Abstract
Symbiotic nitrogen fixation (SNF) in legume root nodules requires differentiation and integration of both plant and bacterial metabolism. Classical approaches of biochemistry, molecular biology, and genetics have revealed many aspects of primary metabolism in legume nodules that underpin SNF. Functional genomics approaches, especially transcriptomics and proteomics, are beginning to provide a more holistic picture of the metabolic potential of nodules in model legumes like Medicago truncatula and Lotus japonicus. To extend these approaches, we have established protocols for nonbiased measurement and analysis of hundreds of metabolites from L. japonicus, using gas chromatography coupled with mass spectrometry. Following creation of mass spectral tag libraries, which represent both known and unknown metabolites, we measured and compared relative metabolite levels in nodules, roots, leaves, and flowers of symbiotic plants. Principal component analysis of the data revealed distinct metabolic phenotypes for the different organs and led to the identification of marker metabolites for each. Metabolites that were enriched in nodules included: octadecanoic acid, asparagine, glutamate, homoserine, cysteine, putrescine, mannitol, threonic acid, gluconic acid, glyceric acid-3-P, and glycerol-3-P. Hierarchical cluster analysis enabled discrimination of 10 groups of metabolites, based on distribution patterns in diverse Lotus organs. The resources and tools described here, together with ongoing efforts in the areas of genome sequencing, and transcriptome and proteome analysis of L. japonicus and Mesorhizobium loti, should lead to a better understanding of nodule metabolism that underpins SNF.
The legume family comprises approximately 700 genera with more than 18,000 species, which occupy niches in almost every environment on earth (Polhill et al., 1981; Doyle and Luckow, 2003). A key to the success of this family was the evolution of mutualistic symbioses with bacteria of the family Rhizobiaceae, which enabled early legumes to utilize atmospheric N2 as a source of nitrogen, especially when colonizing soils lacking mineral or organic nitrogen. Today, symbiotic nitrogen fixation (SNF) by rhizobia in legumes takes place in specialized plant organs called nodules. Nodules develop from cortical cells of the root or stem after contact with rhizobia in the soil (Brewin, 1991). Mature, nitrogen-fixing nodules consist of several layers of uninfected plant cells surrounding a central zone of infected and noninfected plants cells. Infected plant cells typically contain thousands of differentiated, nitrogen-fixing rhizobia, called bacteroids, which are separated from the cytoplasm, either individually or in small groups, by a unique plant membrane called the peribacteroid or symbiosome membrane (SM; Roth et al., 1988; Udvardi and Day, 1997). Microaerobic conditions within legume nodules result from restricted oxygen influx across the outer cell layers of nodules, binding and transport of oxygen by leghemoglobin in the cytoplasm of plant cells, and high rates of respiration by bacteroids and mitochondria in these cells (Appleby, 1984). Low steady-state oxygen concentrations within nodules (in the nanomolar range) have profound effects on plant and bacterial metabolism in nodules. For instance, microaerobiosis is a prerequisite for activity of the oxygen-labile bacteroid enzyme, nitrogenase (Robson and Postgate, 1980).
SNF involves the mutually beneficial exchange of reduced carbon from the plant for reduced nitrogen from the bacteria (Udvardi and Day, 1997), which requires metabolic differentiation of both organisms. Suc, delivered via the phloem, is the primary source of carbon and energy for nodule metabolism (Gordon et al., 1999). However, genetic studies with rhizobial mutants, together with biochemical studies of metabolite transport across the SM and bacteroid membranes, indicate that dicarboxylic acids, especially malate, rather than sugars, are the main source of carbon supplied to bacteroids for SNF (Ronson et al., 1981; Gardiol et al., 1987; Udvardi et al., 1988). An important aspect of plant differentiation during nodule development is the induction of genes and proteins that convert sugars to malate via glycolysis and carbon fixation (Pathirana et al., 1992; Miller et al., 1998; Colebatch et al., 2002, 2004). At about the same time, decreasing oxygen within nodules triggers induction of rhizobial genes for nitrogenase and high-affinity oxidases, which enable bacteroid nitrogen fixation and respiration under these conditions (Batut and Boistard, 1994; Fischer, 1996; Sciotti et al., 2003). Finally, induction of plant genes for ammonium assimilation facilitates rapid incorporation of nitrogen into amino acids and other nitrogen compounds for export to the rest of the plant (Vance et al., 1994; Colebatch et al., 2004). These are some of the principal metabolic changes that occur during nodule development and differentiation, and most studies of nodule metabolism have focused on one or more of these aspects in a variety of different legumes. Few studies have attempted to look more broadly at nodule metabolism in a single, model species. To facilitate such studies, we have developed resources for transcriptome (Colebatch et al., 2002, 2004) and metabolome analyses in the model legume, Lotus japonicus.
In the past, most studies on legume metabolites analyzed a few compounds from preselected classes such as sugars, amino and organic acids, thiols, saponins, and phenolics, using a range of instrumentation, including HPLC (Streeter, 1987; Matamoros et al., 1999; Chen et al., 2003), thin layer chromatography (Khalil and Eladawy, 1994; Steele et al., 1999), and gas chromatography (GC; Streeter and Bosler, 1976; Streeter, 1980; Karoutis et al., 1992). Such studies can best be described as targeted metabolite analysis (Fiehn, 2002), where analysis concentrates on a few, well-defined metabolites, which are often part of well-characterized metabolic pathways. Relatively little attention has been focused on unknown compounds representing potentially novel metabolism. Recent development or refinement of a variety of analytical platforms, including GC-mass spectrometry (GC-MS; Fiehn et al., 2000; Roessner et al., 2000; Wagner et al., 2003) and liquid chromatography-MS (Huhman and Sumner, 2002; Tolstikov and Fiehn, 2002; Chen et al., 2003; Tolstikov et al., 2003), together with software developments, such as GC-MS chromatogram alignment tools (Duran et al., 2003) and deconvolution programs to extricate MS data from overlapping chromatographic peaks (Stein, 1999), have enabled high-throughput, nonbiased analysis of thousands of metabolites from plants and other organisms. These tools afford not only a much broader view of metabolites and metabolism but also the opportunity to discover novel metabolites and previously unknown aspects of metabolism (Fiehn et al., 2000; Sumner et al., 2003). Here, we describe the use of GC-MS to characterize the metabolome of the model legume, L. japonicus. Following the creation of mass spectral tag (MST) libraries, which represent both known and unknown metabolites, we measured and compared relative metabolite levels in nodules, roots, leaves, and flowers of symbiotic plants. Principal component analysis (PCA) and hierarchical cluster analysis (HCA), revealed discrete metabolic phenotypes for the different organs and led to the identification of metabolite markers for each. A large number of novel metabolites, including 2-methylcitrate and many still unidentified metabolites, were uncovered, which alludes to previously unknown aspects of metabolism in nodules and other organs.
RESULTS
GC-MS Chromatograms of L. japonicus Organs and Establishment of MST Libraries
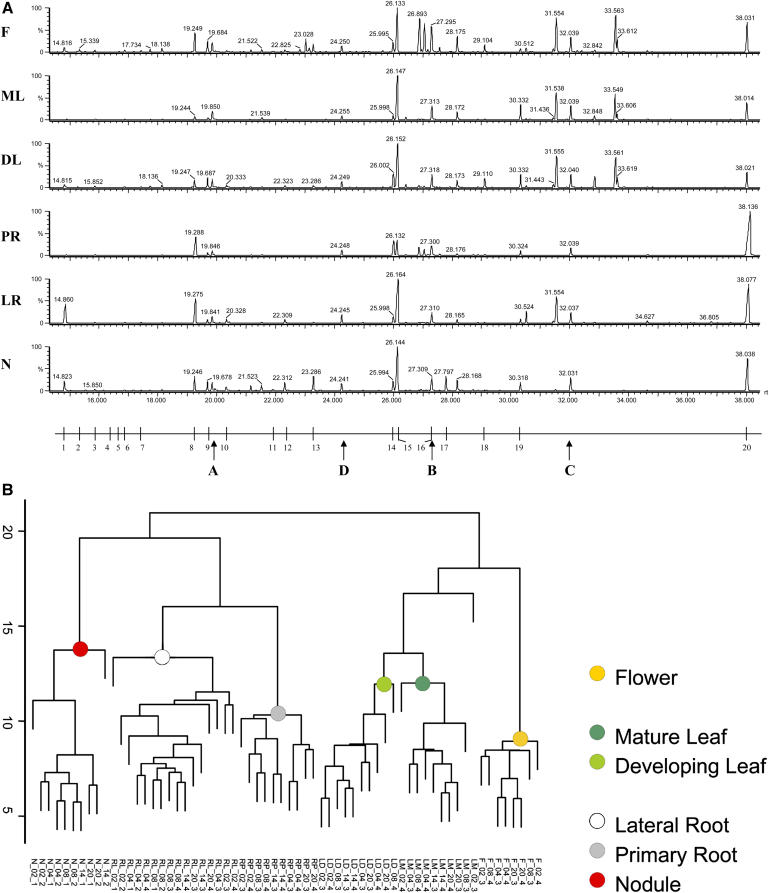
Gas chromatograms of nodules, lateral and primary roots, developing and mature leaves, and flowers from L. japonicus plants, harvested 12 weeks after germination and inoculation with Mesorhizobium loti strain R7A, revealed reproducible and organ-specific features (Fig 1). About 40 major polar metabolite derivatives were detectable by eye from the GC traces, together with a multitude of minor constituents, which are barely or not at all visible on the scale shown in Figure 1.
Figure 1.
Typical GC-MS profiles (A) and hierarchical cluster analysis (B) of polar extracts from different organs of L. japonicus. Profiles were generated from nodules (N), lateral roots (LR), primary roots (PR), developing leaves (DL), mature leaves (ML), and flowers (F). Typical major MSTs represent: (1) phosphate, (2) Pro, (3) succinate, (4) glycerate, (5) fumarate, (6) Ser, (7) threonate, (8) citramalate, (9) malate, (10) Asp, (11) Asn (MST with 4 trimethylsilyl groups), (12) Gln, (13) Asn (MST with 3 trimethylsilyl groups), (14) citrate, (15) pinitol, (16) Fru, (17) ononitol, (18) saccharic acid, (19) myoinositol, and (20) Suc. Arrows indicate internal standard substances, (A) n-pentadecane, (B) n-nonadecane, (C) n-docosane, and (D) ribitol.
GC separates complex mixtures of metabolite derivatives into a series of compounds that enter the mass spectrometer and are subsequently ionized, fragmented, and detected. Each metabolite is, therefore, represented by one or more ionic fragments of precise mass, which together can serve as a tag for that metabolite. We have termed these MST, by analogy to expressed sequence tags of genes. Each MST has properties that facilitate unequivocal identification of the parent metabolite, following comparison to the pure reference compound (Wagner et al., 2003). The properties of an MST are: (1) gas chromatographic retention, which is best characterized by a retention time index (RI), and (2) a specific composition of fragments, which are each characterized by a mass-to-charge ratio (m/z). A library of MSTs was derived from a set of L. japonicus organs using the automated mass spectral deconvolution and identification system, AMDIS (Stein, 1999).
Mass fragments that belong to one MST have the same RI and occur in fixed relative abundance, independent of metabolite concentration. Therefore, any single fragment or set of fragments with identical RI can be used for the quantification of metabolites. As a rule, choice of mass fragments for quantitative purposes must be selective, i.e. only those fragments that are unique to an MST can be used. Mass fragments that are common to coeluting MSTs, i.e. fragments with similar RIs and identical m/z, must be avoided for quantification purposes.
In this work, fragments used for metabolite quantification were identified by m/z, RI, and name of MST to which the fragment belongs. If the MST represents a known or identified metabolite, we add the name of the respective metabolite derivative. We used the following nomenclature: m/z of the selected GC-MS fragments followed by RI and MST name both separated by the underline character; for example, mass fragment 292_2014_glucaric acid (6TMS) or 333_2014_glucaric acid (6TMS; e.g. see Fig. 4). MSTs that remain unidentified were classified tentatively by best matching mass spectra from a custom and a commercial NIST02 library (Institute of Standards and Technology, Gaithersburg, MD). A tentative match required a score >600 on a scale of 0 to 1,000. To address fragments that belong to unidentified MSTs, we use the following nomenclature: match value, and substance name of best fit, separated by a semicolon and set into brackets, e.g. 243_1930_[802; Methylcitric acid (4TMS)] (e.g. Fig. 4).
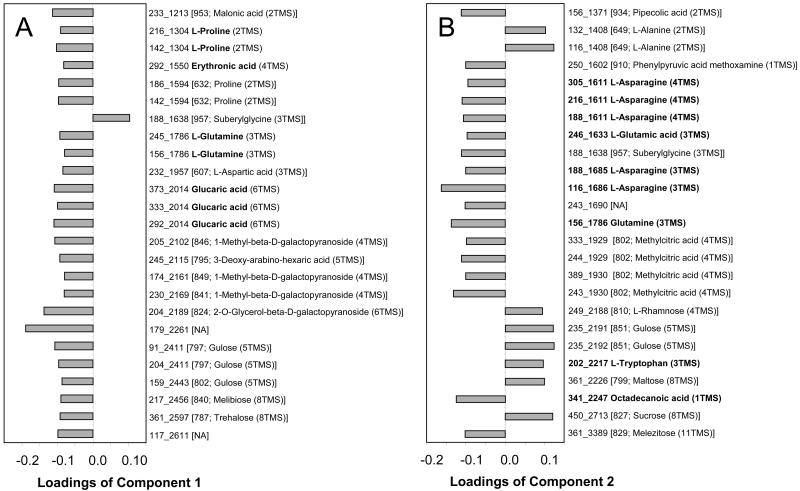
Figure 4.
The 25 most influential fragment masses, which contribute to component 1 (A) and 2 (B) of Figure 2A. The top ranking fragment masses were sorted according to RI, and MSTs as well as metabolite names were manually assigned. MSTs representing metabolites and unidentified MSTs were represented by more than one fragment mass for the purpose of validating metabolite influence. MSTs representing identified metabolites were labeled by fragment mass, RI, and name of metabolite derivative. For example, Asn was found to influence component 2 with 2 MSTs, resulting from different degree of chemical derivatization, namely l-Asn (4TMS) and l-Asn (3TMS). These MSTs were measured by fragment masses 188, 216, 305 at RI = 1,611, and masses 116, 188 at RI = 1,686. Unidentified MSTs (brackets) are characterized by mass spectral match value, name of best match, and degree of silylation (parentheses).
MST-Based Identification of Metabolites in Lotus
Comparison of MSTs derived from Lotus organs with those of pure reference compounds enabled the identification of 87 compounds among the hundreds represented on GC-MS chromatograms (Table I). These included most of the common amino acids as well as polyamines; many organic acids, including TCA cycle intermediates; aromatic acids; sugars and sugar phosphates; and polyols (Table I). A number of likely chemical contaminants, from human or laboratory sources, or reagent impurities, including lactic acid, benzoic acid, and oligomethyl-cyclosiloxanes, were also identified.
Table I.
Identified and unidentified metabolites of L. japonicus organs, GC-MS characteristics (fragment mass, RI), influence in principal components of Figure 2, differential distribution, and metabolite class membership as shown in Figure 6.
Differences in metabolite levels that were significant at P ≤ 0.01 are indicated in bold format.
| m/z | RI, Median | RI, sd | Influence in PCA Component (Metabolite Lists among the Top 25 of Loadings Values) | Response Ratio (Nodule/Plant) | Response Ratio (Nodule/Root) | Response Ratio (Root, Nodule/Shoot) | Response Ratio (Root/Leaf) | Response Ratio (Lateral Root, Primary Root) | Response Ratio (Developing Leaf, Mature Leaf) | Response Ratio (Flower/Other Organs) | Cluster Membershipc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acids | ||||||||||||
| Gly | 248 | 1,313 | 1.4 | 2.6 | 3.3 | 1.1 | 1.4 | 2.8 | 6.5 | 2.3 | 2 | |
| l-Asn | 116 | 1,686 | 3.3 | 2, 3 | 17.6 | 21.4 | 9.1 | 2.3 | 0.6 | 47.7 | 0.5 | 2 |
| l-Homoserine | 218 | 1,455 | 1.9 | 5.9 | 4.9 | 3.5 | 2.1 | 0.2 | 4.4 | 0.7 | 2 | |
| l-Glu | 246 | 1,633 | 2.9 | 2 | 5.2 | 11.8 | 1.4 | 0.4 | 0.2 | 2.5 | 1.3 | 2 |
| l-Cys | 220 | 1,561 | 3.2 | 5.6 | 21.0 | 1.1 | 0.2 | 0.9 | 0.2 | 1.0 | 2 | |
| l-Ala | 116 | 1,095 | 3.1 | 1.5 | 2.7 | 0.6 | 0.5 | 2.0 | 1.7 | 1.8 | 5 | |
| Pyroglutamic acid, l-Gln, l-Glua | 258 | 1,528 | 1.9 | 1.4 | 2.2 | 0.8 | 0.6 | 0.2 | 4.2 | 1.4 | 5 | |
| 2-Aminoadipic acid | 260 | 1,728 | 1.7 | 2.0 | 18.8 | 0.9 | 1.8 | 0.5 | 18.4 | 2.1 | 5 | |
| l-Val | 144 | 1,221 | 1.7 | 0.9 | 1.9 | 0.5 | 0.8 | 1.1 | 0.7 | 5.3 | 6 | |
| l-Tyr | 218 | 1,941 | 2.0 | 0.9 | 4.5 | 0.3 | 0.4 | 1.7 | 1.5 | 9.7 | 6 | |
| l-Met | 176 | 1,523 | 2.5 | 1.0 | 3.0 | 0.5 | 0.9 | 0.3 | 2.9 | 6.7 | 6 | |
| l-Gln | 156 | 1,786 | 3.3 | 1, 2, 3 | 1.4 | 291.0 | 0.3 | 0.1 | 1.0 | 12.7 | 14.6 | 6 |
| l-Phe | 192 | 1,637 | 2.8 | 1.1 | 2.6 | 0.5 | 1.1 | 0.1 | 1.7 | 6.1 | 6 | |
| l-Leu | 232 | 1,279 | 1.8 | 3 | 0.4 | 1.6 | 0.2 | 3.2 | 0.3 | 0.7 | 19.5 | 6 |
| l-Pro | 142 | 1,304 | 1.4 | 1 | 0.1 | 6.2 | 0.0 | 0.1 | 0.5 | 3.7 | 27.7 | 6 |
| l-Trp | 202 | 2,217 | 3.9 | 2 | 0.0 | 12.3 | 0.0 | 0.2 | 0.0 | - | 67.2 | 6 |
| l-Ser | 204 | 1,371 | 1.2 | 0.5 | 3.2 | 0.1 | 0.1 | 0.5 | 1.2 | 5.7 | 6 | |
| l-Thr | 219 | 1,395 | 1.3 | 0.4 | 1.3 | 0.2 | 0.3 | 0.6 | 2.5 | 4.7 | 6 | |
| β-Ala | 174 | 1,432 | 0.8 | 0.3 | 0.3 | 0.8 | 1.9 | 0.7 | 2.7 | 3.0 | 6 | |
| l-Orn, l-Arg, l-Citrullinea | 142 | 1,822 | 2.0 | 3, 4 | 0.4 | 0.6 | 0.4 | 4.4 | 0.0 | 12.8 | 8.3 | 6 |
| l-Lys | 156 | 1,922 | 2.9 | 1.6 | 7.1 | 0.7 | 0.9 | 0.1 | 4.3 | 3.9 | 6 | |
| l-Ile | 158 | 1,302 | 1.9 | 0.7 | 3.3 | 0.2 | 0.1 | 0.4 | 4.3 | 2.7 | 7 | |
| l-Asp | 232 | 1,526 | 1.8 | 4 | 0.9 | 1.0 | 0.9 | 1.4 | 0.1 | 3.0 | 2.1 | 9 |
| 4-Aminobutyric acid | 304 | 1,531 | 1.4 | 0.6 | 0.8 | 0.6 | 0.7 | 0.3 | 1.0 | 1.5 | 9 | |
| Organic Acids | ||||||||||||
| Lactic acidb | 219 | 1,049 | 1.3 | 1.0 | 0.8 | 1.5 | 1.6 | 1.7 | 0.9 | 0.8 | 1 | |
| Octadecanoic acid | 341 | 2,247 | 2.9 | 2 | 52.1 | 110.9 | 14.1 | 0.6 | 0.1 | 1.2 | 0.3 | 2 |
| 2,3,4-Trihydroxybutyric acid (threonic acid) | 292 | 1,570 | 1.2 | 6.7 | 18.4 | 1.6 | 0.4 | 2.1 | 1.5 | 1.5 | 2 | |
| Gluconic acid | 333 | 2,003 | 1.8 | 4.4 | 3.7 | 2.1 | 3.2 | 4.4 | 1.2 | 1.4 | 2 | |
| 2-Ketoglutaric acid | 198 | 1,593 | 1.9 | 1.9 | 3.4 | 0.8 | 0.3 | 2.0 | 1.9 | 0.2 | 3 | |
| DL-2-methylcitric acid | 287 | 1,842 | 0.4 | 2.3 | 4.4 | 0.7 | 0.5 | 0.5 | 0.8 | 2.1 | 5 | |
| Hexadecanoic acid | 313 | 2,052 | 1.1 | 1.4 | 1.6 | 0.9 | 0.9 | 1.0 | 1.0 | 1.5 | 5 | |
| Glucuronic acid | 160 | 1,938 | 1.2 | 0.2 | 1.9 | 0.1 | 0.2 | 0.8 | 1.1 | 13.8 | 6 | |
| Malic acid | 335 | 1,493 | 1.8 | 0.9 | 1.8 | 0.5 | 0.5 | 1.1 | 1.4 | 2.3 | 6 | |
| Glutaric acid | 158 | 1,416 | 1.7 | 1.7 | 4.3 | 0.5 | 0.4 | 1.7 | 2.4 | 3.5 | 6 | |
| Shikimic acid | 204 | 1,822 | 0.9 | 0.8 | 1.8 | 0.4 | 0.3 | 1.0 | 2.3 | 2.1 | 7 | |
| Maleic acid | 245 | 1,313 | 2.3 | 1.1 | 1.5 | 0.8 | 0.6 | 0.0 | 0.4 | 0.9 | 8 | |
| 2,3,4-Trihydroxybutyric acid (erythronic acid) | 292 | 1,550 | 1.3 | 1 | 0.1 | 1.6 | 0.0 | 0.0 | 0.6 | 0.5 | 1.6 | 8 |
| Succinic acid | 147 | 1,318 | 2.0 | 0.2 | 0.4 | 0.2 | 0.3 | 1.7 | 0.4 | 3.1 | 8 | |
| Fumaric acid | 245 | 1,363 | 0.9 | 0.6 | 2.8 | 0.2 | 0.2 | 0.9 | 0.3 | 3.9 | 8 | |
| d-(-)-Quinic acid | 345 | 1,862 | 1.1 | 0.3 | 2.0 | 0.1 | 0.1 | 1.3 | 1.1 | 4.0 | 8 | |
| Gulonic acid | 333 | 1,965 | 1.4 | 0.2 | 1.1 | 0.1 | 0.1 | 1.2 | 0.8 | 3.5 | 8 | |
| cis-Aconitic acid | 229 | 1,763 | 1.7 | 0.5 | 1.1 | 0.3 | 0.4 | 0.9 | 1.2 | 2.2 | 8 | |
| Glucaric acid | 333 | 2,014 | 0.9 | 1 | 0.1 | 1.3 | 0.0 | 0.0 | 0.6 | 1.2 | 2.8 | 8 |
| Citramalic acid | 349 | 1,474 | 0.5 | 0.3 | 0.2 | 1.4 | 2.5 | 1.0 | 0.5 | 1.6 | 9 | |
| Citric acid | 375 | 1,829 | 0.8 | 0.2 | 0.2 | 0.6 | 0.8 | 2.4 | 2.1 | 1.2 | 10 | |
| Isocitric acid | 245 | 1,832 | 2.2 | 0.2 | 0.2 | 0.7 | 1.0 | – | 3.2 | 1.6 | 10 | |
| Galactonic acid | 333 | 1,999 | 1.4 | 0.7 | 1.8 | 0.3 | 0.2 | 0.2 | 0.3 | 1.2 | 11 | |
| Glyceric acid | 292 | 1,341 | 2.0 | 0.1 | 2.0 | 0.1 | 0.0 | 2.7 | 0.9 | 0.6 | 12 | |
| Threonic acid-1,4-lactone | 247 | 1,385 | 1.7 | 0.2 | 2.8 | 0.1 | 0.0 | 0.6 | 0.9 | 0.7 | 12 | |
| Dehydroascorbic acid | 316 | 1,852 | 0.5 | 0.4 | 2.2 | 0.1 | 0.1 | 1.8 | 0.9 | 0.3 | 12 | |
| Aromatic Acids | ||||||||||||
| Benzoic acidb | 179 | 1,253 | 1.9 | 1.7 | 1.6 | 2.4 | 2.8 | 1.9 | – | 0.5 | 1 | |
| Salicylic acid | 267 | 1,514 | 3.1 | 1.0 | 1.7 | 0.6 | 0.4 | 0.9 | 3.1 | 1.1 | 7 | |
| p-Coumaric acid | 293 | 1,948 | 4.7 | 5 | – | – | – | – | – | 0.2 | 2.1 | 8 |
| N-Containing Compounds | ||||||||||||
| Putrescine | 174 | 1,741 | 0.4 | 3.3 | 2.4 | 2.9 | 2.4 | 3.6 | 1.2 | 0.6 | 2 | |
| Urea | 189 | 1,270 | 2.5 | 0.5 | 0.9 | 0.4 | 1.5 | 2.6 | 0.9 | 8.3 | 6 | |
| Allantoin | 331 | 1,888 | 4.2 | 3, 4, 5 | 0.1 | 2.6 | 0.0 | 0.2 | 0.0 | 7.6 | 30.7 | 6 |
| Uric acid | 441 | 2,111 | 0.9 | 0.0 | – | 0.0 | – | – | 1.3 | 69.4 | 6 | |
| Sugars | ||||||||||||
| Rib | 160 | 1,691 | 1.8 | 2.3 | 2.6 | 1.2 | 1.1 | 4.0 | 0.7 | 1.5 | 2 | |
| Raffinose | 217 | 3,402 | 4.4 | 1.8 | 1.7 | 1.4 | 0.9 | 0.3 | 2.4 | 0.1 | 3 | |
| Xyl | 160 | 1,670 | 1.3 | 3 | 0.0 | 0.2 | 0.0 | 0.7 | 2.7 | 0.8 | 97.0 | 6 |
| Ara | 160 | 1,676 | 2.1 | 3 | 0.1 | 0.5 | 0.1 | 0.7 | 1.0 | 0.6 | 40.2 | 6 |
| Gal | 160 | 1,892 | 0.5 | 0.1 | 0.5 | 0.1 | 0.5 | 1.0 | 0.4 | 12.6 | 6 | |
| Fru | 307 | 1,885 | 0.6 | 3 | 0.0 | 0.0 | 0.3 | 2.9 | 8.3 | 0.9 | 11.4 | 6 |
| Glc | 160 | 1,916 | 0.8 | 0.2 | 0.3 | 0.5 | 1.9 | 8.7 | 1.0 | 6.5 | 6 | |
| Trehalose | 191 | 2,751 | 2.2 | 5 | 0.2 | 0.4 | 0.2 | 0.7 | 2.5 | 3.3 | 7.0 | 6 |
| Man | 160 | 1,888 | 0.5 | 0.6 | 0.8 | 0.6 | 0.8 | 0.8 | 1.0 | 2.6 | 6 | |
| Fuc | 117 | 1,747 | 0.4 | 1.3 | 1.4 | 1.0 | 1.7 | 1.3 | 1.0 | 2.6 | 6 | |
| Rha | 160 | 1,728 | 0.3 | 0.5 | 1.4 | 0.3 | 0.3 | 1.3 | 0.5 | 2.7 | 8 | |
| Suc | 451 | 2,653 | 1.0 | 0.6 | 0.6 | 1.0 | 1.3 | 0.7 | 1.1 | 1.5 | 9 | |
| Maltose | 160 | 2,747 | 1.8 | 1.3 | 2.0 | 0.8 | 0.4 | 5.7 | 0.4 | 0.4 | 11 | |
| Polyols | ||||||||||||
| Sorbitol | 319 | 1,937 | 1.2 | 2.1 | 1.8 | 1.8 | 1.6 | 2.9 | 0.8 | 0.8 | 2 | |
| Mannitol | 319 | 1,929 | 1.2 | 3.3 | 2.9 | 2.0 | 1.3 | 7.1 | 0.5 | 0.7 | 2 | |
| Threitol | 217 | 1,503 | 3.9 | 2.5 | 1.0 | 3.0 | 2.9 | – | 0.1 | 0.5 | 2 | |
| Glycerol | 205 | 1,278 | 2.8 | 0.9 | 1.0 | 0.8 | 0.9 | 1.6 | 1.2 | 1.6 | 6 | |
| 4-O-Methyl-myo-inositol, Ononitol | 318 | 1,955 | 0.6 | 1.3 | 5.0 | 0.3 | 0.2 | 1.2 | 0.8 | 3.6 | 6 | |
| Galactitol | 307 | 1,941 | 2.6 | 0.4 | 0.8 | 0.4 | 0.6 | 20.0 | 1.5 | 3.9 | 6 | |
| myo-Inositol | 305 | 2,091 | 0.4 | 0.3 | 1.2 | 0.2 | 0.1 | 0.9 | 0.8 | 1.4 | 8 | |
| Erythritol | 205 | 1,511 | 3.1 | 0.1 | 0.7 | 0.1 | 0.2 | 0.6 | 1.8 | 3.6 | 8 | |
| Galactinol | 191 | 2,995 | 2.2 | 0.4 | 0.5 | 0.5 | 0.7 | 0.5 | 0.9 | 2.2 | 9 | |
| 3-O-Methyl-d-chiro-inositol, d-Pinitol | 231 | 1,835 | 2.2 | 0.4 | 0.8 | 0.3 | 0.3 | 0.3 | 0.7 | 2.2 | 9 | |
| Phosphates | ||||||||||||
| Glyceric acid-3-P | 357 | 1,822 | 1.5 | 7.1 | 19.8 | 2.1 | 0.2 | 0.4 | 7.6 | 0.6 | 2 | |
| Man-6-P | 160 | 2,324 | 2.2 | 1.1 | 5.0 | 0.5 | 0.2 | 0.6 | 2.2 | 2.0 | 5 | |
| Fru-6-P | 315 | 2,324 | 2.4 | 1.1 | 4.6 | 0.6 | 0.2 | 0.4 | 5.4 | 1.5 | 5 | |
| Glc-6-P | 387 | 2,337 | 1.7 | 5 | 1.5 | 5.6 | 0.6 | 0.2 | 0.4 | 5.3 | 1.8 | 5 |
| Glycerol-3-P | 299 | 1,777 | 3.8 | 4 | 3.1 | 31.6 | 1.3 | 0.1 | 0.1 | 108.0 | 1.8 | 5 |
| myo-Inositol-P | 318 | 2,430 | 1.2 | 0.6 | 1.0 | 0.5 | 0.5 | 0.2 | 2.5 | 1.5 | 7 | |
| Phosphoric acid | 314 | 1,282 | 1.0 | 0.6 | 0.6 | 0.9 | 1.4 | 0.3 | 4.9 | 1.9 | 9 | |
| Unidentified | ||||||||||||
| – | 216 | 1,763 | 1.9 | 4 | 2.4 | 1.3 | 4.5 | 5.6 | 0.1 | 3.2 | 0.5 | 1 |
| – | 71 | 1,601 | 3.5 | 3 | 1.8 | 2.2 | 1.3 | 1.2 | 1.4 | 0.8 | 1.6 | 2 |
| [934; Pipecolic acid (2TMS)] | 156 | 1,371 | 1.3 | 2 | 14.5 | 52.4 | 2.8 | 0.4 | 3.0 | 2.9 | 1.4 | 2 |
| [910; Phenylpyruvic acid methoxamine (1TMS)] | 250 | 1,602 | 2.0 | 2, 4 | 40.3 | 16.4 | 46.2 | 4.0 | 32.1 | 1.5 | 0.0 | 2 |
| [824; 2-O-Glycerol-β-d-galactopyranoside (6TMS)] | 263 | 2,190 | 3.1 | 5 | 13.6 | 7.8 | 32.8 | 7.2 | 1.0 | 3.6 | 0.0 | 2 |
| [829; Melezitose (11TMS)] | 361 | 3,389 | 3.0 | 2 | 23.3 | 12.6 | 31.5 | 5.2 | 11.0 | 1.2 | 0.0 | 2 |
| [957; Suberylglycine (3TMS]] | 188 | 1,638 | 4.4 | 1, 2, 3 | 202.5 | 1138.6 | 129.2 | 0.7 | 2.0 | 9.4 | 0.0 | 2 |
| [802; Methylcitric acid (4TMS)] | 243 | 1,930 | 2.0 | 2, 4 | 59.2 | 42.0 | 34.0 | 2.0 | 7.8 | 0.7 | 0.0 | 2 |
| – | 243 | 1,690 | 4.2 | 2 | 38.0 | 75.0 | 7.2 | 1.3 | 0.5 | 0.8 | 0.3 | 2 |
| – | 312 | 1,803 | 3.1 | 4 | 2.8 | 16.3 | 1.2 | 0.1 | 0.6 | 0.1 | 0.8 | 2 |
| [630; DL-2-Methylcitric acid (4TMS)] | 361 | 1,890 | 1.8 | 5 | 2.8 | 2.2 | 2.3 | 1.5 | 0.1 | 0.3 | 0.6 | 2 |
| – | 281 | 1,837 | 3.0 | 2.0 | 2.0 | – | – | – | – | – | 4 | |
| [877; Tetracosamethyl-cyclododecasiloxane]b | 279 | 2,758 | 4.4 | 0.7 | 0.2 | 10.3 | 32.3 | – | 0.2 | 0.2 | 4 | |
| [795; 3-Deoxy-arabino-hexaric acid (5TMS)] | 245 | 2,115 | 1.6 | 1 | 2.2 | 6.4 | 0.8 | 0.3 | 0.0 | 0.9 | 2.3 | 5 |
| [816; Hydroquinone-β-d-glucopyranoside (5TMS)] | 254 | 2,607 | 0.9 | 5 | 2.3 | 4.5 | 0.7 | 0.4 | 1.1 | 0.2 | 1.8 | 5 |
| – | 142 | 1,624 | 3.6 | 5 | 1.1 | 15.9 | 0.4 | 1.6 | 2.7 | 2.8 | 7.3 | 6 |
| [632; Pro (2TMS)] | 186 | 1,594 | 2.6 | 1 | 0.0 | 0.8 | 0.0 | 0.1 | 0.8 | 3.9 | 49.7 | 6 |
| [910; 4-O-d-Glc-β-d-glucopyranoside (8TMS)] | 169 | 3,068 | 7.1 | 5 | – | – | 0.1 | 0.5 | 0.3 | 0.2 | 21.3 | 6 |
| [607; l-Asp (3TMS)] | 232 | 1,957 | 1.8 | 1, 4 | 0.3 | 1.0 | 0.2 | 0.3 | 0.0 | 10.4 | 6.7 | 6 |
| [799; Maltose (8TMS)] | 361 | 2,226 | 2.6 | 2 | 0.0 | – | 0.0 | – | – | 2.0 | 6.4 | 6 |
| [846; 1-Methyl-β-d-galactopyranoside (4TMS)] | 205 | 2,102 | 4.9 | 1, 4 | 0.3 | 0.7 | 0.3 | 0.5 | 0.0 | 1.8 | 4.2 | 6 |
| [674; Gln (4TMS)] | 301 | 1,597 | 3.8 | 4 | 0.9 | 16.4 | 0.5 | – | 0.0 | – | 2.2 | 6 |
| [817; Glc-6-P methoxyamine (6TMS)] | 299 | 2,569 | 3.7 | 5 | 0.3 | 2.3 | 0.2 | 0.1 | 0.4 | 3.8 | 0.9 | 7 |
| [787; Trehalose (8TMS)] | 361 | 2,597 | 3.1 | 1, 5 | 0.1 | 5.0 | 0.0 | 0.0 | 1.0 | 3.1 | 1.1 | 7 |
| [866; Gulose (5TMS)] | 364 | 2,169 | 2.3 | 4 | 0.1 | 0.4 | 0.1 | 0.1 | 0.2 | 2.6 | 1.1 | 7 |
| [746; Gulose (5TMS)] | 204 | 2,431 | 4.4 | 4 | - | - | 0.1 | 0.1 | 0.2 | 0.7 | 1.6 | 8 |
| [810; l-Rha (4TMS)] | 249 | 2,188 | 2.4 | 2 | 0.0 | 1.6 | 0.0 | 0.0 | 0.0 | 0.2 | 2.6 | 8 |
| [802; Gulose (5TMS)] | 159 | 2,443 | 2.5 | 1 | 0.1 | 2.2 | 0.0 | 0.0 | 1.7 | 0.4 | 4.2 | 8 |
| [814; Ribonic acid (5TMS)] | 333 | 1,762 | 0.4 | 0.1 | 0.5 | 0.2 | 0.2 | 1.6 | 0.7 | 3.1 | 8 | |
| [914; Ribonic acid (5TMS)] | 333 | 1,774 | 0.5 | 0.2 | 0.6 | 0.2 | 0.2 | 1.3 | 0.7 | 2.6 | 8 | |
| [849; 1-Methyl-β-d-galactopyranoside (4TMS)] | 174 | 2,161 | 3.1 | 1 | 0.1 | 0.9 | 0.1 | 0.1 | 0.1 | 1.6 | 2.3 | 8 |
| [797; Gulose (5TMS)] | 91 | 2,411 | 2.4 | 1 | 0.1 | 2.4 | 0.0 | 0.0 | 0.8 | 1.5 | 3.7 | 8 |
| [841; 1-Methyl-β-d-galactopyranoside (4TMS)] | 230 | 2,169 | 2.5 | 1 | 0.1 | 0.3 | 0.2 | 0.3 | 0.0 | 1.3 | 2.5 | 8 |
| [649; L-Ala (2TMS)] | 132 | 1,408 | 2.3 | 2, 4 | 0.0 | 0.0 | 0.4 | 0.6 | 0.3 | 1.9 | 2.5 | 9 |
| [953; Malonic acid (2TMS)] | 233 | 1,213 | 4.4 | 1, 3, 4, 5 | 0.0 | 17.2 | 0.0 | 0.0 | 5.9 | 0.3 | 0.4 | 11 |
| [624; Xylulose (4TMS)] | 306 | 1,590 | 1.2 | 5 | 0.2 | 2.6 | 0.1 | 0.0 | 2.4 | 0.1 | 0.9 | 11 |
| [827; Gulose (5TMS)] | 204 | 2,678 | 5.5 | 4 | - | - | 0.0 | 0.0 | 1.3 | 0.0 | 0.2 | 11 |
| [840; Melibiose (8TMS)] | 217 | 2,456 | 1.9 | 1 | 0.0 | 0.7 | 0.0 | 0.0 | 0.4 | 0.1 | 1.6 | 11 |
| [791; 4-O-d-Glc-β-d-galactopyranoside (8TMS)] | 247 | 2,510 | 5.3 | 5 | 0.1 | 0.7 | 0.1 | 0.1 | 2.2 | 0.1 | 0.4 | 11 |
| [716; 4-O-d-Glc-β-d-galactopyranoside (8TMS)] | 361 | 2,950 | 4.0 | 4 | 0.0 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 11 |
| – | 204 | 2,189 | 2.0 | 1, 4 | 0.6 | 94.9 | 0.1 | 0.0 | 0.2 | 0.3 | 1.6 | 11 |
| [851; Gulose (5TMS)] | 235 | 2,191 | 1.2 | 2 | 0.0 | 1.4 | 0.0 | 0.0 | 1.3 | 0.2 | 1.0 | 11 |
| – | 117 | 2,611 | 2.2 | 1 | 0.2 | 2.0 | 0.1 | 0.1 | 0.0 | 0.2 | 1.2 | 11 |
| [690; Raffinose (11TMS)] | 361 | 2,525 | 2.5 | 5 | 0.8 | 4.2 | 0.6 | 0.1 | - | 0.1 | 0.2 | 11 |
| [752; 1-Methyl-6-deoxygalactoopyranoside (3TMS)] | 204 | 2,071 | 2.3 | 5 | 2.0 | 4.7 | 0.7 | 0.3 | 0.2 | 0.1 | 0.5 | 11 |
| [625; 2,2,7,7-Tetramethyl-4,5-diphenyl-3,6-dioxa-2,7-disilaoctane]a | 179 | 2,261 | 2.7 | 1, 3, 5 | 0.0 | 9.9 | 0.0 | 0.0 | 0.8 | 1.2 | 0.7 | 12 |
| [919; Arabino-Hexos-2-ulose-bis(dimethyl-acetal) (4TMS)] | 234 | 1,485 | 2.1 | 3, 5 | 0.0 | 0.8 | 0.0 | 0.0 | 0.6 | 1.5 | 0.1 | 12 |
| [827; Suc (8TMS)] | 450 | 2,713 | 3.5 | 2 | 0.0 | 0.9 | 0.0 | 0.0 | - | 0.5 | 0.3 | 12 |
| [849; 4-O-d-Glucopyranose-β-d-galactopyranoside (8TMS)] | 204 | 2,726 | 4.7 | 3 | 0.1 | 0.5 | 0.1 | 0.1 | 0.1 | 0.6 | 0.3 | 12 |
A small set of MSTs was found to represent more than one metabolite. For example, pyroglutamic acid is formed from Gln, and, to a lesser extent, from Glu during extraction and derivatization of metabolites. However, the classification of Gln, Glu, and pyroglutamic acid into different clusters (see Fig. 6) indicated minimal cross-contamination in this analysis. Arg and citrulline may be converted completely into Orn during chemical derivatization. In our analyses, no specific derivatives of Arg or citrulline were found. Thus, the MST of Orn represented the sum of endogenous Arg, citrulline, and Orn.
Figure 6.
HCA analysis of metabolite and MST distribution in L. japonicus organs. HCA sorts and classifies metabolites according to their distribution pattern. Twelve classes were identified manually (broken lines). Classes 1 and 4 comprised typical laboratory contaminations and allowed to exclude one MST, fragment 281 at RI = 1,837, from further analysis. Class membership and descriptions may be found in Table I. Typical examples of metabolite distributions are presented in Figure 7. Cluster descriptions are as follows.2: N (high), F (low-high), others (low)3: N, DL, ML (high), others (low-high)5: N, F (high), others (low-high)6: F (high), others (low-medium)7: Root (low), DL, F (high), ML (medium)8: Root (low), Shoot (high)9: N (low), LR (low), others (medium-high)10: N (low), others (medium-high)11: ML (high), others (low-medium)12: ML (high), DL (medium-high), F, Root (low-medium)1: Test for solvent contamination4: test for reagent artifact.
Numerical and PCA Analysis of Organ Metabolic Phenotypes
Manual inspection of GC-MS chromatograms indicated major similarities in metabolism of developing and mature leaves, as well as similarities between lateral and primary roots (Fig. 1). To analyze similarities and differences numerically, we performed automated peak integration using 1,046 mass spectral fragments, representative of about 500 MSTs. MSTs representing known or unknown metabolites were analyzed, as a rule, using one to four specific mass spectral fragments within the respective retention time window (see “Materials and Methods” section “Generation of a Metabolite Response Matrix”). Choice of fragment mass and retention time window was performed manually and was facilitated by nonsupervised collection of MSTs (Colebatch et al., 2004). Thus, a large matrix of 1,046 fragment responses, which describe 64 samples from 6 organs of L. japonicus, was generated. PCA (Joliffe, 1986) was applied to gain insight into the nature of the above multivariate data. PCA identifies and ranks major sources of variance within data sets and allows clustering of biological samples into both expected and unexpected groups based on similarities and differences in the measured parameters. PCA also identifies those data elements, e.g. MSTs representing known or unknown metabolites, which contribute most to each of the principal components that describe the variance in metabolite profiling data sets (Roessner et al., 2001a, 2001b). Finally, if sample classes can be clearly distinguished when projected onto any of the principal components, PCA enables identification of those MSTs and metabolites that distinguish the sample classes.
The first 5 principal components derived from the above data matrix encompassed 77.3% of the total variance from this data set (Fig. 2). The first component accounted for 37.1% of the variance and allowed distinction of shoot organs from root organs (Fig. 2A). Nodules exhibited more similarity to roots than to shoot organs according to the first component. However, the second component, which comprised 17.8% of the variance, demonstrated that the data set contained metabolite measurements that distinguished between nodule and root profiles (Fig. 2A). Subsequent principal components revealed other differences between the various organs. Thus, the third and fourth components, encompassing 11.2% and 7.3% of the variance, respectively, indicated that general markers for flowers and primary roots exist (Fig. 2B). The fifth component clearly separated developing leaves from other organs (Fig. 2C). No subsequent components allowed a clear discrimination between sample types (e.g. Fig. 2C, component 6).
Figure 2.
PCA analysis of GC-MS profiles, which represent polar metabolites of L. japonicus organs harvested in the course of 1 d at 12 weeks after germination. Samples were projected into three bi-plots of principal components that were arranged in descending order of variance. Each of the first five principal components allowed clear distinction of metabolite profiles from specific organs. Component 1 separated root from shoot organs, component 2 described the difference of nodules as compared to plant organs, and components 3 to 5 described the distinction of flowers, primary roots, and developing leaves from the remainder of the samples. LR, Lateral roots (n = 15); PR, primary roots (n = 10); F, flowers (n = 8); DL, developing leaves (n = 10); ML, mature leaves (n = 10); and N, nodules (n = 10).
The organ samples described above were harvested at one developmental stage, but at different times of a single day/night cycle. No principal component was found that reflected diurnal changes in metabolism. This finding indicated that diurnal changes resulted in only minor changes in metabolite profiles compared to those resulting from organ development and differentiation. PCA analysis of leaf samples only indicated some diurnal changes in this organ (data not shown). However, the small number of samples from each time point prohibited identification of significant shifts in leaf metabolism during the diurnal cycle.
To test the robustness of PCA in distinguishing between different organs, we analyzed GC-MS data from plants harvested over a 10-week period (7–17 weeks after germination), following growth under different culture conditions in different seasons (Fig. 3A). This data set was expected to be more variable than the first. In fact, this appeared to be the case, and PCA analysis proved less successful in distinguishing between samples of different organs (Fig. 3A). Nonetheless, nodule and root samples were mostly separated from shoot organs by component 1 of PCA, which accounted for 22.9% of the total variance. The 2 subsequent components covered a sum of 14.6% of variance but did not yield distinctions between the samples that could be linked to organ age or plant growth conditions. However, the fourth component, which encompassed 5.5% of the variance, separated nodule samples from those of other organs (Fig. 3A).
Figure 3.
PCA analysis of a second set of GC-MS profiles, which represent polar metabolites of L. japonicus organs that were harvested in the course of a 6-month period at random stages 7 to 17 weeks after germination. PCA analysis of this data set confirmed dominating influence of the root-to-shoot differentiation on the variance observed in GC-MS profiles. In addition, nodule-to-plant differences were detectable in the sample scores plot (A). Loadings analysis (B) confirmed importance of glucaric acid, represented by fragment masses 292, 333, 373 at RI = 2,014 for component 1 and importance of MST [802; Methylcitric acid (4TMS)], represented by fragment mass 243 at RI = 2,014, for component 4. Use of fragments may change from data set to data set, because changes in metabolite levels cause fragments of low abundance to drop below detection limits.
PCA Analysis Reveals Specific Metabolites That Distinguish Different Organs
The contribution of each metabolite to a specific component is reflected by the loading value derived from PCA analysis. Those metabolites with highest loading values are indicated to have the strongest influence on the respective characteristics of a component. We focused on the loading values of components 1 and 2 of experiment 1 (Fig. 2). The 25 most influential fragment masses of each component were analyzed (Fig. 4, A and B). The first component, which described the root to shoot difference, was influenced most by Pro, Gln, erythronic acid, and glucaric acid. The second component, which revealed differences between nodules and other organs, was influenced most by Asn, Gln, Glu, Trp, and octadecanoic acid. Multiple MSTs of unidentified compounds were also found to contribute substantially to components 1 and 2. We selected MST [802; Methylcitric acid (4TMS)] and an identified metabolite, glucaric acid, to demonstrate the possibility of gaining biological insight about a compound, even if its chemical identity is unknown. The choice of these two compounds was made with reference to data from the second experiment described above (Fig. 3). Glucaric acid was found to be important for root and shoot distinction: fragment masses 292, 333, and 373 at RI 2,014 (Fig. 3B). The unknown MST [802; Methylcitric acid (4TMS)] was also found to be a reproducible marker of nodules: fragment 243 at RI 1,929 (Fig. 3B).
Further Analysis of an Unidentified Metabolite
MST [802; Methylcitric acid (4TMS)], as the nomenclature indicates, was found to be highly similar to a typical bacterial metabolite, 2-methylcitric acid. This match was found in the commercially available NIST02 mass spectral library (Ausloos et al., 1999). In contrast, glucaric acid was immediately identified by mass spectral match with a custom set of replicate mass spectra (match value = 944 to 989) and by low RI deviation (ΔRI = 0.3 to 0.5), as described in (Wagner et al., 2003). Typical mass spectra of both compounds contained fragment masses that were used for quantification (Fig. 5A). In an attempt to identify the unknown MST, standard addition experiments were performed with commercially available DL-2-methylcitric acid. This compound generated an MST with mass spectral similarity (match value = 779) but high RI deviation (ΔRI = 89.0). Thus, we confirmed by similarity that MST [802; Methylcitric acid (4TMS)] belongs to the class of methylcitric acids, but we cannot currently define the specific structural position of the methyl group. In addition, we were able to identify true DL-2-methylcitric acid by RI and mass spectral match (700 to 842 and ΔRI = 0.2 to 0.6) with a hitherto unknown MST from the custom MS and RI library of L. japonicus (Colebatch et al., 2004).
Figure 5.
Mass spectra (A) and distribution (B) of MST [802; Methylcitric acid (4TMS)] (black circles), DL-2-methylcitric acid (white circles), and glucaric acid (black circles at bottom). Metabolites were identified by mass spectral match and chromatographic retention. Glucaric acid had match values 944 to 989 (ΔRI = 0.3 to 0.5). MST [802; Methylcitric acid (4TMS)] was similar to DL-2-methylcitric acid but had high RI deviation, match value 779 (ΔRI = 85). The match of DL-2-methyl citric acid was 700 to 842 (ΔRI = 0.2 to 0.6).
Despite the lack of a specific structure for MST [802; Methylcitric acid (4TMS)], we investigated the distribution pattern of the underlying metabolite in Lotus organs (Fig. 5B). MST [802; Methylcitric acid (4TMS)] was found at high levels in nodule samples, while all other organs contained only traces of this compound. In contrast, DL-2-methylcitric acid was relatively high in nodules and flowers but low in lateral roots (Fig. 5B). These results indicate MST [802; Methylcitric acid (4TMS)] is a good marker substance for nodules, while DL-2-methylcitric acid is more evenly distributed throughout the plant. Furthermore, we found glucaric acid to be low in roots and nodules but high in leaves and flowers. Thus, this compound was confirmed as a good marker for shoot organs.
Analysis of Metabolite Distribution Patterns
As illustrated in Figure 5B, metabolites were found to exhibit specific distribution patterns in the organs of Lotus. We applied HCA to the MST distribution of all 87 identified and 49 unidentified compounds that were among the top 25 most discriminatory from PCA analysis (Figs. 1 and 4). Only one mass fragment was used for each MST in this analysis. Following HCA, we grouped the MSTs into 12 classes (Fig. 6). Two of the classes, 1 and 4, were occupied by known laboratory contaminants and were excluded from further analysis. The properties of the remaining 10 classes were investigated further (Fig. 7).
Figure 7.
Typical distribution patterns of representative metabolites from HCA classes. Metabolites were range normalized to minimum 0 and maximum 1 for comparative purpose; error bars represent sds. Class membership is indicated in top right corner. 246_1633_l-Glu, 116_1686_l-Asn, 156_1786_l-Gln, 142_11304_l-Pro, 174_1741_putrescine (Put), 319_1929_mannitol (Mannit), 318_1955_ononitol (Ono), 144_1221_l-Val, 188_1638_[957; SuberylGly (3TMS)] ([957; Sub]), 387_2337_Glc-6-P, 260_1728_2-aminoadipic acid (2-AaA), 202_2217_l-Trp, 217_3402_raffinose (Raff), 198_1593_2-ketoglutaric acid (2-KG), 204_1822_shikimic acid (ShA), 345_1862_D-(-)-quinic acid (QuA), 231_1835_d-pinitol (Pin), 375_1829_citric acid (CitA), 160_2747_maltose (Malt), and 247_1385_threonic acid-1,4-lactone (Thr-L). The nomenclature of fragment mass, RI, and MST names is defined in paragraph “Analysis and Nomenclature of MST.”
Class 2 contained metabolites that were present at relatively high levels in nodules and at low or intermediate levels in other organs. MST [802; Methylcitric acid (4TMS)], like Glu, Asn, putrescine, and mannitol, were characteristic members of this class. Class 3 compounds, with relatively high levels in nodules and leaves, had only 2 members, raffinose and 2-ketoglutaric acid. Class 5, which had high levels in nodules and flowers, comprised DL-2-methylcitric acid and 10 other compounds, including Glc-6-P and 2-aminoadipic acid. Class 6 was the dominant metabolite class and comprised metabolites with high levels in flowers only, for example Pro, Val, Trp, ononitol, and Gln. Classes 7 and 8 were similar and contained metabolites enriched in shoot organs, such as glucaric acid. However, class 8 contained metabolites that had high levels in all above-ground organs, while class 7 metabolites had reduced levels in mature leaves. Class 9 metabolites exhibited relatively high levels in shoot organs and in primary roots. Class 10 contained metabolites with low levels in nodules only. Class 11 and 12 metabolites exhibited high levels in mature leaves. While metabolites of class 12 were also present at high levels in developing leaves, class 11 metabolites had only low or medium levels in other organs. Detailed information on the class membership of each MST, together with short descriptions of each class, is included in Table I.
ANOVA
PCA analysis pointed to metabolites that may be important for organ differentiation. Metabolite clustering by HCA resulted in a rough overview of general metabolite distribution patterns. As an extension to these analyses, ANOVA was used to assess the statistical significance of differences in the distribution of each metabolite. Seven comparisons of organs and groups of organs were performed. These comparisons were motivated by sample classifications made evident by PCA analysis: (1) comparison of nodule with all other plant samples; (2) comparison of nodule with root samples; (3) comparison of below-ground with above-ground samples; (4) comparison of root samples with shoot, including flower samples; (5) comparison of flower samples with all other samples; (6) comparison of lateral and primary roots; and (7) comparison of developing and mature leaves. Differences in metabolite levels were calculated as ratios and compiled in Table I. Differences in metabolite levels that were significant at P ≤ 0.01 are indicated in the table.
Amino acids exhibited two major sites of accumulation: nodules and flowers. Asn, homoSer, Glu, and Cys levels were significantly higher in nodules than in other organs (Table I). In contrast, most other amino acids, especially Trp, Pro, Leu, Val and Gln, were enriched in flowers. Most amino acids were present at higher levels in leaves than in roots. Developing leaves contained higher concentrations of most amino acids than mature leaves, although most differences were not significant.
Only 4 identified organic acids accumulated significantly in nodules compared to all other organs: octadecanoic acid, threonic acid, gluconic acid, and 2-methylcitric acid. Like amino acids, most organic acids were present at significantly lower levels in roots than in leaves. Some organic acids, for example GlcUA and quinic acid, were highly enriched in flowers. Massive accumulation of the N-containing compounds, uric acid, allantoin, and urea was also found in flowers. In contrast, putrescine levels were higher in nodules and roots than in other organs (Table I).
Sugars exhibited variable organ distribution patterns. Most striking was the accumulation of Xyl, Ara, and Gal in flowers. Nodules exhibited significant accumulation of Rib and raffinose compared to other organs. Whereas most sugars were present at similar levels in roots and leaves, Glc and Fru were significantly higher in roots and especially in lateral roots. Sugar alcohols and sugar phosphates were distributed more evenly throughout the plant but exhibited a tendency to be low in roots. Only mannitol, glyceric acid-3-P, and glycerol-3-P exhibited significant accumulation in nodules. Ononitol, pinitol, and galactitol were relatively high in flowers. Developing leaves accumulated a range of phosphorylated compounds, especially phosphoric acid and Glc-6-P.
Most of the MSTs of unidentified compounds, which were selected from the top-ranking loading values of PCA components 1 to 5, exhibited significant differences that substantiated their importance as markers for nodules, roots, or shoot organs. Among these were MSTs that showed the most extreme changes between organs, such as MSTs [957; Suberylglycine (3TMS)] and [802; Methylcitric acid (4TMS)] (Figs. 5 and 7).
DISCUSSION
Current methods of metabolome analysis are far from comprehensive. We selected the GC-MS based method reported earlier (Fiehn et al., 2000; Roessner et al., 2000), which allows analysis of the low to medium Mr, soluble, polar metabolic complement and comprises primary metabolites, such as 24 major and minor amino acids; 29 hydroxylated, nonhydroxylated and aromatic organic acids; 4 amines and amides; 13 mono-, di-, and tri-saccharides; 10 polyols; and 7 phosphorylated compounds (Table I). The choice of this specific GC-MS approach was motivated by the almost complete coverage of those metabolite classes that have received attention in past studies of SNF, namely amino acids, carbohydrates, and organic acids. This enabled validation of some of the GC-MS data by comparison with published data.
Although not as comprehensive as transcript profiles derived from whole-genome oligonucleotide arrays, metabolite profiles allow similar insights at the metabolic level. Biological samples can be classified according to their metabolic phenotype, e.g. the quantitative and qualitative make-up of the metabolome (Figs. 2 and 3); and metabolites, like gene transcripts, can be classified according to their distribution within the various samples under investigation (Figs. 6 and 7). Thus, sets of metabolites can be identified that are not only of diagnostic value but also may indicate the function of metabolites in certain organs or under certain conditions. Below, we discuss three aspects of metabolite profiling: (1) analytical and technical aspects; (2) the data mining strategy applied in this study; and (3) the potential for new insight into a biological process such as SNF that is afforded by comparative metabolomics.
Analytical and Technical Issues
The metabolome of an organism is subject to rapid, enzyme-catalyzed change in response to environmental as well as endogenous factors. Photosynthetic organisms like plants, for instance, undergo profound diurnal changes in metabolism that are related to the light-dark cycle. It is important to take this into account by standardizing growth conditions and synchronizing harvesting times. Equally important is the fact that enzymatic and nonenzymatic conversion of a metabolite to one or more products does not necessarily cease at the time of harvest. Precautions must be taken to avoid such conversions during harvesting, storage, extraction, and derivatization of metabolites (Kopka et al., 2004). For instance, we rapidly harvested plant material in liquid nitrogen, stored at −80 C, and extracted in a mixture of solvents that avoids as much as possible metabolite degradation or modification. To judge the effects of uncontrolled growth conditions and harvest times on metabolite profiles, we compared our well-controlled experiment (Fig. 2) with a less controlled one (Fig. 3). Variance due to experimental intervention is generally much greater than the purely analytical variation, which in the case of metabolite profiling typically ranges from 10% to 25% relative sd, but can be as low as 1% to 5% (Fiehn et al., 2000; Roessner-Tunali et al., 2003).
Data Mining and Analysis
Metabolite profiling data from GC-MS are highly complex, which presents challenges for identification and quantification of metabolites. Unfortunately, bioinformatics tools for automated processing of data are less well-developed for metabolomics than for proteomics and transcriptomics. Currently, the full nonredundant inventory of metabolites from one set of GC-MS profiles cannot be assessed without time consuming manual curation. For this reason, we produced so-called nonsupervised collections of redundant mass spectra from automatically generated mass spectral deconvolution of representative chromatograms (Wagner et al., 2003) for cross-referencing of results and MST identity from Lotus (Colebatch et al., 2004). Known metabolites can be identified by comparing RI and mass spectra of pure standard compounds with mass spectra found in preparations of plant samples. Unidentified MSTs can be retrieved from these libraries for further analysis if there are indications that they may be important. We briefly discuss our current approach to extract useful information from metabolic data matrices below.
A preliminary overview of general similarities and differences between samples is a useful first step in data analysis. Visual inspection of chromatograms is insufficient for this purpose (Fig. 1A). HCA and PCA have been widely applied for data reduction to avoid the need to check each single metabolite for relevant changes (e.g. Roessner et al., 2001a, 2001b). HCA sorts and classifies according to the degree of similarity between metabolite profiles. HCA analysis of our profiles confirmed that each organ of L. japonicus had a characteristic metabolic phenotype (Fig. 1B). PCA of the same data set confirmed HCA results but proved to be superior to HCA in allowing determination not only of differences between sample classes, but also of ranking these differences according to the portion of comprised variance (Fig. 2). Thus, we were able to demonstrate that root-to-shoot differences were responsible for a major part of the variance in the combined data set, followed by differences between nodules and other organs, etc. In the same way, we were able to conclude that intraorgan diurnal changes were relatively small compared to interorgan differences. Nonetheless, diurnal changes were evident in a focused analysis of leaf samples only, which was not presented here because the low number of replications per time point prohibited statistical rigor. Another advantage of PCA over HCA is the possibility to derive a list of metabolites that contribute to each principal component. If principal components separate different sample groups, e.g. components 1 and 2 that separate nodules from all other organs (Fig. 2), a rank-ordered list of MSTs representing known and unknown compounds that distinguish between the groups can be obtained (Fig. 4). In this way, we identified 136 MSTs from among the initial 500 MSTs, representing 87 known and 49 unknown compounds, which distinguished between sample groups. Obviously, such a list also provides insight into the metabolic differences between distinct sample groups such as organs. Thus, by identifying the most striking features of a profiling data set, PCA is an efficient first step of the data mining process.
From the set of 136 MSTs that were identified by PCA to have potential diagnostic properties, we selected 2 MSTs representing a known (glucaric acid) and an unknown compound [802; Methylcitric acid (4TMS)] for further analysis (Fig. 5A). Validation of the diagnostic value of these metabolites was performed via PCA analysis of a second experiment, in which plants were grown under more variable conditions. Once again, both compounds were among the most influential metabolites separating root from shoot, and root from nodule samples, respectively (Fig. 3B). A subsequent analysis of the distribution pattern (Fig. 5) clearly supported the diagnostic properties of the selected compounds. After having established compound relevance we manually confirmed compound identity for glucaric acid and found mass spectral similarity of MST [802; Methylcitric acid (4TMS)] to methylcitric acid. Subsequent standard addition experiments with commercially available DL-2-methylcitric acid confirmed this similarity. As a by-product of this work, we discovered true DL-2-methylcitric acid among the MSTs of unidentified compounds and were able to add this novel identification and its distribution pattern to the set of fully characterized MSTs (Fig. 5B). In the absence of additional commercially available candidate reference substances, further attempts to identify MST [802; Methylcitric acid (4TMS)] will require time-consuming chemical purification of the compound and structural characterization, by NMR analysis, for example.
HCA analysis was applied to all 87 MSTs representing known metabolites and to the 49 representing unidentified compounds, which were found to be interesting from PCA analysis (Fig. 6). HCA demonstrated that the distribution patterns of glucaric acid and MST, [802; Methylcitric acid (4TMS)] were not unique but that both metabolites belonged to groups of metabolites with similar distribution, namely class 2 comprising 24 nodule-enriched metabolites, which were enriched in this organ by between 2- and 200-fold, and class 8 comprising 20 shoot enriched metabolites, which were depleted in roots and nodules by between 2- and 50-fold (Table I). Moreover, HCA supported distribution patterns that were indicated by PCA; e.g. 13 flower-enriched metabolites of class 6 accumulated by more than 10-fold (Table I; Fig. 7). Although HCA allowed rough classification, ANOVA was required to distinguish between those metabolites that were significantly enriched and those that were not. Table I comprises all relevant comparisons that were performed and demonstrates a multitude of significant metabolite enrichments with factors >10-fold or <0.1-fold.
Nodule Metabolism: Some Insights from GC-MS Analysis
PCA analysis revealed that many compounds were enriched in nodules compared to other plant organs, including Asn, Glu, Gln, homoSer, Cys, putrescine, mannitol, threonic acid, gluconic acid, glyceric acid-3-P, glycerol-3-P, and octadecanoic acid. Some of these differences were expected and confirm what is known about nodule metabolism. For instance, SNF is a source of ammonium for amino acid biosynthesis and many legumes, including Lotus export fixed nitrogen in the form of amines, especially Asn and Gln (Vance et al., 1987). Therefore, it was reassuring to find these amino acids at higher levels in nodules than in roots or in the plant as a whole (Table I). In a similar vein, it is known that glycolysis is enhanced in nodules compared to roots (Copeland et al., 1989; Day and Copeland, 1991), and this was reflected by the ratio of hexoses to hexose-phosphates in these organs. The relative abundance of both Fru-6-P and Glc-6-P were about 5 times higher in nodules than in roots, while Fru and Glc were much lower in nodules than in roots. These changes are indicative of increased flux through glycolysis (Roessner et al., 2001; Fernie et al., 2002), even though metabolite levels per se are not a direct measure of flux.
A number of compatible solutes, which typically accumulate in plants in response to osmotic stress, were found to be elevated in nodules compared to roots and other organs. These included the polyols, ononitol, mannitol, and sorbitol; the amino acid, Pro; and the polyamine, putrescine (Table I). Accumulation of osmoprotectants in nodules may indicate that cells in this organ are subject to osmotic stress. Hypoxia, which can cause osmotic stress in plant cells via effects on water uptake and loss (Nuccio et al., 1999), could be responsible for this build-up of compatible solutes. Interestingly, genes encoding putative mannitol transporters are among those induced during nodule development (Fedorova et al., 2002; Colebatch et al., 2004), and these may be involved in importing polyols derived from photosynthesis in the shoot (Noiraud et al., 2001). On the other hand, a proteomic study of the Lotus identified a putative mannitol transporter on isolated peribacteroid membrane/SM (Wienkoop and Saalbach, 2003), which indicates that polyols may be transported between the plant and bacteroids. Sorbitol dehydrogenase, which interconverts d-Fru and d-sorbitol, is induced in nodules (Colebatch et al., 2004), indicating that de novo synthesis of polyols may also occur in this organ. Genes involved in Pro and polyamine biosynthesis are also induced during nodule development, which could account for accumulation of these compounds (Colebatch et al., 2004; Flemetakis et al., 2004). While this data may indicate that osmotic stress is a normal aspect of nodule physiology, a more trivial explanation would be that our sand-grown plants were generally water-stressed at the time of harvest. However, this explanation is at odds with the observation that roots contained significantly lower levels of specific compatible solutes than did nodules of the same plants.
Relatively high levels of Cys were found in nodules (Table I), which is unusual for plant tissues. Two genes encoding Cys synthases were found to be expressed at higher levels in nodules than in roots of Lotus (Colebatch et al., 2004), which could contribute to elevated Cys levels. It is also noteworthy that several genes for sulfate transporters, which presumably deliver substrate for sulfur metabolism, are highly induced during Lotus nodule development (Colebatch et al., 2002, 2004).
While it is not possible to gauge from our GC-MS data the separate contribution that bacteroids make to most metabolite pools, some of the unusual and unidentified compounds that accumulate in nodules, e.g. [802; Methylcitric acid (4TMS)], may be exclusively bacterial products (Table I). Elucidation of the structures of these compounds and their biosynthetic origin will undoubtedly lead to a better understanding of nodule metabolism and the metabolic interactions between legumes and rhizobia. Another important area for future work is metabolic flux determination in nodules. The data presented here give a static picture of metabolite levels averaged over whole organs and provide little insight into metabolic compartmentation or flux though specific pathways. Nonetheless, the resources developed during this project, e.g. MST libraries, will provide a firm basis upon which to build such studies in the future.
MATERIAL AND METHODS
Biological Material, Plant Growth, and Harvesting
Lotus japonicus cv. GIFU seeds were scarified 3 × 10 s in liquid nitrogen, sterilized 10 min in 2% bleach solution, rinsed 5 times with sterile distillated water, and moved to a petri dish with filter paper soaked in B&D medium (Broughton and Dilworth, 1971). After germination in a phytotron set to 25°C and a 16-/8-h day/night cycle, 3-d-old seedlings were transferred to pots, 5 plants each pot, containing coarse quartz sand. Inoculation was performed with Mesorhizobium loti strain R7A. Inoculated plants were grown in a greenhouse with a 16-/8-h day/night cycle, 60% relative humidity, a 21°C/17°C day/night temperature regime, and 1 watering/d with B&D medium.
Two sets of experiments were performed. The first set comprised plant material harvested 12 weeks after germination in the course of 1 diurnal cycle, at 2, 8, and 14 h within the light cycle and at 2, 4, and 6 h during the dark period, respectively. While the diurnal changes were not a topic of this investigation, an equal representation of all diurnal stages was generated for a nonbiased organ-to-organ comparison. A second set of experiments was performed in the course of 6 months, early summer to winter. Samples were taken randomly in the middle of the light cycle at different developmental stages, 7 to 17 weeks after germination. Plants were cultivated either in an open pot or a closed glass jar. This experimental set was expected to be highly variable but allowed to verify persistent metabolic features of L. japonicus organs. At each harvest, plants were carefully pulled from the quartz sand and a complete set of six organ samples prepared by immediate shock freezing in liquid nitrogen, i.e. nodules, lateral and primary root, mature and developing leaves, and flowers. Leaves were separated according to morphological criteria into a group of young expanding leaves from the apex of the plant and a group of mature fully expanded leaves from the middle of the plant shoot. Senescent leaves were discarded. Whole flowers were prepared including all floral organs, petals, sepals, carpels, stamen, and pollen. Lateral roots without visible nodule primordia were collected, followed by pink nodules sampled in a representative range of various sizes. The harvest was completed by preparing the primary root, i.e. 2 cm of the main root directly below the hypocotyl. Only samples without nodules and lateral roots were collected. Primary root material had to be sliced before shock freezing to improve subsequent grinding under liquid nitrogen. Samples were stored for a maximum of 4 weeks at −80°C until GC-MS analysis.
GC-MS Metabolite Profiling of Polar Metabolites
Frozen samples of 25 to 50 mg fresh weight were ground for 2 min in 2-mL micro vials with a clean stainless steel metal ball (5-mm diameter) using a ball mill grinder (MM200, Retsch, Haan, Germany) set to 30 cycles/s. All material was thoroughly precooled in liquid nitrogen. Frozen powder was extracted with hot MeOH/CHCl3 and the fraction of polar metabolites prepared by liquid partitioning into water as described earlier (Wagner et al., 2003; Colebatch et al., 2004). Samples were analyzed by GC-MS using a quadrupole type GC-MS system (MD800, ThermoQuest, Manchester, UK). Ribitol, isoascorbic acid, and deuterated Ala were added for internal standardization. Metabolite samples were derivatized by methoxyamination using a 20- mg/mL solution of methoxyamine hydrochloride in pyridine and subsequent trimethylsilylation with n-methyl-n-(trimethylsilyl)-trifluoroacetamide (Fiehn et al., 2000; Roessner et al., 2000). A C12, C15, C19, C22, C28, C32, C36 n-alkane mixture was used for the determination of RIs. Details of GC-MS analysis were published previously (Colebatch et al., 2004).
MST Definition and Concept
MSTs are defined as full mass spectra obtained from GC-MS chromatograms. MSTs are described by chromatographic retention, for example RI, and mass spectrum, i.e. a set of fragments that are characterized by m/z and relative fragment intensity and normalized to the most abundant fragment of the MS. MSTs represent chemical derivatives of metabolites or metabolites that are not derivatized. MSTs of unidentified compounds can be identified in later experiments by exploiting the above characteristics in standard addition experiments of pure reference substances to the complex biological matrix of interest.
Each mass fragment that belongs to one MST can be used for quantification, and we name these fragments through combination of a single m/z and RI from the MST: for example, fragment 279_2758 below. The best fragment for quantification is generally the most abundant one. However, since metabolite profiles comprise hundreds of MSTs of identified and unidentified compounds, mass fragments need to be highly selective. Because metabolite profiles may contain unexpected, novel MSTs, we analyze multiple fragments per MSTs. If all fragments of one MST exhibit the same relative change, we automatically select the most abundant fragment for quantification. If fragment ratios exhibit discrepancies, we manually overrule the automatic choice and select the next best specific fragment for quantification.
MST Identifications and Test for Artifacts
Reference substances for standard addition experiments were from Sigma-Aldrich (Schnelldorf, Germany) except for DL-2-methylcitric acid, which was obtained from C/D/N Isotopes (Pointe-Claire, Quebec, Canada). Lactic acid and benzoic acid were laboratory contaminations as was oligomethyl-cyclosiloxane, monitored by mass fragment 279_2758. This compound was a chemical artifact caused by the n-methyl-n-(trimethylsilyl)-trifluoroacetamide reagent. Mass spectra were analyzed by AMDIS software (http://chemdata.nist.gov/mass-spc/amdis/; National Institute of Standards and Technology) and compared with commercial and user libraries in NIST02 format (http://chemdata.nist.gov/mass-spc/Srch_v1.7/index.html; National Institute of Standards and Technology). L. japonicus MSTs are made available via the Internet at the CSB.DB resource (http://csbdb.mpimp-golm.mpg.de/gmd.html).
Generation of a Metabolite Response Matrix
We manually selected one or more specific mass fragments and corresponding retention time windows for identified and still unidentified MSTs from L. japonicus. The find algorithm of the MassLab 1.4v software (ThermoQuest, Manchester, UK) was used to automatically retrieve peak areas and chromatographic retention from GC-MS metabolite profiles. Peak identification and area integration was manually supervised as described above. Peak areas with low intensity were rejected.
In accordance with Colebatch et al. (2004) peak areas, X, were defined to represent the fragment responses (Xi of fragment i). Fragment responses were normalized by fresh weight of the sample and response of the internal standard, ribitol, (Ni = Xi × Xribitol−1 × fresh weight−1). This procedure corrects pipette errors and slight differences in sample amount. The relative response of a fragment is defined relative to the average normalized response of all tissue samples (Ri = Ni × avgN−1), namely the average response of flower, nodule, developing leaf, mature leaf, primary root, and lateral root samples.
Statistical Analysis
PCA was performed after log10 transformation of the relative responses, log10(Ri). Missing values were either manually replaced, in the case of identified MSTs, or defined as average of the respective sample group after log10 transformation. If no response was retrievable for any of the samples of a specific organ, log10(Ri) = 0 was substituted for PCA analysis. HCA was applied to classify MSTs, which represented identified metabolites and a selection of unidentified metabolites, according to their relative abundance in different organs. For this purpose, average normalized responses (avgNi) were calculated of each MST and organ. Missing data were substituted by the normalized response at detection limit. HCA was performed after range normalization using Euclidian distance and average linkage. All procedures including ANOVA and visualization were performed with EXCEL software and the S-Plus 2000 software package standard edition release 3 (Insightful, Berlin Germany), and multivariate and cluster analysis was essentially as reported earlier (Colebatch et al., 2004).
Acknowledgments
The authors thank Nicole Gatzke, Cornelia Wagner, and Katrin Bieberich for their patient assistance and technical expertise. The authors greatly appreciate Dr. Andreas Richter (Institute of Ecology and Conservation Biology, Vienna, Austria) for making pinitol and ononitol available for GC-MS standard addition experiments.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.054957.
References
- Appleby CA (1984) Leghemoglobin and rhizobium respiration. Annu Rev Plant Physiol Plant Mol Biol 35: 443–478 [Google Scholar]
- Ausloos P, Clifton CL, Lias SG, Mikaya AI, Stein SE, Tchekhovskoi DV, Sparkman OD, Zaikin V, Zhu D (1999) The critical evaluation of a comprehensive mass spectral library. J Am Soc Mass Spectrom 10: 287–299 [DOI] [PubMed] [Google Scholar]
- Batut J, Boistard P (1994) Oxygen control in rhizobium. Antonie Leeuwenhoek J Microbiol Serol 66: 129–150 [DOI] [PubMed] [Google Scholar]
- Brewin NJ (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth M (1971) Control of leghemoglobin synthesis in snakes beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Duran AL, Blount JW, Sumner LW, Dixon RA (2003) Profiling phenolic metabolites in transgenic alfalfa modified in lignin biosynthesis. Phytochemistry 64: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell T, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15: 411–420 [DOI] [PubMed] [Google Scholar]
- Copeland L, Vella J, Hong ZQ (1989) Enzymes of carbohydrate-metabolism in soybean nodules. Phytochemistry 28: 57–61 [Google Scholar]
- Day DA, Copeland L (1991) Carbon metabolism and compartmentation in nitrogen-fixing legume nodules. Plant Physiol Biochem 29: 185–201 [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131: 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran AL, Yang J, Wang LJ, Sumner LW (2003) Metabolomics spectral formatting, alignment and conversion tools (MSFACTs). Bioinformatics 19: 2283–2293 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Tiessen A, Stitt M, Willmitzer L, Geigenberger P (2002) Altered metabolic fluxes result from shifts in metabolite levels in sucrose phosphorylase-expressing potato tubers. Plant Cell Environ 25: 1219–1232 [Google Scholar]
- Fiehn O (2002) Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol 48: 155–171 [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fischer HM (1996) Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol 4: 317–320 [DOI] [PubMed] [Google Scholar]
- Flemetakis E, Efrose RC, Desbrosses G, Dimou M, Delis C, Aivalakis G, Udvardi MK, Katinakis P (2004) Induction and spatial organization of polyamine biosynthesis during nodule development in Lotus japonicus. Mol Plant Microbe Interact 17: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Gardiol AE, Truchet GL, Dazzo FB (1987) Requirement of succinate dehydrogenase activity for symbiotic bacteroid differentiation of Rhizobium meliloti in alfalfa nodules. Appl Environ Microbiol 53: 1947–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Joliffe IT (1986) Principal Component Analysis. Springer-Verlag, New York
- Karoutis AI, Tyler RT, Slater GP (1992) Analysis of legume oligosaccharides by high-resolution gas-chromatography. J Chromatogr 623: 186–190 [DOI] [PubMed] [Google Scholar]
- Khalil AH, Eladawy TA (1994) Isolation, identification and toxicity of saponin from different legumes. Food Chem 50: 197–201 [Google Scholar]
- Kopka J, Fernie A, Weckwerth W, Gibon Y, Stitt M (2004) Metabolite profiling in plant biology: platforms and destinations. Genome Biology 5: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M (1999) Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol 121: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Driscoll BT, Gregerson RG, Gantt JS, Vance CP (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J 15: 173–184 [DOI] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R (2001) Transport of polyols in higher plants. Plant Physiol Biochem 39: 717–728 [Google Scholar]
- Nuccio ML, Rhodes D, McNeil SD, Hanson AD (1999) Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol 2: 128–134 [DOI] [PubMed] [Google Scholar]
- Pathirana SM, Vance CP, Miller SS, Gantt JS (1992) Alfalfa root nodule phosphoenolpyruvate carboxylase: characterization of the cDNA and expression in effective and plant-controlled ineffective nodules. Plant Mol Biol 20: 437–450 [DOI] [PubMed] [Google Scholar]
- Polhill RM, Raven PH, Stirton CH (1981) Evolution and systematics of the Leguminosae. In RM Polhill, PH Raven, eds, Advances in Legume Systematics Part 1. Royal Botanic Gardens, Kew, UK pp 1–26
- Robson RL, Postgate JR (1980) Oxygen and hydrogen in biological nitrogen-fixation. Annu Rev Microbiol 34: 183–207 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001. a) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR (2001. b) High-resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems. Identification of phenocopies. Plant Physiol 127: 749–764 [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson CW, Lyttleton P, Robertson JG (1981) C4-dicarboxylate transport mutants of rhizobium trifolii form ineffective nodules on trifolium repens. Proc Natl Acad Sci USA 78: 4284–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth E, Jeon K, Stacey G (1988) Homology in endosymbiotic systems: the term ‘symbiosome’. In R Palacios, DPS Verma, eds, Molecular Genetics of Plant-Microbe Interactions. American Phytopathological Society Press, St. Paul, pp 220–225
- Sciotti MA, Chanfon A, Hennecke H, Fischer HM (2003) Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum. J Bacteriol 185: 5639–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele HL, Werner D, Cooper JE (1999) Flavonoids in seed and root exudates of Lotus pedunculatus and their biotransformation by Mesorhizobium loti. Physiol Plant 107: 251–258 [Google Scholar]
- Stein SE (1999) An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J Am Soc Mass Spectrom 10: 770–781 [Google Scholar]
- Streeter JG (1980) Carbohydrates in soybean nodules. II. Distribution of compounds in seedlings during the onset of nitrogen fixation. Plant Physiol 66: 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter JG (1987) Carbohydrate, organic acid and amino acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol 85: 768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter JG, Bosler ME (1976) Carbohydrates in soybean nodules: identification of compounds and possible relationships to nitrogen fixation. Plant Sci Lett 7: 321–329 [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62: 817–836 [DOI] [PubMed] [Google Scholar]
- Tolstikov VV, Fiehn O (2002) Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal Biochem 301: 298–307 [DOI] [PubMed] [Google Scholar]
- Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O (2003) Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal Chem 75: 6737–6740 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membrane of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett 231: 36–40 [Google Scholar]
- Vance CP, Gregerson RG, Robinson DL, Miller SS, Gantt JS (1994) Primary assimilation of nitrogen in alfalfa nodules: molecular-features of the enzymes involved. Plant Sci 101: 51–64 [Google Scholar]
- Vance CP, Reibach PH, Pankhurst CE (1987) Symbiotic properties of Lotus-pedunculatus root-nodules induced by Rhizobium-loti and Bradyrhizobium sp (Lotus). Physiol Plant 69: 435–442 [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887–900 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Saalbach G (2003) Proteome analysis. novel proteins identified at the Peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]