Abstract
Biochemical studies have demonstrated decreased binding of various proteins to DNA in nucleosome cores as their cognate sites are moved from the edge of the nucleosome to the pseudodyad (center). However, to date no study has addressed whether this structural characteristic of nucleosomes modulates the function of a transcription factor in living cells, where processes of DNA replication and chromatin modification or remodeling could significantly affect factor binding. Using a sensitive, high-resolution methyltransferase assay, we have monitored the ability of Gal4p in vivo to interact with a nucleosome at positions that are known to be inaccessible in nucleosome cores in vitro. Gal4p efficiently bound a single cognate site (UASG) centered at 41 bp from the edge of a positioned nucleosome, perturbing chromatin structure and inducing transcription. DNA binding and chromatin perturbation accompanying this interaction also occurred in the presence of hydroxyurea, indicating that DNA replication is not necessary for Gal4p-mediated nucleosome disruption. These data extend previous studies, which demonstrated DNA replication-independent chromatin remodeling, by showing that a single dimer of Gal4p, without the benefit of cooperative interactions that occur at complex wild-type promoters, is competent for invasion of a preestablished nucleosome. When the UASG was localized at the nucleosomal pseudodyad, relative occupancy by Gal4p, nucleosome disruption, and transcriptional activation were substantially compromised. Therefore, despite the increased nucleosome binding capability of Gal4p in cells, the precise translational position of a factor binding site in one nucleosome in an array can affect the ability of a transcriptional regulator to overcome the repressive influence of chromatin.
The interaction of transcription factors with chromatin is an important issue in understanding gene regulation. Transcriptional activation upon factor binding often results in the disruption or displacement of one or more nucleosomes within promoter regulatory regions (1, 53). However, the mechanisms by which transcriptional regulators gain access to their target DNA sequences in chromatin remain unclear. Biochemical studies in vitro have disclosed that factor binding to nucleosome cores depends on variables such as the type of factor, the number and location of binding sites, and the status of the histone amino termini (reviewed in reference 44). For example, Gal4p derivatives (e.g., Gal4-AH) can bind to reconstituted nucleosomes containing single or multiple GAL4 binding sites to form a ternary complex (15, 68, 72, 78). Binding to multiple nucleosomal GAL4 binding sites initiates at the edge of nucleosome cores and also occurs when a single binding site is centered at 21 bp from the edge, whereas single sites centered at 40 or 74 bp (at the pseudodyad) from the edge were not accessible in unmodified nucleosomes (72). These data and other physical studies (34, 41, 60, 75) indicate that the two helical turns of DNA at each end of the nucleosome core are not associated as strongly with the histone octamer as sequences more internal. Because nucleosomes occur within arrays in cells, whether a similar structure pertains in vivo and its potential relevance to gene activation are highly controversial (19, 47) and have not been addressed.
A previous structural study suggested that Gal4p led to perturbation of chromatin when UASG was localized near the pseudodyad of a positioned nucleosome in yeast (40). The fractional occupancy of UASG by Gal4p and the degree of chromatin reconfiguration were not determined. The mechanism by which Gal4p gained access to and perturbed this nucleosome is also currently unknown. Finally, nucleosome positioning is known to occur by several different mechanisms, making generalizations from study of a single positioned nucleosome hazardous.
One model to explain how transcription factors achieve increased access to their regulatory sequences in vivo suggests that removal of histones, presumed to occur transiently during replication, provides a window of opportunity that enables factor binding (6, 65, 74). For instance, it was reported that Gal4-VP16 can potentiate transcription of a template assembled into chromatin in vitro if added during, but not after, DNA replication (27). Likewise, DNA replication in the presence of specific erythroid factors is also required for transcriptional activation of nucleosome-repressed β-globin templates in synthetic nuclei (4). On the other hand, Gal4p derivatives can potentiate replication-independent transcription in vitro from preassembled chromatin templates (11, 48). DNA replication is also not required for chromatin reconfiguration of the rat tyrosine aminotransferase, mouse mammary tumor virus, or yeast PHO5 promoters in vivo (2, 52, 57). As these promoters are complex, cooperative binding of multiple upstream factors and the basal transcription machinery could be directly responsible for relieving the requirement for DNA replication in chromatin remodeling or could be indirectly responsible through recruitment of a chromatin remodeling-modification activity (8, 28). The absence of such an activity in protein extracts employed for in vitro studies could affect conclusions regarding the role of DNA replication in effecting factor access to chromatin in vivo.
To gain insight into the mechanism by which transcription factor binding and chromatin remodeling occur in vivo, we have employed a simple minichromosome system in which the interaction of a single factor with a template having a well-defined chromatin structure can be studied (40, 42, 61). We have monitored factor binding and changes in chromatin structure in this system by a novel in vivo footprinting strategy utilizing SssI DNA methyltransferase (MTase) (31). Insertion of a single GAL4 binding site at 41 bp from either edge of a positioned nucleosome, a location that is refractory to binding in nucleosomes cores in vitro (72), and growth under activating conditions led to substantial binding of Gal4p and perturbation of chromatin structure in both replicating and nonreplicating cells. In contrast, placing UASG at the nucleosomal pseudodyad significantly inhibited chromatin disruption and transcriptional activation, as only partial occupancy and nucleosome perturbation were seen, even under conditions of Gal4p overexpression. These data demonstrate that the ability of endogenous Gal4p to invade a positioned nucleosome in vivo does not derive from transient disruption of DNA-histone contacts by the DNA replication machinery and may involve chromatin modification or remodeling activities. Despite a potential role of such activities in Gal4p-mediated chromatin disruption and their abundance in cells, our results also demonstrate that the precise translational positioning of a regulatory element within a single nucleosome contained in an array influences factor accessibility. Thus, the influence of translational positioning on factor accessibility is not unique to nucleosome cores and appears to be a bona fide physical property of nucleosomes in vivo which exerts biological consequences.
MATERIALS AND METHODS
Plasmid construction and yeast strains.
Yeast plasmid TALS4 was created by modifying pTALS (55) (containing TALS inserted into the HindIII site of pUC19) by PCR site-directed mutagenesis (26) as described by Kladde et al. (31); five additional CG reporter sites were inserted, and UASgl (5′-CGGTCCACTGTGTGCCG-3′), centered at map unit (m.u.) 1415 within GAL3 sequences (3), was eliminated by changing its sequence to 5′-CGtTCCACgaTcTGaCG-3′ (altered positions are indicated in lowercase). In the present study, site-directed mutagenesis by PCR (26) was used to change sequences of the parent pTALS4 (31) from m.u. 1400 to 1431, 1432 to 1463, and 1464 to 1495 to 5′-CGATCACCGGAAGACTCTCCTCCGCGAGCTCG-3′ (the near-consensus UASG is underlined) (22), creating pTALS4-17L, pTALS4-17C, and pTALS4-17R, respectively. The positions of the C residues in each CG on the lower strand relative to the operator-distal or left edge of the nucleosome in this invariant sequence in the resulting minichromosomes (see below) are as follows: TALS4-17L, 27, 34, 49, 51, and 57 bp; TALS4-17C, 59, 66, 81, 83, and 89 bp; and TALS4-17R, 91, 98, 113, 115, and 121 bp. The accuracy of all base substitutions was verified by double-stranded DNA sequencing. TALS4 and its UASG-containing derivatives were excised from pUC19 by digestion with HindIII and religated prior to transformation by electroporation (54) into Saccharomyces cerevisiae YPH500ΔL.19-2 (MATα ade2-101ochre his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ1::LYS2-GAL1 promoter/SssI) and the isogenic MATa strain, YPH499ΔL.19-2 (31). Propagation of each minichromosome as a monomeric species and the absence of genetic rearrangements were verified by Southern blotting. Where indicated, cells were also transformed with the 2μm-based shuttle vector pRS426 (13) or pRS426-GAL4, which comprises the 3.6-kbp BamHI fragment from pG525 (33), encompassing the entire GAL4 promoter and coding sequence, inserted into the BamHI site of pRS426.
The corresponding series of β-galactosidase reporter plasmids, YCpTALS4 and its derivatives (see Fig. 6), were constructed by using primers 5′-GCATCGCTGCAGGTCG-3′ and 5′-GAAGATCTCGAGCGTTGCCTCATCATCAATGC-3′ to PCR amplify the region from m.u. 1347 to 1619 (see Fig. 1) in TALS4 and its versions harboring UASG. The PCR product was digested with PstI and XhoI, and the resulting 261-bp fragment was ligated into the same sites upstream of the minimal CYC1 promoter driving expression of lacIZ in YCpCSG1 (66). The cloned region in each construct was verified by double-stranded DNA sequencing. YCp reporter plasmids were directly transformed into isogenic strains YPH500ΔL.19-2 and YPH499ΔL.19-2, which express wild-type levels of Gal4p.
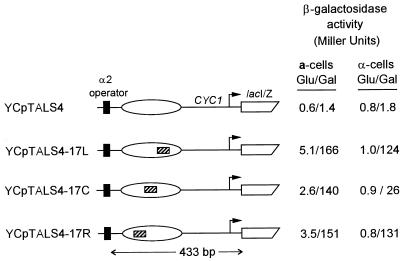
FIG. 6.
The location of UASG in a positioned nucleosome modulates transcriptional activation by Gal4p. The region indicated in Fig. 1 encompassing the α2 operator and nucleosome (ellipse) from TALS4 and its derivatives (UASG is indicated by hatched bar) were subcloned upstream of lacIZ to create the four indicated YCp constructs. Reporter plasmids were transformed into cells expressing wild-type levels of Gal4p, and levels of β-galactosidase activity were determined after growth in medium containing glucose (Glu) or galactose (Gal). The expression levels of β-galactosidase shown are the averages obtained from two different yeast transformants from two representative, independent experiments that included all of the samples. In addition, although the absolute levels of expression varied slightly between experiments, the fold inductions from glucose to galactose were very similar for YCpTALS4-17C versus YCpTALS4-17L and YCpTALS4-17R in α-cells for two independent transformants in two additional experiments.
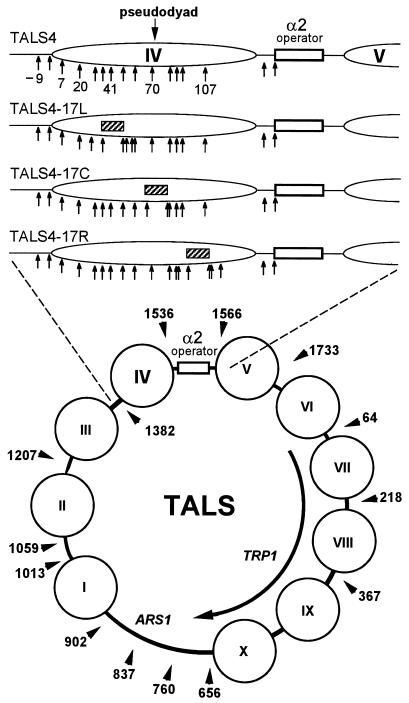
FIG. 1.
Chromatin structure of the TALS4 minichromosome and its derivatives in α-cells. The lower diagram shows the locations of preferential cleavages by MNase (arrowheads), in m.u., as determined at low resolution (10 to 20 bp) by indirect end labeling, and inferred positions of nucleosomes (circles) in TALS minichromosomes (55). The α2 operator is indicated by the open rectangle. The upper diagram shows a to-scale enlargement of the region from m.u. 1347 to 1619 (left to right) from the minichromosome TALS4 and its derivatives. This region was amplified by PCR and cloned in the reverse orientation to construct the lacIZ reporter plasmids of Fig. 6 and 7. The position of nucleosome IV (ellipse) (m.u. 1375 to 1520), determined by mapping of MNase cutting sites at high resolution (59) (Fig. 2), and locations of target CG sites (arrows) relative to the left edge (arbitrarily assigned position 1) of the nucleosome are given. A 32-bp sequence (see Materials and Methods) containing a near-consensus 17-bp binding site for Gal4p (UASG; hatched bar) was inserted at three different positions into TALS4 (no UASG) to create the three UASG-containing plasmids, TALS4-17L, TALS4-17C, and TALS4-17R. In α-cells, nucleosome IV is precisely positioned next to the α2 operator, incorporating UASG at three different translational positions, centered 41 bp from the left (17L) and right (17R) edges or at the center (17C; pseudodyad), of the nucleosome.
Primer extension analysis of chromatin.
High-resolution mapping of micrococcal nuclease (MNase) cleavage sites was performed as described by Shimizu et al. (59) and modified by Weiss and Simpson (76). Briefly, yeast nuclei, prepared from cells grown in 1 liter of either 2% glucose or 2% galactose complete synthetic medium (CSM) (Bio101, La Jolla, Calif.) lacking tryptophan (to select for TALS4-17L) and uracil (to select for pRS426-GAL4), were treated with various concentrations of MNase. Following termination of digestion, DNA was purified and MNase-cut sites were detected by multiple rounds of primer extension with 32P-end-labeled primer 5′-CTCAAGTCGTCAAGTAAAGATTTCGTGTTC-3′ followed by electrophoresis on a 6% polyacrylamide (acrylamide-bisacrylamide, 19:1)–50% urea gel buffered by an electrolyte gradient (58). DNA was also isolated from nuclei, deproteinized, and subsequently treated with MNase to determine cleavage preferences in naked DNA.
Mobility shift assay and in vitro SssI footprinting.
Primers 5′-GCATAAACACCATCAGCCTC-3′ and 5′-CAGATATCAAAACTGTTGCATTATT-3′ were used to PCR amplify a 242-bp probe (m.u. 1267 to 1508) from pTALS4-17L containing a single UASG which was subsequently gel purified.
The Gal4p derivative Gal4-AH was purified from Escherichia coli as described by Lin et al. (35). Serial dilutions of the Gal4-AH stock (monomer concentration of 5 μM) were made in G4D buffer (100 mM KCl, 10 mM HEPES [pH 7.4], 10 μM ZnCl2, 0.2 mM phenylmethylsulfonyl fluoride). Dilutions of Gal4-AH (2 μl) were added to duplicate 18-μl reaction mixtures containing the following: 1× SW binding buffer (2.5 mM HEPES [pH 7.9], 0.05 mM EDTA, 5% glycerol, 0.01 mg of bovine serum albumin per ml, 0.1 mM dithiothreitol, 0.02 mM phenylmethylsulfonyl fluoride), 55.6 mM NaCl, 3.33 mM MgCl2, 5.6 ng of sheared calf thymus DNA per μl, and 0.18 mM S-adenosylmethionine. In addition, reaction mixtures contained 1 ng of either 32P-end-labeled (7,500 cpm; for mobility shift gel) or nonradioactive (for SssI footprinting) probe (final concentration of 0.056 ng/μl).
The radioactive binding reaction mixtures were incubated at 30°C for 42 min and then electrophoresed on a mobility shift gel as described previously (72). Following incubation for 40 min at 30°C, 2 μl of purified SssI (New England Biolabs), diluted from a concentration of 2 U/μl to 0.5 U/μl with 1× SW binding buffer, was added to each duplicate, nonradioactive sample. After an additional 2 min at 30°C, 10 μl of freshly made deamination denaturation buffer (0.9 N NaOH [dissolved from solid], 25 mM EDTA, 0.2 mg of sheared calf thymus DNA per ml) was added. Following 5 min at 98°C, deamination was initiated by adding 200 μl of saturated sodium metabisulfite, and the samples were subsequently processed as described by Kladde et al. (31). The primers used to amplify from the purified deaminated probe DNA were MK1b1T4 (31) and MK1b2T4 (5′-GAAAATTGTTGTATTATTGTGTTTTGTATTTT-3′).
Cell culturing and in vivo SssI footprinting.
Yeast cells (10 ml) for Fig. 4 and 5 were grown with shaking at 30°C in 2% galactose CSM lacking tryptophan (to select for TALS4 and derivatives) and uracil (to select for pRS426 or pRS426-GAL4) to an optical density at 600 nm (OD600) of approximately 1. The cells were then centrifuged, resuspended in fresh selective medium containing 2 to 4% galactose, grown for an additional 16 h, and then treated to identify modified cytosines on the lower DNA strand as described previously (14, 20, 31). The primers used were MK1b1T4 and MS6b2T4 (31) for TALS4, TALS4-17L, TALS4-17C, and TALS4-17R and YCpb1T4 (5′-TATATATATCAACACTAAAATTCCTAACCATCC-3′) and YCpb2T4 (5′-TAATTTTTATTAAAGGGAATAAAAGTTGGG-3′) for the YCp plasmids used for Fig. 7.
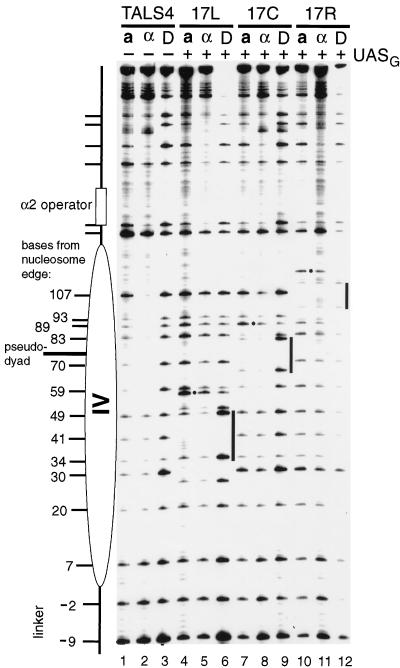
FIG. 4.
Remodeling of a positioned nucleosome by Gal4p binding is dependent on the position of UASG. The chromatin structures in living yeast cells grown in galactose of the four different plasmids depicted in Fig. 1, TALS4 (lanes 1 to 3), TALS4-17L (lanes 4 to 6), TALS4-17C (lanes 7 to 9), and TALS4-17R (lanes 10 to 12), were analyzed in the presence of pRS426-GAL4, which overexpresses Gal4p from the wild-type GAL4 promoter. Protein-free DNA for each plasmid (D) was methylated by SssI in vitro to identify target CG sequences and site preferences of the enzyme (lanes 3, 6, 9, and 12). For analysis of chromatin, DNA was rapidly isolated from isogenic, SssI-expressing a-cells (lanes 1, 4, 7, and 10) and α-cells (lanes 2, 5, 8, and 11), and modified cytosines were identified as described previously (31). The number of nucleotides that the C residue in each target CG site is away from the operator-distal edge (i.e., left edge) of the positioned nucleosome in TALS4 in α-cells is indicated. Where present, each UASG is marked by a bar at the immediate right of the samples that were methylated in vitro (D), and the position of the hypermethylated residue next to each binding site is indicated by a dot.
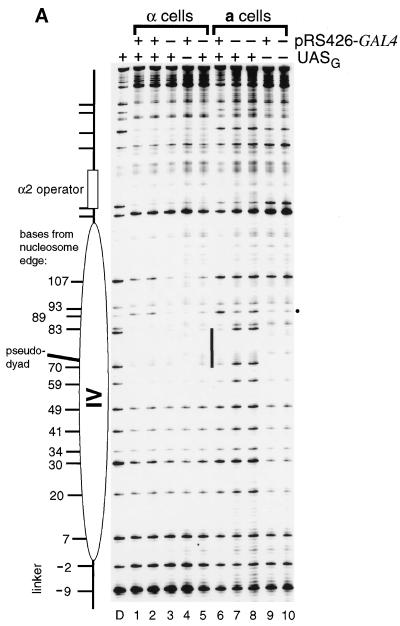
FIG. 5.
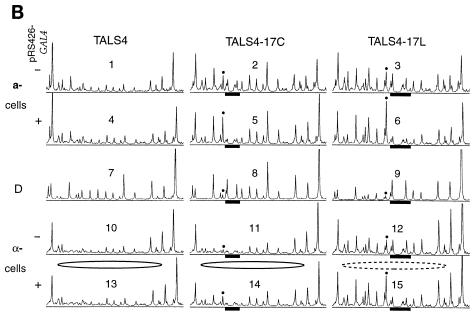
Chromatin remodeling and Gal4p occupancy in a- and α-cells at endogenous and overexpressed levels of Gal4p. (A) TALS4-17C (+UASG) and TALS4 (−UASG) were introduced into SssI+ a- and α-cells containing endogenous levels of Gal4p (−pRS426-GAL4) (i.e., transformed with pRS426 vector only) or overexpressing Gal4p (+pRS426-GAL4) and probed with SssI MTase by growing the cells in galactose. Positions of CG sites were identified in protein-free DNA (D) as indicated in the legend to Fig. 4. Endogenous levels of Gal4p in a-cells led to marked disruption of chromatin in the region of nucleosome IV (compare lanes 7 and 8 to lane 10), whereas no remodeling was visible in α-cells (compare lanes 3 and 5 to lane 4). Overexpression of Gal4p increased disruption of nucleosome IV in α-cells only slightly at sites 107, 89, and 30 (compare lanes 1 and 2 to lanes 3 and 5). Note that overexpression of Gal4p in cells harboring TALS4 (no UASG) had no effect on chromatin structure (compare lanes 9 and 10). (B) Scans of in vivo SssI probing data. Phosphorimager scans from an experiment completely independent from that presented in panel A are shown, analyzing disruption of TALS4, TALS4-17C, and TALS4-17L in a- and α-cells grown in galactose containing endogenous (transformed with pRS426 vector) (scans 1 to 3 and 10 to 12) or overexpressed (transformed with pRS426-GAL4) (scans 4 to 6 and 13 to 15) levels of Gal4p. UASG is indicated by filled bars, and the site that becomes hypermethylated upon Gal4p binding is marked by dots. The CG within the α2 operator is located at the leftmost side of each scan, and the linker is at the right. Perturbation of control a-cell chromatin, seen as increases in methylation at many CG sites in the region (except sites within UASG that are protected by bound Gal4p in the presence of pRS426-GAL4), occurs equally well in TALS4-17C and TALS4-17L (compare scans 2 and 3 to scan 1 and compare scans 5 and 6 to scan 4). The precisely positioned nucleosome in TALS4 and TALS4-17C in α-cells is depicted as a solid ellipse. Perturbation of this nucleosome in α-cells in TALS4-17L (compare scans 12 and 10 and compare scans 15 and 13) is indicated by the dashed ellipse. Endogenous levels of Gal4p efficiently remodel UASG-containing a-cell chromatin (compare scans 2 and 3 to scan 1) but disrupt chromatin in α-cells only when UASG is removed from the pseudodyad in TALS4-17L (i.e., lack of chromatin perturbation in scan 11 versus scan 10 but clear disruption in scan 12 versus scan 10 or 11). Note that the level of disruption in TALS4-17L at endogenous levels of Gal4p (scan 12) is greater than that in TALS4-17C even when Gal4p is overexpressed (scan 14).
FIG. 7.
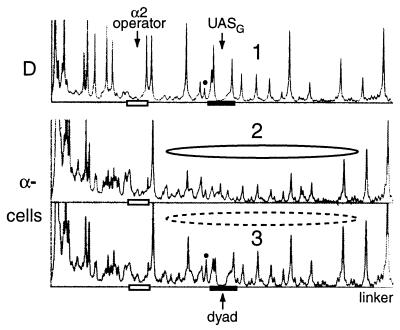
A nucleosome is positioned next to the α2 operator in the lacIZ reporter plasmids in α-cells. Linear phosphorimager scans of SssI methylation of YCpTALS4 (no UASG) and YCpTALS4-17C (UASG is demarcated by the filled bar) in SssI-expressing cells which express endogenous levels of Gal4p are shown. The open bar indicates the α2 operator. Cells were grown in galactose and then processed to identify methylated cytosines. For YCpTALS4 (scan 2), note the protection by the nucleosome (solid ellipse) (compare scans 2 and 1). Insertion of UASG (i.e., YCpTALS4-17C [scan 3]) leads to partial disruption of the nucleosome (depicted by the dashed ellipse) in α-cells (compare scans 3 and 2). The increase in signal at the hypermethylated site (marked by the dot) also indicates partial occupancy of UASG by Gal4p.
For experiments in which DNA replication was inhibited (see Fig. 9), initially, to repress Gal4p and SssI expression (i.e., SssI is transcribed from the GAL1 promoter), yeast cells (10 ml) were grown in 2% glucose CSM lacking tryptophan and uracil until they reached stationary phase (OD600, ∼4). The cells were then centrifuged, washed once with medium containing 2% galactose, resuspended in 2% galactose selective medium plus 200 mM hydroxyurea, and incubated with shaking for another 12 h at 30°C to achieve synthesis of Gal4p and SssI. In vivo-methylated cytosines were identified as described previously (14, 20, 31) by using primers MK1b1T4 and MS6b2T4 (31). Under the conditions employed, as reported previously (23, 62), the cell number did not increase throughout the time course of the experiment. In other studies, to ensure that cells had stopped replicating far in advance of accumulating significant SssI activity, we determined that a minimum of 8 to 10 h of incubation is required to accumulate detectable methylation following a shift from glucose to galactose. An additional control was performed in which logarithmically growing cells were first synchronized in late G1 phase by addition of 6 μg of α-factor per ml. The cells were then released from the cell cycle block by washing once with yeast extract-peptone-dextrose (YPD) medium (54) and preincubation in YPD for 30 min at 30°C. Following preincubation in YPD, one-half of the cells were centrifuged and resuspended in YPD containing 200 mM hydroxyurea. No budding was observed, even after 3 h at 30°C, indicating that hydroxyurea had completely inhibited DNA replication and arrested cells in early S phase. The remaining cells, resuspended in YPD lacking hydroxyurea, exhibited significant budding after 30 min.
FIG. 9.
Gal4p binds 41 bp from the edge of a positioned nucleosome and causes disruption in the absence of DNA replication. Cells overexpressing Gal4p and harboring TALS4-17L (lanes 1–3; UASG is indicated by the bar at right of lane 3) or TALS4 (lanes 4 to 6; no UASG) were initially grown in glucose-containing medium to repress synthesis of SssI MTase and Gal4p and were then transferred to galactose medium that also contained hydroxyurea. Following the induction of Gal4p and SssI expression, identification of lower-strand cytosines methylated in chromatin in vivo was performed as described (31). Positions of CG sites were identified in protein-free DNA (lanes D) as indicated in the legend to Fig. 4. Note the increase in methylation in chromatin of both cell types in the presence of UASG (for α-cells, compare lanes 1 and 4; for a-cells, compare lanes 3 and 6), which is indicative of chromatin remodeling.
β-Galactosidase expression assays.
Cells for Fig. 6 were grown at 30°C in CSM lacking leucine and containing 2% glucose or 2% galactose for 16 to 20 h to an OD600 of about 2 to 3 and were assayed for β-galactosidase activity (39, 54), which is expressed as 1,000 × A420/(OD600 × time [minutes] × volume [milliliters]), at 30°C. For Fig. 8, following growth in 200 ml of 2% glucose CSM lacking leucine to an OD600 of 0.2, cells were washed extensively with sterile, distilled water to remove the glucose, resuspended in 200 ml of 2% galactose CSM lacking leucine, and incubated at 30°C with shaking. Aliquots of cells (6 OD600 units, 10 to 20 ml) were removed at 1-h intervals following resuspension in galactose-containing medium and assayed for β-galactosidase activity.
FIG. 8.
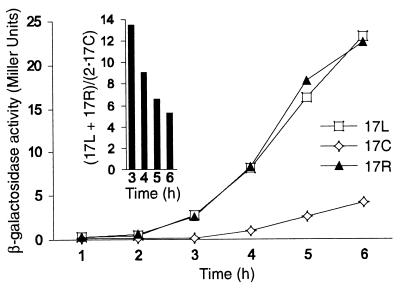
Transcriptional activation of the lacIZ reporter plasmids in α-cells at limiting levels of Gal4p expression. Cells containing YCpTALS4, YCpTALS4-17C (17C), YCpTALS4-17L (17L), and YCpTALS4-17R (17R) were initially cultured in medium containing glucose then transferred to galactose and assayed at 1-h intervals for expression of β-galactosidase. The activity of YCpTALS4 lacking UASG remained below 0.1 Miller unit at all time points. The inset shows the relative activities of 17L and 17R versus 17C at several time points.
RESULTS
Strategy for assessing Gal4p-mediated chromatin disruption in living cells.
We have employed derivatives of a yeast minichromosome, TALS4 (31), to study the interaction of Gal4p within two different chromatin environments that are well characterized. In α-cells, we assessed the ability of Gal4p to compete with histones within a nucleosome that is precisely positioned by the Mcm1p-Matα2p complex (Fig. 1). The assignment of a nucleosome and its position adjacent to the α2 operator in α-cells is based on a considerable body of earlier mapping studies utilizing MNase (21, 46, 55, 56, 59, 76), MTases (Dam, Sau3A1, and SssI) (29–31), and DNase I (21, 46, 56, 59). In sum, these studies demonstrate that, abutting the α2 operator, a nucleosome is positioned by Mcm1p-Matα2p over the promoters of a-cell-specific genes (21, 56), the recombination enhancer (76), minichromosomes (29, 55, 59), and heterologous random sequence in a lacZ-containing reporter plasmid (46). Nucleosome positioning over such disparate sequences reinforces the likelihood that the chromatin structures of closely related minichromosomes would be the same. Indeed, protection against MNase (29) (see Fig. 2) and MTases (29–31) of an extended region next to the operator, inferred to be a nucleosome, persists in α-cells when the original TALS plasmid (55) is modified by the introduction of MTase target sites in the nucleosome (e.g., TALS4). Most conclusively, mutations in the globular region of histone H4 which produce Sin− phenotypes lead to increases in accessibility adjacent to the α2 operator in TALS4 in α-cells (73), demonstrating that a nucleosome is responsible for the observed protection. In a-cells, which do not synthesize Matα2p, Mcm1p remains bound and nucleosomes are present adjacent to the α2 operator but are relatively disordered. The minichromosomes offer an additional advantage in that the interaction of a single factor within a well-defined chromatin context can be monitored and changes in chromatin structure are unlikely to be due to transcription. This derives from the fact that the minichromosomes lack a natural promoter (i.e., even the promoter of the TRP1 selectable marker is defective such that only a low percentage of the templates are transcribed, which is sufficient to confer growth in medium lacking tryptophan [16]).
FIG. 2.
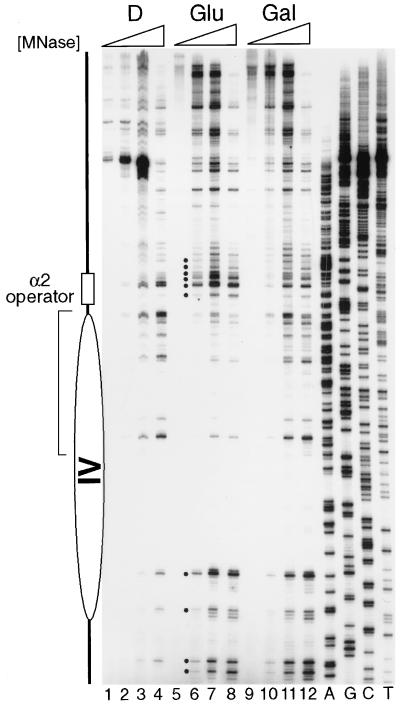
Primer extension analysis of MNase-cut sites in TALS4-17L. Nuclei (lanes 5 to 12) and protein-free DNA (D) (lanes 1 to 4) from α-cells containing TALS4-17L and pRS426-GAL4 grown in glucose (Glu) (lanes 5 to 8) or galactose (Gal) (lanes 9 to 12) were isolated and treated with increasing concentrations of MNase as follows: 0.05, 0.25, 0.5, and 1 U/ml, respectively, in lanes 1 to 4; 0, 2.5, 5, and 10 U/ml in lanes 5 to 8; and 0, 1.25, 2.5, and 5 U/ml in lanes 9 to 12. The higher concentrations used to digest cells from glucose were to compensate for increased cell numbers. The dots mark cleavage sites that are enhanced in glucose-grown cells, and the bracket delineates those sites that are protected relative to control DNA. MNase cleavages and the inferred nucleosome position (ellipse) are as those observed in the propositus TALS plasmid by Shimizu et al. (59). Loss of protection due to binding of activated Gal4p, indicating disruption of the nucleosome, is observed when cells are grown in galactose (40, 63).
In this study, a near-consensus 17-mer binding site for Gal4p was introduced at three different translational positions within the region occupied by nucleosome IV of TALS4 (Fig. 1). Plasmids TALS4-17C, TALS4-17L, and TALS4-17R contain UASG at the pseudodyad (center) and 41 bp from the left and right edges of the nucleosome, respectively. Although Gal4p is apparently insensitive to the rotational orientation of its binding site on the surface of a nucleosome (68), we kept the rotational setting of UASG virtually identical and 7 bp flanking each element constant to permit an accurate test of the variable of translational position.
In view of the above-described results regarding the dominance of nucleosome positioning mediated by Mcm1p-Matα2p, it was unlikely that insertion of a Gal4p binding site would simply destabilize the nucleosome in α-cells during growth in glucose, where Gal4p synthesis is repressed and Gal4p does not bind nucleosomal DNA (40, 63). For example, TALS, which contains UASgl from the GAL3 promoter at the identical location as the near-consensus site in TALS4-17L, positions a nucleosome in α-cells grown in glucose (55, 59, 63). To control for other minor sequence differences between TALS and TALS4-17L that formally could contribute to nucleosome destabilization, we analyzed the chromatin structure of the region next to the α2 operator in TALS4-17L in cells grown in glucose versus those grown in galactose (Fig. 2). Nuclei were isolated from cells and digested with various concentrations of MNase, and nuclease cleavages were mapped by primer extension (59). Relative to deproteinized DNA (Fig. 2, lanes 1 to 4), chromatin in nuclei isolated from cells grown in glucose (lanes 5 to 8) exhibited protection against and enhancement of MNase cleavage at several sites, indicating the presence of a positioned nucleosome flanked by histone-free linker regions that are nuclease hypersensitive. Compared to when cells were grown in glucose, in galactose (Fig. 2, lanes 9 to 12) the pattern of MNase cleavage in TALS4-17L chromatin was very similar to that observed for protein-free DNA, consistent with disruption of the nucleosome concomitant with binding of activated Gal4p (40, 63). While this high-resolution experiment yields information about the status of DNA-histone contacts in cells grown in galactose, it yields no indication of the degree of Gal4p binding.
As a probe for chromatin structure in living cells, we utilized the SssI DNA MTase, stably integrated as a single copy under GAL1 control (31). This sensitive strategy offers the advantage over conventional methodologies that it can simultaneously map both DNA-histone and -non-histone protein interactions. In addition, as lengthy or harsh treatments (cell permeabilization, preparation of nuclei, or DNA alkylation damage) are not employed, it avoids potential rearrangement or loss of chromatin constituents (as seen above for Gal4p), allowing an accurate assessment of in vivo states. The MTase modifies accessible CG sites at a low level without altering the growth characteristics of cells. Following rapid isolation of DNA from cells, 5-methylcytosines are identified (14, 20), and footprints are visualized by comparison of modifications in chromatin with those in control, protein-free DNA, methylated in vitro (31).
Nucleosome translational positioning affects Gal4p-mediated chromatin reconfiguration in vivo.
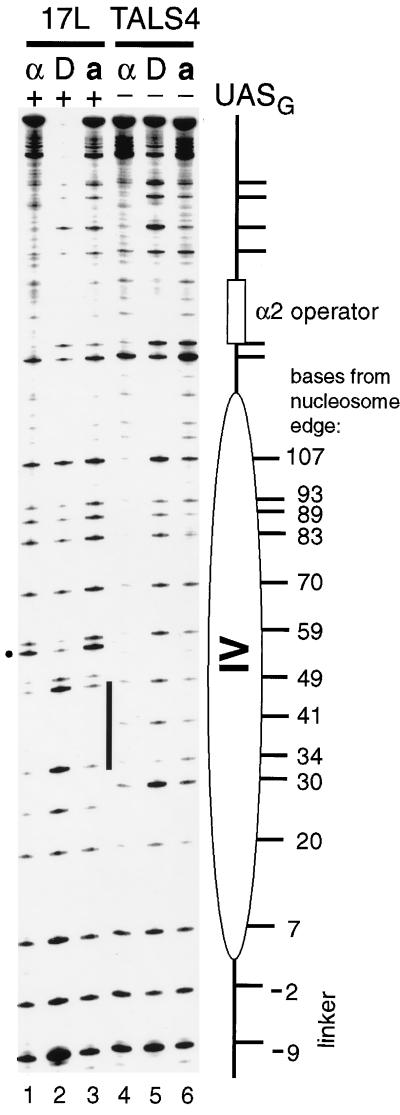
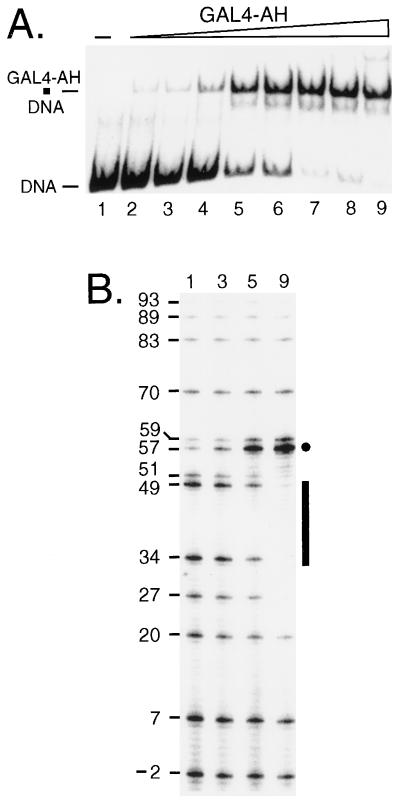
We first analyzed the interaction of the Gal4p derivative Gal4-AH by footprinting with SssI in vitro to provide a benchmark for comparison with in vivo experiments in chromatin (Fig. 3). A 242-bp fragment was amplified by PCR from TALS4-17L DNA (Fig. 1) containing a single UASG. The mobility shift gel of this fragment in the presence of increasing concentrations of Gal4-AH is shown in Fig. 3A. In parallel, identical binding reactions, purified SssI enzyme was added to footprint the interaction of Gal4-AH on protein-free DNA. As more Gal4-AH was added to the binding reactions, a footprint became apparent as evidenced by strong protection of cytosines (sites 34 and 49) within the major groove of each half-site of UASG. In addition, adjacent to UASG, there was protection at sites 27 and 51 and striking enhancement of methylation (sites 57 and 59). Accessibility at other CG sites was relatively unaffected by bound Gal4-AH. A change in the ratio of the modification at the hypermethylated cytosine to that at either of those protected against methylation in each major groove of the binding site could be detected at only 10% occupancy of the probe (Fig. 3, lanes 3). This ratio increased dramatically as additional Gal4-AH was included in the binding reaction. Thus, SssI footprinting should be a sensitive and effective means for detecting Gal4p binding in vivo. As each 17-bp UASG (17L, 17C, and 17R) is flanked by identical nearest-neighbor sequences (i.e., the UASG was contained within a constant 32-bp sequence that was inserted into the original TALS plasmid), the same characteristic protections and enhancements would be expected to occur upon occupancy of UASG in each plasmid.
FIG. 3.
In vitro SssI footprinting of Gal4-AH bound to a single site. (A) Mobility shift gel showing resolution of naked DNA probe containing sequences from m.u. 1267 to 1508 of TALS4-17L (DNA) from the complex of probe bound by Gal4-AH (Gal4-AH · DNA). The concentrations of Gal4-AH monomer added to each sample and the resultant percent occupancies (quantified with a phosphorimager) of the probe are as follows: lane 1, 0 nM; lane 2, 0.21 nM, 7.1%; lane 3, 0.42 nM, 10%; lane 4, 0.85 nM, 17%; lane 5, 1.7 nM, 59%; lane 6, 3.3 nM, 70%; lane 7, 6.6 nM, 90%; lane 8, 13.3 nM, 90%; and lane 9, 27.5 nM, 93%. (B) Duplicate binding reaction mixtures were incubated with purified SssI DNA MTase, and selected reaction mixtures (numerals indicated above gel correspond to samples in panel A) were treated to identify methylated cytosines as described previously (31). The signal intensity of a given band corresponds directly to the amount of methylation at that cytosine. UASG is indicated by the bar, and the location of each CG site in bases from the left edge of the positioned nucleosome in α-cells is given to facilitate comparison to data in Fig. 4, 5, and 9. Protection against SssI of two CG sites, one in each UASG half-site (sites 34 and 49), as well as sites just outside UASG (sites 27 and 51), and an accompanying enhancement of methylation (marked by dot) occur with increasing concentrations of Gal4-AH. As the 17-bp UASG is contained in the same 32-bp sequence within each of the plasmids, these sites can be used to assess protections and enhancement of SssI methylation upon Gal4p binding within chromatin in vivo. The corresponding positions of CG sites from the left edge of nucleosome IV within TALS4-17C and TALS4-17R can be obtained by adding 32 and 64 bp, respectively, to each of the above locations.
To analyze the chromatin structure of each multicopy minichromosome, the TALS4 series of plasmids was transformed into SssI-producing a- and α-cells which also overexpress Gal4p (Fig. 4). All samples were cultured in galactose to induce synthesis of Gal4p and SssI and subsequently processed to identify methylated cytosine residues. TALS4, which lacks a UASG, was methylated by SssI in α-cells in the linker (bottom of gel) between two positioned nucleosomes and the adjacent 20-bp DNA in the nucleosome edge; methylation was inhibited substantially by DNA-histone contacts nearer to the nucleosomal pseudodyad (Fig. 4, compare lanes 2 and 3). This pattern of modification, as demonstrated previously with SssI (31) as well as Dam (29) and Sau3A1 (30) MTases, is diagnostic of a positioned nucleosome. We have further verified by indirect end labeling that nucleosome IV in TALS4 protects against cleavage by MNase (data not shown). Levels of SssI methylation in TALS4 in a-cells (Fig. 4, lane 1), particularly at sites 59 to 107, were significantly greater than those in α-cell chromatin (lane 2) but less than those of naked DNA (lane 3), suggesting the presence of nucleosomes that are positioned less precisely (31, 55, 59). This conclusion is supported further by the reproducible decrease in modification at sites 7 to −9, the linker region in α-cells, indicating that the central, inaccessible region of some nucleosomes occupies these sites in a significant fraction of a-cell minichromosomes.
The patterns of SssI methylation in the region of nucleosome IV were changed to different extents depending on the translational setting of UASG and the cell type (Fig. 4). There are four possible means by which methylation at a particular CG site may be altered in this experiment: protection due to bound Gal4p or histones and increases in methylation either directly due to Gal4p binding (i.e., hypermethylation adjacent to UASG as seen in Fig. 3B) or indirectly through disruption of DNA-histone interactions. First, we consider changes in methylation at CG targets which are more remote from UASG and thus are not likely to be influenced by bound Gal4p (Fig. 3B). In a-cells, where the nucleosomes are less positioned than in α-cells, substantial changes in methylation were seen at several CG sites in TALS4-17L (sites 70 to 107), TALS4-17C (sites 30 to 49 and 107), and TALS4-17R (sites 30 to 83) relative to the control TALS4 plasmid lacking UASG (Fig. 4, compare lanes 4, 7, and 10 to lane 1; for normalization of methylation intensity, it is recommended to use site 7 because it is localized within the accessible periphery of the nucleosome in α-cells and is distant from each UASG). Since Gal4p can bind nucleosomes in vivo (40, 63) (Fig. 2) and SssI detects differences in chromatin structure (e.g., between a- and α-cells [Fig. 4, lanes 1 and 2] [31]), we conclude that in a-cells, overexpressed levels of Gal4p can efficiently gain access to UASG 17L, 17C, and 17R and lead to disruption of chromatin. Increased accessibility to SssI was particularly striking in the region between the α2 operator and UASG, likely reflecting exclusion of nucleosomes from this region with simultaneous binding of Gal4p and Mcm1p (or Mcm1p-Matα2p in α-cells [see below]). Hence, assessment of methylation between the α2 operator and UASG appears to provide the most informative estimate of the degree of Gal4p-mediated chromatin disruption. The term disruption is used only to refer to the apparent changes in DNA-histone contacts sensed by SssI, and no mechanism regarding displacement of histones in trans is implied.
Evaluation of the changes in methylation within and in the vicinity of UASG in a-cells allows an estimate of UASG occupancy. As the 17-bp UASG is embedded within the same 32-nucleotide sequence in each construct, we can compare the methylation in chromatin in each plasmid to that seen in vitro in the presence of bound Gal4-AH (Fig. 3B). In contrast to the increased accessibility of SssI to sites remote from Gal4p, relative to protein-free DNA (Fig. 4, compare lanes 4 and 6, lanes 7 and 9, and lanes 10 and 12), there was clear protection against SssI of the CG in each half-site of UASG (17L, sites 34 and 49; 17C, sites 66 and 81; 17R, sites 98 and 113) and of sites near the binding site (17L, sites 27 and 51; 17C, sites 59 and 83; 17R, sites 91 and 115). Although sites within each UASG are strongly protected in the classical sense of a footprint (comparing methylation in chromatin to that in protein-free DNA), by themselves these data do not formally allow protection due to bound Gal4p versus histones to be ascribed. Nonetheless, given that Gal4p is overexpressed in this experiment and the significant disruption of DNA-histone interactions flanking each UASG, it is likely that most of protection within the binding site can be attributed to Gal4p. In addition, as in naked DNA occupied by Gal4-AH (Fig. 3B), the hypermethylation near UASG was also evident in each construct (Fig. 4) (17L, sites 57 and 59; 17C, site 89; 17R, site 121). Since complete loss of nucleosomes would lead to the methylation levels of naked DNA, it is probable that the pronounced hypermethylation indicates substantial occupancy of UASG by Gal4p in a-cells. In addition, we are confident that the local enhancements in methylation are due to Gal4p binding because (i) they are absent from the control (TALS4) which does not contain UASG, (ii) they are seen when Gal4-AH is bound to DNA in vitro (Fig. 3B), (iii) Gal4p can access nucleosomal DNA in vivo (40, 63) (Fig. 2), and (iv) they vary with the level of expression of Gal4p (i.e., endogenous versus overexpression) (see Fig. 5).
To determine the influence of translational location of UASG in a nucleosome on Gal4p binding and chromatin disruption, we analyzed SssI accessibility of the same series of multicopy plasmids in α-cells in the presence of Gal4p overexpression. Relative to that of TALS4, increases in methylation occurred in TALS4-17L, and to a lesser extent in TALS4-17R, concomitant with protection of UASG and hypermethylation at sites 57 to 59 and 121, respectively, suggesting that Gal4p binding occurred at a site centered 41 bp from either edge of the nucleosome and led to disruption (Fig. 4, compare lanes 5 and 11 to lane 2). Again, as observed above in a-cell chromatin, the most pronounced changes in methylation occurred between UASG and the α2 operator. The absence of such sites in TALS4-17R might contribute to the seemingly lesser disruption compared to that in TALS4-17L; however, the similar extents of methylation of TALS4-17R in a- and α-cells suggests that Gal4p efficiently accessed UASG 17R (Fig. 4, compare lanes 10 and 11). When UASG was inserted at the pseudodyad of nucleosome IV (17C), in α-cell chromatin, methylation was increased relative to that with TALS4 at positions 107, 93, 89, and 30 (Fig. 4, compare lanes 8 and 2), suggesting that Gal4p was able to bind UASG 17C in the presence of a high-copy-number expression plasmid. Modification at these CG sites, however, was reproducibly weaker than that in a-cell chromatin (Fig. 4, compare lanes 8 and 7), suggesting that the UASG 17C was less accessible to Gal4p than UASG 17R or 17L. Reduced hypermethylation at position 89 adjacent to UASG 17C in α-cells (Fig. 4, lane 8) relative to that in a-cells (lane 7) also suggests less binding by Gal4p in the former.
Relation of extent of chromatin disruption to UASG fractional occupancy.
We wanted to investigate further the relative efficacy with which Gal4p can access different positions in a nucleosome (Fig. 5). It should be possible to determine this under conditions in which Gal4p is more limiting, in the absence of the Gal4p expression vector at endogenous or wild-type levels of Gal4p (i.e., TALS4 and its derivatives are multicopy minichromosomes). At this reduced level of Gal4p, in contrast to when Gal4p was overexpressed (Fig. 4), methylation in TALS4-17C in α-cells was indistinguishable from that in TALS4 (Fig. 5A, compare lanes 3 and 5 to lane 4; Fig. 5B, compare scans 10 and 11). Strikingly, methylation in α-cells of TALS4-17L at endogenous levels of Gal4p expression (Fig. 5B, scan 12) was significantly more than that in TALS4 (scan 10) or TALS4-17C at both endogenous (scan 11) and overexpressed (scan 14) levels of the factor. Endogenous levels of Gal4p led to substantial methylation throughout the region of nucleosome IV in both TALS4-17C and TALS4-17L in a-cells (Fig. 5A, compare lanes 7 and 8 to lane 10; Fig. 5B, compare scans 2 and 3 to scan 1), indicating that chromatin in a-cells is significantly more susceptible to disruption than that in α-cells. This susceptibility is also evident in that wild-type levels of Gal4p expression led to more disruption of a-cell chromatin than was observed in TALS4-17C upon overexpression of Gal4p in α-cells (Fig. 5A, compare lanes 7 and 8 to lanes 1 and 2; Fig. 5B, compare scans 2 and 14). Overexpression of Gal4p did not alter methylation in the nucleosome IV region in either cell type when UASG was absent (i.e., in TALS4) (Fig. 5A, compare lane 4 to lanes 3 and 5 and compare lane 9 to lane 10; Fig. 5B, compare scans 4 and 1 and scans 13 and 10). Thus, the SssI methylation assay suggests that the ability of endogenous or overexpressed levels of Gal4p to outcompete histones for DNA occupancy in vivo is affected by the translational position of UASG in a nucleosome and the overall chromatin organization of a region (i.e., a- versus α-cells).
Since the intensity of SssI hypermethylation adjacent to each UASG is a sensitive indicator of Gal4p binding (Fig. 3B), this gave us a unique opportunity to relate the degree of chromatin disruption to the fractional occupancy of UASG in different chromatin contexts. First, in α-cells, there was excellent concordance between the severity of chromatin disruption and the intensity of hypermethylation adjacent to UASG (Fig. 4 and 5; see Fig. 7). For example, hypermethylation at position 89 in TALS4-17C increased with Gal4p overexpression and was UASG dependent (Fig. 5A, compare band marked by dot in lanes 1 and 2 to that in lanes 3 to 5; Fig. 5B, compare peak marked by dot in scan 14 to that in scan 11). Similar findings were also seen at positions 57 and 59 abutting UASG 17L (Fig. 5B, compare scans 15 and 12). In addition, in α-cells at endogenous levels of Gal4p, while hypermethylation was not detectable next to UASG 17C it was apparent at UASG 17L (Fig. 5B, compare peak indicated by dot in scans 11 and 12). Therefore, as the intensity of SssI hypermethylation in α-cells appears to reflect accurately the degree of UASG saturation and the extent of chromatin disruption, we conclude that accessibility of Gal4p at the pseudodyad of a positioned nucleosome is substantially restricted relative to that 41 bp from the nucleosome edge.
In contrast to the differences between hypermethylation at UASG 17C and 17L in α-cells, in a-cells the hypermethylation was greater at each level of Gal4p expression and was observed next to both UASs (Fig. 5A, compare band marked by dot in lanes 3 and 5 to that in lanes 7 and 8 and compare lanes 1 and 2 to lane 6; Fig. 5B, compare peak indicated with dot in scans 2 and 11, 3 and 12, 5 and 14, and 6 and 15). Interestingly, protection of cytosines in each binding site was not readily apparent at UASG 17C and 17L in a-cells at endogenous levels of Gal4p and could be detected only by quantitative scanning with a phosphorimager (Fig. 5A, compare lanes 7 and 8 to lane D; Fig. 5B, compare scans 2 and 8 and scans 3 and 9), suggesting a low level of fractional occupancy by Gal4p. It is intriguing that the apparently low occupancy at wild-type levels of Gal4p is sufficient to lead to extensive chromatin perturbation in a-cells (see Discussion). From the comparison of the scans in Fig. 5B, binding and chromatin remodeling by Gal4p were apparently most effective in the following order: a-cell chromatin > 41 bp from nucleosome edge in α-cell chromatin > nucleosome pseudodyad in α-cell chromatin.
Translational positioning of UASG affects Gal4p-induced gene expression.
The fragment encompassing the α2 operator and region of nucleosome IV from TALS4 and each of its derivatives (Fig. 1) was cloned upstream of the minimal CYC1 promoter driving expression of lacIZ to generate the corresponding series of single-copy YCp plasmids (Fig. 6). In each plasmid, the α2 operator was intentionally placed at a relatively far distance from the promoter in order to abrogate both activation by Mcm1p (46) bound at the operator in a-cells (i.e., in the absence of MATα2p) (Fig. 6, YCpTALS4 data) and repression of RNA polymerase II transcription by MATα2p in α-cells (51). In addition, this excludes all CYC1 TATA elements, which are constitutively bound by TATA-binding protein (12), from the positioned nucleosome. Although the presence of a β-galactosidase reporter was previously shown not to interfere with the positioning of a nucleosome adjacent to the α2 operator in α-cells (46), we confirmed the presence of the nucleosome in YCpTALS4 (no UASG) by SssI (Fig. 7). In α-cells, YCpTALS4 exhibited protection against SssI and a defined linker (compare scans 2 and 1), demonstrating the positioning of nucleosomes adjacent to the operator in a predominant translational setting. In addition, Gal4p occupancy and chromatin disruption required the presence of UASG (Fig. 7, compare scans 2 and 3).
As shown in Fig. 6, compared to the low level of β-galactosidase activity of cells grown in glucose, transcription was stimulated in galactose in all cells harboring a minichromosome containing UASG. This stimulation was clearly Gal4p and UASG dependent, because galactose did not stimulate transcription in cells harboring YCpTALS4 which lacks UASG. In a-cells, Gal4p binding led to high levels of β-galactosidase expression from all three UASG-containing episomes. The increase in basal-level transcription in glucose may result from minor synergism between Mcm1p, bound at the α2 operator, and residual Gal4p binding at UASG. In contrast, when UASG was incorporated 41 bp from the edge of the positioned nucleosome (YCpTALS4-17L and YCpTALS4-17R in α-cells), Gal4p stimulated β-galactosidase expression in galactose to more than 120 U of activity. In contrast, α-cells harboring YCpTALS4-17C showed a much lower level of transcription (26 U). In this and other experiments, the inhibition of activated transcription exerted at the nucleosome center was reproducibly four- to sixfold. The occurrence of two transcriptional maxima (17L and 17R) flanking a minimum (17C) strongly implies that repression of UASG at 17C in α-cells is due to restricted access of Gal4p mediated by a positioned nucleosome. These data rule out the possibility that our results can be attributed to different distances between UASG and either the α2 operator or CYC1 promoter. The four- to sixfold transcriptional repression is expected to be an underestimate of the ability of a positioned nucleosome to exclude Gal4p, because the presence of a functional promoter is known to increase fractional occupancy of a single Gal4p binding site in vivo (see Discussion) (70). Likewise, it is not known whether this repression of Gal4p, which has a strong acidic activation domain, would be improved with a weaker activator.
As the data in Fig. 6 were obtained under conditions used for probing with SssI, where expression of Gal4p was maximal, we hypothesized that reduced levels of Gal4p expression might lead to a greater level of repression at 17C in α-cells. Cells containing each YCp construct were initially grown in glucose-containing medium to repress synthesis of Gal4p. Following transfer to galactose, aliquots of cells were removed and assayed for β-galactosidase activity. At each time assayed, the activity mediated by UASG 17L and 17R (removed from the nucleosomal dyad) was greater than that exerted by UASG 17C located at the dyad (Fig. 8). The inset of Fig. 8 shows that the relative activity of UASG 17C versus those of 17L and 17R was 13.5-fold reduced immediately following the lag in induction (at 3 h) and sequentially diminished to ∼5-fold as observed in Fig. 6 as levels of Gal4p expression increased. This result is consistent with preferential binding of limiting levels of Gal4p to each off-dyad site and subsequent occupancy at the dyad driven by mass action. In conclusion, the functional data correlate well with the structural results: Gal4p stimulated transcription more efficiently when UASG was centered 41 bp from the edge of the positioned nucleosome (α-cells) or when the chromatin was not as precisely organized (a-cells).
Gal4p is capable of invading and reconfiguring a preexisting nucleosome.
Considering that the UASG elements in TALS4-17C and TALS4-17L differ in location by only 32 bp, it seemed unlikely that transient removal of histones by DNA replication would favor Gal4p binding in one minichromosome over the other. To investigate the influence of replication on Gal4p binding, we checked the methylation patterns of TALS4 and TALS4-17L when DNA replication was inhibited by hydroxyurea. Under the experimental conditions used, SssI was not expressed until DNA replication was completely blocked (see Materials and Methods). As shown in Fig. 9, in the presence of hydroxyurea, protection of methylation by nucleosome IV was still evident in TALS4 chromatin in α-cells (compare lanes 4 and 5 and lanes 4 and 6). Significant increases in methylation near the pseudodyad as well as a Gal4p footprint were observed in TALS4-17L chromatin in both cell types (Fig. 9, compare lanes 1 and 4 and lanes 3 and 6). Therefore, without prior removal of histones by DNA replication, Gal4p can directly access UASG when located 41 bp from the edge of a preexisting nucleosome and can effect destabilization or disruption.
DISCUSSION
We have utilized derivatives of TRP1ARS1 minichromosomes, which have a well-characterized chromatin structure, to assess the structural and functional consequences of introducing cis-acting elements at various locations in positioned nucleosomes (40, 42, 61, 63). In any such study in vivo, it is important to dissociate structural changes which precede transcription from those which are consequences of transcription. In our study, transcription should not affect the structural results. While the TRP1 gene is expressed, the level of expression from the 102 bp of the 5′ flanking sequence is 3 to 4% of that from the wild-type promoter (16). This absence of a natural promoter within these minichromosomes permits study of the interaction of a single transcription factor in chromatin with a greatly reduced, if not eliminated, influence of the basal transcriptional machinery. In the present study, we have introduced a single, near-consensus Gal4p binding site, UASG, at various locations in the region incorporated within a nucleosome positioned by Mcm1p-Matα2p, allowing us to assess Gal4p accessibility in two different, cell-type-specific chromatin environments (55, 59).
Location of UASG within a nucleosome affects occupancy by Gal4p in vivo.
Evaluation of the SssI footprinting and reporter expression studies leads us to conclude that, in vivo, relative to binding at the pseudodyad, Gal4p disrupts a precisely positioned nucleosome (in α-cells) and activates transcription more efficiently when UASG is centered 41 bp from a nucleosome edge. Thus, as is the case for SssI MTase (Fig. 4 and 5) (31), Dam MTase (29), and the DNA replication machinery (61), the accessibility in vivo of Gal4p to DNA in a positioned nucleosome is greater at translational positions removed from the nucleosomal pseudodyad. In vitro, binding of the Gal4p derivative Gal4-AH (72), elongation of transcription by bacteriophage SP6 RNA polymerase (64), and digestion by restriction endonucleases (49) also occur more readily within the two helical turns entering and exiting a nucleosome, a region where DNA-histone interactions are less stable than those at the pseudodyad (34, 41, 60, 75). Accessibility to sites more internal was compromised. While it is likely that the increased repression further into the nucleosome is primarily due to more robust DNA-histone interactions, it is also possible that Gal4p binding is impaired by underwinding of the DNA helix and/or steric occlusion of UASG by the neighboring gyres of DNA at the pseudodyad (24, 25). Thus, our data provide evidence that the general physical property of increased accessibility further from the pseudodyad is retained in vivo and translates to functional consequences in gene expression.
What is the mechanism by which Gal4p exhibits increased accessibility at the periphery of nucleosomes? It is possible that transient disruption of DNA-histone contacts near the edges of nucleosomes, a “nucleosome breathing,” exposes sites for protein binding. According to one model (49, 50), disruption of DNA-histone contacts initiates at nucleosome termini and proceeds sequentially into the nucleosome, leading to dissociation of the DNA helix from the histone octamer surface. This dynamic structure of nucleosomes would obviate the need for replication to precede factor binding and predicts less occupancy of a target sequence when it is located at the pseudodyad versus the nucleosome periphery, as we have observed. It should be noted, however, that in these previous studies, the region of relatively increased accessibility to restriction endonucleases in nucleosome cores was confined to the terminal 20 to 25 bp of DNA. For example, the rate of cleavage at a site 7 to 11 bp from the end of nucleosome cores decreased at least 10-fold when the site was 43 to 52 bp from the end (49). In vitro, Gal4-AH also binds preferentially 7 to 21 bp from the edge of nucleosome cores, but binding was very poor when UASG was centered at 26 to 40 or 60 to 74 bp from the nucleosome core edge (the 14-bp uncertainty is due to potential heterogeneity of the position of the reconstituted nucleosome on the probe DNA [72]). Thus, these in vitro studies demonstrate that, energetically, the cumulative disruption of DNA-histone contacts beyond approximately 25 bp into the nucleosome is very unfavorable.
Accessibility of Gal4p to nucleosomes is enhanced in vivo relative to in vitro.
In contrast to these in vitro results, in vivo we have observed more occupancy of single Gal4p sites that were localized 33 to 49 bp internal (i.e., centered 41 bp from the edge) to a positioned nucleosome than of one that was positioned at the pseudodyad. Morse and colleagues have reported disruption of a nucleosome in vivo in a TRP1ARS1 derivative containing UASG near the pseudodyad (40) and in TALS (63), which has a natural Gal4p binding site centered at 41 bp from the nucleosome edge. A comparison of the degrees of chromatin disruption and relative occupancies of these binding sites was not made. In any case, Gal4p has the capability to access a nucleosome in vivo in a region that is quite refractory to binding in nucleosome cores in vitro.
The enhanced ability of Gal4p to bind at sites more internal in the nucleosome in vivo compared to in vitro may be due to a difference from Gal4-AH, altered nucleosome structure (e.g., nucleosome cores versus minichromosomes [19, 47]), an influence of DNA replication, or the presence of nucleosome modification activities, etc. In comparison to that of Gal4p in vivo, several observations make it unlikely that the decreased capability of Gal4-AH to access nucleosomes in vitro can be ascribed simply to a trivial deficiency. First, since the DNA-binding and dimerization domains of Gal4p are contained within amino acids 1 to 94, all GAL4 derivatives containing these residues and wild-type Gal4p, purified from yeast as a Gal80p complex, exhibit similar specific affinities for protein-free DNA (10, 45, 68, 78). Second, activation domains are not required for the binding of GAL4 derivatives to nucleosomes or for displacement of histones after binding in vitro (45, 68), making it doubtful that wild-type Gal4p would behave differently in such assays. Third, as mentioned above, in addition to that of Gal4-AH, the accessibility of several different proteins to nucleosome cores is similarly restricted to nucleosome termini (29, 31, 49, 61, 64, 72). Interestingly, the enhanced ability of Gal4p to bind nucleosomes in cells also does not appear to be solely due to dissimilar nucleosome structures in vivo as opposed to in vitro; in vivo accessibility of both the Dam and SssI MTases is also restricted beyond 20 to 29 bp (i.e., sites at ≥30 bp internal are protected from methylation) into a positioned nucleosome in both replicating (29, 31) (Fig. 4, lane 2; Fig. 5A, lane 4) and nonreplicating (Fig. 9, lane 4) cells. These observations suggest that the prokaryotic enzymatic probes, foreign to the environment of chromatin in S. cerevisiae, reflect the state of accessibility of a particular region, whereas Gal4p has additional means for invading chromatin structure.
Disruption of chromatin architecture accompanying passage of the DNA replication fork could provide a greatly enhanced opportunity for activators to access regulatory sites, with the outcome being disruption of nucleosomes through the exclusion of the core histones (6, 74, 77). It has been suggested that this is a primary mechanism by which the majority of factors access their binding sites in chromatin (65). On the other hand, replication-independent activation of transcription by Gal4p derivatives has been seen in vitro in templates preassembled into chromatin by Drosophila or Xenopus oocyte extracts (11, 48). While these experiments utilized arrays of physiologically spaced nucleosomes (5), the arrays in the population are heterogeneous in that they are not precisely positioned with respect to the underlying sequence. Since ∼40% of the DNA in any given molecule would be accessible to an activator due to localization of a single binding site in an internucleosomal linker or in the periphery of a nucleosome, an accurate test of the effect of nucleosome positioning in this system is precluded. This is a conservative estimate of the percentage of accessible DNA, as multiple factor binding sites (48) are usually employed and most extracts contain ATP-dependent remodeling activities (48, 69) which may facilitate factor binding in chromatin.
In some instances, particularly at genes that rapidly respond to environmental signals, chromatin reconfiguration and transcriptional activation in vivo are replication independent (2, 52, 57). It is possible that this replication-independent activation of these genes is accomplished through cooperativity between the multiple bound factors and/or the basal transcription machinery (18, 44, 70, 79). At the PHO5 promoter, for example, a low-affinity binding site for Pho4p is located between two positioned nucleosomes, one of which (nucleosome −2) contains additional sites for Pho4p and Pho2p (17, 71). During induction in low-phosphate medium, these factors bind their sites and reconfigure chromatin in the absence of replication (57). Consistent with these studies, we find that the trans-activator Gal4p is also able to effect chromatin disruption in nonreplicating cells (Fig. 9). In studying binding to a single UASG, in the absence of a known promoter, our data extend previous observations indicating that a single Gal4p dimer does not require DNA replication to facilitate binding in chromatin. This suggests a two-step process in which, following passage of the DNA replication fork, chromatin assembly precedes Gal4p binding (and methylation by SssI) and subsequently the activator invades and disrupts preformed nucleosomes. Such a model is consistent with the ability of Gal4p derivatives to form a ternary complex with nucleosomes containing a single binding site in vitro (15, 72) and, furthermore, is in agreement with investigations which have revealed that an active chromatin configuration is not inherited directly but must be continually reestablished following each successive round of replication (36).
Binding of Gal4p to a preformed nucleosome in vivo is striking because the affinity of the activator for a single site in reconstituted chromatin is approximately 100-fold lower than that in DNA (68). Our data obtained from a-cells expressing wild-type levels of Gal4p may give an indication of how the activator overcomes this impediment in vivo. In this situation, although only a partial occupancy of UASG by Gal4p was observed, disruption of the chromatin structure was apparently fairly extensive (Fig. 5). The simplest interpretation of this phenomenon is that transient binding of Gal4p leads to recruitment of a nucleosome modification-remodeling activity which irreversibly disrupts DNA-histone contacts. For example, the activation domain of Gal4p interacts in vitro with Ada2p (38), which is part of a larger yeast complex that contains the histone acetyltransferase, Gcn5p (7, 8). A mechanism in which a recruited nucleosome-remodeling complex is only transiently required to reconfigure chromatin can also be envisaged (43). However, despite the presence of numerous histone modification and nucleosome disruption activities within the cell (28), moving UASG to the pseudodyad significantly inhibited disruption and transcriptional activation.
In conclusion, our data suggest a role for nucleosome positioning in modulating DNA occupancy of specific upstream activators in addition to factors within the basal transcription machinery (32, 37, 79). Our results also indicate that DNA occupancy is a primary element in effecting chromatin disruption and fine-tuning levels of gene expression (9, 67, 70, 80). While we have not determined the exact nature of the Gal4p-mediated nucleosome reconfiguration, our data clearly demonstrate that the activator substantially perturbs DNA-histone contacts in intact cells without requiring DNA replication. Furthermore, our demonstration that the surface along a nucleosome is not uniformly repressive implicates another level of regulation in factor binding that can be utilized to achieve appropriate physiological responses.
ACKNOWLEDGMENTS
We are particularly grateful to Randy Morse for many valuable discussions throughout the course of this study, for editing an earlier version of the manuscript, and for communicating results before publication. We also thank H.-G. Patterton, J. L. Workman, and other members of the Simpson and Workman laboratories for many helpful suggestions, Randy Morse for providing pRS426-GAL4, and Tom Owen-Hughes and Jerry Workman for supplying the purified Gal4-AH.
This work was supported by NIH grant GM52908 awarded to R.T.S.
REFERENCES
- 1.Almer A, Rudolph H, Hinner A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer T K, Cordingley M G, Marsaud V, Richard-Foy H, Hager G L. Steroid transactivation at a promoter organized in a specifically-positioned array of nucleosomes. In: Gustafsson J A, Eriksson H, Carlstedt-Duke J, editors. Steroid/thyroid hormone receptor family and gene regulation. Berlin, Germany: Berkhauser Verlag, AG; 1989. pp. 221–238. [Google Scholar]
- 3.Bajwa W, Torchia T E, Hopper J E. Yeast regulatory gene GAL3: carbon regulation; UASGal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol Cell Biol. 1988;8:3439–3447. doi: 10.1128/mcb.8.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton M C, Emerson B M. Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev. 1994;8:2453–2465. doi: 10.1101/gad.8.20.2453. [DOI] [PubMed] [Google Scholar]
- 5.Becker P B, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown D D. The role of stable complexes that repress and activate eukaryotic genes. Cell. 1984;37:359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Zhou J X, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Bunker C A, Kingston R E. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc Natl Acad Sci USA. 1996;93:10820–10825. doi: 10.1073/pnas.93.20.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey M, Kakidani H, Leatherwood J, Mostashari R, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 11.Chang C B, Gralla J D. A critical role for chromatin in mounting a synergistic transcriptional response to GAL4-VP16. Mol Cell Biol. 1994;14:5175–5181. doi: 10.1128/mcb.14.8.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J J, Ding M, Pederson D S. Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences. Proc Natl Acad Sci USA. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 14.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 16.Dobson M J, Tuite M F, Mellor J, Roberts N A, King R M, Burke D C, Kingsman A J, Kingsman S M. Expression in Saccharomyces cerevisiae of human interferon-alpha directed by the TRP1 5′ region. Nucleic Acids Res. 1983;11:2287–2302. doi: 10.1093/nar/11.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fascher K D, Schmitz J, Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher T M, Hansen J C. The nucleosomal array: structure/function relationships. Crit Rev Eukaryot Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 20.Frommer M, MacDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganter B, Tan S, Richmond T J. Genomic footprinting of the promoter regions of STE2 and STE3 genes in the yeast Saccharomyces cerevisiae. J Mol Biol. 1993;234:975–987. doi: 10.1006/jmbi.1993.1652. [DOI] [PubMed] [Google Scholar]
- 22.Giniger E, Varnum S M, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell L H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- 24.Hayes J J, Tullius T D, Wolffe A P. The structure of DNA in a nucleosome. Proc Natl Acad Sci USA. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes J J, Wolffe A P. The interaction of transcription factors with nucleosomal DNA. Bioessays. 1992;14:597–603. doi: 10.1002/bies.950140905. [DOI] [PubMed] [Google Scholar]
- 26.Higuchi R, Krummel B, Saiki R K. A general method for in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase-II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 28.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 29.Kladde M P, Simpson R T. Positioned nucleosomes inhibit dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1360–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kladde M P, Simpson R T. Chromatin structure mapping in vivo using methyltransferases. Methods Enzymol. 1996;274:214–233. doi: 10.1016/s0076-6879(96)74019-3. [DOI] [PubMed] [Google Scholar]
- 31.Kladde M P, Xu M, Simpson R T. Direct probing of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 1996;15:6190–6200. [PMC free article] [PubMed] [Google Scholar]
- 32.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1988;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 33.Laughon A, Gesteland R F. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci USA. 1982;79:6827–6831. doi: 10.1073/pnas.79.22.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K P, Baxter H J, Guillemette J G, Lawford H G, Lewis P N. Structural studies on yeast nucleosomes. Can J Biochem. 1982;60:379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y-S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vivo. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 36.Lucchini R, Sogo J M. Replication of transcriptionally active chromatin. Nature. 1995;374:276–280. doi: 10.1038/374276a0. [DOI] [PubMed] [Google Scholar]
- 37.Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987;7:1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melcher K, Johnston S A. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Morse R H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1562–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 41.Morse R H, Pederson D S, Dean A, Simpson R T. Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res. 1987;15:10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morse R H, Roth S Y, Simpson R T. A transcriptionally active transfer RNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol. 1992;12:4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen-Hughes T, Utley R T, Côté J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 44.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 45.Parthun M, Jaehning J A. Purification and characterization of the yeast transcriptional activator GAL4. J Biol Chem. 1990;265:209–213. [PubMed] [Google Scholar]
- 46.Patterton H G, Simpson R T. Nucleosomal location of the STE6 TATA-box- and Matα2p-mediated repression. Mol Cell Biol. 1994;14:4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 48.Pazin M J, Kamakaka R T, Kadonaga J T. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- 49.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 50.Polach K J, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 51.Redd M J, Stark M R, Johnson A D. Accessibility of α2-repressed promoters to the activator Gal4. Mol Cell Biol. 1996;16:2865–2869. doi: 10.1128/mcb.16.6.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reik A, Schütz G, Stewart A F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991;10:2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richard-Foy H, Hager G L. Sequence specific positioning of nucleosomes over the steroid inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 55.Roth S Y, Dean A, Simpson R T. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Stable nucleosome positioning and complete repression by the yeast α2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 57.Schmid A, Fascher K D, Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 58.Sheen J-Y, Seed B. Electrolyte gradient gels for DNA sequencing. BioTechniques. 1988;6:942–944. [PubMed] [Google Scholar]
- 59.Shimizu M, Roth S Y, Szent-Gyorgyi C, Simpson R T. Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simpson R T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979;254:10123–10127. [PubMed] [Google Scholar]
- 61.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 62.Slater M. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stafford G A, Morse R H. Chromatin remodeling by transcriptional activation domains in a yeast episome. J Biol Chem. 1997;272:11526–11534. doi: 10.1074/jbc.272.17.11526. [DOI] [PubMed] [Google Scholar]
- 64.Studitsky V M, Clark D J, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 65.Svaren J, Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990;6:52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- 66.Szent-Gyorgyi C. A bipartite operator interacts with a heat shock element to mediate early meiotic induction of Saccharomyces cerevisiae HSP82. Mol Cell Biol. 1995;15:6754–6769. doi: 10.1128/mcb.15.12.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka M. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor I C A, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 69.Varga-Weisz P D, Blank T A, Becker P B. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vashee S, Kodadek T. The activation domain of GAL4 protein mediates cooperative promoter binding with general transcription factors in vivo. Proc Natl Acad Sci USA. 1995;92:10683–10687. doi: 10.1073/pnas.92.23.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vettese-Dadey M, Walter P, Chen H, Juan L J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, Gal4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wechser M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weintraub H. Assembly and propagation of repressed and derepressed chromosomal states. Cell. 1985;42:705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- 75.Weischet W O, Tatchell K, van Holde K E, Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978;5:139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiss K, Simpson R T. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolffe A P. The role of transcription factors, chromatin structure and DNA replication in 5S RNA gene regulation. J Cell Sci. 1994;107:2055–2063. doi: 10.1242/jcs.107.8.2055. [DOI] [PubMed] [Google Scholar]
- 78.Workman J L, Kingston R E. Nucleosome core displacement in vitro via a metastable transcription factor nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 79.Workman J L, Roeder R G, Kingston R E. An upstream transcription factor, USF (MLTF), facilitates the formation of preinitiation complexes during in vitro chromatin assembly. EMBO J. 1990;9:1299–1308. doi: 10.1002/j.1460-2075.1990.tb08239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu H E, Kodadek T, Johnston S A. A single GAL4 dimer can maximally activate transcription under physiological conditions. Proc Natl Acad Sci USA. 1995;92:7677–7680. doi: 10.1073/pnas.92.17.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]