, Oxford, pp 773–823
- Dixon RA, Sumner LW (2003) Legume natural products. Understanding and manipulating complex pathways for human and animal health. Plant Physiol 131: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll J, Hause B, Demchenko K, Pawlowski K, Krajinski F (2003) A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene. Plant Cell Physiol 44: 1208–1214 [DOI] [PubMed] [Google Scholar]
- Dondrup M, Goesmann A, Bartels D, Kalinowski J, Krause L, Linke B, Rupp O, Szyrba A, Pühler A, Meyer F (2003) EMMA: a platform for consistent storage and efficient analysis of microarray data. J Biotechnol 106: 135–146 [DOI] [PubMed] [Google Scholar]
- Dumas-Gaudot E, Guillaume P, Tahiri-Alaoui A, Gianinazzi-Pearson V, Gianinazzi S (1994) Changes in polypeptide patterns in tobacco roots colonized by two Glomus species. Mycorrhiza 4: 215–221 [Google Scholar]
- El Ghachtouli N, Martin-Tanguy J, Paynot M, Gianinazzi S (1996) First report of the inhibition of arbuscular mycorrhizal infection of Pisum sativum by specific and irreversible inhibition of polyamine biosynthesis or by gibberellic acid treatment. FEBS Lett 385: 189–192 [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136: 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fester T, Maier W, Strack D (1999) Accumulation of secondary compounds in barley and wheat roots in response to inoculation with an arbuscular mycorrhizal fungus and co-inoculation with rhizosphere bacteria. Mycorrhiza 8: 241–246 [Google Scholar]
- Fester T, Schmidt T, Lohse S, Walter MH, Giuliano G, Bramley PM, Fraser PD, Hause B, Strack D (2002) Stimulation of carotenoid metabolism in arbuscular mycorrhizal roots. Planta 216: 148–154 [DOI] [PubMed] [Google Scholar]
- Fester T, Strack D, Hause B (2001) Reorganization of tobacco root plastids during arbuscule development. Planta 213: 864–868 [DOI] [PubMed] [Google Scholar]
- Franken P, Requena N (2001) Analysis of gene expression in arbuscular mycorrhiza: new approaches and challenges. New Phytol 150: 431–439 [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P, Dedieu A, Journet E-P, Huguet T, Barker DG (1991) Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen-fixing root nodules development and response to exogenously supplied nitrate. Plant Mol Biol 17: 335–349 [DOI] [PubMed] [Google Scholar]
- Gamas P, de Carvalho-Niebel F, Lescure N, Cullimore J (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Geil RD, Guinel FC (2002) Effects of elevated substrate-ethylene on colonization of leek (Allium porrum) by the arbuscular mycorrhizal fungus Glomus aggregatum. Can J Bot 80: 114–119 [Google Scholar]
- Gianinazzi-Pearson V (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Arnould C, Oufattole M, Arango M, Gianinazzi S (2000) Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211: 609–613 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Brechenmacher L (2004) Functional genomics of arbuscular mycorrhiza: decoding the symbiotic cell programme. Can J Bot 82: 1228–1234 [Google Scholar]
- Gianinazzi-Pearson V, Dumas-Gaudot E, Golotte A, Tahiri-Alaoui A, Gianinazzi S (1996) Cellular and molecular defense-related root response to invasion by arbuscular mycorrhizal fungi. New Phytol 133: 45–57 [Google Scholar]
- Ginzberg I, Stein H, Kapulnik Y, Szabados L, Strizhov N, Schell J, Koncz C, Zilberstein A (1998) Isolation and characterization of two different cDNAs of delta1-pyrroline-5-carboxylate synthase in alfalfa, transcriptionally induced upon salt stress. Plant Mol Biol 38: 755–764 [DOI] [PubMed] [Google Scholar]
- Harrison MJ (1996) A sugar transporter from Medicago truncatula: altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J 9: 491–503 [DOI] [PubMed] [Google Scholar]
- Harrison MJ (1997) The arbuscular mycorrhizal symbiosis: an underground association. Trends Plant Sci 2: 54–60 [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dixon RA (1993) Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microbe Interact 6: 643–654 [Google Scholar]
- Hawker JS (1985) Sucrose. In PM Dey, RA Dixon, eds, Biochemistry of Storage Carbohydrates in Green Plants. Academic Press, London, pp 1–51
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Hildebrandt U, Schmelzer E, Bothe H (2002) Expression of nitrate transporter genes in tomato colonized by an arbuscular mycorrhizal fungus. Physiol Plant 115: 125–136 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16: 903–915 [DOI] [PubMed] [Google Scholar]
- Johnson C, Glover G, Arias J (2001) Regulation of DNA binding and trans-activation by a xenobiotic stress-activated plant transcription factor. J Biol Chem 276: 172–178 [DOI] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring the root symbiotic programs of the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldorf M, Ludwig-Müller J (2000) AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiol Plant 109: 58–67 [Google Scholar]
- Klironomos JN, Hart MM (2002) Colonization of roots by arbuscular mycorrhizal fungi using different sources of inoculum. Mycorrhiza 12: 181–184 [DOI] [PubMed] [Google Scholar]
- Koltai H, Dhandaydham M, Oppermann C, Thomas J, Bird D (2001) Overlapping plant signal transduction pathways induced by a parasitic nematode and a rhizobial endosymbiont. Mol Plant Microbe Interact 14: 1168–1177 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker DG, Bécard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu G-J, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aiko T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11: 263–274 [DOI] [PubMed] [Google Scholar]
- Krajinski F, Hause B, Gianinazzi-Pearson V, Franken P (2002) Mtha1 a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-induced expression. Plant Biol 4: 754–761 [Google Scholar]
- Küster H, Hohnjec N, Krajinski F, El Yahyaoui F, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al (2004) Construction and validation of cDNA-based Mt6k-RIT macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108: 95–113 [DOI] [PubMed] [Google Scholar]
- Lee H, Hur C-G, Oh CJ, Kim HB, Park S-Y, An CS (2004) Analysis of the root nodule-enhanced transcriptome in soybean. Mol Cells 18: 53–62 [PubMed] [Google Scholar]
- Lerat S, Lapointe L, Piché Y, Vierheilig H (2003) Variable carbon-sink strength of different Glomus mosseae strains colonizing barley roots. Can J Bot 81: 886–889 [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Liu C-J, Huhmann D, Sumner LW, Dixon RA (2003. b) Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula. Plant J 36: 471–484 [DOI] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003. a) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán A-F, Ellis DR, Grusak MA (2004) Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula. Plant Mol Biol 54: 583–596 [DOI] [PubMed] [Google Scholar]
- Manthey M, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Schultze M (2001) Analysis of arbuscular mycorrhizas using symbiosis-defective plants mutants. New Phytol 150: 525–532 [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 5572: 1470–1472 [DOI] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Galbraith DW, Nelson T, Agrawal V (2004) Methods for transcriptional profiling in plants. Be fruitful and replicate. Plant Physiol 135: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I (2004) High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol 164: 357–364 [DOI] [PubMed] [Google Scholar]
- Ng TB (2004) Antifungal proteins and peptides of leguminous and non-leguminous origins. Peptides 25: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JN, Jakobsen I (1993) The relative contribution of hyphae and roots to phosphorus uptake by arbusculoar mycorrhizal plants, measured by dual labeling with P-32 and P-33. New Phytol 124: 489–494 [Google Scholar]
- Peretto P, Bettini V, Favaron F, Alghisi P, Bonfante P (1995) Polygalacturonase activity and location in arbuscular mycorrhizal roots of Allium porrum L. Mycorrhiza 5: 157–163 [Google Scholar]
- Pfeffer PE, Douds DD, Becard G, Shachar-Hill Y (1999) Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol 120: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provorov NA, Borisov AY, Tikhonovich IA (2002) Developmental genetics and evolution of symbiotic structures in nitrogen-fixing nodules and arbuscular mycorrhiza. J Theor Biol 214: 215–232 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M (2001) A phosphate transporter expressed in arbuscule-containing cells of potato. Nature 414: 462–466 [DOI] [PubMed] [Google Scholar]
- Ravnskov S, Wu Y, Graham JH (2003) Arbuscular mycorrhizal fungi differentially affect expression of genes coding for sucrose synthase in maize roots. New Phytol 157: 539–545 [DOI] [PubMed] [Google Scholar]
- Repp A, Mikami K, Mittmann F, Hartmann E (2004) Phosphoinositide-specific phospholipase C is involved in cytokinin and gravity responses in the moss Physcomitrella patens. Plant J 40: 250–259 [DOI] [PubMed] [Google Scholar]
- Requena N, Breuninger M, Franken P, Ocon A (2003) Symbiotic status, phosphate, and sucrose regulate the expression of two plasma membrane H+-ATPase genes from the mycorrhizal fungus Glomus mosseae. Plant Physiol 132: 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewarne GM, Barker SJ, Smith SE, Smith FA, Schachtman DP (1999) A Lycopersicon esculentum phosphate transporter (LePT1) involved in phosphorus uptake from a vesicular-arbuscular mycorrhizal fungus. New Phytol 144: 507–516 [DOI] [PubMed] [Google Scholar]
- Salzer P, Bonanomi A, Beyer K, Vögeli-Lange R, Aeschbacher RA, Lange JAW, Kim D, Cook DR, Boller T (2000) Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection. Mol Plant Microbe Interact 13: 763–777 [DOI] [PubMed] [Google Scholar]
- Sanchez L, Weidmann S, Brechenmacher L, Batoux M, van Tuinen D, Lemanceau P, Gianinazzi S, Gianinazzi-Pearson V (2004) Common gene expression in Medicago truncatula roots in response to Pseudomonas fluorescens colonization, mycorrhiza development and nodulation. New Phytol 161: 855–863 [DOI] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32: 33–57 [DOI] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG (1995) Partitioning of intermediate carbon metabolism in VAM colonized leek. Plant Physiol 108: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Smith FW, Mudge SR, Rae AL, Glassop D (2003. a) Phosphate transport in plants. Plant Soil 248: 71–83 [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis. Academic Press, London
- Smith SE, Smith FA, Jakobsen I (2003. b) Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol 133: 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D, Fester T, Hause B, Schliemann W, Walter MH (2003) Arbuscular mycorrhiza: biological, chemical, and molecular aspects. J Chem Ecol 29: 1955–1979 [DOI] [PubMed] [Google Scholar]
- van Buuren ML, Maldonado-Mendoza IE, Trieu AT, Blaylock LA, Harrison MJ (1999) Novel genes induced during an arbuscular mycorrhizal (AM) symbiosis formed between Medicago truncatula and Glomus versiforme. Mol Plant Microbe Interact 12: 171–181 [DOI] [PubMed] [Google Scholar]
- van Rhijn P, Goldberg RB, Hirsch AM (1998) Lotus corniculatus nodulation specificity is changed by the presence of a soybean lectin gene. Plant Cell 10: 1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch K, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanRhijn P, Fang Y, Galili S, Shaul O, Atzmon N, Wininger S, Eshed Y, Lum M, Li Y, To V, et al (1997) Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc Natl Acad Sci USA 94: 5467–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Schwitzguebel JP, Thewys T, Van Der Lelie D, Vangronsveld J (2004) The use of plants for remediation of metal-contaminated soils. Sci World J 16: 9–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Chiou T-J, Harrison MJ (2002) Phosphate transporters of Medicago truncatula and arbuscular mycorrhizal fungi. Plant Soil 244: 239–245 [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piche Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieweg MF, Frühling M, Quandt H-J, Heim U, Bäumlein H, Pühler A, Küster H, Perlick AM (2004) The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe Interact 17: 62–69 [DOI] [PubMed] [Google Scholar]
- Walter MH, Hans J, Strack D (2002) Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J 31: 243–254 [DOI] [PubMed] [Google Scholar]
- Weidmann S, Sanchez L, Descombin J, Chatagnier O, Gianinazzi S, Gianinazzi-Pearson V (2004) Fungal elicitation of signal transduction-related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula. Mol Plant Microbe Interact 17: 1385–1393 [DOI] [PubMed] [Google Scholar]
- Weidner S, Pühler A, Küster H (2003) Genomics insights into symbiotic nitrogen fixation. Curr Opin Biotechnol 14: 200–205 [DOI] [PubMed] [Google Scholar]
- Winzer T, Bairl A, Linder M, Linder D, Werner D, Müller P (1999) A novel 53-kDa nodulin of the symbiosome membrane of soybean nodules, controlled by Bradyrhizobium japonicum. Mol Plant Microbe Interact 12: 218–226 [DOI] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A (2004) POPP the question: What do LEA proteins do? Trends Plant Sci 9: 13–17 [DOI] [PubMed] [Google Scholar]
- Wright DP, Read DJ, Scholes JD (1998) Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ 21: 881–891 [Google Scholar]
- Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Küster H, Krajinski F (2003) Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact 16: 306–314 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.