Abstract
Aphids and related insects feed from a single cell type in plants: the phloem sieve element. Genetic resistance to Acyrthosiphon kondoi Shinji (bluegreen aphid or blue alfalfa aphid) has been identified in Medicago truncatula Gaert. (barrel medic) and backcrossed into susceptible cultivars. The status of M. truncatula as a model legume allows an in-depth study of defense against this aphid at physiological, biochemical, and molecular levels. In this study, two closely related resistant and susceptible genotypes were used to characterize the aphid-resistance phenotype. Resistance conditions antixenosis since migratory aphids were deterred from settling on resistant plants within 6 h of release, preferring to settle on susceptible plants. Analysis of feeding behavior revealed the trait affects A. kondoi at the level of the phloem sieve element. Aphid reproduction on excised shoots demonstrated that resistance requires an intact plant. Antibiosis against A. kondoi is enhanced by prior infestation, indicating induction of this phloem-specific defense. Resistance segregates as a single dominant gene, AKR (Acyrthosiphon kondoi resistance), in two mapping populations, which have been used to map the locus to a region flanked by resistance gene analogs predicted to encode the CC-NBS-LRR subfamily of resistance proteins. This work provides the basis for future molecular analysis of defense against phloem parasitism in a plant model system.
Parasitism by phloem-feeding insects, such as aphids and whiteflies, is a widespread and often serious constraint on plant production. Aphids have been especially successful in exploiting a broad range of vascular plants. In temperate regions, approximately one in four plant species can be colonized by at least one species of aphid (Dixon, 1998). Phloem feeders may harm plants by direct feeding damage and by vectoring microbial pathogens. These insects are exquisitely adapted to their hosts, feeding from a single cell type, the sieve element, at the plant interior. This cell-specific mode of herbivory presents both a technical challenge and an opportunity for plant biologists to elucidate ways in which plants defend against parasitism of the translocation stream.
Despite the ubiquity of phloem feeding, basic knowledge of its relation to plant physiology and, in particular, to plant defense has lagged behind knowledge of plant-microbe interactions. This imbalance is starting to change, however, as molecular tools are applied to the study of induced responses to phloem feeding and to mechanisms of genetic resistance (for review, see Walling, 2000; Kessler and Baldwin, 2002; Moran et al., 2002). Studies with Arabidopsis (Arabidopsis thaliana) and cultivated species have identified changes in gene expression when plants are challenged with phloem feeders (Walling, 2000; Moran and Thompson, 2001; Moran et al., 2002; de Ilarduya et al., 2003; Zhu-Salzman et al., 2004). Considering the intimate and enduring contact of insect stylets with the host tissue, it is not surprising that these and other studies have found remarkable similarities between plant responses to phloem feeders and pathogens. As with pathogens, it is difficult to distinguish factors that are specific and proximate defenses against insects from those that merely coincide with a general immune or stress response.
Understanding of the molecular basis of resistance to phloem feeding was greatly advanced with the cloning and characterization of the Mi gene from tomato (Lycopersicon esculentum), which confers resistance to root-knot nematodes (Meloidogyne spp.), potato aphid (Macrosiphum euphorbiae), and sweetpotato whitefly biotypes B and Q (Bemisia tabaci; Milligan et al., 1998; Rossi et al., 1998; Vos et al., 1998; Nombela et al., 2003). The gene encodes a classical resistance (R) protein homologous to proteins conferring resistance against viruses, bacteria, and fungi (Milligan et al., 1998). Arabidopsis has been used to make impressive inroads toward the molecular dissection of R gene-mediated pathogen resistance (Holt et al., 2003). Unfortunately, naturally occurring and simply inherited aphid resistance has not been reported in Arabidopsis.
Medicago truncatula Gaert. (barrel medic), an annual pasture species of economic importance in Australia, has attained the status of a model legume. Resources for M. truncatula are well developed, including expressed sequence tag (EST) databases, genetic and physical maps, and a genome sequencing project (http://medicago.org/genome/). Since legumes comprise a major portion of the world's agricultural systems, the study of resistance to phloem feeding in a model legume could have important ramifications in a broad range of crop settings.
Acyrthosiphon kondoi Shinji (bluegreen aphid or blue alfalfa aphid) is an important pest of pasture legumes, particularly Medicago spp. such as alfalfa/lucerne (Medicago sativa; Blackman and Eastop, 1984). Breeders at the South Australian Research and Development Institute (SARDI) screened a large collection of germplasm from around the world and identified two M. truncatula accessions, SA1499 and SA10419, with resistance to A. kondoi. These accessions were used as donor parents in a backcrossing program conducted at the institute for the introgression of A. kondoi resistance into widely grown M. truncatula cultivars (Crawford et al., 1989). For example, resistance in SA1499 was repeatedly backcrossed into susceptible cultivar Jemalong to create aphid-resistant Jester (Hill, 2000). Aphid resistance and susceptibility in two closely related cultivars, such as Jester and Jemalong, provides the opportunity to identify the mechanism by which one of A. kondoi's many host species defends itself against attack.
One significant advantage of this system is that a derivative of Jemalong, genotype A17, has been adopted as a reference genotype by M. truncatula researchers worldwide. The genome of A17 is being sequenced, and most EST libraries and a large collection of molecular markers were generated from this genotype. These resources facilitate the molecular-genetic analysis of aphid resistance in the A17 genetic background. This paper reports a study of the aphid-resistance phenotype and its genetic control in Jemalong/A17's closely related line Jester. The trait is characterized at multiple levels including field performance and feeding behavior from single cells. Results show that A. kondoi resistance exerts its effect at the level of the phloem sieve element. The trait is conditioned by a single dominant gene flanked by classical-resistance gene analogs. These results lay the groundwork for extensive molecular and biochemical elucidation of this agriculturally important trait in a well-developed plant model system.
RESULTS
Jester Is Resistant to Aphids in the Field
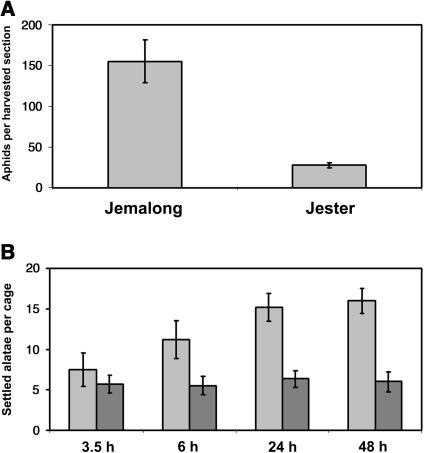
A. kondoi resistance was identified by South Australian plant breeders in M. truncatula accession SA1499 and backcrossed into cv Jemalong to create the closely related, aphid-resistant cv Jester (Hill, 2000). We confirmed and quantified a high degree of A. kondoi resistance in the field in Jester compared to the susceptible cv Jemalong. A. kondoi were first observed on plots in mid-July, 1 month after sowing. By 8 weeks after sowing, when plants were sampled for aphid numbers, a pronounced difference in plant damage was observed between Jester and Jemalong, with mean damage scores of 2.3 and 9.8, respectively. Damage assessments were based on symptoms including shortened internodes, small and deformed leaves, and leaf chlorosis. No signs of a hypersensitive response (HR) appeared on resistant line Jester. More than 95% of aphids on stem sections were identified as A. kondoi. The average number of A. kondoi per stem was significantly lower on Jester than on Jemalong by 5.6-fold (Fig. 1A; F1,6 = 61; P = 0.0002). Because the damage on Jemalong plants was so severe, it is likely that aphid numbers were limited by the deterioration of host quality; hence, aphid numbers alone do not reflect the actual severity of the infestation on this susceptible cultivar. These results indicate that, by late in the growing season, the A. kondoi resistance trait in Jester had prevented the substantial colonization and plant damage observed in Jemalong.
Figure 1.
Resistance phenotype in Jester as measured by A. kondoi performance in the field (A) and settling of A. kondoi alatae in a choice-test conducted in a growth chamber (B). Each bar represents the mean of four replicates in A and six replicates in B. Error bars represent ±1 se. In B, light gray bars, A17; dark gray bars, Jester.
Jester Resists Stunting and Leaf Damage by Aphids under Controlled Conditions
The field results prompted a closer analysis of resistance to feeding damage in Jester, in which aphids could choose between hosts in a controlled environment. Following 5 weeks of infestation by A. kondoi, the fresh weight of above-ground tissue was higher on potted Jester than A17 by 6.8-fold: 7.5 g ± 0.5 g se for Jester compared with 1.1 g ± 0.2 g se for A17 (F1,20 = 161; P < 0.0001). Jester had an average of 10 pods/plant, while no pods were found on any plants of A17. All infested A17 plants had white, necrotic patches of 1- to 2-mm diameter on many of their trifoliate leaves, often surrounded by a ring of red pigment extending approximately 0.5 mm from the patch. The petioles of these damaged leaves were often sharply bent with a darkened area at the bend. These small patches of necrotic tissue appeared similar to HR symptoms in response to pathogens. Some infested leaves of A17 were also chlorotic and deformed. Interestingly, no HR-like flecks, chlorosis, or deformation were observed on any leaves of resistant line Jester, even though aphids had colonized all Jester plants.
Alatae Prefer Susceptible Line A17 over Resistant Line Jester
Observation of host choice by alatae (the winged, migratory morph) can reveal clues to mechanisms of aphid resistance, such as whether antixenotic (deterrent) factors are present and the speed with which they influence behavior of a foraging aphid. In the host-choice test, alatae quickly dispersed from the point of release, and most flew to the tops of cages before settling on a plant. The average number of settled alatae remained relatively constant on Jester plants over the 48 h of observation, while the average number on A17 increased (Fig. 1B). Pooled chi-square tests indicate that alatae showed no significant preference between genotypes at 3.5 h after release (χ2 = 1.3; P = 0.26; degrees of freedom [df] = 1), while alatae at each subsequent time point showed a highly significant preference for susceptible line A17 (χ2 > 10; P < 0.001 at 6, 24, and 48 h). This choice test was repeated in a glasshouse, where similar results were obtained (data not shown).
Resistance in Jester Is Phloem Specific
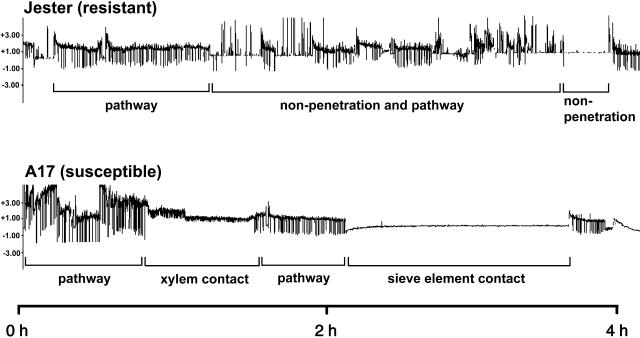
The electrical penetration graph (EPG) method is a powerful means of discerning, in real time, the locations and activities of aphid stylets during probing, including their salivation into sieve elements and passive uptake of phloem sap (Walker, 2000). Representative EPG traces produced by A. kondoi probing A17 and Jester are shown in Figure 2. We tested for the possibility that resistance in Jester is enhanced by prior infestation by comparing probing activities of single aphids on both genotypes, with and without a preinfestation treatment. The proportions of time that tethered apterae spent outside the cuticle (nonpenetration), penetrating between cells en route to the vascular tissue (pathway phase), contacting xylem, salivating into phloem sieve elements, or briefly puncturing cells (of unknown cell types) did not differ significantly between A17 and Jester, for either infestation treatment, as measured by Kruskal-Wallis tests (Table I). Transitions between most of these activities represent the process by which aphids penetrate the plant cuticle, navigate to the phloem, and prepare to ingest sap from sieve elements. Aphids spent the same proportion of time, on average, negotiating through leaf peripheral tissues to the phloem of both genotypes, regardless of whether plants had been previously infested. The similarities between the behavior of the aphids for these activity durations suggest that neither surface features (e.g. epicuticular waxes or trichomes) nor cell wall properties play a role in Jester's resistance mechanism.
Figure 2.
EPG showing representative waveform patterns produced when A. kondoi apterae feed on resistant line Jester (top) or susceptible line A17 (bottom). Plants used for these traces had not been preinfested with A. kondoi. The horizontal axis represents a 4-h time period; the vertical axes represent voltage. Histological studies of plant-aphid interactions have correlated stylet positions in plant tissues with specific EPG waveforms (Walker, 2000). “Sieve element contact,” consisting primarily of sap ingestion with short periods of salivation into sieve elements, was frequently seen with plants of A17 and only rarely seen with plants of Jester. “Pathway” indicates mostly intramural probing activities between mesophyll or parenchyma cells. Sharp, downward spikes indicate cell puncture events by stylets, each lasting approximately 5 s. “Xylem contact” indicates stylet penetration of tracheary elements. “Nonpenetration” indicates stylets are outside the plant.
Table I.
Feeding activities of aphids on control or previously infested plants
Numbers indicate the percentage of time aphids spent in various activities on A17 or Jester during 16-h exposure to the host plants. Numbers in parentheses are ses.
| Feeding Activity
|
A17
|
Jester
|
||
|---|---|---|---|---|
| Control n = 10 | Preinfested n = 9 | Control n = 10 | Preinfested n = 8 | |
| Nonpenetration | 37.8 (8.9) | 38.1 (7.8) | 58.3 (8.4) | 51.0 (11.0) |
| Pathway phase | 25.0 (4.6) | 22.5 (4.2) | 17.7 (2.6) | 31.2 (8.0) |
| Cell puncture | 3.8 (0.7) | 4.0 (0.9) | 2.2 (0.3) | 2.6 (0.6) |
| Sieve element salivation | 3.0 (1.5) | 3.7 (1.7) | 1.5 (0.8) | 0.8 (0.4) |
| Phloem sap ingestiona | 22.6 (7.9) | 18.3 (7.1) | 11.8 (7.4) | 0.04 (0.04) |
| Xylem contact | 7.8 (1.8) | 13.3 (2.6) | 8.5 (3.1) | 14.3 (5.8) |
A significant difference exists for this activity among the four genotype-treatment combinations (P < 0.05).
In contrast to these preingestion activities, the proportion of time aphids spent ingesting phloem sap (E2 phase) was dramatically reduced for aphids on previously infested Jester plants (Table I). Sap ingestion occupied an average of less than 0.05% of total recorded activity on preinfested Jester plants. The mean duration spent on individual bouts of phloem ingestion was also reduced on these plants, lasting an average of only 12 s compared to at least 3,000 s for the other genotype-treatment combinations (data not shown). The disparity between the preinfested Jester treatment and the other genotype-infestation combinations was likely the cause of a significant difference in sap ingestion among the four genotype-treatment combinations (Table I; H = 8.0; df = 3; P = 0.046). There appeared to be a trend toward less sap ingestion in both types of Jester plants compared to both types of A17 plants, although unequal variances and unequal sample sizes prevented statistical comparisons between each genotype-treatment combination. Only 2 of 8 aphids on previously infested Jester plants registered any bouts of phloem ingestion during the experiment, one lasting 85 s and the other only 8 s. The absence of significant differences in the proportions of time spent in feeding-related activities, outside of phloem ingestion, indicates the resistance mechanism in Jester exerts a major effect on A. kondoi at the level of the phloem sieve element. Moreover, the results suggest resistance in Jester is enhanced by prior aphid infestation.
The following are the proportions of aphids that achieved at least one bout of sap ingestion for each of the treatments: 8 of 10 on nonpreinfested A17; 7 of 9 on preinfested A17; 4 of 10 on nonpreinfested Jester; and 2 of 8 on preinfested Jester. These figures reveal a significant effect of genotype-treatment combination on the probability of ingesting phloem sap during the experiment, with a trend toward reduced sap ingestion on Jester plants (χ2 = 8.3; df = 3; P = 0.041). In general, aphids were less likely to ingest sap from Jester than A17, regardless of infestation treatment (χ2 = 7.8; df = 1; P = 0.0051). These results offer additional support for phloem-specific resistance in Jester, suggesting the trait is due, at least in part, to reduced sap ingestion from sieve elements. The results also suggest the antixenotic effect of resistance, observed in the choice test, may derive from the inhibition of sap uptake by alatae as they probe alternative host plants.
Prior Infestation Reduces Aphid Performance on Jester
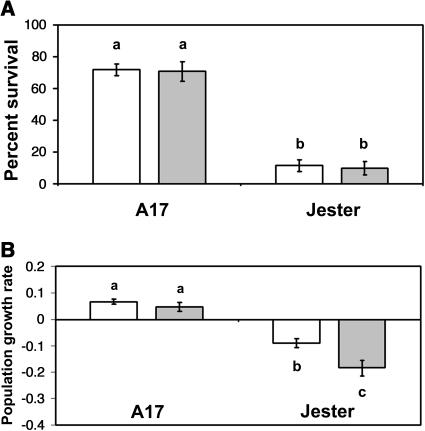
The reduced sap ingestion caused by prior infestation suggests this induced defense has an effect on aphid performance. To determine whether aphid feeding causes systemic effects on colony development, we compared aphid survival and population growth rate (PGR) either with or without prior infestation of A17 and Jester. By 4 d after infestation the resistance trait in Jester caused a 7-fold reduction in survival of A. kondoi nymphs compared to A17 (Fig. 3A; F1,53 = 173; P < 0.001). Prior infestation with aphids had no significant effect on survival relative to the no-aphid control treatment, for either genotype, over this 4-d period (F1,53 = 0.07; P = 0.80). On A17 prior infestation did not affect PGR; in contrast, PGR of aphids on Jester was significantly reduced by prior infestation (Fig. 3B; F1,37 = 124; P < 0.001). The reduction was greater for preinfested Jester plants, leading to a significant genotype-by-treatment interaction (F1,37 = 5.43; P = 0.026). The negative values of PGR on Jester plants reflect both decreased survival and slower development of surviving aphids during 4 d of confinement on these plants. The significant difference in PGR between naïve and preinfested Jester plants indicates that resistance in Jester involves a systemic reduction of host suitability for A. kondoi.
Figure 3.
Effects of plant genotype and prior infestation on A. kondoi survival (A) and population growth rate (B). White bars, no prior infestation; gray bars, prior infestation. Population growth rate in B was measured as (log[mg]aphid−1) d−1. Bars represent means of 14 replicates. Error bars represent ±1 se. Means labeled with the same letter are not significantly different (P < 0.05).
Resistance in Jester Requires an Intact Plant
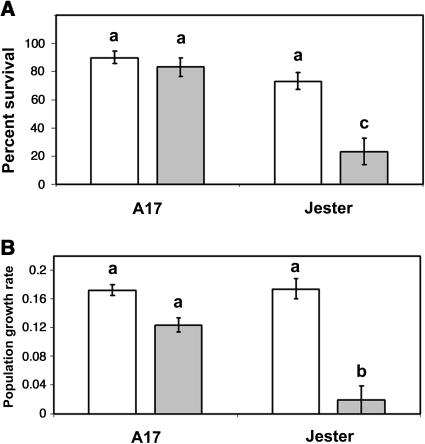
One possible mechanism for phloem-specific aphid resistance is the importation of a phloem-mobile resistance factor(s) to the site of stylet insertion. We tested this possibility by measuring aphid performance on shoots excised from the host plant. Excision and maintenance of shoots on nutrient-supplemented agar did not cause any visible wilting or other signs of damage during the 7-d assay. Aphids settled on excised shoots, deposited honeydew, and produced nymphs as they would on an intact plant. Despite a lack of obvious change in tissue quality, shoot excision in Jester caused a striking enhancement in the survival and growth of aphids, compared to their performance on intact plants of this genotype (Fig. 4). As expected, aphid survival was significantly lower on intact plants of resistant line Jester than on A17 (Fig. 4A; F1,20 = 8.4; P = 0.0009). Interestingly, excision abolished this difference, causing aphids to survive as well on excised shoots of Jester as on excised shoots of A17 (F1,20 = 19; P = 0.0003). Excision did not significantly affect survival on A17. Colony performance, as measured by PGR, increased by 9-fold on excised shoots of Jester, compared to shoots of A17 (F1,20 = 58; P < 0.0001). Excision on A17 caused a nonsignificant increase of 39% in PGR. These contrasting effects of excision between the two genotypes led to a significant genotype-by-treatment interaction for PGR (F1,20 = 16; P = 0.0008).
Figure 4.
Effects of plant genotype and shoot excision on A. kondoi survival (A) and population growth rate (B). Population growth rate in B was measured as described in “Materials and Methods.” White bars, excised shoots; gray bars, shoots of an intact plant. Each bar represents the mean of six replicates. Error bars represent ±1 se. Means labeled with the same letter are not significantly different (P < 0.05).
Resistance Is Controlled by a Single Dominant Gene
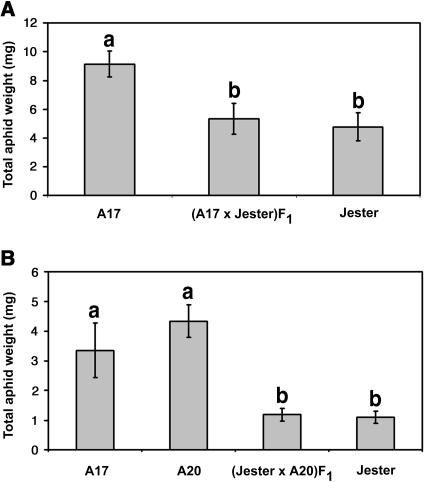
For genetic analysis of A. kondoi resistance, we used F1 hybrids and F2 populations from two crosses in which Jester was the aphid-resistant parent. The aphid-susceptible genotype A20 was chosen as an additional parent for crossing with Jester because A17 and A20 were parents of the mapping population used by the M. truncatula Consortium to produce a reference map of the genome (http://medicago.org/genome/) and by Zhu et al. (2002) to map resistance gene analogs. The aphid performance phenotypes of F1 plants from A17 × Jester, and from Jester × A20, were indistinguishable from that of Jester, indicating resistance is dominant in both crosses (Fig. 5). Genotype A20 had a nonsignificant trend toward supporting higher aphid population densities than A17 (Fig. 5B; this was later borne out by the dramatically higher aphid densities that accrued on A20 compared with A17 during F2 screening). After being infested and phenotyped for aphid resistance, the hybrid plants were grown to produce F2 seed. The F2 phenotyping method, in which aphids could move freely among host plants during colonization, was designed to combine the effects of antixenosis, antibiosis, and plant tolerance. By the end of the third week of infestation, all plants, including the Jester controls, had at least some aphid colonization. Plants of the F2 population from A17 × Jester were either extremely stunted with necrotic lesions (like A17) or were much larger with few aphids and no visible necrosis (like Jester). Plants of the F2 population from Jester × A20 population either had very abundant aphids with moderate stunting and no necrosis (like A20) or looked like the much larger Jester plants. A relatively small number of plants in either population had intermediate phenotypes and could not be scored with confidence (0.7% of the F2 population from A17 × Jester and 12% in the F2 population from Jester × A20). These were excluded from all further analyses. A total of 1,278 plants were assigned phenotypes in the F2 population from A17 × Jester, and 202 plants in the F2 population from Jester × A20. For both F2 populations, the segregation ratios for aphid-resistance phenotypes strongly support the model of a single, dominant, nuclear gene controlling resistance to A. kondoi in Jester (Table II). We propose the name AKR (Acyrthosiphon kondoi resistance) for this gene.
Figure 5.
Performance of A. kondoi on F1 and parent plants from the cross A17 × Jester (A) and Jester × A20 (B). A and B show results of two separate experiments in which different numbers of aphids were used for different lengths of time (see “Materials and Methods”). In A, bars represent means of 10 to 12 replicate plants; in B, bars represent means of 7 to 9 plants. Error bars are ±1 se. Means labeled with the same letter are not significantly different (P < 0.01).
Table II.
Segregation of aphid resistance in F2 populations indicates dominant, Mendelian inheritance
| Population | Observed Resistant:Susceptible | Expected Resistant:Susceptible | χ2 | P |
|---|---|---|---|---|
| A17 × Jester | 959:319 | 958.5:319.5 | 0.00104 | 0.974 |
| Jester × A20 | 148:54 | 151.5:50.5 | 0.323 | 0.570 |
AKR Is Flanked by Resistance Gene Analogs
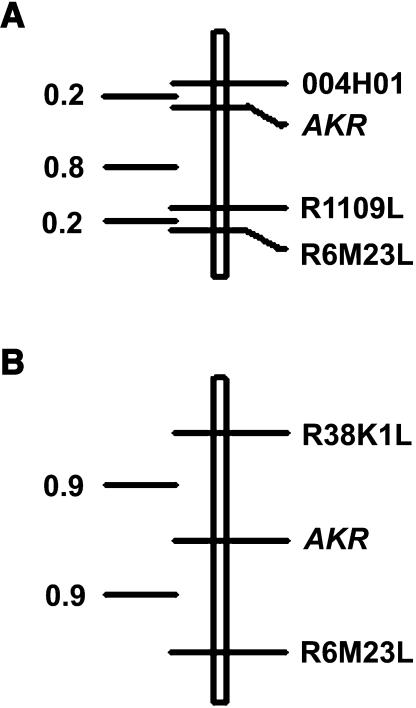
A selection of cleavable amplified polymorphic sequence (CAPS) markers, mapped in the M. truncatula genome by the Medicago truncatula Consortium, was tested for polymorphism between A17 and Jester (maps and marker information are posted at http://medicago.org/genome). The markers were selected to span all eight linkage groups (LG), with a maximum distance of about 10 cM between each marker, and with at least 5 and up to 21 markers tested per LG. A total of 72 CAPS markers were tested and only 5 of these, all on LG 3, were polymorphic between the 2 lines. The markers were R1109L, R6M23L, DK417L, R-EST-BE187590, and DK202R. Four of these were tested and found to be linked to AKR in the first group of phenotyped F2 plants from the A17 × Jester population. From this population, 672 plants were genotyped for markers R1109L and R6M23L. These plants were also scored with a simple-sequence repeat marker, 004H01, since it was found by the Medicago truncatula Consortium to map to this same region of LG 3. CAPS marker R38K1L was found to be polymorphic between Jester and A20, and was tested for linkage to AKR in the F2 population derived from these lines.
We identified tight genetic linkage between AKR and markers 004H01, R1109L, and R6M23L (Fig. 6A). R6M23L and the linked marker R38K1L were tested on 181 plants from the F2 population from Jester × A20, again showing tight linkage between AKR and R6M23L (Fig. 6B). Since R1109L and 004H01 were monomorphic between Jester and A20, we were unable to map these markers with respect to AKR in the F2 population from these lines. Similarly, R38K1L was monomorphic between A17 and Jester and therefore does not appear on the map in Figure 6A. Plants of seven F3 families of the A17 × Jester cross were infested with aphids to determine their F2 progenitors' genotypes at the AKR locus; these results added to the resolution of the map from the population from A17 × Jester. The two maps in Figure 6 are consistent with a map of LG 3 (known to represent chromosome 3) produced by the Medicago truncatula Consortium, in which markers R6M23L and R38K1L flank R1109L, and R38K1L is tightly linked to 004H01.
Figure 6.
A. kondoi resistance locus, AKR, on maps derived from two F2 populations of M. truncatula. A shows the map based on the A17 × Jester population; B shows the map based on the Jester × A20 population. Numbers on the left are interval distances in centiMorgans.
DISCUSSION
Greater knowledge of defense mechanisms against phloem-feeding insects may enhance the exploitation of genetic resistance to manage these agricultural pests. This knowledge may also shed light on fundamental processes such as defense signaling within the phloem and intercellular trafficking of macromolecules, as within the sieve element-companion cell complex. We have characterized the interaction between M. truncatula and A. kondoi at several levels. At each of these levels, the advantages of this model system offer the prospect of substantial elaboration on the mechanism of aphid defense.
Host selection by alatae is normally the first stage of colonization and plays a major role in determining aphid populations in the field (Klingauf, 1987). Resistant line Jester exhibits antixenosis, i.e. deters alatae from settling, within 6 h of aphid release. It is possible that aphids used plant cues such as volatiles or surface waxes and did not require stylet penetration of plants to choose A17, the more susceptible host. However, EPG recordings of feeding behavior do not support this possibility and suggest instead that A. kondoi makes no distinction between surface features of the two closely related lines. Kennedy and Kishaba (1977) compared settling behavior of alate cotton-melon aphid (Aphis gossypii) between unrelated, resistant and susceptible lines of melon (Cucumis melo). Interestingly, they found no preference for either melon genotype at 4 h after release, but a highly significant preference for the susceptible genotype at 24 and 48 h after release. Like AKR-mediated resistance, the resistance mechanism in melon was later shown to be phloem localized (Kennedy et al., 1978; Chen et al., 1997; Klingler et al., 1998), suggesting that time was spent probing phloem tissue before alatae chose between alternative hosts.
The field evaluation of aphid performance on Jester supported laboratory studies of population growth rate, showing that aphid reproduction is possible on this resistant genotype. This contrasts with Mi-mediated resistance against Macrosiphum euphorbiae in tomato, which caused 100% mortality within 10 d (Kaloshian et al., 1997). Since AKR-mediated resistance in M. truncatula permits a low level of reproduction, it should impose a relatively moderate selection pressure on A. kondoi and thereby retain durability in the field. As in the laboratory, the field results can be explained by a combination of antixenosis (reduced settling behavior by alatae), antibiosis (reduced longevity, growth, and fecundity of apterae), and tolerance (relatively more growth of the host plant in the presence of aphids). The enhancement of these effects by prior infestation gives further insight on the dynamics of Jester's multiple modes of resistance. Within 4 d of an aphid's exposure to a mature leaf of Jester, it had a 6.3-fold higher probability of death than an aphid on A17 (Fig. 3A), probably from the lower rate of sap ingestion that occurred as resistance was induced by colonization. Despite this pronounced antibiotic effect on caged A. kondoi, uncaged aphids are able to colonize Jester (i.e. the plant is not lethal). This may be due to a lower level of resistance in shoot tips (the preferred tissue of A. kondoi) compared to mature leaves.
Comparative analysis of aphid feeding behavior between resistant and susceptible plants, using the EPG technique, allows the identification of host tissues most likely to play a role in the resistance mechanism. In two cases, results have indicated that physical or chemical features outside the phloem were involved in aphid resistance (Dreyer and Campbell, 1987; Givovich and Niemeyer, 1991). Plant properties outside of the phloem were reported to affect feeding behavior of another phloem feeder, sweetpotato whitefly (B. tabaci), when the insect was exposed to Mi-containing tomato plants (Jiang et al., 2001). However, phloem-specific resistance similar to that of AKR has been reported for several aphid-plant interactions, some of which are mediated by a single dominant gene (van Helden and Tjallingii, 1993; Chen et al., 1997; Klingler et al., 1998; Kaloshian et al., 2000). Sauge et al. (2002) tested the effect of prior infestation on feeding behavior of Myzus persicae on a resistant genotype of peach (Prunus persicae), in which resistance is also conditioned by a single, dominant gene (Pascal et al., 2002). As in our study, they observed a significant reduction in phloem sap ingestion on the preinfested, resistant cultivar. Unlike our study, however, preinfestation of a susceptible cultivar (unrelated to the resistant cultivar) caused an increase in sap ingestion, suggesting that aphids induced changes in the host plant's physiology leading to enhanced susceptibility. Our results and those of Sauge et al. contrast with those of Chen et al. (1997), who observed no effect of prior infestation on the feeding behavior of A. gossypii on melon near-isogenic lines, with or without the Vat gene.
It is important to note that our tests for antibiosis and altered feeding behavior were conducted on different time scales. In the former case, aphids infested plants for 4 d; in the latter case they probed for only 16 h. The decreased survival of aphids on nonpreinfested Jester, compared to nonpreinfested A17, is likely due to the accumulation of a small but significant, deleterious effect of resistant plants on aphid biology, which may have been induced locally by the aphid cohort itself. In contrast, individual aphids, whose feeding behavior was monitored on uninfested plants for 16 h, may not have been able to induce sufficient levels of a resistance factor in Jester to create a measurable effect on the duration of sap ingestion.
If A. kondoi resistance is based on phloem properties in Jester, the causal factor may be produced locally, i.e. within infested tissue. One possible mechanism for local and phloem-specific resistance is the physical blockage of sap uptake, at the feeding site, through rapid polymerization and deposition of macromolecules such as phloem proteins or callose. Another possible mechanism is the biosynthesis of resistance factors in the vicinity of aphid feeding sites. Even sieve elements themselves can produce allelochemicals in their parietal cytoplasm, as reported by Bird et al. (2003) for alkaloid biosynthesis in opium poppy (Papaver somniferum). However, our finding that shoot excision eliminates A. kondoi resistance in M. truncatula raises the possibility that a resistance factor(s) is imported to the feeding site and that resistance may not be tissue-autonomous. Reciprocal grafting experiments between Jester and A17 will be necessary to confirm this hypothesis.
Aphid-induced necrosis and plant growth inhibition are clearly correlated in genotype A17. Experiments with spotted alfalfa aphid (Therioaphis trifolii f. maculata) on M. sativa (Miles, 1999) suggest that an interaction exists between oxidative enzymes of the aphid saliva and the reduction-oxidation system of sieve elements, which may lead to an uncontrolled production of reactive oxygen species and tissue necrosis. A similar process may occur when A. kondoi parasitizes A17. It is possible that prolonged sieve element contact by aphids on A17 provokes necrotic lesions and the concomitant reduction in plant growth, while the inhibition of phloem feeding in Jester prevents these symptoms.
Aphid resistance can be mediated by classical R genes, as illustrated by Mi in tomato (Rossi et al., 1998). The Mi gene is a member of the non-TIR (or coiled-coil, CC) NBS-LRR subfamily and resides in a cluster of NBS-LRR homologs (Milligan et al., 1998). The Vat gene in melon, which confers aphid and virus resistance (Pitrat and Lecoq, 1982), has been mapped to a cluster of NBS-LRR associated sequences, or RGAs (Klingler et al., 2001; Brotman et al., 2002). Strong genetic evidence supports the Vat gene's membership in the CC-NBS-LRR subfamily (Dogimont et al., 2003). Molecular markers we identified as linked to AKR (R1109L, R6M23L, and R38K1L) also represent RGAs highly similar to the CC-NBS-LRR subfamily (Zhu et al., 2002). Marker 004H01, derived from bacterial artificial chromosome sequence AC138014, lies in close physical proximity to CC-NBS-LRR homologs (http://mtgenome.ucdavis.edu; http://www.genome.ou.edu/medicago_totals.html). Thus, our results raise the possibility that AKR resides in a cluster of such genes. High-resolution mapping of the locus is under way to positionally clone and characterize AKR, which will determine whether it also encodes a classical R protein.
CONCLUSION
We have demonstrated the effectiveness of AKR-mediated resistance in the field and have characterized the trait at multiple temporal and spatial levels. Our results indicate aphids have unrestricted access to the phloem on resistant plants, but that an inhibition of phloem sap ingestion is a likely cause of the nonpreference behavior by aphids in choice tests and of antibiosis in leaf cages. The inducibility of the trait suggests that long distance signaling plays an integral role in its expression. We have identified a single gene controlling the trait and shown its linkage to classical resistance gene-like sequences. This study has established the framework for molecular-genetic and biochemical dissection of aphid resistance in an agricultural context.
MATERIALS AND METHODS
Plants
Plant genotypes used in this study included Medicago truncatula Gaertn. cv Jemalong and the closely related, aphid-resistant cv Jester. Jester was developed from three successive backcrosses to aphid susceptible Jemalong after incorporation of resistance to Acyrthosiphon kondoi Shinji (bluegreen aphid) derived from M. truncatula accession SA1499 (Hill, 2000). Based on its breeding pedigree, Jester has 91% of its genome derived from Jemalong (S. Hughes, personal communication). A derivative of Jemalong, genotype A17, was also used in this study. A17 and two other derivatives from Jemalong were monomorphic for 4,000 molecular markers (Thoquet et al., 2002), suggesting a very low level of heterogeneity in Jemalong. Thus, in our studies the aphid susceptible genotypes A17 and Jemalong are considered equivalent and closely related to Jester. M. truncatula genotype A20, described by Penmetsa and Cook (2000), was used in genetic analysis of A. kondoi resistance in Jester. Prior to laboratory or glasshouse experiments, seeds of A17 and Jester were scarified and germinated in the dark on moist filter paper at 4°C for 10 to 14 d to synchronize radical growth before transfer to soil.
Aphids
A single aphid isolate (an asexual clone) of A. kondoi Shinji, collected from narrow-leaf lupin (Lupinus angustifolius) near Kelleberrin, Western Australia, founded the colony used for most experiments in this study. The colony reproduced on subterranean clover (Trifolium subterraneum) L. cv Dalkeith with 14 h light (23°C)/10 h dark (20°C) under high pressure sodium and fluorescent light at 280 μE m−2 s−1. Under these conditions aphids were asexual females with parthenogenetic, viviparous reproduction. Most aphids were apterae when mature (the wingless, sedentary morph); alatae (the winged, migratory morph) were generally produced at a frequency of less than 1%. An additional colony of A. kondoi was collected from alfalfa/lucerne (Medicago sativa) in South Australia and maintained on this same plant species under controlled conditions. This aphid colony was used in most of the F2 phenotyping for genetic analysis of resistance. Aphids were transferred to experimental plants with a fine paintbrush.
Aphid Colonization in the Field
Plants from cultivars Jemalong and Jester were planted in a single field at Mullewa, Western Australia, on June 15, 1999. These cultivars were evaluated as part of a large trial including 49 genotypes from 26 pasture legume species (Berlandier et al., 1999). Plants were grown in 2-m-long plot rows in a randomized block design, with 4 plots/genotype. Naturally occurring aphids were allowed to infest the plots over the growing season. Eight weeks after sowing, aphid damage in each plot was visually rated using a graduated scale of 0 to 10, where 0 = no visual damage, through to 10 = death of all plants. After damage scoring, a 15-cm section of the 2-m row was randomly selected, and all above-ground plant material in this section was excised and placed in a bag, including any aphids present on the material. The total number of aphids in each sample was counted in the laboratory. Due to inequality of variances, aphid counts were log-transformed before a one-way ANOVA was performed with Statview 5.0.1 (SAS Institute, Cary, NC).
Plant Growth under Infestation in Controlled Conditions
Twelve plants of each genotype were grown individually in 1.2-L pots in a controlled temperature glasshouse in March 2003 in Perth, Western Australia, under natural light. Temperature was 17°C at night and 23°C during the day. Pots were placed in contact with one another in a completely randomized design. Four weeks after sowing, 3 adult apterae (the wingless, sedentary morph) of A. kondoi were placed on each plant and were allowed to reproduce and move among plants for 5 weeks. The above-ground fresh weight of each plant and number of pods per plant were then recorded. Due to inequality of variances, fresh weights were transformed as log (x + 1), and one-way ANOVA was performed with Statview 5.0.1 (SAS Institute).
Host Selection Behavior
Twelve plants each of A17 and Jester were grown in separate 1.2-L pots in a growth chamber with 14 h light at 23°C and 10 h dark at 18°C under high pressure sodium and incandescent light at 250 to 300 μE m−2 s−1. Nineteen days after sowing, two plants of A17 and two plants of Jester were placed in each of six insect-proof cages (38 cm length × 28 cm width × 46 cm height) covered with fine, light-transmitting mesh on the top and on three sides, and a sliding Perspex cover on the remaining side. Two plants of each genotype were randomly placed in the cage so that one plant occupied each of the four corners. Pots were spaced so that no leaves touched other plants. A 5-cm petri dish was placed in the center of the cage, suspended at a height of approximately 10 cm above the soil level of each pot. Twenty-four A. kondoi alatae were placed on the platform in each cage and allowed to choose host plants on which to feed and reproduce over the next 48 h. Settling of aphids on each plant was observed at 3.5, 6, 24, and 48 h after release. Goodness-of-fit to the null hypothesis of equal preference for the two genotypes was tested for settled alatae at each time point, using chi-square tests with the Yates correction for continuity (Zar, 1998). An experiment with a similar design was also conducted in a glasshouse in Perth, Western Australia, in November, 2003, where temperatures ranged from 12°C at night to 30°C during the day. A major difference between this and the growth chamber test was that only 12 aphids were released per cage, and a 3.5 h observation was not included.
Aphid Feeding Behavior
Aphid feeding behavior on preinfested and control plants of A17 and Jester was studied using the EPG technique (Tjallingii, 1987). Plants were grown with 16 h light (20°C)/8 h dark (15°C) under metal halide and incandescent lamps producing 300 μE m−2 s−1. When plants were 3 to 5 weeks old, a single cage was placed on a stem node of each plant. The cage was a 35-mm diameter, 80-mm length, clear plastic cylinder, with the stem passing through slotted, gas-permeable sponge discs at each end of the cylinder so that the distal end of the stem was uncaged. A wooden stake supported the stem and cage. Plants were randomly placed into one of two treatments: preinfested and control. Plants in the preinfested treatment had 20 alatae (the winged morph) inside the cage for 2 d. Aphids had access to the stem, a single trifoliate leaf, and its petiole. Plants that were not preinfested had cages without aphids. At the end of the 2-d preinfestation period, a single apterous adult was placed on a single trifoliate leave outside and distal to the cage. The feeding behavior of this test aphid was monitored while the original, caged aphids remained on preinfested plants.
This monitoring protocol involved starving the test aphids for about 1 h while a 2-to 4-cm length of 20-μm diameter gold wire was attached to the dorsal surface of each aphid's abdomen using silver conductive paint (Ladd, Burlington, VT). The other end of the wire was connected to a Giga-4 direct current amplifier with four channels and 109-Ω input resistance (Wageningen Agricultural University, Wageningen, The Netherlands) in an electrical circuit that also included the host plant, via an electrode placed in the soil. The behavior of individual aphids was monitored for 16 h, most of which occurred during the dark period. All plants and insects were held inside a Faraday cage during recording at an ambient temperature of 23°C. Use of a four-channel amplifier enabled simultaneous recording from four individual aphids on four plants with each treatment combination (Jester, with and without preinfestation, and Jemalong, with and without preinfestation). Voltage waveforms were digitized at 100 Hz with a Metrabyte DAS-8 A/D card (Keithley Instruments, Cleveland, OH). Waveform recordings were analyzed with the EPG analysis software MacStylet 2.0 (Febvay et al., 1996). Histological studies of plant-aphid interactions have correlated specific electrical waveforms with specific positions of insect stylets in plant tissues (Walker, 2000). Waveform patterns in this study were scored according to categories described by Tjallingii and Esch (1993): nonprobing; pooled pathway phase activities; salivary secretion into sieve elements; phloem sap ingestion; xylem ingestion; and cell puncture events of several seconds duration. Both the mean and proportional time spent in each behavior on preinfested and control plants of the two cultivars were analyzed with Kruskall-Wallis tests using Genstat 6.2 (Lawes Agricultural Trust, Rothamsted Experimental Station, Harpenden, Hertfordshire, UK). Numbers of aphids that achieved phloem sap ingestion on different genotypes and under different treatments were analyzed with contingency table analysis using Statview 5.0.1 (SAS Institute).
Aphid Development on Potted Plants
Aphid survival and growth were measured after 4 d on preinfested and control plants of A17 and Jester using cohorts of 10 preweighed, early-instar nymphs. Plants were grown in individual 0.9-L pots in a growth chamber with 16 h light/8 h dark under fluorescent light at 100 to 120 μE m−2 s−1 and a constant temperature of 22°C. Four weeks after sowing, one-half of the plants were preinfested with caged aphids as for the EPG analysis, except that a single trifoliate leaf was caged instead of a stem length as described above. The other half had caged leaves without aphids. Fourteen replicate plants were set up for each genotype-treatment combination. The cage was placed on either the fourth or fifth trifoliate leaf to emerge on the primary stem of each plant. At the end of the 2-d preinfestation treatment, a mesh cage was placed on the next trifoliate leaf distal to (younger than) the original caged leaf on the same stem. A cohort of 10 preweighed, early-instar nymphs was placed inside this second cage, while the original aphids remained in their cage on the other leaf. Four days after the second infestation, the number and weight of surviving aphids in the second cage were recorded. The PGR of surviving nymphs was calculated as the per diem difference between the logarithm of the initial mean weight of aphids placed on the plant (Worig) and the logarithm of the final, total weight of living aphids removed per aphid originally placed on the plant (Wtotal), according to Edwards (2001) and Leather and Dixon (1984):
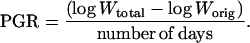 |
This statistic combines effects of aphid growth and survival, providing an estimate of colonization potential on the host plant (Edwards, 2001). The proportion of aphids that survived and PGR were analyzed by two-way ANOVA (genotype, A17 and Jester; treatment, preinfestation and no preinfestation) and compared by the LSD test at a 5% significance level using GenStat 6.2 (Lawes Agricultural Trust, Rothamsted Experimental Station).
Aphid Development on Excised Shoots
Plants were grown with 16 h light (20°C)/8 h dark (15°C) under metal halide and incandescent lamps producing 300 μE m−2 s−1. Five weeks after planting, a stem tip with three nodes was excised from each plant and inserted into agar supplemented with soluble fertilizer in an inverted 90-mm diameter petri dish according to Milner (1982). Each dish contained filter paper to absorb condensate and aphid honeydew. From the remaining stems on the same plant a stem tip, also with three nodes, was caged to receive test aphids. Groups of 5 preweighed, early-instar nymphs were placed onto each excised stem and each caged, intact stem. There were 6 replicate excised stems and intact stems per plant genotype. The plants and petri dishes were then placed in a controlled environment chamber with 12 h light (22°C)/12 h dark (18°C), with 100 to 130 μE m−2 s−1 from fluorescent lamps. Seven days later, the number and weight of surviving aphids and new nymphs produced were recorded to calculate and analyze PGR as above. The proportion of aphids that survived and PRG were analyzed by two-way ANOVA (genotype, A17 and Jester; treatment, shoot excision and no shoot excision) and compared by the Bonferroni/Dunn test at the 5% significance level using Statview 5.0.1 (SAS Institute).
Genetic Analysis of Resistance
Flowers of A17 were emasculated and fertilized with pollen from Jester to produce F1 plants based on the method of Pathipanawat et al. (1994). To determine their aphid performance phenotypes, hybrids were infested with 8 early-instar aphids in leaf cages for 10 d, after which the final weights of aphid cohorts were recorded. Since a genetic map was already available from a cross between A17 and the unrelated line A20 (http://medicago.org/genome/), an additional cross was made between Jester and A20. F1 plants from Jester × A20 were produced and tested as in the A17 × Jester cross, except that Jester was the female parent, and only 6 early-instar aphids were tested in leaf cages for 8 d. Due to inequality of variances, aphid cohort weights were log-transformed for one-way ANOVA, and means were compared by the Bonferroni/Dunn test at the 5% significance level using Statview 5.0.1 (SAS Institute).
Both types of hybrid plants were self-fertilized to produce seed for two populations of F2 plants, which were used for genetic analysis of the A. kondoi resistance trait. F2 individuals were phenotyped for aphid resistance by assessing the amount of feeding damage caused by aphids on plants grown in separate pots in a glasshouse. Phenotyping experiments were performed repeatedly throughout the year under natural light in southern Australia, with temperatures ranging from around 10°C to 30°C. Two weeks after sowing, 2 apterous aphids were placed on each seedling and were allowed to develop, reproduce, and move freely among plants for a period of 3 weeks. Parental lines for each F2 population were randomly distributed among the F2 plants as controls. At the end of 3 weeks, each F2 plant was given a subjective score for the amount of aphid-induced stunting and leaf damage, using a scale of either 1 to 5 or 1 to 10. Low values indicated little or no visible damage while high values indicated severe stunting and necrosis. The appearance of parental plants was used to standardize the damage scales. Each round of phenotyping had between 50 and 350 F2 plants tested at one time. After scoring, plants were chemically treated to remove aphids and grown to maturity to produce leaf tissue (for DNA analysis) and F3 seed.
The related lines A17 and Jester were tested for molecular polymorphisms using CAPS markers (also known as PCR-RFLP markers) and a simple sequence repeat marker developed and mapped in a population of 93 F2 plants from A17 × A20 by the Medicago truncatula Consortium (http://medicago.org/genome/). DNA was isolated from 5 mg freeze dried leaves using a Puregene mini-prep kit (Gentra Systems, Minneapolis). The PCR was used with primers for molecular markers known to reveal polymorphisms between parental genotypes. PCR solutions had 10-μL volumes and consisted of the following components: approximately 50 ng DNA, 0.025 units Taq DNA polymerase (from either Qiagen, Valencia, CA, or Invitrogen, Carlsbad, CA), the recommended dilution of PCR buffer from the manufacturer of Taq DNA polymerase, 2.5 mm MgCl2, 0.25 mm each dNTP, 0.25 μm each primer. After an initial denaturing step at 95°C for 3 min, products were amplified for 38 cycles using the following conditions: 94°C, 30 s; 55°C, 30 s; 72°C, 90 s. Amplification was concluded with a final elongation step at 72°C for 5 min. PCR products for CAPS markers were digested with the appropriate restriction enzyme; products for all markers were separated on agarose gels and visualized with ethidium bromide to identify molecular polymorphisms.
DNA was isolated from each F2 leaf sample as described above, and genotyped using molecular markers identified as polymorphic between the parental lines. Genetic distances between markers and the aphid-resistance phenotype were determined by the Kosambi function using Mapmaker (Lander et al., 1987).
Acknowledgments
We thank Steve Hughes and Dr. Doug Cook for providing seed, Darryl McClements for advice on crossing M. truncatula, Dr. Carol Andersson for helpful discussions in the early stages of the project, Ross Ballard for use of a freeze drier facility, and Steve Robinson, Louisa Bell, Caroline Wielinga, Rick Horbury, Stephanie Whitehand, and Jay Patterson for technical support. We thank Drs. Danny Llewellyn and David Tattersall for helpful comments on the manuscript.
This work was supported in part by the Centre for Legumes in Mediterranean Agriculture and by the Grains Research and Development Corporation (an Honours Scholarship to R.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051243.
References
- Berlandier FA, Edwards OR, Nichols PGH, Blake A (1999) Aphid resistance in annual pasture legumes. In JN Matthiessen, ed, Proceedings 7th Australasian Grassland Invertebrate Ecology Conference, October 4–6, 1999. CSIRO Entomology, Perth, Australia, pp 299–304
- Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: Alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman RL, Eastop VF (1984) Aphids on the World's Crops. Wiley-Interscience, Chichester, UK
- Brotman Y, Silberstein L, Kovalski I, Perin C, Dogimont C, Pitrat M, Klingler J, Thompson GA, Perl-Treves R (2002) Resistance gene homologues in melon are linked to genetic loci conferring disease and pest resistance. Theor Appl Genet 104: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Chen JQ, Rahbe Y, Delobel B, Sauvion N, Guillaud J, Febvay G (1997) Melon resistance to the aphid Aphis gossypii: behavioural analysis and chemical correlations with nitrogenous compounds. Entomol Exp Appl 85: 33–44 [Google Scholar]
- Crawford E, Lake A, Boyce K (1989) Breeding annual Medicago species for semiarid conditions in Southern Australia. Adv Agron 42: 399–437 [Google Scholar]
- de Ilarduya OM, Xie QG, Kaloshian I (2003) Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol Plant Microbe Interact 16: 699–708 [DOI] [PubMed] [Google Scholar]
- Dixon AFG (1998) Aphid Ecology: An Optimization Approach, Ed 2. Chapman and Hall, London
- Dogimont C, Bendahmane A, Pauquet J, Burget E, Desloire S, Hagen L, Caboch M, Pitrat M (2003) Map-based cloning of the Vat melon gene that confers resistance to both aphid colonization and virus transmission. In 11th International Congress on Molecular Plant-Microbe Interactions, July 18–25, 2003, St. Petersburg, Russia
- Dreyer DL, Campbell BC (1987) Chemical basis of host-plant resistance to aphids. Plant Cell Environ 10: 353–361 [Google Scholar]
- Edwards OR (2001) Interspecific and intraspecific variation in the performance of three pest aphid species on five grain legume hosts. Entomol Exp Appl 100: 21–30 [Google Scholar]
- Febvay G, Rahbe Y, vanHelden M (1996) MacStylet, software to analyse electrical penetration graph data on the Macintosh. Entomol Exp Appl 80: 105–108 [Google Scholar]
- Givovich A, Niemeyer HM (1991) Hydroxamic acids affecting barley yellow dwarf virus transmission by the aphid Rhopalosiphum padi. Entomol Exp Appl 59: 79–85 [Google Scholar]
- Hill JR (2000) Jester. Plant Varieties J 13: 40 [Google Scholar]
- Holt BF, Hubert DA, Dangl JL (2003) Resistance gene signaling in plants: complex similarities to animal innate immunity. Curr Opin Immunol 15: 20–25 [DOI] [PubMed] [Google Scholar]
- Jiang YX, Nombela G, Muniz M (2001) Analysis by DC-EPG of the resistance to Bemisia tabaci on an Mi-tomato line. Entomol Exp Appl 99: 295–302 [Google Scholar]
- Kaloshian I, Kinsey MG, Ullman DE, Williamson VM (1997) The impact of Meu1-mediated resistance in tomato on longevity, fecundity and behavior of the potato aphid, Macrosiphum euphorbiae. Entomol Exp Appl 83: 181–187 [Google Scholar]
- Kaloshian I, Kinsey MG, Williamson VM, Ullman DE (2000) Mi-mediated resistance against the potato aphid Macrosiphum euphorbiae (Hemiptera: Aphididae) limits sieve element ingestion. Environ Entomol 29: 690–695 [Google Scholar]
- Kennedy GG, Kishaba AN (1977) Response of alate melon aphids to resistant and susceptible muskmelon lines. J Econ Entomol 70: 407–410 [Google Scholar]
- Kennedy GG, McLean DL, Kinsey MG (1978) Probing behavior of Aphis gossypii on resistant and susceptible muskmelon. J Econ Entomol 71: 13–16 [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53: 299–328 [DOI] [PubMed] [Google Scholar]
- Klingauf FA (1987) Host plant finding and acceptance. In AK Minks, P Harrewijn, eds, Aphids: Their Biology, Natural Enemies and Control, Vol 2A. Elsevier, Amsterdam, pp 209–223
- Klingler J, Kovalski I, Silberstein L, Thompson GA, Perl-Treves R (2001) Mapping of cotton-melon aphid resistance in melon. J Am Soc Hortic Sci 126: 56–63 [Google Scholar]
- Klingler J, Powell G, Thompson GA, Isaacs R (1998) Phloem specific aphid resistance in Cucumis melo line AR 5: effects on feeding behaviour and performance of Aphis gossypii. Entomol Exp Appl 86: 79–88 [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Leather SR, Dixon AFG (1984) Aphid growth and reproductive rates. Entomol Exp Appl 35: 137–140 [Google Scholar]
- Miles PW (1999) Aphid saliva. Biol Rev Camb Philos Soc 74: 41–85 [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10: 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner RJ (1982) On the occurrence of pea aphids, Acyrthosiphon pisum, resistant to isolates of the fungal pathogen, Erynia neoaphis. Entomol Exp Appl 31: 23–27 [Google Scholar]
- Moran PJ, Cheng YF, Cassell JL, Thompson GA (2002) Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch Insect Biochem Physiol 51: 182–203 [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125: 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela G, Williamson VM, Muniz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16: 645–649 [DOI] [PubMed] [Google Scholar]
- Pascal T, Pfeiffer F, Kervella J, Lacroze JP, Sauge MH (2002) Inheritance of green peach aphid resistance in the peach cultivar ‘Rubira’. Plant Breed 121: 459–461 [Google Scholar]
- Pathipanawat W, Jones RAC, Sivasithamparam K (1994) An improved method for artificial hybridization in annual Medicago species. Aust J Agric Res 45: 1329–1335 [Google Scholar]
- Penmetsa RV, Cook DR (2000) Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol 123: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitrat M, Lecoq H (1982) Relations génétiques entre les résistances par non-acceptation et par antibiose de melon Aphis gossypii. Recherche de liaisons avec d'autres gènes. Agronomie 2: 503–508 [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95: 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauge MH, Lacroze JP, Poessel JL, Pascal T, Kervella J (2002) Induced resistance by Myzus persicae in the peach cultivar ‘Rubira’. Entomol Exp Appl 102: 29–37 [Google Scholar]
- Thoquet P, Gherardi M, Journet E-P, Kereszt A, Ane J-M, Prosperi J-M, Huguet T (2002) The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii WF (1987) Electrical recording of stylet penetration activities. In AK Minks, P Harrewijn, eds, Aphids: Their Biology, Natural Enemies and Control, Vol 2B. Elsevier, Amsterdam, pp 95–108
- Tjallingii WF, Esch TH (1993) Fine-structure of aphid stylet routes in plant-tissues in correlation with EPG signals. Physiol Entomol 18: 317–328 [Google Scholar]
- van Helden M, Tjallingii WF (1993) Tissue localisation of lettuce resistance to the aphid Nasonovia ribisnigri using electrical penetration graphs. Entomol Exp Appl 68: 269–278 [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, et al (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16: 1365–1369 [DOI] [PubMed] [Google Scholar]
- Walker GP (2000) A beginner's guide to electronic monitoring of homopteran probing behavior. In GP Walker, EA Backus, eds, Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior. Thomas Say Publications in Entomology, Entomological Society of America, Lanham, MD, pp 14–40
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19: 195–216 [DOI] [PubMed] [Google Scholar]
- Zar JH (1998) Biostatistical Analysis, Ed 4. Pearson Education, Upper Saddle River, NJ
- Zhu HY, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15: 529–539 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn J-E, Koiwa H (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]