Abstract
Orellana et al. (2021) and Dai et al. (2021) demonstrate that increased m7G modification of a subset of tRNAs by the METTL1/WDR4 complex stabilizes these mRNAs against decay, increases translation efficiency, reduces ribosome pausing, is associated with poor survival in human cancers, and is directly transforming.
The first RNA modification was discovered in 1957 when pseudouridine was identified, found in the transfer RNA (tRNA) dihydrouridine (D) and anticodon stem-loops, and now known to be the most abundant modification in cellular RNA. Since that time, the emerging “epitranscriptomics” field has uncovered over 100 modified nucleosides and more than 160 modification enzymes that covalently alter nucleosides in tRNAs, ribosomal RNAs (rRNAs), mRNAs, long-noncoding (lnc)RNAs, small nuclear (sn) RNAs, small nucleolar (sno)RNAs, and microRNAs. By far, tRNAs contain the greatest number and variety of modifications than any other cellular RNA. While the chemical identities of tRNA nucleoside modifications have now been well established, understanding their functions has proven to be more difficult, limited not just by our technological tools but also by our conceptual understanding of the complex biology of tRNA functions.
Despite more than 60 years of study, one of the most prevalent modifications, N7-methyguanosine (m7G) at nucleotide position 46 in the variable loop of a subset of tRNAs (Alexandrov et al., 2005; Lin et al., 2018), continues to reveal exciting new functional secrets. In this issue of Molecular Cell, Dai et al. (2021) and Orellana et al. (2021) demonstrate that m7G modification of a subset of tRNAs, catalyzed by the RNA methyltransferase complex METTL1/WDR4 (Alexandrov et al., 2005; Lin et al., 2018), is upregulated in certain cancers. m7G was found by both groups to selectively promote the translation of certain cell cycle regulatory and oncogenic mRNAs that are enriched in corresponding m7G-tRNA cognate codons, which buffers against ribosome pausing, which can cause ribosome collision-mediated translation inhibition (Figure 1). Collectively, these two studies comprise a tour-de-force that connect the remarkably complex molecular functions of tRNA modifications to selective mRNA translational regulation and human cancer. While many studies have associated dysregulated tRNA modifications or tRNA expression with a wide variety of human diseases (Suzuki, 2021), there are surprisingly few well-characterized molecular mechanisms that functionally link specific tRNA modifications to human cancer. These two studies provide a highly detailed molecular understanding by which m7G-tRNA modifications can drive cellular transformation and cancer progression.
Figure 1. m7G-tRNA modification prevents ribosome stalling at m7G-tRNA decoded codons.
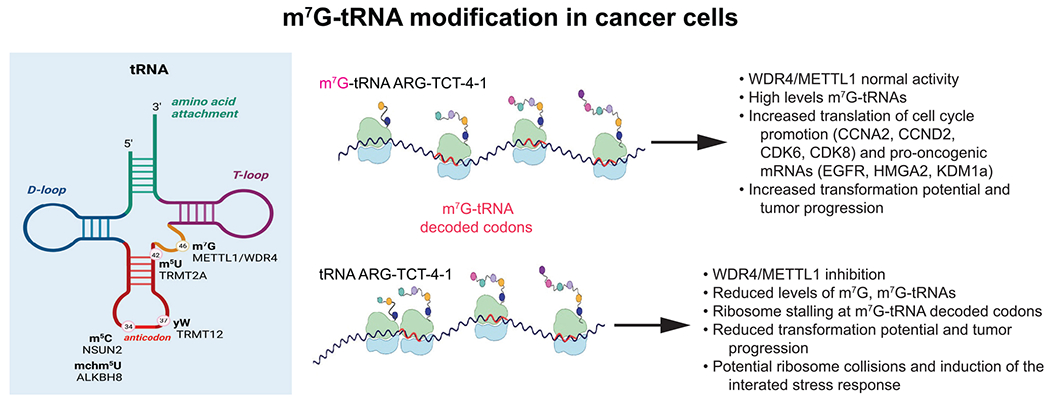
Shown is an overview of tRNA modifications associated with increased oncogenic activity, including m7G modification of nucleotide position 46. The METTL1/WDR4 enzymatic complex carries out m7G modification of a subset of tRNAs that decode m7G-tRNA-dependent codons, thereby promoting normal ribosome elongation without stalling. Certain cell cycle and pro-oncogenic mRNAs that are enriched in m7G-tRNA-dependent codons are increased in their translation, thereby increasing oncogenic potential. Reduction in METTL1/WDR4 activity results in ribosome stalling, shown specifically for the m7G-modified ARG-TCT-4-1 tRNA, resulting in reduced translation of these same mRNAs, resulting in reduction in transformation and cancer progression.
It is well understood that nucleoside modifications in the body and structural loops of tRNAs are dynamic and can change with different physiological conditions and stimuli, conferring regulation of tRNA folding, stability, anticodon function, and aminoacyl-tRNA synthetase amino acid charging specificity of cognate tRNAs, among other functions (Kirchner and Ignatova, 2015; Suzuki, 2021). tRNA modifications therefore ultimately impact not only on the ability of tRNAs to function as codon decoders that provide amino acids for the growing peptide chain during protein synthesis, but also tRNA control of translation fidelity, rates of protein synthesis, and contextually link active ongoing mRNA translation to different cell physiological settings, such as translation-limiting stress responses.
One of the most heavily modified tRNA sites is the anticodon “wobble” nucleotide (position 34), where a number of different modifications confer different codon-anticodon pairing possibilities and codon interaction strengths, and adjacent anticodon position 37, modification of which impacts on decoding and reading frame accuracy (Kirchner and Ignatova, 2015; Suzuki, 2021). Previous studies established that engineered METTL1 knockout or WDR4 mutation impairs m7G tRNA modification and results in a variety of different disorders, including abnormal embryonic growth and differentiation (Lin et al., 2018), microcephalic primordial dwarfism, Galloway-Mowat syndrome (Braun et al., 2018), and increased sensitivity to genotoxic chemotherapy (Okamoto et al., 2014), associated with rapid tRNA degradation (Alexandrov et al., 2006).
Dai et al. (2021) found that levels of METTL1/WDR4 and m7G tRNA modifications are increased in human intrahepatic cholangiocarcinomas (ICCs), a primary hepatic tumor, and is associated with poor survival. Silencing METTL1 or WDR4 in human ICC cell lines resulted in loss of typical markers of malignant transformation, including cell proliferation arrest, decreased colony formation and cell migration, increased apoptosis, and reduced tumor formation in xenografted mice. By characterizing tRNA m7G modifications at single-nucleotide resolution, they identified 22 tRNAs that contain m7G modification in the variable loop, which was shown to stabilize these tRNAs against rapid decay. Non-m7G modified tRNAs were unaffected. Ribosome profiling studies found that METTL1 silencing and reduced m7G tRNA modification caused increased ribosome pausing at m7G-tRNA decoded codons. Identification of the mRNAs most translationally impaired by reduced levels of m7G tRNAs showed enrichment in cell cycle promoting mRNAs such as those encoding cyclin A2, cyclin D2, CDK6, CDK8, and pro-oncogenic mRNAs including epidermal growth factor receptor (EGFR). To seal the deal, the authors overexpressed wild-type or enzymatically mutant METTL1 and showed that it increased m7G tRNA levels, while promoting parameters of transformation and tumorigenesis.
Orellana et al. (2021) also connected increased expression of the METTL1/WDR4 complex and increased m7G tRNA levels to increased malignancy and poor survival in certain human cancers, including breast cancers, glioblastomas, certain sarcomas, acute myelogenous leukemias (AML), and others, which they link mechanistically to the increased abundance of a specific cognate m7G-containing tRNA, Arg-TCT-4-1. Knockout or silencing of METTL1 or WDR4 in highly transformed cell lines impaired their growth and tumorigenic properties in culture and in animals. In murine AML cells, loss of METTL1 also impaired oncogenic capacity but had no effect on normal non-leukemic hematopoietic stem and progenitor cells (HSPCs), suggesting that the METTL1/WDR4 complex may comprise another potential target for therapeutic intervention. While certain human cancers show increased abundance of m7G-tRNAs, the findings of Orellana et al. and Dai et al. converge, with both showing that cell cycle progression mRNAs such as Cdk4, and certain oncogenic mRNAs such as Hmga2, Ash2l, Setdb1, Ube2t, are surprisingly enriched in AGA codons that correspond to the Arg-TCT-4-1 cognate tRNA, and therefore increased in their translation. In fact, Orellana et al. (2021) show that merely overexpressing m7G-tRNA Arg-TCT-4-1 in the absence of increased activity or levels of METTL1/WDR4, phenocopies METTL1/WDR4 overexpression and is itself malignantly transforming. These findings add an important new conceptual layer to the understanding of the complex and often under-appreciated role of translational regulation as a driver and not just a participant in oncogenesis.
Both the Orellana and Dai studies show that m7G modification of tRNAs, and particularly of the Arg-TCT-4-1 tRNA, promotes its stabilization and therefore its ability to participate in mRNA translation and reduce or eliminate ribosome pausing. This leads to a key question that now needs to be addressed in future studies that might directly link m7G tRNA modification to stress-mediated ribosome pausing as a mechanism by which translational regulation drives transformation and cancer progression. Ribosome collisions during translation elongation due to increased pausing has been shown to mediate a general stress response that increases eIF2α serine-51 phosphorylation, which in turn inhibits overall protein synthesis (Wu et al., 2020). There are hints of ribosome collision-mediated translation inhibition in both the Dai and Orellana studies. Studies now need to determine whether m7G modification of certain tRNAs serves as an integrator between selectively increasing translation of cell cycle promoting mRNAs while suppressing inhibition of global protein synthesis due to ribosome pausing, by eliminating ribosome collision-mediated translation inhibition.
REFERENCES
- Alexandrov A, Grayhack EJ, and Phizicky EM (2005). tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, and Phizicky EM (2006). Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96. [DOI] [PubMed] [Google Scholar]
- Braun DA, Shril S, Sinha A, Schneider R, Tan W, Ashraf S, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, et al. (2018). Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 176, 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Liu H, Huang LC, Ren X, Zhu W, Zhu S, Peng B, Li S, Lai J, Liang L, et al. (2021). N7-methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 81, 3339–3355. [DOI] [PubMed] [Google Scholar]
- Kirchner S, and Ignatova Z (2015). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet 16, 98–112. [DOI] [PubMed] [Google Scholar]
- Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, and Gregory RI (2018). Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol. Cell 71, 244–255.e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H, Xiao Y, Qi G, Shimamoto F, Ota T, et al. (2014). tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 10, e1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana E, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W, Lim J, Aspris D, Sendinc E, Garyfallos D, et al. (2021). METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 81, 3323–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. (2021). The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol 22, 375–392. [DOI] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, and Green R (2020). Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 182, 404–416.e414. [DOI] [PMC free article] [PubMed] [Google Scholar]


