Abstract
Introduction
The application of artificial intelligence to facial aesthetics has been limited by the inability to discern facial zones of interest, as defined by complex facial musculature and underlying structures. Although semantic segmentation models (SSMs) could potentially overcome this limitation, existing facial SSMs distinguish only three to nine facial zones of interest.
Methods
We developed a new supervised SSM, trained on 669 high‐resolution clinical‐grade facial images; a subset of these images was used in an iterative process between facial aesthetics experts and manual annotators that defined and labeled 33 facial zones of interest.
Results
Because some zones overlap, some pixels are included in multiple zones, violating the one‐to‐one relationship between a given pixel and a specific class (zone) required for SSMs. The full facial zone model was therefore used to create three sub‐models, each with completely non‐overlapping zones, generating three outputs for each input image that can be treated as standalone models. For each facial zone, the output demonstrating the best Intersection Over Union (IOU) value was selected as the winning prediction.
Conclusions
The new SSM demonstrates mean IOU values superior to manual annotation and landmark analyses, and it is more robust than landmark methods in handling variances in facial shape and structure.
Keywords: computer vision, face and gesture recognition, image processing and computer vision, pixel classification, segmentation
Abbreviations
- CVAT
Computer Vision Annotation Tool
- HRNetv2
High‐Resolution Network modification 2
- IOU
Intersection over union
- JSON
JavaScript Object Notation
- SSMs
semantic segmentation models
1. INTRODUCTION
The use of artificial intelligence in medicine has greatly increased in recent years. In the field of facial aesthetics, artificial intelligence has been used for diagnosis, prognosis, and preoperative planning, as well as in cosmetology. 1 , 2 , 3 A number of these approaches have demonstrated their value by showing greater accuracy than experienced aesthetic surgeons in many areas, including surgical burn treatment, congenital or acquired facial deformities, and cosmetic surgery. 1
Early machine learning approaches to facial aesthetics were based on landmark analyses, which interpret individual fixed points on the face using pattern recognition models to detect and evaluate facial characteristics. This technique has been very effective in facial recognition, 2 , 3 and in developing arbitrary measures of attractiveness, as well as in some clinical settings. 4 , 5 A limitation of these models in facial aesthetics, however, has been their inability to distinguish discrete facial zones of interest. 6 The face is made up of different structures rather than individual points, with extensive overlap existing between some regions (Figure 1). In fact, any individual pixel may lie within multiple different facial regions, for example, overlap of the upper perioral region with the nasolabial folds and the submalar regions. Furthermore, regions such as the forehead and cheeks are not well represented in landmark analyses. In order to apply artificial intelligence techniques to specific regions of the face (e.g., the infraorbital or glabellar areas), it is necessary to first accurately identify these regions.
FIGURE 1.
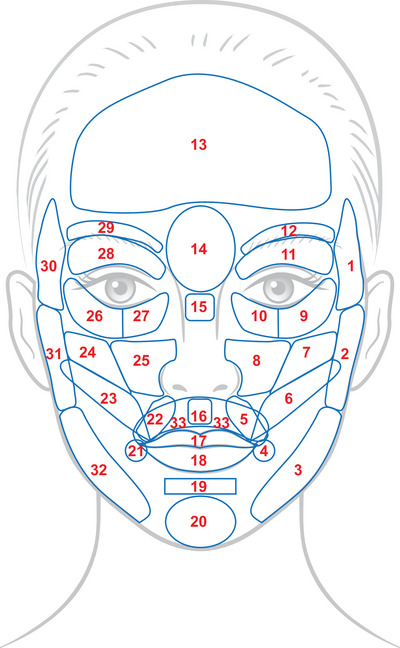
Complete facial zone map with 33 annotated zones of interest, the result of multiple iterations between manual annotation and review/adjustment by facial aesthetics experts. Note the overlapping zones in the lower half of the face, which required the separation of the full model into three sub‐models with no overlap between zones.
Semantic segmentation models (SSMs) have been developed to overcome the limitations of landmark analysis and identify discrete regions of the face. 7 However, to date, these models have been only able to differentiate three to nine facial zones. 7 , 8 , 9 , 10 , 11 , 12 SSMs have seen little clinical utility to date, but have demonstrated potential applications in measuring skin surface temperature for identifying individuals with COVID‐19 and in ophthalmology for assessment of eyelid and periorbital soft tissue position. 10 , 11
There are over 30 distinct muscles in the face, all of which help to define unique regions of the face. 13 An SSM, therefore, needs to precisely define and accurately differentiate the numerous regions of the face currently targeted by aesthetic procedures if it is to be of value for accurate clinical outcomes assessment when paired with digital diagnostics. 13 Here we describe a supervised machine learning model designed to identify 33 distinct regions of the face, as defined by a panel of facial aesthetic surgeons. Accurate facial segmentation will in turn allow for development of other artificial intelligence models with clinical utility for directing specific facial aesthetic procedures targeted at individual regions or groups of regions.
2. METHODS
2.1. Facial zone detection
The facial SSM described here was designed to take standardized clinical‐grade facial images as inputs and to output a prediction on 33 facial zones defined by clinicians. The process is composed of several sections: data, domain expertise, annotation, preprocessing, model training, and evaluation.
2.1.1. Data
The model was trained using high‐resolution clinical‐grade images taken from 53 studies, with data selection being agnostic as to any specific treatment or study phase. In other words, the only criteria when selecting studies are those that include photographic capture as part of their study design. All images were captured using the VISIA‐CR imaging system, manufactured by Canfield Scientific (Parsippany‐Troy Hills, NJ, USA), an industry standard for generating high‐quality, reproducible facial imaging for clinical research. In addition, a number of images captured via mobile device as part of a clinical study were used to provide additional data and introduce lower‐quality images to the model. The purpose of including these lower‐quality images was to further reinforce the robustness of the model by exposing it to unideal inputs during training so that it is capable in some capacity to handle a variety of image qualities.
2.1.2. Domain expertise and annotation
In order to maximize the validity of the model, we collaborated with three clinical experts in the field of facial aesthetics. To ensure model accuracy and consistency, a total of 33 distinct zones were identified, which were manually annotated by the experts on a total of 10 clinical‐grade images. These annotations were reviewed and refined until all the experts confirmed their clinical accuracy. Subsequently, nonclinical personnel were trained on identification of the respective facial zones, referencing the clinically accurate annotations confirmed by the clinicians (Figure 1). These raters manually annotated images for input to the training set for the development of the supervised SSM. Annotators drew each area on a given image, provided it was visible within the image. The open‐source Computer Vision Annotation Tool (CVAT; Intel Corporation, Santa Clara, CA, USA) was used to complete the manual annotation. A total of 33 labels, corresponding to the 33 facial segmentation zones, were employed. An example of a complete annotation is shown in Figure 2.
FIGURE 2.
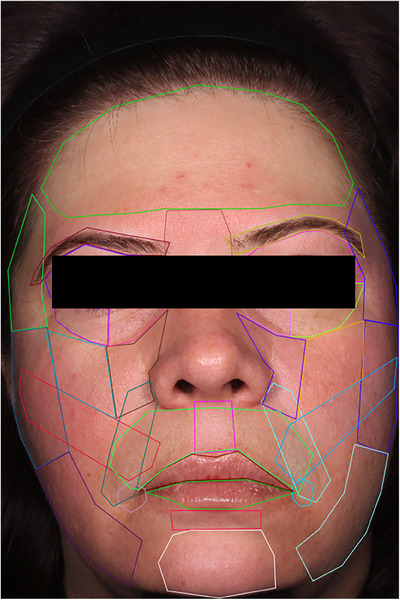
Manual annotation of a high‐resolution facial image based on the standardized facial map. Line colors are arbitrary; they are added to distinguish individual markups.
For measuring the consistency of image annotators, we adopted the Intersection Over Union (IOU) metric to calculate inter‐annotator correlations. This metric also served to set quantitative expectations toward what the model can achieve, as well as to make a comparison to a landmark analysis. Once annotations were complete, a final data quality check was conducted to ensure the accuracy of annotations and labels.
2.1.3. Preprocessing
Images were stored separately from their respective annotation masks. A mask indicated which pixels in an image belong to which object or class. As such, part of our preprocessing included data organization in a similar fashion to the scene‐parsing ADE20K dataset (Figure 3). In addition, ODGT files were created, whereby each line is a JavaScript Object Notation (JSON). Each JSON details the dataset, splitting them into training and validation sets along with their respective annotations. With the requirement that all pixels must be labeled for semantic segmentation, we conformed to this standard by turning the background to its own class. This step brought focus to the areas that needed attention. Our preprocessing pipeline ensured our data were ready for model ingestion.
FIGURE 3.
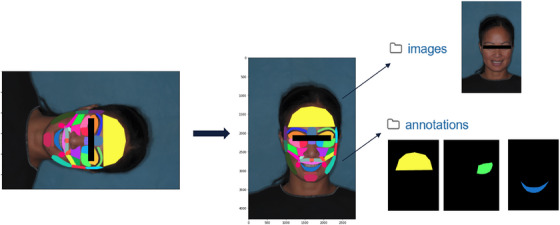
Schematic illustration of the preprocess pipeline, showing (left to right) the annotated high‐resolution image, the same image properly oriented, and the separation of image and annotation mask data.
2.1.4. Model training
Given that the images used were primarily clinical grade, the model was built to manage high‐resolution images, in order to preserve detail and granularity. To achieve this, Integrated High‐Resolution Network modification 2 (HRNetv2), which maintains high‐resolution representations through the entirety of the pipeline (Figure 4), 14 , 15 was included in the model architecture.
FIGURE 4.
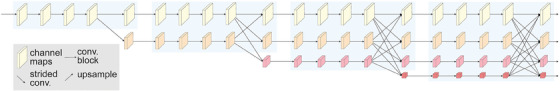
An example of a HRNetv2. HRNetv2 maintains high‐resolution images by connecting high‐to‐low convolution streams in parallel. Stage 1 consists of high‐resolution convolutions. High‐to‐low resolution streams are gradually added individually and connect the multiresolution streams in parallel. Stages 2−4 are formed by repeating modularized 2‐resolution (3‐resolution, 4‐resolution) blocks. Reproduced with permission from Wang et al. 15 HRNetv2, High‐Resolution Network modification 2.
2.1.5. Inference
To avoid overlapping pixels, three separate strategically mapped facial zone sets were created, each of which acted as a standalone model for predicting its assigned zones (Figure 5). For the inference process, an image was fed into each respective model. This means that there was a total of three outputs of each input image. Based on prior IOU analysis, the best zones for each model were chosen to be the winning prediction. For example, given that model 1 and model 2 both have the forehead area in their zone sets, the model that generated the best IOU for final forehead prediction was chosen. This logic ensued for all zones until a full image prediction with 33 zones annotated was achieved.
FIGURE 5.
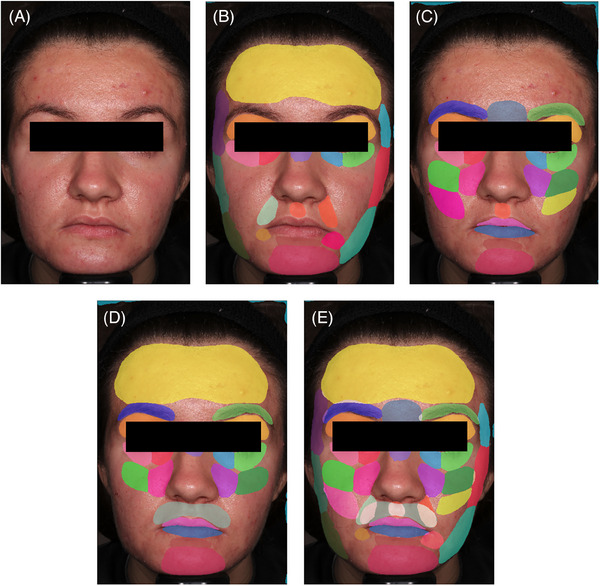
The full inference process, beginning with high‐resolution input (A), continuing through annotation based on the three sub‐models with non‐overlapping zones (B), (C), (D), and re‐aggregation into a final image prediction with all 33 zones annotated (E).
3. RESULTS
The model was trained on a total of 669 high‐resolution clinical‐grade images taken from 53 studies and 59 images captured via mobile phone. Figure 6 describes the available demographic distribution of the image dataset. The dataset was well distributed, but most patients were 40−60 years of age and female, and White race was most common. The training hyperparameters that yielded the best IOU results are detailed in Table 1.
FIGURE 6.
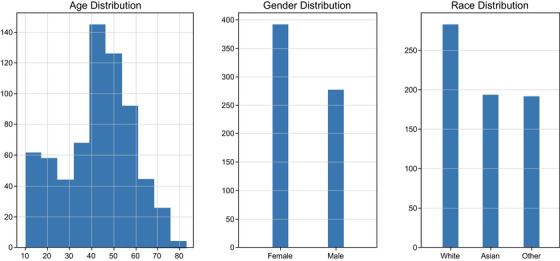
Data breakdown describing demographic distributions separated by age, gender, and race for 53 studies used in model generation.
TABLE 1.
Hyperparameter settings used for model training.
| Hyperparameter | Value |
|---|---|
| Batch size per GPU | 2 |
| Epochs | 30 |
| Iteration of each epoch | 30 |
| Optimizer | SGD |
| Learning rate for encoder | 0.02 |
| Learning rate for decoder | 0.02 |
| Power in poly to drop learning rate | 0.9 |
| Momentum for SGD | 0.9 |
| Weights regularizer | 0.0001 |
| Weighting of deep supervision loss | 0.4 |
Abrreviations: GPU, graphics processing unit; SGD, stochastic gradient descent.
Each image was rated by three annotators. Inter‐annotator mean IOU scores across three separate images ranged from 0.3534 to 0.4018. A comparison of inter‐rater, landmark model, and SSM yielded mean IOU scores of 0.3584, 0.3952, and 0.5538, respectively. The mean IOU for the SSM represents a 55% improvement in accuracy compared with human annotators, and a 40% improvement relative to the landmark model, indicating a superior performance.
The model performance was quite variable across the 33 different zones, with mean IOU scores from the SSM ranging from 0.2119 for LTA (zone 1) to 0.8357 for the forehead (FFA; zone 13). IOU values for all facial segmentation zones are shown in Table 2.
TABLE 2.
Mean Intersection Over Union (IOU) values for 33 facial zones.
| Zone | Abbreviation | IOU Score |
|---|---|---|
| 1 | LTA | 0.2119 |
| 2 | LPA | 0.5792 |
| 3 | LJA | 0.5858 |
| 4 | LOC | 0.5078 |
| 5 | LNF | 0.4314 |
| 6 | LSA | 0.4918 |
| 7 | LZA | 0.5728 |
| 8 | LAA | 0.6229 |
| 9 | LLI | 0.5728 |
| 10 | LMI | 0.6815 |
| 11 | LUL | 0.4768 |
| 12 | LBA | 0.2498 |
| 13 | FFA | 0.8357 |
| 14 | CGA | 0.6536 |
| 15 | NBA | 0.3531 |
| 16 | FPA | 0.6534 |
| 17 | FUL | 0.6596 |
| 18 | FLW | 0.7629 |
| 19 | LMS | 0.5583 |
| 20 | FCS | 0.7111 |
| 21 | ROC | 0.4661 |
| 22 | RNF | 0.2519 |
| 23 | RSA | 0.4917 |
| 24 | RZA | 0.6014 |
| 25 | RAA | 0.6543 |
| 26 | RLI | 0.6081 |
| 27 | RMI | 0.6303 |
| 28 | RUL | 0.5025 |
| 29 | RBA | 0.3718 |
| 30 | RTA | 0.5332 |
| 31 | RPA | 0.5808 |
| 32 | RJA | 0.5827 |
| 33 | UPA | 0.8280 |
4. DISCUSSION
In order to create an artificial intelligence tool that is meaningful and can be properly utilized, we must understand the regions of interest, which means that these regions must have clinical significance. Because there are numerous zones in a relatively small canvas, precisely defining each zone is of utmost importance. We identified a total of 33 distinct zones that effectively defined meaningful facial regions.
The performance of any machine learning model is underpinned by the quality of the imputation data. Because our goal was to develop a robust, clinically applicable model, it was imperative that the training dataset be as balanced and diverse as possible. Given that our dataset included a range of images from subjects of different age, gender, and race, our model should perform as expected on nearly all individuals. Two major characteristics that further define quality data are accuracy and consistency. By collaborating with experts in facial aesthetic medicine, we were able to develop clinically relevant facial zone annotations for inputting to the model. The quality of these data was further improved by using an iterative process whereby the experts reviewed a subset of images following annotation by image raters, adjusting accordingly until they accepted annotations as clinically accurate.
Consistency is also critical for training of the model, requiring accurate annotations during the training phase. This metric is especially important considering that these compartmentalized facial zones are novel and have no clear boundaries. In this context, the IOU metric showed good inter‐annotator correlation, demonstrating good consistency from annotator to annotator. Another key factor determining model accuracy is noise taking the form of artifacts that may obstruct the face and can misguide the model and hinder its performance. This notion is critical when we consider a supervised learning model where the model directly learns from target data. Data preprocessing utilized in our model helps to eliminate these artifacts and directly helps to improve the model's ability to learn. Furthermore, HRNet models are exceptionally apt for visual recognition, including semantic segmentation, and have demonstrated high performances across a range of applications. 16 , 17 , 18 , 19 Taken together, it is evident that the inputs for our model provide a solid foundation for an accurate and useful tool.
An inherent issue of semantic segmentation is overlapping pixels because pixels are meant to have a one‐to‐one relationship with a class, that is, a single pixel can belong to only one class. Whereas it is not uncommon to have 33 separate classes, the face is a relatively small region and sectioning it anatomically can violate this rule. Indeed, many areas overlap with one another, and a pixel may belong to more than three classes. SSMs disallow this behavior because we want to map each pixel to a single class. We were able to overcome this limitation by creating separate facial zone sets, strategically mapped to avoid overlap.
Both the landmark model and SSM had higher IOUs than the inter‐rater IOU, suggesting that automation of facial zone annotation is valid and favorable, at least in the context of annotation by nonclinical raters, as may occur in a clinical trial setting. Both models also outperformed human annotations compared with one another. Additionally, because the model is discrete, it guarantees consistency and will always predict the same outcome on a given input, thus minimizing the latent issue of human error and variability. Between the two machine learning models, the SSM exceeds the performance of a landmark detection approach. With quantitative results to confirm our initial hypothesis, the SSM is more robust to manage facial differences, especially in terms of varying facial structures, while the landmark detection model is rigid in nature due to having to essentially “connect the dots.”
Although our model provides promising results, there are certain standards that are assumed. That is, the input must be of clinical‐grade quality. Whereas some mobile‐capture images are present in the training set, the model is trained primarily on high‐resolution images; therefore, it will perform more poorly on lower‐quality images it has not adequately learned from before. The input should also be cropped to the face in the image and oriented correctly with a yaw, pitch, and roll equal to zero. Another assumption is that the background is labeled as its own class. Due to the nature of semantic segmentation, this is still a class that needs to be predicted. Whereas background prediction is > 97% accurate, this leaves erroneous predictions of some facial zones to be labeled incorrectly as the background. Overall, the IOU scores measured were poor relative to usual standards for both the human annotators and machine learning models, reflecting the complexity of the human face. However, the improvement seen with the SSM is encouraging. The model provided robust results for larger, more prominent regions of the face, including the forehead, with a mean IOU score of 0.8357. However, scores for some smaller, more‐complex regions were poor, with IOU scores for some segmental zones < 0.3.
Whereas the model described here has demonstrated a novel application in the field of facial aesthetics, there are several paths for improvement and additional utility. The model is trained only for front‐facing images. However, there are several treatment areas on the face that are assessed at different angles. Thus, front‐facing images would not be suitable in these cases. Future studies should expand this functionality to other angles, including 45‐degree angle captures. Another improvement could be the possible additions of new zones. Though we have exercised due diligence in creating the 33 facial zones, there are still uncaptured areas of the face that may prove to be important in the future. New discoveries within facial aesthetics are inevitable and may carry their own unique treatment areas.
With the surge of telehealth, a major opportunity lies in integrating edge devices into our model. Whereas our model is robust on clinical‐grade images, we would like to extend this performance to mobile‐capture images as well. There are several differences between VISIA‐CR and mobile‐capture images, namely resolution and a lack of standardization. Because of hardware differences between VISIA‐CR and edge devices, there is a ceiling to the degree that imaging techniques and software in general can achieve. Sensors on current edge devices pale in comparison to those in VISIA‐CR devices, exemplifying the latent issue of hardware limitations, leading to a stark difference in quality. Another key difference that will need to be accounted for is the introduction of human error through mobile self‐capture. Whereas the VISIA‐CR has forehead rests, chin rests, and precisely controlled lighting, mobile users carry the burden of ensuring their environment is controlled and consistent with regard to lighting, distance from their camera, framing, and steady movement. Training a model to manage a variation that comes with mobile‐capture images is an arduous but potentially fruitful next step.
5. CONCLUSION
The development of an ensemble SSM framework for facial zones of interest, as described in this paper, paves the way for clinical‐grade machine analysis of individual faces, and ultimately for the routine use of artificial intelligence–assisted diagnostic, prognostic, and treatment‐related applications across multiple clinical settings. The iterative interaction between key opinion leaders in facial aesthetics and experienced annotators, in the creation and refinement of the facial zone map, also provides a model for developers facing similar image‐analysis challenges for other medical and nonmedical applications. In a broader sense, the ability to create valid SSMs despite the existence of overlapping zones/classes of interest by generating multiple non‐overlapping zone sets, and to select optimal zone boundaries based on comparison of IOU values across sets, opens a wide range of image‐analysis scenarios characterized by overlapping or diffuse zone boundaries to the semantic segmentation approach. Results from the SSM described here demonstrate that automatic labeling of facial zones through semantic segmentation is viable and proven to be a direction we may confidently strive toward. The model can be used as a utility to extract any facial zone of interest and expedite further computer vision work in facial aesthetics.
CONFLICT OF INTEREST STATEMENT
R.T. and S.M. are full‐time employees of AbbVie.
ETHICS APPROVAL
Due to the nature of this publication, no ethics approval was necessary.
ACKNOWLEDGMENTS
The authors thank Damon Caiazza, M.H.I. and Vivian Luong, B.S. for their contributions to the manuscript. Medical writing support was provided by Stephen Collins, PhD and James Street, MD of Peloton Advantage, LLC (an OPEN Health company), and funded by Allergan Aesthetics, an AbbVie Company. Allergan Aesthetics, an AbbVie Company, funded this study and participated in the study design, research, analysis, data collection, interpretation of data, reviewing, and approval of the publication. All authors had access to relevant data and participated in the drafting, review, and approval of this publication. No honoraria or payments were made for authorship.
Tuazon R, Mortezavi S. Automatic labeling of facial zones for digital clinical application: An ensemble of semantic segmentation models. Skin Res Technol. 2024;30:e13625. 10.1111/srt.13625
DATA AVAILABILITY STATEMENT
AbbVie is committed to responsible data sharing regarding the clinical trials we sponsor. This includes access to anonymized, individual, and trial‐level data (analysis data sets), as well as other information (e.g., protocols, clinical study reports, or analysis plans), as long as the trials are not part of an ongoing or planned regulatory submission. This includes requests for clinical trial data for unlicensed products and indications. These clinical trial data can be requested by any qualified researchers who engage in rigorous, independent, scientific research, and will be provided following review and approval of a research proposal, Statistical Analysis Plan (SAP), and execution of a Data Sharing Agreement (DSA). Data requests can be submitted at any time after approval in the US and Europe and after acceptance of this manuscript for publication. The data will be accessible for 12 months, with possible extensions considered. For more information on the process or to submit a request, visit the following link: https://vivli.org/ourmember/abbvie/ then select “Home.”
REFERENCES
- 1. Mantelakis A, Assael Y, Sorooshian P, Khajuria A. Machine learning demonstrates high accuracy for disease diagnosis and prognosis in plastic surgery. Plast Reconstr Surg Glob Open. 2021;9(6):e3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarvis T, Thornburg D, Rebecca AM, Teven CM. Artificial intelligence in plastic surgery: current applications, future directions, and ethical implications. Plast Reconstr Surg Glob Open. 2020;8(10):e3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hennocq Q, Khonsari RH, Benoît V, Rio M, Garcelon N. Computational diagnostic methods on 2D photographs: a review of the literature. J Stomatol Oral Maxillofac Surg. 2021;122(4):e71‐e75. [DOI] [PubMed] [Google Scholar]
- 4. Wei W, Ho ESL, McCay KD, Damaševičius R, Maskeliūnas R, Esposito A. Assessing facial symmetry and attractiveness using augmented reality. Pattern Anal Appl. 2022;25:635–65. [Google Scholar]
- 5. Hong YJ, Nam GP, Choi H, Cho J, Kim I‐J. A novel framework for assessing facial attractiveness based on facial proportions. Symmetry. 2017;9(12):294. [Google Scholar]
- 6. Wan J, Lai Z, Shen L, et al. Robust facial landmark detection by cross‐order cross‐semantic deep network. Neural Netw. 2021;136:233‐243. [DOI] [PubMed] [Google Scholar]
- 7. Huang P, Han J, Zhang D, Xu M. CLRNet: component‐level refinement network for deep face parsing. IEEE Trans Neural Netw Learn Syst. 2023;34(3):1439‐1453. [DOI] [PubMed] [Google Scholar]
- 8. Kim H, Park J, Kim H, Hwang E. Facial landmark extraction scheme based on semantic segmentation. In: International Conference on Platform Technology and Service (PlatCon). IEEE; 2018. [Google Scholar]
- 9. Benini S, Khan K, Leonardi R, Mauro M, Migliorati P. FASSEG: A FAce semantic SEGmentation repository for face image analysis. Data Brief. 2019;24:103881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller D, Ehlen A, Valeske B. Convolutional neural metworks for semantic segmentation as a tool for multiclass face analysis in thermal infrared. J Nondestr Eval. 2021;40(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Brummen A, Owen JP, Spaide T, et al. PeriorbitAI: artificial intelligence automation of eyelid and periorbital measurements. Am J Ophthalmol. 2021;230:285‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan K, Attique M, Syed I, Sarwar G, Irfan MA, Khan RU. A unified framework for head pose, age and gender classification through end‐to‐end face segmentation. Entropy. 2019;21(7):647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu WT, Chang KV, Chang HC, et al. Ultrasound imaging of the facial muscles and relevance with botulinum toxin injections: a pictorial essay and narrative review. Toxins. 2022;14(2):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun K, Zhao Y, Jiang B, et al. High‐resolution representations for labeling pixels and regions. 2019. Accessed November 7, 2022. https://arxiv.org/abs/1904.04514
- 15. Wang J, Sun K, Cheng T, et al. Deep high‐resolution representation learning for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2021;43(10):3349‐3364. [DOI] [PubMed] [Google Scholar]
- 16. Cheng TW, Chua YW, Huang CC, Chang J, Kuo C, Cheng YC. Feature‐enhanced adversarial semi‐supervised semantic segmentation network for pulmonary embolism annotation. Heliyon. 2023;9(5):e16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Zheng JC, Zhao SJ. An improved lightweight high‐resolution network based on multi‐dimensional weighting for human pose estimation. Sci Rep. 2023;13(1):7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bao W, Niu T, Wang N, Yang X. Pose estimation and motion analysis of ski jumpers based on ECA‐HRNet. Sci Rep. 2023;13(1):6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan Y, Zhang X, Meng Y, et al. Sagittal intervertebral rotational motion: a deep learning‐based measurement on flexion‐neutral‐extension cervical lateral radiographs. BMC Musculoskelet Disord. 2022;23(1):967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
AbbVie is committed to responsible data sharing regarding the clinical trials we sponsor. This includes access to anonymized, individual, and trial‐level data (analysis data sets), as well as other information (e.g., protocols, clinical study reports, or analysis plans), as long as the trials are not part of an ongoing or planned regulatory submission. This includes requests for clinical trial data for unlicensed products and indications. These clinical trial data can be requested by any qualified researchers who engage in rigorous, independent, scientific research, and will be provided following review and approval of a research proposal, Statistical Analysis Plan (SAP), and execution of a Data Sharing Agreement (DSA). Data requests can be submitted at any time after approval in the US and Europe and after acceptance of this manuscript for publication. The data will be accessible for 12 months, with possible extensions considered. For more information on the process or to submit a request, visit the following link: https://vivli.org/ourmember/abbvie/ then select “Home.”


