Abstract
The soybean apyrase, GS52, was previously characterized as an early nodulin that is expressed in roots and localized to the plasma membrane. Transgenic Lotus japonicus plants were constructed constitutively expressing the GS52 apyrase. Segregation and Southern-blot analysis identified four single-copy sense lines, several double-copy sense lines, and one double-copy antisense line for further analysis. The single- and double-copy sense gs52 L. japonicus lines had enhanced nodulation that correlated with expression of the transgene. The sense transgenic lines were also found to have increased infection thread formation and enhanced infection zone length when infected by Mesorhizobium loti, the natural symbiont of L. japonicus. The data presented show that expression of the GS52 apyrase can enhance nodulation in L. japonicus and points to an important role for this group of enzymes in nodulation.
Apyrases (nucleotide phosphohydrolases; EC 3.6.1.15) are nonenergy-coupled NTPases that have been observed to play diverse roles, as might be expected of enzymes that can change the ratios of key energy carriers (e.g. ATP), inorganic phosphorus, and signaling molecules (e.g. GMP, cAMP). The apyrase NTPase catalytic domain can be located both cytoplasmically (endoapyrases) or extracellularly (ectoapyrases; Komoszynski and Wojtczak, 1996; Day et al., 2000). In animals, the roles of apyrases include modulation of neurotransmission (Edwards and Gibb, 1993) and blood platelet aggregation (Marcus and Safier, 1993). In yeast (Saccharomyces cerevisiae), two apyrases facilitate the glycosylation of N- and O-linked oligosaccharides in the Golgi lumen (Abeijon et al., 1993; Gao et al., 1999).
In plants, apyrases were shown to play a role in phosphate transport and mobilization (Thomas et al., 1999). Transgenic expression of a pea apyrase in Arabidopsis (Arabidopsis thaliana) increased growth and phosphate transport (using either inorganic phosphate or ATP). During animal neurotransmission, ectoapyrases recycle and diminish the hormonal activity of extracellular ATP/ADP (for review, see Clark et al., 2001). There is now a growing realization that extracellular ATP may exist in plants and ectoapyrases may be involved in catalysis of this ATP. Lew and Dearnaley (2000) showed that extracellular ATP (approximately 1 mm) would depolarize the membrane potential of growing Arabidopsis root hairs. Thomas et al. (2000) showed that transgenic expression of either PGP1 (a multidrug resistance transporter) or ectoapyrase resulted in enhanced resistance to cycloheximide. The authors suggested that symport-mediated extrusion of this antibiotic could make use of an ATP gradient across the plasma membrane. Tang et al. (2003) also showed that extracellular ATP (>1 mm) inhibited root gravitropism and polar auxin transport. Extracellular ATP increased the sensitivity of roots to exogenous auxin. Demidchik et al. (2003) showed that ATP could trigger an increase in intracellular calcium levels. More recently, Jeter et al. (2004) showed that ATP-induced increases in cytoplasmic calcium levels were coupled to downstream gene expression, implying a complete signaling pathway responsive to ATP.
Membrane depolarization, hormonal activity, calcium oscillations, and signal transduction are all relevant to legume nodulation in response to inoculation with rhizobia (for review, see Cohn et al., 1997). Therefore, ectoapyrases, perhaps through action on extracellular ATP, could play an important role in nodulation. Indeed, Etzler et al. (1999) reported that an ectoapyrase from the roots of the legume Dolichos biflorus could bind the lipo-chitin Nod signal produced by rhizobia. The Nod signal is essential for the induction of a nodule structure in response to rhizobial infection. The ATPase activity of the Dolichos apyrase was stimulated upon binding the Nod signal, as well as other related ligands. Etzler et al. (1999) proposed that the D. biflorus apyrase could be the Nod signal receptor postulated to be essential to nodulation. However, this now seems unlikely due to the identification of the LysM domain receptor kinases as the mostly likely receptors for the Nod signal (Limpens and Bisseling, 2003; Madsen et al., 2003; Radutoiu et al., 2003). However, interaction of the Nod signal with an ectoapyrase on the root hair surface could still be relevant to nodulation.
Indeed, a direct role for apyrases in nodulation was suggested by the fact that treatment of roots with antibody directed against either the D. biflorus apyrase (Etzler et al., 1999) or the soybean (Glycine max) GS52 apyrase (Day et al., 2000) inhibited nodulation. These data suggested that the apyrase was localized to the root hair (i.e. the site of rhizobial invasion) and, indeed, Kalsi and Etzler (2000) were able to confirm this by immunolocalization of the apyrase to Dolichos root hairs. Inoculation with compatible, but not with incompatible, rhizobia led to a redistribution of the D. biflorus apyrase to the root hair tips. Day et al. (2000) were also able to localize the soybean GS52 apyrase to root plasma membrane. In addition, both Cohn et al. (2001), working with Medicago truncatula, and Day et al. (2000), working with soybean (L. Merr.), showed that inoculation with the respective, compatible rhizobium resulted in induction of specific apyrase gene expression.
An open question is what unique features are found in legumes, which allow them to be nodulated by rhizobia, compared to other angiosperms. Cannon et al. (2003) compared apyrase gene sequences from various legumes (i.e. M. truncatula, soybean, and Lotus japonicus) to one another and to those from nonlegumes (e.g. Arabidopsis). This phylogenetic analysis identified a potential legume-specific clade, which included the soybean GS52 apyrase and the Nod signal-binding D. biflorus apyrase. Comparisons of rates of change at synonymous and nonsynonymous sites in the GS52 apyrase and sister clades showed rapid evolution in the potential legume-specific clade. The authors suggested that local apyrase gene duplication in an ancestor of the legumes, followed by functional diversification and increased rates of change in the new genes, resulted in the formation of a legume-specific apyrase gene subfamily. This analysis, as well as earlier analyses (Roberts et al., 1999; Cohn et al., 2001), is consistent with the hypothesis that this legume apyrase subfamily plays a unique role in legume biology, and perhaps a critical role in nodulation.
Consistent with this hypothesis, we now report that transgenic expression of the soybean GS52 apyrase in L. japonicus plants significantly increased nodule number upon inoculation with the compatible Mesorhizobium loti. This increase in nodule number was correlated with an increased number of root hair infections and an expansion in the root zone infected.
RESULTS
Construction of gs52 Transgenic L. japonicus
At the time we instigated these studies, the gs52 gene was the only apyrase gene in our possession. Given the difficulties with soybean transformation, we opted to express this gene in L. japonicus, which is more amenable to genetic transformation. The gs52 gene and orthologous LjLNP gene of L. japonicus, isolated by Roberts et al. (1999), share approximately 80% sequence identity, suggesting that the expression of gs52 could result in silencing of the endogenous L. japonicus apyrase genes. Therefore, transgenic L. japonicus plants were constructed in which the Glycine soja gs52 gene (Day et al., 2000) was constitutively expressed from the strong cauliflower mosaic virus 35S promoter in either the sense or antisense orientation. Transformation was via Agrobacterium-mediated hypocotyl transformation as described by Stiller et al. (1997).
Analysis of gs52 Copy Number in Transgenic Plants
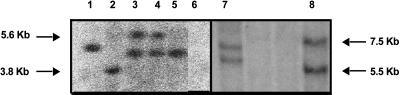
Primary (T1) transgenic plants were selfed and segregation of the transgene was analyzed in T2- and T3-generation seeds from individual transformed plants (Table I). Segregation of the transgene was scored based on a comparison of seedling growth on medium with and without G418 antibiotic. Segregation analysis suggested that numerous lines were either single copy (segregating 3:1) or homozygous (copy number unknown) for the transgene. Copy number was subsequently determined by Southern blotting (Fig. 1). Three single-copy sense lines, 45W T2, 45B1K T3, and 45D5 T3, and one double-copy sense line, 45B77 T3, were identified and used for further analysis. In addition, a double-copy antisense line, 44HH T3, was identified and used as a control in further analysis. With the exception of the nodulation assay, these lines were used exclusively for further analyses.
Table I.
Segregation analysis of gs52 L. japonicus plants
Plants were germinated on filter paper and then transferred to selection medium (see “Materials and Methods”) and scored for resistance or sensitivity to G418. Transgene copy number was determined by Southern blotting and is listed in column 4 of the table. S, Sense copy of gs52; AS, antisense copy of gs52.
| Line | Resistant:Sensitive | Segregation | Southern Results |
|---|---|---|---|
| 45 W T2 | 34:9 | 3.8:1 | 1 Copy |
| 45D5 T3 | 68:26 | 2.8:1 | 1S |
| 45B1K T3 | 51:15 | 3.4:1 | 1S |
| 45B77 T3 | 62:0 | Homozygousa | 2S |
| 45A7 T3 | 75:0 | Homozygous | 2S |
| 45B38 T3 | 64:0 | Homozygous | 2S |
| 45B1.2 T3 | 92:0 | Homozygous | 2S |
| 45B1H T3 | 60:0 | Homozygous | 2S |
| 45B35 T3 | 22:0 | –b | 2S |
| 45B1I T3 | 100:0 | Homozygous | 2S |
| 45B16 T3 | 53:0 | Homozygous | 2S |
| 45B37 T3 | 73:0 | Homozygous | 2S |
| 44HH T2 | 99:0 | Homozygous | 2AS |
All plants were G418 resistant and were homozygous for at least one of the two inserts.
Inconclusive.
Figure 1.
gs52 genomic Southern blot. Eight-microgram genomic DNA was digested with HindIII and separated by agarose gel electrophoresis. A 1.4-kb PCR product complementary to the gs52 cDNA was used as a probe. 1, 45W T2 (single-copy line); 2, 45D5 T3 (single-copy line); 3, 45B1.2 T3; 4, 45B1H T3; 5, 45B1K T3; 6, wild type; 7, 44HH T2 (double-copy antisense line); and 8, 45B77 T3 (double-copy sense line). The enzyme HindIII cuts once within the T-DNA of pGA941 (but not within the gs52 sequence); therefore, each band represents an independent insertion into the plant genome.
gs52 Expression Analysis of Transgenic L. japonicus Lines
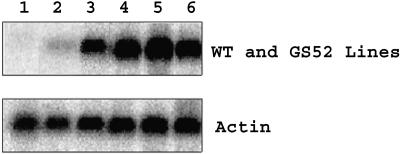
Southern-blot analysis indicated the presence of gs52 integrated into the genome at independent locations in each of the lines. Northern-blot analysis was carried out to determine whether the gs52 gene was successfully expressed in these transgenic lines. As shown in Figure 2, 3 single-copy lines had varying levels of gs52 expression. The double-copy gs52 line, 45B77 T3, had strong gs52 expression. In addition, the northern blot indicated that neither the wild-type L. japonicus nor the antisense gs52 line, 44HH T2, had detectable expression of gs52 mRNA.
Figure 2.
Northern-blot analysis of wild-type and gs52 transgenic L. japonicus. A 1.4-kb PCR product complementary to gs52 cDNA was used to detect gs52 mRNA expression levels (top). Lanes 1 to 6 represent the results of 20 μg total RNA from each sample. 1, Wild type; 2, 44HH T2 (double-copy antisense line); 3, 45D5 T3 (single-copy sense line); 4, 45W T2 (single-copy sense line); 5, 45B1K T3 (single-copy sense line); and 6, 45B77 T3 (double-copy sense line). A soybean actin probe was used as a RNA loading control (bottom).
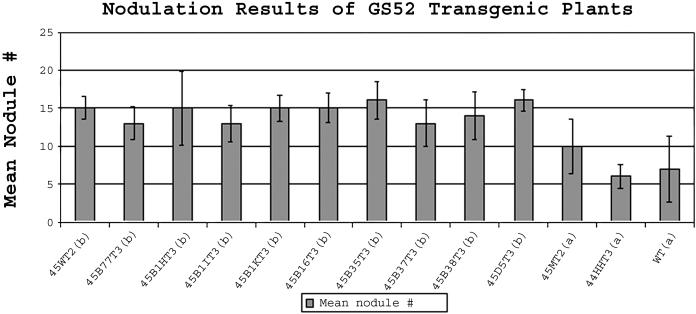
Figure 3.
Nodulation results of gs52 transgenic L. japonicus plants. Compared to wild-type and line 44HH T3 plants, sense gs52 plants had enhanced nodulation. Seeds from each line were sterilized, germinated, and grown as described in “Materials and Methods.” Three-week-old plants were equally inoculated with M. loti NZP2235 and then grown for an additional 4 weeks. The plants were scored for their nodulation phenotype 4 weeks postinoculation. Bars indicated ±se. Groups marked (a) and (b) are statistically different from one another (α = 0.05) by Student's t test. The data shown are from 1 experiment, but are representative of the 3 independent trials that were conducted (n ≥ 30 for each treatment in each trial). Line 45M T2 (not listed in Table I) shown in this figure was a sense gs52 line that exhibited greater variation with regard to nodulation and, therefore, did not differ from the wild type. It is included here to indicate that not all sense lines showed greater nodulation, although most did. The differences are probably due to positional effects of the various T-DNA insertions.
Although antisense expression of a transgene can result in gene silencing (e.g. Escobar et al., 2003), significant silencing was not apparent in the case of line 44HH T2 (as evidenced by northern blots; data not shown), consistent with the lack of antisense gs52 mRNA expression in this line (Fig. 2). It is interesting that very few antisense GS52 lines were regenerated, perhaps suggesting that significant silencing of apyrase expression is lethal. Studies of Arabidopsis T-DNA mutants showed that a functional apyrase gene was essential for pollen germination (Steinebrunner et al., 2003). Therefore, apyrase function for plant regeneration during the transformation process may be essential. For the purposes of subsequent experiments, the genotype of line 44HH T2 allowed it to serve as a useful control.
gs52 Sense Plants Showed Enhanced Nodulation
Figure 3 shows the nodulation results of 12 lines of gs52 transgenic L. japonicus in comparison to wild-type plants. In every case, the mean nodule number represents the mean results from at least 30 individual plants (n = 30) and data are representative results from 3 independent experiments. Sense gs52 lines exhibited a 30% to 50% increase in nodulation compared to wild-type controls. In contrast, the control antisense line, 44HH T2, displayed a wild-type nodulation phenotype.
gs52 Sense Plants Have Enhanced InfectionThread Formation
The single-copy transgenic L. japonicus plants had significantly enhanced nodulation compared to the control wild-type and 44HH T2 L. japonicus plants. To explore the mechanism of the enhanced nodulation, the number of infection threads visible on each of the plants was counted. Each of the plants was inoculated with M. loti NZP2235 harboring a hemA-lacZ fusion (Boivin et al., 1990). This strain carries a marker (constitutive lacZ fusion) allowing detection of bacterial infection events via staining with X-gal. Visualization of the infection threads demonstrated that the enhanced nodulation phenotype of the gs52 plants correlated with a marked enhancement in infection thread number (Table II). Indeed, there was a direct correlation between the level of gs52 mRNA expression (as measured by northern analysis) and the increase in infection threads formed. As shown in Table II, infection by the antisense 44HH T2 appeared to be slightly inhibited in comparison to the wild type. It is tempting to speculate that these results are due to a slight reduction in endogenous apyrase levels due to gene silencing. However, as reported above, such silencing was not confirmed by northern blots. Therefore, if present, gene silencing was limited to only critical tissues or occurred at such a low level as to remain undetectable by northern blotting.
Table II.
Infection thread analysis of GS52 L. japonicus plants
Groups marked (a), (b), (c), (d), or (e) were statistically significant at α = 0.05 by Student's t test. WT, Wild type; Avg, average.
| Data | WT | 44HH T2 | 45D5 T3 | 45W T2 | 45B1K T3 | 45B77 T3 |
|---|---|---|---|---|---|---|
| Copy no. | 0 | 2 antisense | 1 sense | 1 sense | 1 sense | 2 sense |
| Avg total root length (mm) | 210.2 (a) | 144.4 (b) | 231.8 (a) | 195.2 (a) | 185.4 (c) | 182.3 (c) |
| Avg nodule no./plant | 8 (a) | 7 (a) | 12 (c) | 13 (c) | 11 (c) | 9 (c) |
| Avg tap root infection threads/plant | 83 (a) | 50 (b) | 144 (c) | 229 (d) | 316 (e) | 207 (d) |
| Avg lateral root infection threads/plant | 55 (a) | 42 (b) | 137 (c) | 164 (d) | 270 (e) | 123 (c) |
| Avg infection threads/plant | 138 (a) | 92 (b) | 281 (c) | 393 (d) | 586 (e) | 330 (c) |
| Avg root infection zone length (mm) | 28.3 (a) | 30 (a) | 39 (c) | 59 (d) | 85 (e) | 57 (c) |
| Avg root infection threads/mm in the avg total root infection zone length (mm) | 11 (a) | 7 (b) | 17 (c) | 20 (d) | 20 (d) | 13 (c) |
| Avg root infection threads/mm of avg total root length (mm) | 0.65 (a) | 0.64 (a) | 1.21 (c) | 2.01 (d) | 3.16 (e) | 1.81 (d) |
It is well accepted that legume roots have defined regions (i.e. infection zones) that are most susceptible to infection (Calvert et al., 1985). The infection zone length of the GS52-expressing plants was significantly larger than those found on control plants (Table II). Further, the GS52 transgenic plants had greater infection thread numbers per unit infection zone length (millimeters). In other words, the GS52 plants had enhanced infection thread formation; i.e. infection threads were more abundant inside the infection zone.
DISCUSSION
Transgenic L. japonicus plants were created to express the soybean apyrase, GS52. This approach was utilized because L. japonicus can be easily transformed and manipulated. This plant has also been adopted as a model for the study of nodulation and nitrogen fixation (Handberg and Stougaard, 1992). A variety of transgenic lines were generated, varying in transgene copy number. To simplify further analysis, only genetically independent lines containing a single insertion of the gs52 transgene were analyzed in detail. Initially, we also attempted to silence the endogenous L. japonicus apyrase by antisense expression of gs52. Although several transformations were attempted, we recovered only 1 line, 44HH T2, which exhibited no detectable antisense expression of gs52, had normal endogenous apyrase expression (as measured by northern blotting), but showed a normal nodulation phenotype. Although not definitive, the difficulty in recovering antisense gs52 lines may suggest that silencing of the apyrase could be detrimental to plant growth and/or regeneration. This hypothesis would be consistent with the findings of Steinebrunner et al. (2003), who showed that a functional apyrase is required for pollen germination in Arabidopsis.
Northern-blot analysis revealed that the 3 single-copy lines (expressing sense gs52) had various levels of gs52 mRNA expression. 45W T2 strongly expressed gs52, but had the lowest level of expression compared to the other single-copy sense lines. The other single-copy sense lines, 45D5 T3 and 45B1K T3, had medium and maximal levels of gs52 expression, respectively. The level of gs52 expression in the transgenic lines directly correlated with the infection and nodulation phenotypes observed. These data, coupled with analysis of segregation, lead us to conclude that GS52 expression is responsible for the phenotypes seen.
Expression of GS52 in L. japonicus enhanced nodulation by M. loti from 30% to 50%, compared to control plants. Such a result could be explained by either an increase in the number of infections or by an increase in the percentage of infections that can lead to nodule formation. It is well known that not all infections actually result in nodule formation (Calvert et al., 1985). We tested these two possibilities by directly counting the number of infections, as visualized using a M. loti strain constitutively expressing β-galactosidase. The results showed a significant enhancement in infection thread formation when the GS52 sense plants were inoculated with M. loti.
The number of nodules formed on legume roots is under autoregulatory control (for review, see Caetano-Anollés and Gresshoff, 1991) The net result is that only a small segment of the developing root is susceptible to nodulation. This is termed the infection zone. We measured this area by counting the number of infections as a function of root length. The GS52-expressing L. japonicus plants had significantly larger infection zones. Taken together, the infection thread and infection zone data suggest that the GS52 apyrase plays a role in early nodulation at the stage of infection thread formation. This would be consistent with earlier reports of Nod signal enhancement of apyrase enzyme activity (Etzler et al., 1999).
Recently, LysM receptor-like kinases were identified as the likely receptors for the Nod signal. These proteins were identified by positional cloning of their corresponding genes starting with plant mutants blocked in the earliest steps in nodulation (Limpens and Bisseling, 2003; Madsen et al., 2003; Radutoiu et al., 2003). The LysM domain was first found in bacterial proteins involved in binding murein (peptidoglycan) found in bacterial cell walls (Bateman and Bycroft, 2000). Peptidoglycan, a polymer of N-acetylmuramic acid and GlcNAc, is structurally similar to chitin and, therefore, the presence of the LysM domain in the nodulation-related receptor-like kinases suggests a role in direct binding of the lipo-chitin Nod signal. Although direct biochemical evidence for Nod signal binding to these proteins is still lacking, the mutant phenotype supports their role as Nod signal receptors. Given these findings, the earlier suggestion (Etzler et al., 1999) that apyrases, due to Nod signal-binding activity, could be the postulated, essential Nod signal receptor seems unlikely. However, available evidence, including the information reported here, supports a critical role for apyrases in the early infection mechanism.
If GS52 does not play a role as a bona fide Nod signal receptor, then what other possible roles exist for this protein in nodulation? Our results do not allow a definitive answer to this question. However, the work of Demidchik et al. (2003) and Jeter et al. (2004) suggest one possible function. Demidchik et al. (2003) showed that extracellular ATP could increase intracellular calcium levels. It is known that an increase in cytoplasmic calcium is an essential component in the initial phases of rhizobial infection of the root hair (for review, see Cohn et al., 1997). Jeter et al. (2004) recently reported that extracellular ATP triggers an increase in cytoplasmic calcium levels, resulting in enhanced expression of stress-related transcripts, including those involved in ethylene biosynthesis. Ethylene is a known inhibitor of nodulation and, therefore, the ability to control extracellular ATP levels via apyrase activity could allow fine control of cellular responses both beneficial (e.g. calcium oscillations) and detrimental (e.g. ethylene production) to nodulation.
MATERIALS AND METHODS
Chemicals and Enzymes
Na-32phosphate was purchased from ICN (Costa Mesa, CA). The protease inhibitor phenylmethylsulfonyl fluoride was obtained from Sigma Chemical (St. Louis). Unless specified, all other reagents were purchased from Fisher Scientific (Pittsburgh). Most enzymes in this study were purchased from Promega (Madison, WI) or Fisher Scientific.
Bacterial Culture Media and Growth Conditions
Escherichia coli strain JM109 was grown and maintained on Luria-Bretani medium (Sambrook et al., 1989). The antibiotic used for plasmid maintenance and selection in E. coli was 200 μg/mL ampicillin.
Mesorhizobium loti NZP2235 was cultivated on yeast mannitol broth medium (Schauser et al., 1998) at 30°C. The Agrobacterium strain was grown at 30°C on yeast extract peptone medium (Vervliet et al., 1975). Antibiotic concentrations were 2 μg/mL tetracycline and 100 μg/mL carbenicillin for M. loti NZP2235 and Agrobacterium tumefaciens LBA4404, respectively. Antibiotic concentrations for Bradyrhizobium japonicum carrying the constitutive npt-lacZ fusion were 25 μg/mL tetracycline and 50 μg/mL chloramphenicol.
Construction of gs52 Transgenic Lotus japonicus
The gs52 cDNA was isolated and cloned previously as described in Day et al. (2000). Subsequently, the entire GS52 sequence was subcloned into the plant binary vector pGA941 (An et al., 1988). This vector was then electroporated into A. tumefaciens strain LBA4404 by procedures described in Sambrook et al. (1989). Agrobacterium-mediated hypocotyl transformation of L. japonicus was carried out based on the protocol described by Stiller et al. (1997).
Seed Germination and Sterilization
Wild-type and gs52 transgenic L. japonicus ecotype Gifu seeds were first scarified by rubbing briefly between 2 sheets of fine 150-grain sandpaper. This was done until the seed coat was visibly roughened. The scarified seeds were then soaked in 3% hydrogen peroxide (H2O2) and 95% ethanol (ethanol) for 15 min with shaking at room temperature. The seeds were germinated in square petri dishes on a 0.5-cm stack of sterile Whatman No. 1 filter paper soaked in sterile, distilled, deionized water. The petri dishes were sealed with parafilm and placed in a Percival model CU-32 L (Percival Scientific, Boone, IA) incubator for 1 week with a light cycle of 8-h, 22°C day and 16-h, 20°C night. The relative humidity was 70% to 80%.
Segregation Analysis of Transgenic L. japonicus
Progeny from individual T1- or T2-generation plants (equivalent to F2 or F3 seeds) were analyzed. One-week-old germinated seedlings were transferred to 0.5× Gamborg's B5 medium (Gamborg and Shyluk, 1970) agar plates containing 5 μg/mL G418 (Sigma) for segregation analysis. The seedlings were allowed to grow on selection medium for 4 weeks before scoring for antibiotic resistance. Antibiotic resistance was based on growth phenotype compared to wild-type plants that were obviously sensitive to the antibiotic. Resistant plants had enhanced root growth compared to wild-type sensitive plants and, in most cases, the roots of resistant plants grew in a curled and/or wavy pattern. In addition to an obvious root phenotype, resistant plants had healthy, green leaves, while sensitive plants appeared chlorotic.
Nucleic Acid Isolation and Manipulation
Plasmid DNA Isolation
Plasmid DNA was isolated from E. coli and A. tumefaciens using the alkaline lysis method described in Sambrook et al. (1989) or by using the Wizard Plus Miniprep DNA purification system (Promega) for automated DNA sequencing. DNA concentrations were determined using a DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech, San Francisco).
Isolation of DNA Fragments
DNA restriction endonuclease fragments used in cloning were separated by agarose gel electrophoresis, as described by Sambrook et al. (1989). Gel-purified fragments were isolated using the DNA gel extraction kit (Qiagen, Valencia, CA).
Genomic DNA Isolation
Genomic DNA was isolated from L. japonicus using the protocol described by Dellaporta et al. (1983). DNA concentrations were determined as described previously in Sambrook et al. (1989).
Isolation of Total RNA
Total RNA was isolated from L. japonicus plants using the protocol described by Verwoerd et al. (1989). Briefly, plant material (approximately 100 mg) was ground in liquid nitrogen and 500 μL of hot (80°C) extraction buffer were added (phenol 0.1 m LiCl, 100 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS [1:1]) and homogenized by vortexing for 30 s. Next, 250 μL of chloroform:isoamyl alcohol (24:1) were added and the samples were vortexed again. The samples were centrifuged in a microfuge at 14,000g for 10 min at 4°C. The water phase was removed and mixed with 1 volume of 4 m LiCl. RNA was precipitated overnight and collected by centrifugation as described above. The resultant pellet was dissolved in 250 μL of nuclease-free water and 0.1 volume of 3 m NaOAc, pH 5.2, and 2 volumes of ethanol were added. The sample was incubated for 30 min at −80°C and the RNA was collected by centrifugation as described above. Finally, the pellet was washed with 70% ethanol and allowed to air dry for 10 min before dissolving in 50 μL of sterile nuclease-free water. RNA quantity and integrity were determined by monitoring the optical density as 260 nm and by agarose gel electrophoresis, respectively.
DNA Sequencing
Automated DNA sequencing was performed by Dr. Neil Quigley (University of Tennessee). Plasmid DNA sequencing was performed with the ABI prism dye terminator cycle sequencing reaction kit on an ABI 373 DNA sequencer (Perkin-Elmer, Foster City, CA). Both strands of two independent clones were sequenced to ensure the fidelity of sequences.
Southern-Blot Analysis
Eight micrograms of genomic DNA from L. japonicus were digested with HindIII overnight at 37°C, according to the supplier's specifications. Digests were loaded onto a 0.7% agarose gel and separated overnight at 5 volumes/cm. The DNA was then blotted onto ZetaProbe nylon membrane (Bio-Rad Laboratories, Hercules, CA), according to the method of Sambrook et al. (1989). Hybridization and washing were performed according to the manufacturer's protocol. Gene-specific probes for gs52 were labeled to a specificity of 108 cpm/μg DNA using random primer labeling (Promega).
Northern-Blot Analysis
gs52 transgenic L. japonicus plants were sterilized and germinated in the dark. Etiolated seedlings were ground in liquid nitrogen and total RNA was isolated as described below. Total RNA (20 μg) was separated on formaldehyde-denaturing gels in 1% agarose and blotted onto ZetaProbe nylon membrane (Bio-Rad Laboratories), according to the method of Sambrook et al. (1989). Hybridization and washing were performed according to the manufacturer's protocol. In all cases, gene-specific probes were labeled to a specific activity of 108 cpm/μg DNA using random primer labeling (Promega).
Nodulation Assays
Lotus seeds were sterilized and germinated as described above. One-week-old seedlings were planted in 4-inch plastic pots containing sterile vermiculite and given Broughton and Dilworth nutrient solution (Broughton and Dilworth, 1971). After planting, the seedlings were allowed to grow for 2 additional weeks (as described above) and then inoculated with a M. loti culture as described above. The plants were placed back into the growth chamber and allowed to grow for an additional 4 weeks (as described above). At 4 weeks postinoculation, the plants were harvested and scored for nodule number.
Infection Thread Assays
Lotus seeds were sterilized and germinated as described previously. After 1 week, the germinated seedlings were transferred to sterile Leonard jars containing sterile vermiculite for further growth and nodulation assays. To ensure sterility, all containers, vermiculite, and water and/or nutrient solutions used in these experiments were autoclaved. The plants were watered with Broughton and Dilworth nutrient solution (Broughton and Dilworth, 1971) and grown for an additional 2 weeks as described above.
The 3-week-old plants were inoculated with 1 mL of a 3-d-old M. loti (hemA-lacZ) culture washed with sterile water and diluted to an optical density 600 of 0.1. The culture inoculant was manually applied to each plant with a pipette. The plants were allowed to grow for an additional 2 weeks postinoculation before the roots were harvested, fixed, and stained for visualization of infection threads. At the time of harvest, the plants were removed from the Leonard jars by flooding gently with water. The roots were further washed gently in water and subsequently detached from the plant using a razor blade. The detached roots were immediately fixed in glutaraldehyde and stained for LacZ (β-galactosidase) expression as described by Boivin et al. (1990). Roots were stored in the dark in sterile, distilled water at 4°C until use.
The roots were measured and photographed using a stereoscope (Olympus SZX12) equipped with a Nikon DXM1200 digital camera. The infection zone was defined as an area on the root showing the most abundant infections (compare with Calvert et al., 1985). The edges of the infection zone were arbitrarily determined as the point where no additional infections were apparent within 3 mm.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–97ER20260).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055939.
References
- Abeijon C, Yanagisawa K, Mandon EC, Hausler A, Moreman K, Hirschberg CB, Robbins PW (1993) Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J Biol Chem 122: 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, DP Verma, eds, Plant Molecular Biology Manual A3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–19
- Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299: 1113–1119 [DOI] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica C, Truchet G, Rosenburg C (1990) Rhizobium meliloti genes encoding trignoelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G, Gresshoff PM (1991) Plant genetic control of nodulation in legumes. Annu Rev Microbiol 45: 345–382 [DOI] [PubMed] [Google Scholar]
- Calvert HE, Pence MK, Pierce M, Malik NSA, Bauer WD (1985) Anatomical analysis of the development and distribution of rhizobium infections in soybean roots. Can J Bot 62: 2375–2384 [Google Scholar]
- Cannon SB, McCombie WR, Tabata S, Denny R, Palmer L, Katari M, Young ND, Stacey G (2003) Evolution and microsynteny of the apyrase gene family in three legume genomes. Mol Genet Genomics 270: 347–361 [DOI] [PubMed] [Google Scholar]
- Clark G, Thompson G, Roux SJ (2001) Signal transduction mechanisms in plants: an overview. Curr Sci 80: 181–188 [PubMed] [Google Scholar]
- Cohn J, Day RB, Stacey G (1997) Legume nodule organogenesis. Trends Plant Sci 3: 105–110 [Google Scholar]
- Cohn JR, Uhm T, Ramu S, Nam Y-W, Kim D-J, Penmetsa RV, Wood TC, Denny RL, Young ND, Cook DR, et al (2001) Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol 125: 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G (2000) Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant-Microbe Interact 13: 1053–1070 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: version II. Plant Mol Biol Rep 1: 19–21 [Google Scholar]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM (2003) Is ATP a signaling agent in plants? Plant Physiol 133: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards FA, Gibb AJ (1993) ATP: a fast neurotransmitter. FEBS Lett 325: 86–89 [DOI] [PubMed] [Google Scholar]
- Escobar MA, Civerolo EL, Polito VS (2003) Characterization of oncogene silenced transgenic plants: implications for Agrobacterium biology and posttranscriptional gene silencing. Mol Plant Pathol 4: 57–65 [DOI] [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB (1999) A Nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA 96: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Shyluk JP (1970) Culture of plant cells with ammonium salts as sole nitrogen source. Plant Physiol 45: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Kaigorodov V, Jigami Y (1999) YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J Biol Chem 274: 21450–21456 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- Jeter CR, Tang W, Henaff E, Butterfield T, Roux SJ (2004) Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 16: 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi G, Etzler ME (2000) Localization of a Nod factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol 124: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoszynski M, Wojtczak A (1996) Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochim Biophys Acta 1310: 233–241 [DOI] [PubMed] [Google Scholar]
- Lew RR, Dearnaley JDW (2000) Extracellular nucleotide effects on the electrical properties of growing Arabidopsis thaliana root hairs. Plant Sci 153: 1–6 [Google Scholar]
- Limpens E, Bisseling T (2003) Signaling in symbiosis. Curr Opin Plant Biol 6: 343–350 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen L, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Marcus AJ, Safier LB (1993) Thromboregulation: multicellular modulation of platelet reactivity in hemostasis and thrombosis. FASEB J 7: 516–522 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen L, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Philips DA, Etzler ME (1999) A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet 262: 261–267 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TA (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Neilsen A, Stougaard J (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259: 414–423 [DOI] [PubMed] [Google Scholar]
- Steinebrunner I, Wu J, Sung Y, Corbett A, Roux SJ (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J, Martirani L, Túpale SL, Chian R, Chiurazzi M, Gresshoff PM (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48: 1357–1365 [Google Scholar]
- Tang WQ, Brady SR, Sun Y, Corbett A, Roux SJ (2003) Extracellular ATP inhibits root gravitropism at concentrations that inhibit polar auxin transport. Plant Physiol 131: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Rajagopal A, Windsor B, Dudler R, Lloyd A, Roux SJ (2000) A role for ectophosphatase in xenobiotic resistance. Plant Cell 12: 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux SJ (1999) Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol 119: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervliet G, Holsters H, Teuchy H, Van Montagu M, Schell J (1975) Characterization of different plaque-forming and defective temperate phages in Agrobacterium strains. J Gen Virol 26: 33–48 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker MM, Hoeckema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]