Abstract
Sclerotinia minor Jagger is the causal agent of Sclerotinia blight, a highly destructive disease of peanut (Arachis hypogaea). Based on evidence that oxalic acid is involved in the pathogenicity of many Sclerotinia species, our objectives were to recover transgenic peanut plants expressing an oxalic acid-degrading oxalate oxidase and to evaluate them for increased resistance to S. minor. Transformed plants were regenerated from embryogenic cultures of three Virginia peanut cultivars (Wilson, Perry, and NC-7). A colorimetric enzyme assay was used to screen for oxalate oxidase activity in leaf tissue. Candidate plants with a range of expression levels were chosen for further analysis. Integration of the transgene was confirmed by Southern-blot analysis, and gene expression was demonstrated in transformants by northern-blot analysis. A sensitive fluorescent enzyme assay was used to quantify expression levels for comparison to the colorimetric protocol. A detached leaflet assay tested whether transgene expression could limit lesion size resulting from direct application of oxalic acid. Lesion size was significantly reduced in transgenic plants compared to nontransformed controls (65%–89% reduction at high oxalic acid concentrations). A second bioassay examined lesion size after inoculation of leaflets with S. minor mycelia. Lesion size was reduced by 75% to 97% in transformed plants, providing evidence that oxalate oxidase can confer enhanced resistance to Sclerotinia blight in peanut.
Sclerotinia blight of peanut, caused by the necrotrophic fungus Sclerotinia minor, is one of the most devastating diseases of peanut (Arachis hypogaea) in Virginia, northeastern North Carolina, Oklahoma, and Texas. The fungicide fluazinam provides some protection (Smith et al., 1992), but its benefit to peanut producers is offset by the expense of the multiple applications necessary to achieve modest control. Yield losses due to Sclerotinia blight can be significant. For example, prior to the availability of fluazinam, losses to Sclerotinia blight averaged 10% in Virginia, with pod loss exceeding 50% in severely affected fields (Porter and Melouk, 1997).
Oxalic acid is considered a pathogenicity factor in Sclerotinia sclerotiorum and many other fungal pathogens (Maxwell and Lumsden, 1970; Godoy et al., 1990). A wide variety of other fungi also secrete oxalic acid, including Poria placenta (Ritschkoff et al., 1995), Septoria musiva (Liang et al., 2001), Sclerotium rolfsii (Bateman and Beer, 1965), Endothia (Cryphonectria) parasitica (Havir and Anagnostakis, 1985; Bennett and Hindal, 1989), Penicillium oxalicum (Ikotun, 1984), Cristulariella pyramidalis (Kurian and Stelzig, 1979), Sclerotium cepivorum (Stone and Armentrout, 1985), Mycena citricolor (Rao and Tewari, 1987), and S. minor (Hollowell et al., 2001). Direct application of oxalic acid to stem or leaf tissue causes marked tissue injury and wilting, similar to plant responses to fungal infection by S. rolfsii (Bateman and Beer, 1965) and S. sclerotiorum (Noyes and Hancock, 1981). The most compelling evidence for the involvement of oxalic acid in disease initiation was the demonstration that mutant isolates of S. sclerotiorum, deficient in oxalic acid production, were not pathogenic on bean (Phaseolus vulgaris), but revertants became pathogenic once they regained the ability to produce oxalic acid (Godoy et al., 1990). Screening for resistance to oxalic acid has also been studied as an indirect test of physiological resistance to white mold in common bean (Kolkman and Kelly, 2000).
Oxalic acid may aid in infection through a number of proposed routes, including acidification to facilitate cell wall-degrading enzyme activity, through pH-mediated tissue damage, or via sequestration of Ca2+ ions (for review, see Dutton and Evans, 1996). The optimal pH for many cell wall-degrading enzymes lies in the acidic range (Lumsden, 1979). Degradative synergy between oxalic acid and polygalacturonase has been shown for Sclerotinia species and other fungal pathogens (Bateman and Beer, 1965; Kurian and Stelzig, 1979). Rollins (2003) demonstrated that both oxalic acid production and endopolygalacturonase activity are regulated by pH in S. sclerotiorum. The low pH resulting from oxalic acid production may weaken plants to improve access to fungal infection. Oxalic acid is also a strong chelator of divalent cations, and sequestration of calcium may weaken cell walls (Bateman and Beer, 1965). Oxalic acid can directly suppress the oxidative burst associated with the detection of pathogens by the plant (Cessna et al., 2000). Recently, oxalic acid has been reported to disturb guard cell function during infection by S. sclerotiorum by inducing stomatal opening and inhibiting abscisic acid-induced stomatal closure (Guimarães and Stotz, 2004).
Oxalate oxidase belongs to the germin family of proteins and catalyzes the degradation of oxalic acid to produce carbon dioxide and hydrogen peroxide (H2O2; Chiriboga, 1966; Lane et al., 1993). Oxalate oxidase is expressed during germination, where it associates with cell wall components such as glucuronogalactoarabinoxylans (Lane, 2000). Plant oxalate oxidase activity increases the pH at the site of infection following contact with oxalate-secreting pathogens (Rollins, 2003). Oxalate oxidase has received considerable attention for its possible utility in plant defense. It has been proposed that through the production of H2O2, oxalate oxidase or oxalate oxidase-like proteins may cause cross-linking of plant cell wall proteins in papillae at the site of infection (Wei et al., 1998) or play a role in the plant hypersensitive response (Lane, 1994; Zhou et al., 1998), previously shown to be orchestrated by H2O2 from the oxidative burst (Levine et al., 1994). Wheat (Triticum aestivum) oxalate oxidase activity increased following biotic (e.g. fungal pathogens) and abiotic (e.g. salt or heavy-metal ions) stress (Hurkman et al., 1994; Dessalegne et al., 1997; Berna and Bernier, 1999). Barley (Hordeum vulgare) oxalate oxidase is also induced in leaves following challenge with the fungus Erysiphe graminis (Dumas et al., 1995; Zhang et al., 1995).
Reports of oxalate oxidase activity in response to pathogen attack have been restricted to cereals (Pundir, 1991; Lane et al., 1993); however, transgenic technology has been used to successfully transfer oxalate oxidase genes from cereals to other plants. Transgenic oilseed rape (Brassica napus) expressing a barley oxalate oxidase can break down exogenously applied oxalic acid (Thompson et al., 1995). Soybean (Glycine max), tobacco (Nicotiana tabacum), and sunflower (Helianthus annuus) transformed with a wheat oxalate oxidase gene demonstrated increased resistance to S. sclerotiorum (Zaghmout et al., 1997; Donaldson et al., 2001; Hu et al., 2003). Poplar (Populus spp.) expressing the same gene was resistant to S. musiva (Liang et al., 2001). Although not examined in the soybean or poplar studies, it was demonstrated in transformed sunflower that oxalate oxidase expression resulted in the induction of plant defense proteins (Hu et al., 2003). There are also reports of increased resistance to insect predation in corn (Zea mays) expressing a wheat oxalate oxidase (Ramputh et al., 2002). Expression of another oxalate-degrading enzyme, oxalate decarboxylase, isolated from Collybia velutipes, also resulted in resistance of transgenic tomato (Lycopersicon esculentum) and tobacco to S. sclerotiorum (Kesarwani et al., 2000).
Further evidence for the utility of H2O2-generating enzymes in protective plant responses was provided by an examination of Glc oxidase expression in transgenic rice (Kachroo et al., 2003). Expression of Glc oxidase and release of H2O2 led to enhanced resistance to the bacterial pathogen Xanthomonas oryzae and the fungus Magnaporthe grisea. Multiple strategies for engineering oilseed crops for resistance to S. sclerotiorum, including introduction of H2O2-producing enzymes, have recently been reviewed (Lu, 2003). Together, these reports suggest that introduction of an oxalate oxidase to degrade oxalic acid and generate H2O2 can be used in peanut to enhance resistance to oxalic acid-producing fungi, including S. minor.
RESULTS
Assay of Transformants for Oxalate Oxidase Activity
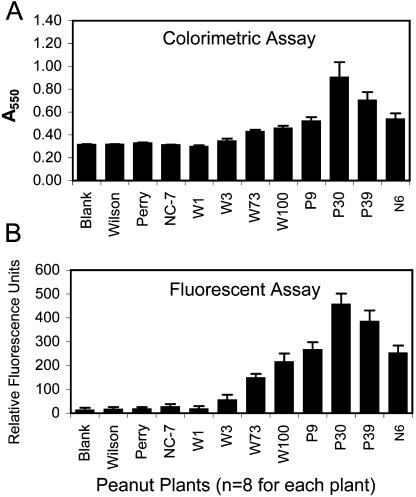
Following establishment of embryogenic tissue culture lines for several Virginia peanut cultivars, embryos were transformed by particle bombardment with a plasmid vector containing the barley oxalate oxidase coding sequence. Selection and regeneration resulted in greater than 200 viable plants from three different cultivars. A simple colorimetric assay was used to screen hygromycin-resistant transformants for oxalate oxidase activity, and 40% to 60% of regenerated T0 plants showed elevated activity levels, depending on cultivar (data not shown). Eight transformed plants were chosen for a closer examination of enzyme activity (Fig. 1). Oxalate oxidase activity levels were higher in seven of the transformants than in all three nontransformed controls (Wilson, Perry, and NC-7). One additional regenerated plant, W1, was included as a nonexpressing control transformant.
Figure 1.
Comparison of two oxalate oxidase activity assays using transgenic and control peanut plants. Transgenic plants were regenerated from embryogenic peanut cultures bombarded with an oxalate oxidase gene under the control of the cauliflower mosaic virus 35S promoter and selected in the presence of hygromycin. Transformants were identified by screening for transgene expression using a leaf disc assay for oxalate oxidase activity. Quantitative measure of enzyme activity was determined for eight transformants and three untransformed controls using two different detection methods. A, The colorimetric detection protocol used 100 μL of the enzyme reaction, and results were measured spectrophotometrically at A550. B, The Amplex Red detection kit used 20 μL of the enzyme reaction, and results were measured with a fluorescence plate reader with an excitation filter of 530 nm and emission filter of 590 nm. (The same reaction was assayed by both methods for each of the eight leaf discs per plant.) Untransformed controls were Perry, Wilson, and NC-7. P9, P30, and P39 were regenerated from bombardment of Perry embryogenic cultures. W1, W3, W73, and W100 were regenerated from bombardment of Wilson embryogenic cultures. N6 is derived from NC-7 embryos, although some caution in making this assignment was described in “Materials and Methods.” “Blank” indicates a reaction performed without plant material (buffer alone). Means and se are presented for eight replicates for each plant.
Figure 1 shows a comparison of results from two different protocols for detection of oxalate oxidase activity. Both the colorimetric assay and the commercially available fluorescent kit relied on the detection of H2O2 produced by the activity of oxalate oxidase on the substrate, oxalic acid. The difference between the two protocols was the greater sensitivity and lower background of the fluorescent assay, which required 5-fold less reaction volume for a comparable response. Both methods are easy to perform in a microtiter format, which allowed for high throughput analysis of transformed material. The assay results were highly repeatable with comparable levels of expression in different leaves of the same plant. One line, W3, gave variable oxalate oxidase activity results at different sampling dates, with some leaflets showing moderate activity and some with only background levels, indicating possible chimerism (data not shown).
Southern and Northern Analysis
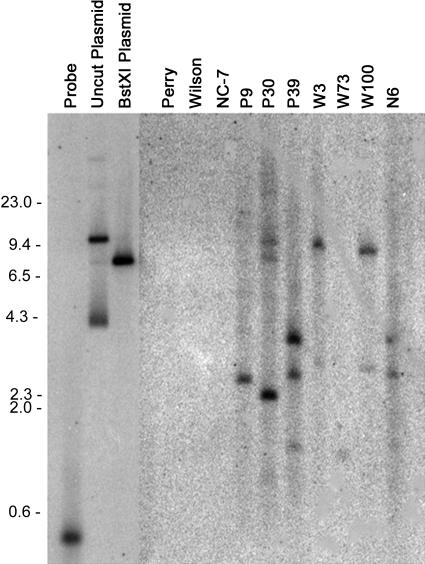
Genomic DNA samples from transformed and control peanut plants were analyzed by Southern-blot hybridization to demonstrate the integration of the oxalate oxidase transgene into the peanut genome. The number of insertion sites varied among transformants, as illustrated in Figure 2. The variable intensity of bands also suggested the possible insertion of multiple copies of the coding sequence in several lines, as is frequently observed for microprojectile bombardment. Hybridization patterns differed for the three Perry transformants, P9, P30, and P39, indicating that they were all independent transformation events. Hybridization patterns for two of the Wilson transformants (W3 and W100) were similar, raising the possibility that they may have originated from the same bombardment event. However, the difference in oxalate oxidase expression and other parameters tested below suggested that W3 and W100 originated from different events and that the patterns appeared similar due to the inability to resolve small size differences for large fragments on the gel. A similar consideration applies to transformant N6 obtained from bombardment of NC-7 cultures. The similarity of its hybridization pattern to P39 called into question whether it could have been inadvertently mislabeled during transfer to the greenhouse. We have retained the N6 designator while recognizing this caveat and will confirm its origin if progeny from this plant are used for any future studies.
Figure 2.
Southern-blot analysis of peanut genomic DNA with the oxalate oxidase probe. BstXI-digested DNA was probed with a 32P-labeled fragment amplified by PCR from the barley oxalate oxidase cDNA. Size markers were HindIII-digested lambda DNA (sizes to the left of the blot are in base pairs). The first three lanes show hybridization of the probe to the PCR product used as the probe (470 bp), to undigested pOxOx plasmid, and to BstXI-digested pOxOx plasmid. Untransformed controls are cultivars Perry, Wilson, and NC-7. P9, P30, and P39 were Perry transformants. W3, W73, and W100 were Wilson transformants, and N6 is believed to be a NC-7 transformant.
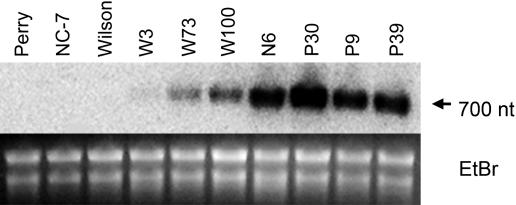
Oxalate oxidase RNA expression was examined by northern-blot analysis to confirm the enzyme activity results. RNA from young leaflets was hybridized with the oxalate oxidase probe (Fig. 3). The seven oxalate oxidase-positive transformants showed the presence of a single 700-nucleotide RNA band that was absent from nontransformed controls. RNA expression levels were variable among lines. Line W3 showed very low levels of oxalate oxidase RNA, while P30 had the highest level of expression, consistent with enzyme activity results.
Figure 3.
Northern-blot analysis of peanut leaflet RNA with the oxalate oxidase probe. A, Total RNA from transformants and nontransformed controls was probed with the same 470-bp probe as described for the Southern blot in Figure 2. Expression in transgenic peanuts is observed by the presence of a 700-nucleotide mRNA band. B, Ethidium bromide (EtBr) staining was performed to demonstrate sample loading.
Resistance to Oxalic Acid
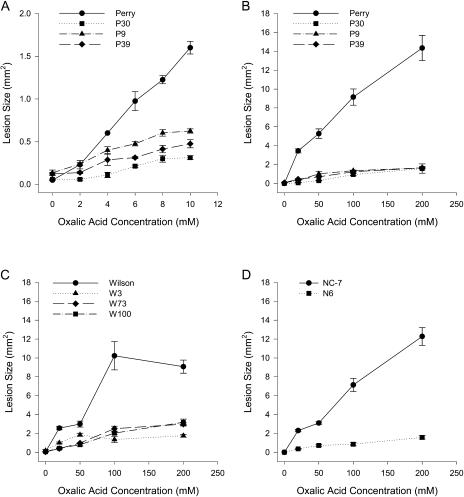
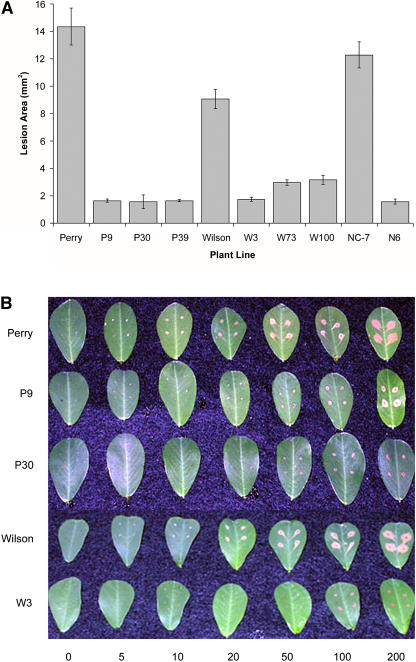
To determine whether transgenic peanut plants expressing oxalate oxidase showed increased resistance to the damaging effects of oxalic acid, varying concentrations of oxalic acid were applied to the surface of detached leaflets. Resistance was measured as a reduction in lesion size on transgenic plant leaflets compared to lesion size on nontransformed controls. Transformed peanut plants showed decreased damage from exogenously applied oxalic acid over a wide range of oxalic acid concentrations, as illustrated in Figures 4 and 5. The assays showed that expression of oxalate oxidase in transgenic peanut was able to neutralize the effects of oxalic acid at pathophysiological concentrations (Fig. 4A) and at concentrations up to 20-fold higher than normally found in infected tissue (Fig. 4, B–D). At high (200 mm) oxalic acid concentrations, lesions were reduced by 65% to 89% compared to the corresponding nontransformed controls (Fig. 5A). Figure 5B shows the appearance of leaflets from several transformants following application of oxalic acid, illustrating the reduced lesion size. Liang et al. (2001) found that lesion area resulting from exposure to 200 mm oxalic acid on leaf discs from oxalate oxidase-expressing poplar transformants was reduced by 18% compared to nontransformed controls. The apparent difference in oxalic acid resistance in our transgenic peanut lines compared to the transgenic poplar may have resulted from differences in parameters such as plant species, expression levels, and/or assay conditions.
Figure 4.
Resistance of transgenic plants to application of oxalic acid. Different concentrations of oxalic acid were applied to detached leaflets from transformants and nontransformed controls. Oxalic acid in A was applied at 0, 2, 4, 6, 8, and 10 mm. Oxalic acid in B to D was applied at 0, 20, 50, 100, and 200 mm. A, Nontransformed Perry and three transformants (P9, P30, and P39) at low oxalic acid concentrations. B, Same samples as in A except for high range of oxalic acid concentrations. C, Nontransformed Wilson and three transformants (W3, W73, and W100). D, Nontransformed NC-7 and transformed N6. Means and se are shown for four replicates.
Figure 5.
Resistance to oxalic acid in control and transformed peanut. A, Bar graph showing lesion size (in mm2) for transformed lines and nontransformed controls following application of 200 mm oxalic acid to peanut lines. B, Photograph of selected peanut lines showing lesion size and appearance on detached leaflets after application of increasing amounts of oxalic acid. Means and se are shown for four replicates.
Resistance to S. minor
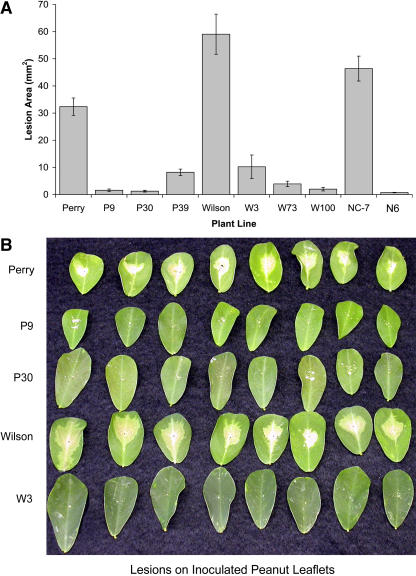
To test for enhanced resistance to S. minor, thin agar plugs from the actively growing edge of a fungal culture were used to inoculate isolated leaflets from both transformed and nontransformed peanut plants. All the transgenic lines showed increased resistance to S. minor as measured by a reduction in lesion area (Fig. 6A). The type of lesion also varied between transgenic and control lines. Controls showed a large, continuous, tan-colored lesion, whereas the transgenic lines typically exhibited numerous smaller lesions resembling a hypersensitive response (Fig. 6B). The average reduction compared to the respective nontransformed control cultivars ranged from 75% for P39 to 97% for W100. The resistance results in peanut correlated well with previously published reports in other plants. Lesion size resulting from inoculation with the oxalate-secreting pathogen S. musiva was reduced by 63% in poplar expressing a wheat oxalate oxidase gene (Liang et al., 2001). Using oxalate decarboxylase to degrade oxalic acid, Kesarwani et al. (2000) showed decreased lesion size of approximately 89% in transgenic tomato inoculated with S. sclerotiorum.
Figure 6.
Resistance to S. minor in oxalate oxidase-expressing peanut lines compared to controls. A, Lesion size (in mm2) on detached leaflets in response to inoculation with S. minor. B, Comparison of selected peanut lines in response to inoculation with S. minor showing lesion size and appearance on detached leaflets. Means and se are shown for 16 replicates.
Fertility of Transgenic Lines
Primary transgenic plants often show reduced fertility and lower yields than nontransformed plants. This can be due to epigenetic effects related to long tissue culture regimens or to the direct effect of the transgene itself, either because of position effects or the toxic nature of some transgene products. For example, Kachroo et al. (2003) showed that ectopic, wound-inducible expression of a Glc oxidase gene in transgenic rice resulted in lesion mimicry. Our T0 peanut transformants did not grow as well as nonregenerated control plants and exhibited variable fertility. T1 progeny plants showed improved growth, and no lesion mimicry has been observed. Line P30 had the highest average enzyme activity, but, although it flowered and produced several pods, all seeds were aborted at an early stage. Lines P9, P39, W73, and N6 produced seed yields similar to the nonexpressing transformant from line W1 (21–54 seeds). Line W100 produced only five viable seeds. Line W3 showed the lowest levels of transgene expression and most variable enzyme activity but also failed to produce any viable seed. From a comparison of RNA expression levels and seed recovery, we concluded that the lack of seed production in several transformants could not be attributed to the expression of oxalate oxidase alone.
DISCUSSION
These studies demonstrated that the expression of a barley oxalate oxidase in transgenic peanut increased resistance to exogenously applied oxalic acid over a wide range of concentrations (up to 20 times pathophysiological levels). Transgenic peanut plants also exhibited enhanced resistance to S. minor as measured by reduced lesion size in detached leaflet assays. Peanut leaf inoculations were previously shown to serve as a reliable indicator of host resistance to S. minor (Hollowell and Shew, 2003).
In addition to meeting the goal of enhancing resistance to S. minor, the project results also demonstrated the potential utility of oxalic acid as a screen for fungal disease resistance. Previous studies used wilting during immersion of cut bean seedlings in 20 mm oxalic acid as a screen for white mold resistance (Kolkman and Kelly, 2000). Our studies quantified the response to oxalic acid over a much wider range of concentrations. In addition, the leaflet assay is much more rapid and simple to perform.
Due to the absence of obvious deleterious effects of the oxalate oxidase transgene in most peanut transformants, we have used it as a sensitive marker for following transformation and regeneration, and as a reporter gene to evaluate expression in transformants. It is inexpensive and easy to assay, allowing screening of many progeny plants for transgene segregation patterns during generation of sufficient peanut seeds for field studies. The utility of oxalate oxidase as a reporter gene and its advantages over luciferase, β-glucuronidase, and green fluorescent protein have been recently described for other dicots (soybean) and monocots (maize) alike (Simmonds et al., 2004).
As encouraging as these results are in support of oxalate oxidase as a useful resistance gene, a limitation to the experiments is that the leaflet assays were performed under highly controlled conditions, including constant temperature, humidity, and use of a single fungal isolate. This will not be the case in field conditions. S. minor is an obligate necrotrophic fungus and requires dead or decaying tissue for successful establishment of infection. Variable growth and environmental conditions will come into play in a field production situation. Oxalic acid is considered a pathogenicity factor for Sclerotinia spp. (Godoy et al., 1990) but may be only one of a number of factors that contribute to successful infection by fungal pathogens.
The oxalate oxidase gene may also play a role in cell wall defense unrelated to its enzyme activity. For example, Schweizer et al. (1999) showed that the expression of a mutant wheat oxalate oxidase sequence lacking enzyme activity conferred increased resistance to penetration by S. sclerotiorum (20%–67% of control levels). Other studies have suggested that additional factors, such as hydrolytic enzymes, are likely to be important in establishing infection and progression of disease caused by S. minor. Based on the observation that coexpression of glucanase and chitinase genes in transgenic tobacco enhanced protection against fungal attack (Zhu et al., 1994), these genes have also been introduced into peanut (Chenault et al., 2002). Expression of multiple resistance genes may offer a better approach for providing greater and long-lasting fungal resistance.
Although there are probably multiple factors involved in the establishment of fungal infection, this study demonstrates the importance of oxalic acid as a pathogenicity factor in infection of peanut by S. minor, and that counteracting the effects of oxalic acid through oxalate oxidase expression can enhance resistance to injury and establishment of fungal infection. Further studies are in progress to assess disease resistance and peanut yields for a number of oxalate oxidase-expressing peanut transformants under field conditions.
MATERIALS AND METHODS
Tissue Culture and Regeneration
Embryos from mature seeds of Virginia-type peanut (Arachis hypogaea) cultivars (NC-7, Wilson, and Perry) were surface sterilized by soaking the seeds in 70% ethanol for 5 min, twice in 1% dichloroisocyanuric acid for 5 min, and finally rinsing the seeds with sterile distilled water. Following sterilization, the embryo was excised and the radicle end removed. Excised embryos were washed in 1% dichloroisocyanuric acid for 30 s and rinsed with three volumes of sterile distilled water. All reagents for tissue culture and subsequent assays were obtained from Sigma (St. Louis) unless otherwise specified.
Embryogenic callus was induced by culturing the mature embryos on MP3 medium, which contained Murashige and Skoog (MS; Murashige and Skoog, 1962) salts and vitamins (Caisson Laboratories, Sugar City, ID), 3% (w/v) Suc, 1 g L−1 Gln, 0.8% (w/v) agar, and 3 mg L−1 picloram. Embryogenic callus was maintained on MP3 medium at 28°C, in the dark, with a subculture period of 3 to 4 weeks.
Maintenance, bombardment, and regeneration of peanut embryogenic cultures were conducted as described previously (Livingstone and Birch, 1999). For regeneration, differentiated embryos with a clearly distinguishable “apex” and “root” bipolarity were transferred to MS medium, which contained MS salts and vitamins, 2% (w/v) Suc, and 0.1% activated charcoal, and allowed to grow for 1 month. The embryos were transferred to a thin layer of MS medium and allowed to dessicate for approximately 1 month before final transfer to fresh MS medium. Any embryos that failed to turn green or show signs of growth and differentiation were discarded. Both regeneration steps were performed at 28°C in the light (80 μmol m−2 s−1) with a 16-h photoperiod.
Vector Construction
RNA was purified from germinating barley seedlings (RNeasy kit; Qiagen, Valencia, CA) for amplification of the oxalate oxidase coding region by reverse transcription-PCR. Oligonucleotide primers were designed from the published sequence for barley oxalate oxidase (accession no. Y14203; Zhou et al., 1998). Upstream (5′GCAAGGTACCATGGGTTACTCTAAAAACCTAG) and downstream (5′GCAAGTCGACTGGGGCTCATGGAAGTTAAG) primers included KpnI and SalI sites, respectively, for cloning into the transformation vector. First-strand synthesis was carried out using Superscript reverse transcriptase according to manufacturer's specifications (Invitrogen, Carlsbad, CA), and amplification was performed using Taq PCR master mix (Qiagen). PCR conditions were denaturation for 30 s at 94°C, annealing for 1 min at 44°C and 50°C for the first and second cycles, respectively, followed by 56°C for the remaining 38 cycles, and extension for 1 min at 72°C. After cloning the 710-bp PCR product into the SmaI site of an intermediate vector pTZ19R and verification of the sequence for oxalate oxidase, cDNA was excised as a KpnI-SalI fragment and used to replace the coding sequence for a fungal phyA gene in a previously described plasmid vector (Li et al., 1997) under control of the dual-enhanced cauliflower mosaic virus 35S promoter. The final oxalate oxidase construct (pOxOx) included a selectable marker for hygromycin resistance.
Plant Transformation and Growth of Regenerated Plants
Embryogenic callus was bombarded with the pOxOx plasmid DNA that had been coated onto 1-μm-diameter tungsten microparticles. Particles were accelerated using a PDS-1000/He Particle Delivery System and an 1,100-psi rupture disc according to manufacturer's recommendations (Bio-Rad, Hercules, CA). Callus was incubated on MP3 medium supplemented with 0.4 m D-mannitol for bombardment. After 2 h of recovery, callus was transferred to MS medium for 2 to 3 d and then plated onto MP3 medium supplemented with filter-sterilized hygromycin B at a concentration of 40 mg L−1 for selection. After 3 to 6 months, transformed somatic embryos were regenerated on antibiotic-free medium as described previously (Livingstone and Birch, 1999). Once plants were transferred from Magenta boxes to the greenhouse, growth conditions included daylight plus supplemental light (sodium halide and high-intensity discharge lighting) for 12 h a day. Plants were treated with Osmocote fertilizer (Scotts, Marysville, OH) and Nitro-Fix (TCI, Pekin, IL) according to manufacturers' recommendations.
DNA and RNA Extraction
DNA was extracted by a modification of the hexadecyltrimethylammonium bromide method described by Murray and Thompson (1980). Unexpanded peanut leaflets (1–1.5 g) were ground in a mortar and pestle with liquid nitrogen and transferred to 10 mL of extraction buffer containing 2% hexadecyltrimethylammonium bromide, 2% polyvinylpyrrolidone (PVP 40; Sigma), 50 mm MES, and 1.4 m NaCl, pH 5.5, which was preheated to 65°C. A volume of 100 μL of 0.2 m β-mercaptoethanol was added to each tube immediately before adding the frozen tissue. The mixture was incubated at 65°C for 10 min with several gentle inversions to ensure mixing, and debris was pelleted at 12,000 rpm in a prewarmed (>15°C) Sorvall SA-600 rotor (Kendro, Asheville, NC) for 10 min. The supernatant was poured through two layers of Miracloth (Calbiochem, La Jolla, CA) into a clean tube containing 10 μL of 10 mg mL−1 RNase A and incubated for 30 min at 37°C. Following RNase digestion, the sample was transferred to a clean tube, and an equal volume of chloroform was added. The tube was gently agitated for 5 min before centrifugation at 12,000 rpm for 10 min. The supernatant was retained and the DNA precipitated with 0.1 volumes of 7.5 m ammonium acetate and 2 volumes of ice-cold ethanol. After 30 min at −20°C, the DNA was spooled into a fresh tube and allowed to stand at room temperature to evaporate the remaining alcohol. The sample was resuspended in 3 mL of TE (10 mm Tris, pH 8, 1 mm EDTA). RNA was extracted from 100 to 150 mg of fresh, tightly folded peanut leaflets using an RNeasy kit (Qiagen).
Southern and Northern Hybridization
To confirm integration of the transgene in peanut transformants, genomic DNA was digested with BstXI. BstXI cuts the pOxOx plasmid once in the oxalate oxidase coding sequence, adjacent to the region amplified as the hybridization probe. DNA fragments were separated by agarose gel electrophoresis and transferred to positively charged nylon membranes (Nytran supercharge; Schleicher and Schuell BioScience, Keene, NH). The membrane was hybridized with a 32P-labeled oxalate oxidase probe. The probe sequence was a 470-bp fragment amplified by PCR using puReTaqReady-To-Go PCR beads (Amersham Biosciences, Piscataway, NJ) with forward primer 5′CCCTCTACAGGACTTCTGCG3′ and reverse primer 5′CTGGCTGTTGAAGGAACACAA3′. The PCR product was purified using a QIAquick PCR purification kit (Qiagen) and labeled using a Prime-It RmT Random Primer labeling kit (Stratagene, La Jolla, CA) and radiolabeled 32P-dCTP (Perkin Elmer, Boston). Hybridization and subsequent washes were performed at 65°C using conditions described by Sambrook et al. (1989) and analyzed on a Storm 850 phosphorimager (Molecular Dynamics, Piscataway, NJ).
RNA was separated by formaldehyde agarose gel electrophoresis and transferred to positively charged nylon membranes as described previously (Sambrook et al., 1989). Membranes were probed with the same oxalate oxidase sequence as described above for Southern blots.
Oxalate Oxidase Assays
To determine oxalate oxidase activity in transgenic peanut plants, leaf discs (5 mm diameter) were placed in 1.5-mL microfuge tubes and assayed using a modification of the procedure by Sugiura et al. (1979). Next, 200 μL of assay buffer (18 mg oxalic acid in 100 mL of 2.5 mm succinic acid, pH 4) were added to tubes and reactions incubated for 15 min at 37°C. After incubation, developing solution (135 μL) was added to the tubes and the reactions allowed to continue at room temperature for 30 min. Developing solution consisted of 6 mg of aminoantipyrene dissolved in 30 μL of N,N-dimethylaniline, which was added to 100 mL of 0.1 m sodium phosphate buffer, pH 7.0, containing 57 μL of a 140 mg mL−1 solution of horseradish peroxidase. The absorbance was measured at a wavelength of 550 nm. For assay in a microtiter plate format, volumes were reduced to one-half the microfuge assay reaction size.
An alternative assay for release of H2O2 as a measure of oxalate oxidase activity was performed with an Amplex Red kit (Molecular Probes, Eugene, OR). Amplex Red in the presence of H2O2 and horseradish peroxidase forms the fluorescent compound resorufin. For the fluorescent assay, leaf discs were incubated in microtiter wells with the same assay buffer as described above. After incubation of leaf discs with oxalic acid substrate, a 20-μL aliquot of each sample was transferred with a multichannel pipettor to a fresh plate and brought to a volume of 50 μL with kit reaction buffer. To start the detection reaction, 50 μL of the Amplex Red/horseradish peroxidase reagent was added and incubated for 30 min in the dark. Fluorescence was detected in a plate reader (Bio-Tek Instruments, Winooski, VT), using a 530-nm excitation filter and 590-nm emission filter. A standard curve allowed calculation of the total H2O2 released. The products of the Amplex Red reaction can also be measured spectrophotometrically at 550 nm due to the high extinction coefficient.
Oxalic Acid Bioassays
Leaflet assays were conducted to assess the ability of oxalate oxidase expression to prevent damage in response to application of oxalic acid to plant tissue. Detached peanut leaflets were arranged on an inverted weigh boat in 15-cm petri dishes containing dampened paper towels. Eight leaflets were used for each plant line, one for each concentration of oxalic acid. Each leaflet was wounded in four locations on the abaxial surface with an 18-gauge needle, and 15 μL of oxalic acid was applied to each wound. For oxalic acid concentrations in the 0 to 200 mm range, the leaflets were incubated for 6 h at room temperature. For concentrations in the 0 to 10 mm range, leaflets were incubated for 48 h at room temperature. To quantify lesion size, leaflets were washed and viewed with a Zeiss Stemi SV 11 dissecting microscope (Carl Zeiss, Jena, Germany). Images were recorded with an attached Scion video camera and imaging software (Scion, Frederick, MD). The lesion area was traced with the computer mouse on the image displayed on the computer monitor. The delimited area was calculated in square pixels by the software package and converted to mm2.
Fungal Bioassays
Fungal assays were performed using detached leaflets inoculated with an agar plug of S. minor mycelia. S. minor cultures were grown on thin potato dextrose agar plates and 5-mm plugs taken from the actively growing edge. Leaflets were wounded with an 18-gauge needle, and plugs were placed on the adaxial surface, near the midvein. Eight leaflets were inoculated for each plant line tested using a minimal quantity of agar in each plug. The plates were incubated for 48 h at 19°C, and lesion area was measured as in the oxalic acid assays above.
Acknowledgments
We thank Dr. Carla Hegeman for assistance in cloning the oxalate oxidase cDNA.
This work was supported by the Virginia Agricultural Council, the Virginia Peanut Growers Association, and the National Peanut Board.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057232.
References
- Bateman DF, Beer SV (1965) Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 55: 204–211 [PubMed] [Google Scholar]
- Bennett AR, Hindal DF (1989) Mycelial growth and oxalate production by five strains of Cryphonectria parasitica in selected liquid culture media. Mycologia 81: 554–560 [Google Scholar]
- Berna A, Bernier F (1999) Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol Biol 39: 539–549 [DOI] [PubMed] [Google Scholar]
- Cessna SG, Sears VE, Dickman MB, Low PS (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12: 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenault KD, Burns JA, Melouk HA, Payton ME (2002) Hydrolase activity in transgenic peanut. Peanut Sci 29: 89–95 [Google Scholar]
- Chiriboga J (1966) Purification and properties of oxalic acid oxidase. Arch Biochem Biophys 116: 516–523 [DOI] [PubMed] [Google Scholar]
- Dessalegne L, Wetten AC, Caligari PDS (1997) Production of transgenic tomatoes expressing oxalate oxidase. Acta Hortic 447: 457–458 [Google Scholar]
- Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotinia sclerotiorum. Physiol Mol Plant Pathol 59: 297–307 [Google Scholar]
- Dumas B, Freyssinet G, Pallett KE (1995) Tissue-specific expression of germin-like oxalate oxidase during development and fungal infection of barley seedlings. Plant Physiol 107: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton MV, Evans CS (1996) Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can J Microbiol 42: 881–895 [Google Scholar]
- Godoy G, Steadman JR, Dickman MB, Dam R (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol Mol Plant Pathol 37: 179–191 [Google Scholar]
- Guimarães RL, Stotz HU (2004) Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol 136: 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havir EA, Anagnostakis SL (1985) Oxaloacetate acetylhydrolase activity in virulent and hypovirulent strains of Endothia (Cryphonectria) parasitica. Physiol Plant Pathol 26: 1–9 [Google Scholar]
- Hollowell JE, Shew BB (2003) Evaluating isolate aggressiveness and host resistance from peanut leaflet inoculations with Sclerotinia minor. Plant Dis 87: 402–406 [DOI] [PubMed] [Google Scholar]
- Hollowell JE, Smith MR, Shew BB (2001) Oxalic acid production by nine isolates of Sclerotinia minor. Proc Am Peanut Res Ed Soc 33: 24 [Google Scholar]
- Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, Lu G (2003) Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol 133: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman WJ, Lane BG, Tanaka CK (1994) Nucleotide sequence of a transcript encoding a germin-like protein that is present in salt-stressed barley (Hordeum vulgare L.) roots. Plant Physiol 104: 803–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikotun T (1984) Production of oxalic acid by Penicillium oxalicum in culture and in infected yam tissue and interaction with macerating enzyme. Mycopathologia 88: 9–14 [Google Scholar]
- Kachroo A, He Z, Patkar R, Zhu Q, Zhong J, Li D, Ronald P, Lamb C, Chattoo BB (2003) Induction of H2O2 in transgenic rice leads to cell death and enhanced resistance to both bacterial and fungal pathogens. Transgenic Res 12: 577–586 [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Azam M, Natarajan K, Mehta A, Datta A (2000) An oxalate decarboxylase from Collybia velutipes: molecular cloning and its overexpression to confer resistance to fungal infection in transgenic tobacco and tomato. J Biol Chem 275: 7230–7238 [DOI] [PubMed] [Google Scholar]
- Kolkman JM, Kelly JD (2000) An indirect test using oxalate to determine physiological resistance to white mold in common bean. Crop Sci 40: 281–285 [Google Scholar]
- Kurian P, Stelzig DA (1979) The synergistic role of oxalic acid and endopolygalacturonase in bean leaves infected by Cristulariella pyramidalis. Phytopathology 69: 1301–1304 [Google Scholar]
- Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC (1993) Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem 268: 12239–12242 [PubMed] [Google Scholar]
- Lane BG (1994) Oxalate, germin, and the extracellular matrix of higher plants. FASEB J 5: 294–301 [DOI] [PubMed] [Google Scholar]
- Lane BG (2000) Oxalate oxidases and differentiating surface structure in wheat: germins. Biochem J 349: 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Li J, Hegeman CE, Hanlon RW, Lacy GH, Denbow DM, Grabau EA (1997) Secretion of active recombinant phytase from soybean cell-suspension cultures. Plant Physiol 114: 1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Maynard CA, Allen RD, Powell WA (2001) Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol Biol 45: 619–629 [DOI] [PubMed] [Google Scholar]
- Livingstone DM, Birch RG (1999) Efficient transformation and regeneration of diverse cultivars of peanut (Arachis hypogaea L.) by particle bombardment into embryogenic callus produced from mature seeds. Mol Breed 5: 43–51 [Google Scholar]
- Lu G (2003) Engineering Sclerotinia sclerotiorum resistance in oilseed crops. Afr J Biotechnol 2: 509–516 [Google Scholar]
- Lumsden RD (1979) Histology and physiology of pathogenesis in plant diseases caused by Sclerotinia species. Phytopathology 69: 890–896 [Google Scholar]
- Maxwell DP, Lumsden RD (1970) Oxalic acid production by Sclerotinia sclerotiorum in infected bean and in culture. Phytopathology 60: 1395–1398 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes RD, Hancock JG (1981) Role of oxalic acid in the Sclerotinia wilt of sunflower. Physiol Plant Pathol 18: 123–132 [Google Scholar]
- Porter DM, Melouk HA (1997) Sclerotinia blight. In N Kokkalis-Burelle, DM Porter, R Rodriguez-Kabana, DH Smith, P Subrahmanyam, eds, Compendium of Peanut Diseases. APS Press, St. Paul, pp 34–36
- Pundir CS (1991) Purification and properties of oxalate oxidase from Sorghum leaves. Phytochemistry 30: 1065–1067 [Google Scholar]
- Ramputh AI, Arnason JT, Cass L, Simmonds JA (2002) Reduced herbivory of the European corn borer (Ostrinia nubilalis) on corn transformed with germin, a wheat oxalate oxidase gene. Plant Sci 162: 431–440 [Google Scholar]
- Rao DV, Tewari JP (1987) Production of oxalic acid by Mycena citricolor, causal agent of American leaf spot of coffee. Phytopathology 77: 780–785 [Google Scholar]
- Ritschkoff A-C, Rättö M, Buchert J, Viikari L (1995) Effect of carbon source on the production of oxalic acid and hydrogen peroxide by brown-rot fungus Poria placenta. J Biotechnol 40: 179–186 [Google Scholar]
- Rollins JA (2003) The Sclerotinia sclerotiorum Pac 1 gene is required for sclerotial development and virulence. Mol Plant-Microbe Interact 16: 785–795 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schweizer P, Christoffel A, Dudler R (1999) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20: 541–552 [DOI] [PubMed] [Google Scholar]
- Simmonds J, Cass L, Routly E, Hubbard K, Donaldson P, Bancroft B, Davidson A, Hubbard S, Simmonds D (2004) Oxalate oxidase: a novel reporter gene for monocot and dicot transformations. Mol Breed 13: 79–91 [Google Scholar]
- Smith FD, Phipps PM, Stipes RJ (1992) Fluazinam: a new fungicide for control of sclerotinia blight and other soil borne diseases of peanut. Peanut Sci 19: 115–120 [Google Scholar]
- Stone HE, Armentrout VN (1985) Production of oxalic acid by Sclerotium cepivorum during infection of onion. Mycologia 77: 526–530 [Google Scholar]
- Sugiura M, Yamamura H, Harano K, Sasaki M, Morikawa M, Tsuboi M (1979) Purification and properties of oxalate oxidase from barley seedlings. Chem Pharm Bull (Tokyo) 27: 2003–2007 [Google Scholar]
- Thompson C, Dunwell JM, Johnstone CE, Lay V, Schmitt M, Watson H, Nisbet G (1995) Degradation of oxalic acid by transgenic oilseed rape plants expressing oxalate oxidase. Euphytica 85: 169–172 [Google Scholar]
- Wei YD, Zhang Z, Anderson CH, Schmelzer E, Gregerson PL, Collinge DB, Smedegaard-Peterson V, Thordal-Christensen H (1998) An epidermis/papilla-specific oxalate oxidase-like protein in the defense response of barley attacked by the powdery mildew fungus. Plant Mol Biol 36: 101–112 [DOI] [PubMed] [Google Scholar]
- Zaghmout OF, Dang PD, Allen RD (1997) Expression of oxalate oxidase in transgenic plants provides resistance to oxalic acid and oxalate-producing fungi (abstract no. 1152). Plant Physiol (Suppl) 114: 227 [Google Scholar]
- Zhang Z, Collinge DB, Thordal-Christensen H (1995) Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J 8: 139–145 [Google Scholar]
- Zhou F, Zhang Z, Gregersen PL, Mikkelsen JD, de Neergaard E, Collinge DB, Thordal-Christensen H (1998) Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol 117: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ (1994) Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Nat Biotechnol 12: 807–812 [Google Scholar]