Abstract
A patient-tailored, ex vivo drug response platform for glioblastoma (GBM) would facilitate therapy planning, provide insights into treatment-induced mechanisms in the immune tumor microenvironment (iTME), and enable the discovery of biomarkers of response. We cultured regionally annotated GBM explants in perfusion bioreactors to assess iTME responses to immunotherapy. Explants were treated with anti-CD47, anti–PD-1, or their combination, and analyzed by multiplexed microscopy [CO-Detection by indEXing (CODEX)], enabling the spatially resolved identification of >850,000 single cells, accompanied by explant secretome interrogation. Center and periphery explants differed in their cell type and soluble factor composition, and responses to immunotherapy. A subset of explants displayed increased interferon-γ levels, which correlated with shifts in immune cell composition within specified tissue compartments. Our study demonstrates that ex vivo immunotherapy of GBM explants enables an active antitumoral immune response within the tumor center and provides a framework for multidimensional personalized assessment of tumor response to immunotherapy.
Immune cells within glioblastoma explants can be re-invigorated by immunotherapies, preferentially in the tumor center.
INTRODUCTION
Glioblastoma (GBM) is a fatal brain tumor without effective treatment options. Current standard of care consists of gross total surgical resection followed by chemoradiation, resulting in a mean overall survival of 14 months (1). Recently, strategies harnessing the non-neoplastic immune tumor microenvironment (iTME) have evolved. A major problem in GBM therapy lies in the inherent immunosuppression exerted by the cell types residing in the iTME. The GBM iTME consists of myeloid-derived macrophages and yolk sac–derived microglia [collectively termed glioma-associated macrophages/microglia (GAMs)] as well as other myeloid cell types and lymphocytes. GAMs infiltrate into GBM tumors and, by interaction with tumor cells, change their functional state toward an immunosuppressive and regenerative phenotype (2). The composition of GAMs within GBM and their origin and phenotypic evolution during tumorigenesis are currently under intense research (3, 4). GBM is a heterogeneous and widely invasive malignancy, and human data on the composition of the iTME and its interaction with neoplastic cells in different tumor regions (contrast medium–enhancing tumor center, peripheral infiltration zone) are scarce (5). Recently, major efforts have been undertaken to describe the GBM iTME on the single-cell level, highlighting the predominance of myeloid cells (6, 7).
Immune checkpoint inhibitor therapies with anti–CTLA-4 and/or anti–PD-1/PD-L1 antibodies have revolutionized the treatment of many solid tumors. However, so far, results from clinical trials of systemic T cell checkpoint blockade in GBM were disappointing (8). Other studies suggest that PD-1 blockade as a neoadjuvant treatment in combination with adjuvant maintenance therapy could increase survival compared to adjuvant PD-1 blockade alone (9). High PD-L1 expression levels are associated with decreased survival in GBM patients (9). The expression of PD-L1 and indoleamine 2,3-dioxygenase (IDO-1), as well as the accumulation of regulatory T cells (Tregs) in response to the presence of CD8+ T cells are known mechanisms of adaptive resistance (10) and might counteract immune responses. Combinatorial immunotherapies addressing both innate and adaptive immune cell types of the iTME may therefore circumvent these resistance mechanisms (11).
Previously, we focused on the disruption of the CD47-Sirpα axis to regain antitumor activity of GAMs against malignant brain tumors. We and others showed that Sirpα itself is a potent modulator of macrophage-mediated phagocytosis (12, 13). Blocking the CD47-Sirpα axis suppresses a “don’t eat me” signal to macrophages and allows efficient phagocytosis of most cancers tested so far (14–16). Preclinical analysis of a humanized anti-CD47 antibody demonstrated potent in vitro and in vivo tumor killing ability against GBM (17, 18). Anti-CD47 treatment induced a macrophage polarization change of GAMs in vivo. Furthermore, both M1- and M2-polarized macrophages displayed a higher GBM cell phagocytosis rate under anti-CD47 treatment, indicating that even M2 macrophages can be rendered phagocytic (18). Moreover, anti-CD47 treatment induced exclusive microglia-mediated GBM cell phagocytosis in a xenograft mouse model, when macrophage influx was impeded (19).
The culture of intact tumor tissues is an attractive strategy to assess the effects of cancer immunotherapies on the iTME (20). To dissect the composition of the human GBM iTME and its response to innate and adaptive immune checkpoint inhibitor therapy ex vivo, we used three-dimensional (3D) tissue perfusion bioreactors, CO-Detection by indEXing (CODEX) highly multiplexed microscopy, soluble protein arrays, and mass spectrometry. Here, we present an in-depth analysis of the GBM iTME of intra-operatively annotated samples from tumor center and periphery at baseline and after 7 days of ex vivo immunotherapy targeting CD47 and/or PD-1. We included the peripheral invasion zone because most tumor recurrences originate from this region. Hence, targeting the periphery after surgical tumor control by immunotherapy would be an important pillar in adjuvant GBM treatment. Our approach mimics a localized application of immunotherapy, which poses a clinically feasible modality with less systemic toxicity. We found that a subset of explants from the tumor center was responsive to the tested immunotherapies. Moreover, we provide evidence of intratumoral reactivation of T cells and relief of immunosuppression using immunotherapies targeting both innate and adaptive immune cell types in a subset of responding tumors. This approach provides a multidimensional proof-of-concept strategy to identify patients who might be amenable to local immunotherapeutic approaches.
RESULTS
Ex vivo culture and immunotherapy of intact GBM explants from tumor center and periphery using perfusion bioreactors
To profile the region-specific GBM iTME by highly multiplexed microscopy and assess treatment responses to immune checkpoint inhibitors, we prospectively collected GBM specimens from seven patients undergoing neuronavigated surgery (Fig. 1A and table S1). From each tumor, contrast medium–enhancing, 5-aminolevulinic acid (5-ALA)–positive, vital tissue from the center as well as 5-ALA–low/negative tissue from the periphery/infiltration zone were acquired. Tissue acquisition was documented by intraoperative imaging (taking into consideration 5-ALA positivity and neuronavigation; fig. S1). After neuropathological examination by two board-certified neuropathologists, all samples were diagnosed as GBM, World Health Organization (WHO) grade IV, isocitrate dehydrogenase 1/2 (IDH) wild type. Furthermore, GBM subtype analysis was performed by whole-genome methylation analysis according to Capper et al. (21), documenting three receptor tyrosine kinase RTK II (classical) and four mesenchymal subtypes (Fig. 1B and table S1). Preoperatively, patients were exposed to varying dexamethasone dosage schemes (average cumulative pre- and intrasurgical steroid dosage in our patients was 73 mg, ranging from 28 to 116 mg; table S1).
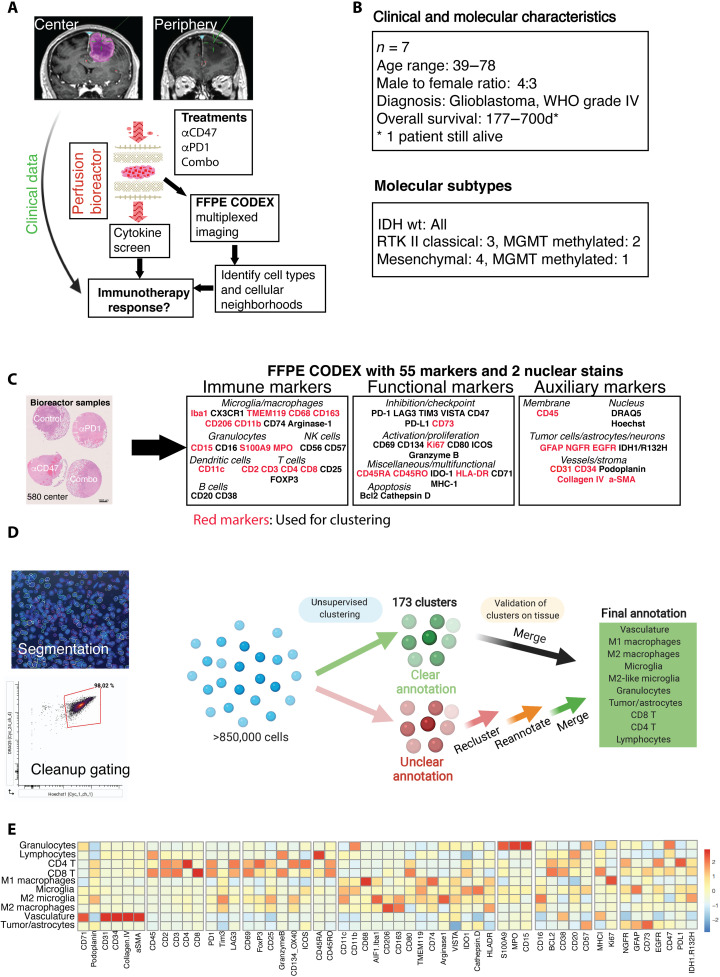
Fig. 1. Experimental setup, clinical characteristics, and CODEX clustering pipeline.
(A) Experimental setup of the proof-of-concept study: Fresh tumor biopsies were taken according to neuronavigation and directly transferred into 3D perfusion bioreactors. Immunomodulatory treatments consisted of innate (anti-CD47) and/or adaptive (anti–PD-1) immune checkpoint inhibition for 7 days. Soluble proteins and metabolites from bioreactor media, and multidimensional CODEX microscopy data were integrated to assess immunotherapy response per patient/tumor region sample. (B) Clinical baseline and genetic data of the included patients (n = 7). (C) Left: Representative arrangement of an FFPE-processed, hematoxylin and eosin (H&E)–stained center biopsy consisting of explants cultured in the bioreactor and treated with different immunotherapies. Scale bar, 1000 μm. Right: FFPE CODEX antibody panel. (D) Clustering strategy for cell type annotation. Cells were segmented on the basis of nuclear DRAQ5 staining, and DRAQ5/Hoechst double-positive cells were gated in CellEngine (bottom left), resulting in >850,000 single cells across all samples. Vortex clustering at first using the markers specified led to 173 clusters, which were subsequently merged/simplified on the basis of manual validation on the tissue. Unclear cells were reclustered, reannotated, and revalidated on each tissue section using the cell finder package. Last, 10 main cell types throughout all samples could be annotated with high confidence (see fig. S3, detailed clustering strategy, cell passports, and flow cytometry–like plots). (E) Heatmap of mean fluorescence intensities of individual markers used in the CODEX pipeline per annotated cell type across all samples.
Intact tumor fragments (explants) were subsequently cultured in 3D tissue perfusion bioreactors (22). This culture system provides flow of the media through the tissue, which enables culturing intact tissues of greater thickness and overcomes typical limitations of static cultures, including limited transport of nutrients and waste removal, particularly in the tissue center. We validated and optimized this technology and found superior tissue preservation in perfused cultures compared to nonperfused/static conditions (fig. S2). Specifically, an intact iTME, tumor cell proliferation, and invasion of GBM cells into the scaffold could be detected in explants cultured for up to 3 weeks (fig. S2).
To simulate a continuous local immunotherapy, we treated the perfused explants for 7 days with central nervous system–adapted doses (23) of an antibody against the microglia-macrophage checkpoint CD47 (14), with the clinically approved PD-1 checkpoint inhibitor nivolumab (24), or a combination of both (Fig. 1A). Control samples were left untreated. To investigate potential differential effects of these immunotherapies on tumor center versus periphery, we treated patient-matched samples from both regions simultaneously. Our results indicate that intact GBM tissues from tumor center and periphery can be cultured alive for multiple days and provide a framework for GBM-tailored immunotherapy testing using multimodal analysis of the iTME.
Formalin-fixed paraffin-embedded CODEX enables interrogating changes in iTME composition and architecture in GBM explants treated with immunotherapy
To analyze changes in immune cell type abundance and localization in the GBM iTME induced by immunotherapies, we performed highly multiplexed microscopy using CODEX on a total of 47 explants (25). We selected and validated a panel of 55 antibodies for GBM iTME–specific protein markers on formalin-fixed paraffin-embedded (FFPE) sections, which included markers for identifying immune cell types, functional markers (costimulatory proteins, immune checkpoints, and molecules involved in apoptosis and proliferation), as well as auxiliary markers for tumor, vascular, and stromal cells (Fig. 1C and data S1).
After thorough validation of staining quality, marker expression, and distribution in the tissue sections, iTME cell types were identified using a combined approach of unsupervised clustering and manual cluster refinement and by overlaying the clusters onto each individual tissue for morphological confirmation (Fig. 1D and fig. S3, page 1). Only markers that had a robust expression (moderate or strong signal intensity) throughout all the samples were used for clustering (see Materials and Methods). To validate the clustering result, we generated “cell passports” for each cell type based on the expression of cell type–defining markers and appropriate negative controls (fig. S3, pages 2 to 5). For all cell types, standardized mean fluorescence intensities across all the markers in the panel are shown in Fig. 1E. In addition, we generated flow cytometry–like plots to visualize the gating strategy (fig. S3, page 6).
Tissue integrity after 7 days of perfusion culture was well preserved, and perfused samples were comparable to fresh tissues based on hematoxylin and eosin (H&E) staining (Fig. 2A and fig. S2). Using composite multicolor overlays derived from the CODEX dataset, we visualized each of the 10 annotated, final cell types in the tissue context based on phenotype-defining markers and morphology (Fig. 2B). Vasculature-defining markers were CD34, CD31, and collagen IV. Tumor cells and astrocytes were positive for GFAP and NGFR, with a subset of proliferating, Ki-67+ tumor cells. CD45, CD68, CD11b, Iba1, and HLA-DR were used to identify macrophages and microglia. CD163 was used as a surrogate marker of an M2-like state for both microglia and macrophages; microglia and M2-like microglia were discriminated from macrophages by TMEM119 positivity. Granulocytes represented another major myeloid population and were characterized by MPO, CD11b, S100A9, and CD15 expression. In line with a previous study (6), the adaptive immune compartment was relatively low abundant in our sample set and was dominated by CD3+CD4+ and CD3+CD8+ T cells and a CD45 high-expressing, morphologically defined lymphocytic population negative for T cell markers, which we called “lymphocytes” (fig. S3).
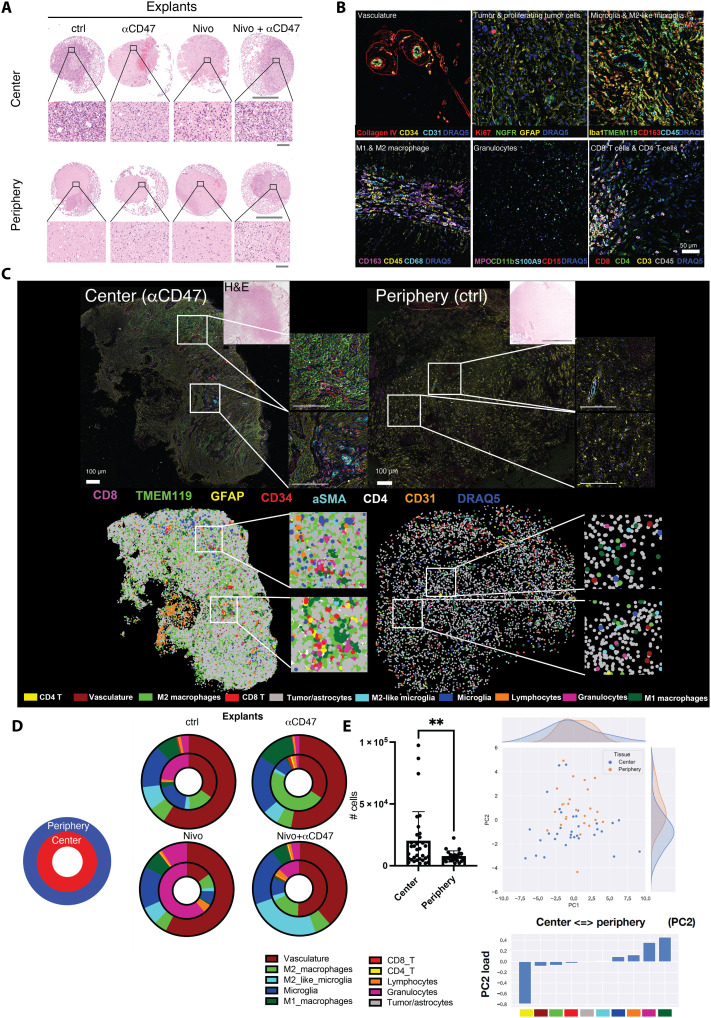
Fig. 2. Differential composition of cellular phenotypes across biopsy locations.
(A) Representative H&E-stained images of FFPE GBM center and periphery explants after 7 days of culture in perfusion bioreactors. Scale bars, 1000 μm (overview) and 50 μm (close-up). (B) Multiplexed immunofluorescence overlay images of the most important cell types and their defining markers after definitive cell type annotation. (C) Top: FFPE-CODEX–stained representative GBM bioreactor sample from center and periphery (tumor ID 588). Seven of 54 markers are depicted indicating microglia (TMEM119+), tumor cells/reactive astrocytes (GFAP+), CD8 T cells (CD8+), CD4 T cells (CD4+), and vasculature (CD31+CD34+ SMA+). Scale bars, 100 μm. Bottom: Assembly of annotated cell types according to x-y coordinates of the corresponding tissue sample in the top. (D) Cell type frequency distribution throughout all center and periphery samples used in the experiment. Inner circle, center samples; outer circle, periphery samples. (E) Left: Overall cellularity among all bioreactor samples in center versus periphery (n = 31 center samples, mean = 20,995 cells per sample; n = 22 periphery samples, mean = 8467 cells per sample; P = 0.0095, unpaired t test with Welch’s correction). Individual cellular abundances are visualized in fig. S6 and Fig. 5G; right: principal components analysis (PCA) correlating abundances in center versus periphery samples (across pooled bioreactor samples). Statistics: (E) Student’s t test; **P < 0.01.
To obtain an overview of the overall tissue composition with respect to the 10 cell type clusters identified, we used a color-coded graphical representation and correlated this to the morphology and multicolor overlays (Fig. 2C). We observed great intratumoral heterogeneity in explants from the tumor center, exemplified by islands of pronounced microglia accumulation in parts of an anti-CD47–treated center explant and a perivascular enrichment of CD4+ and CD8+ T cells (Fig. 2C, bottom left, insets). In an untreated explant from the tumor periphery, we observed the typical stellate morphology of astrocytes with homogeneously intermingled microglia and only few adaptive immune cells (Fig. 2C, right). By spatially arranging the annotated, color-coded clusters based on their x-y coordinates, differences in cellular density and spatial distribution between the exemplary center and periphery explants can be appreciated (Fig. 2C, bottom).
Cell frequencies and composition differ between explants from tumor center and periphery
We observed differences in the cell type composition and density of explants from tumor center and periphery (Fig. 2D and figs. S4 to S6). Consistent with our biopsy selection, the total cell density in explants from the tumor center was significantly higher compared to that in explants from the tumor periphery [mean total cell counts per center explant: 20,779, periphery explant: 8467; P = 0.0095; Fig. 2, C (bottom) and E, and figs. S4 and S5]. Moreover, tumor center explants contained significantly higher numbers of CD4+ T cells (P = 0.004) and lymphocytes (P = 0.0055) and tended toward higher M2 macrophage (P = 0.11) numbers. Conversely, M2-like microglia (P = 0.0014) and M1 macrophages (P = 0.00019) were more abundant in periphery explants (fig. S5). After performing a principal components analysis (PCA) of the relative abundance of the annotated cell types across samples, we observed marked differences between explants from tumor center and periphery (Fig. 2E). The first principal component (PC1) did not significantly differ between the biopsy locations. Notably, the second principal component (PC2), which was more prevalent in tumor center explants (P = 0.0041), had a positive weight for CD4+ T cells, CD8+ T cells, vasculature, and M2 macrophages (Fig. 2E).
Spatial analysis identifies GBM iTME tissue compartments conserved across samples and patients
To further investigate whether immunotherapies affect the spatial localization of cell types in the GBM iTME, and whether this depends on the type of tumor sample (center versus periphery), we identified GBM tissue compartments (TCs) using a raster-scanned radius method (26). This analysis resulted in seven distinct TCs that were conserved across samples (Fig. 3A). The cell type composition of each TC as well as the abundance of the different TCs across all samples are visualized as a heatmap in Fig. 3B, and their frequencies across explants from center and periphery are depicted in Fig. 3C. As expected, the most prevalent TC (TC1) was composed mainly of tumor cells/astrocytes, and therefore corresponded to the “bulk tumor/astrocyte compartment” (Fig. 3, B and C, TC1, dark blue). TC2 was enriched in not only immunosuppressive cell types such as M2-like microglia and granulocytes but also M1-macrophages; we therefore named this TC the “myeloid compartment” (Fig. 3, B and C, TC2, cyan). TC3, the second most abundant compartment in all samples, consisted of a mixture of tumor cells and immune cells; we therefore named this compartment the “tumor-immune interface” (Fig. 3, B and C, TC3, green). TC4 morphologically corresponded to GBM-typical “glomeruloid vascular proliferations” (Fig. 3, B and C, TC4, yellow). CD4+ and CD8+ T cells, lymphocytes, and M2 macrophages were enriched in TC5 (“adaptive immune compartment”; Fig. 3B, TC5, orange), while microglia defined TC6 (“microglia-enriched compartment;” Fig. 3B, TC6, red). Last, TC7 was a vascular compartment with immune cell enrichment, which we termed “vascular-immune interface” (Fig. 3B, TC7, brown).
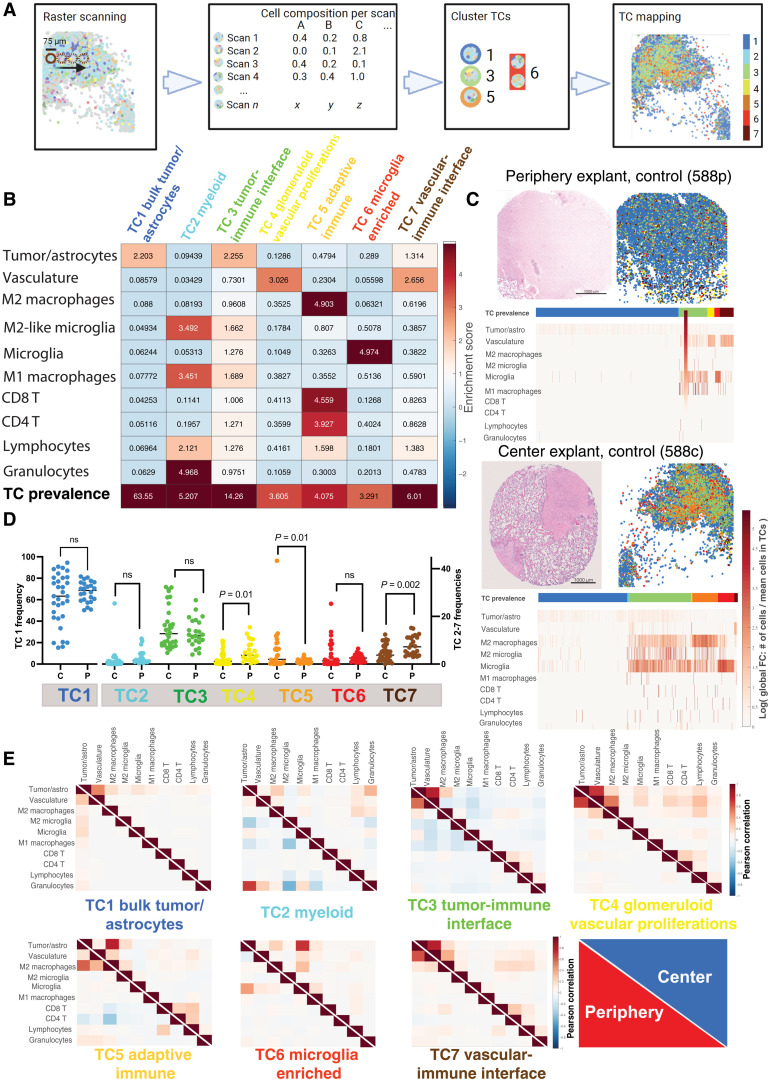
Fig. 3. Glioblastoma explants can be subclassified into distinct TCs that differ depending on biopsy location.
(A) Schematic of neighborhood TC identification: Tissue regions were scanned in cylinders of 75 μm diameter (best focus area). The cellular composition of each scan was recorded and subsequently clustered using the NN self-organizing map (SOM) algorithm and the Davies-Bouldin criterion. This resulted in seven distinct neighborhoods. (B) Region enrichment score depicting the fold change of cellular composition in each region and their overall prevalence throughout all explants. Color code for TCs is used for all subsequent analysis. TCs were named on the basis of their most prevalent cell types and functionality: TC1 (blue), bulk tumor-astrocyte compartment; TC2 (cyan), myeloid compartment; TC3 (green), tumor-immune interface; TC4 (yellow), glomeruloid vascular proliferations; TC5 (orange), adaptive immune compartment; TC6 (red), microglia-enriched compartment; TC7 (brown), vascular-immune interface. (C) Representative TC map on bioreactor tissue from untreated periphery (top image) and center (bottom image). H&E-stained tissue section is supplemented by its corresponding TC overlay. Individual region heatmaps display the prevalence of TCs and the cellular composition of each region (log of cell number/total cells per TC). (D) Frequencies of TCs in all bioreactor samples depending on biopsy location [center (C) versus periphery (P)]. Each data point represents one bioreactor sample, and the horizontal line represents the median of all samples. Statistical analysis: Student’s t test. (E) Normalized cell-cell correlation per TC in periphery (red, lower triangle) and center (blue, upper triangle). Pearson correlation coefficients (−1 to 1) between individual cell types per region are plotted. Reading example: in TC6, microglia and M2 macrophages have a stronger spatial interaction with tumor cells in the center than in the periphery.
GBM iTME TC distribution points toward higher adaptive immune activity in tumor center explants
To visualize the different TCs and their spatial contexts in the tissue, we overlaid them onto the respective tissue samples (Figs. 3C and 4). In samples from the tumor periphery, we observed, as expected, a lower cell density in bioreactor explants examined and a more homogeneous distribution of the TCs (Fig. 3C, top, and fig. S4). In contrast, in samples from the tumor center, higher cell densities and a more heterogeneous TC distribution prevailed, and the region heatmap analyses showed a more pronounced infiltration by adaptive and especially innate immune cells, corresponding mainly to the presence of the respective TCs “M2 macrophages,” “M2-like microglia,” and “microglia” (Fig. 3C, bottom, and fig. S4). We performed this analysis for each patient sample, biopsy location, and treatment condition, cross-comparing H&E stainings, cell density plots, cell type and TC localizations, and cell type enrichments (fig. S6). We then analyzed TC frequencies between all bioreactor center (n = 34) and periphery (n = 24) samples independently of the applied treatment. The frequencies of TC4 (glomeruloid vascular proliferations), TC5 (adaptive immune compartment), and TC7 (vascular-immune interface) significantly differed in their frequency between tumor center and periphery explants (Fig. 3D).
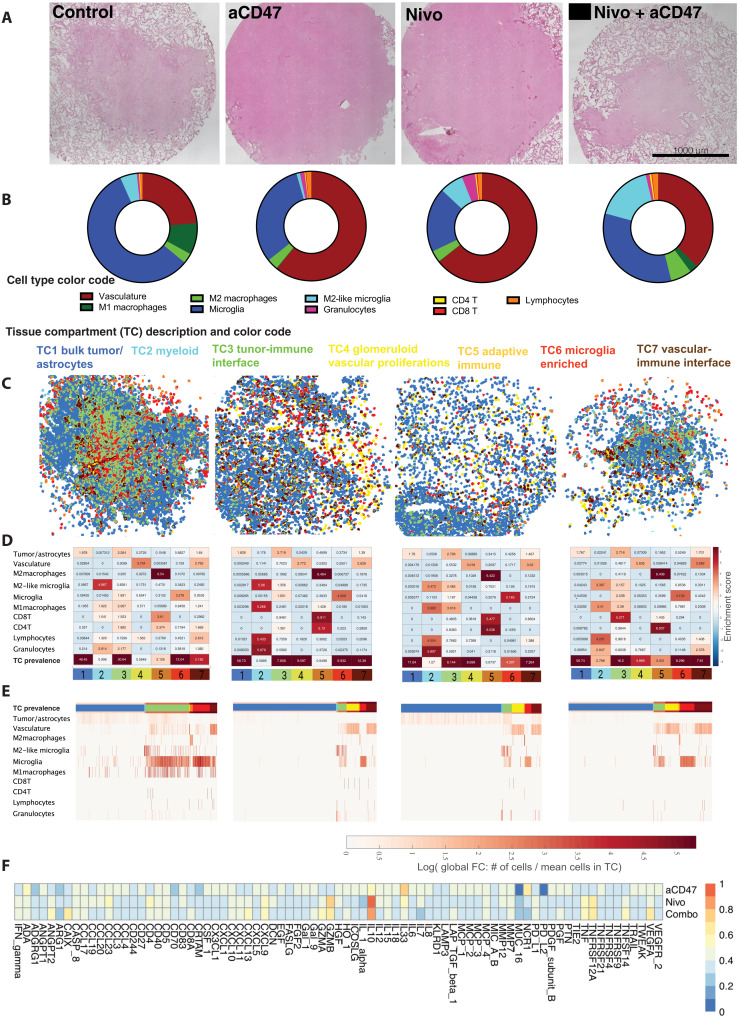
Fig. 4. Personalized local immunotherapy assessments in GBM explants by integrated analysis of soluble proteins and spatial iTME rearrangements.
Representative example of patient-specific treatment assessment toward “local” immunotherapy (patient 583, center explant). (A) H&E-stained tissue sections of bioreactor samples after 7 days of immunomodulatory treatment, after CODEX run. (B) Cell frequency distribution pie chart per specific explant, excluding tumor cells and astrocytes. (C) TC mapping and overlay on tissue sections based on x-y coordinates. (D) Individual TC enrichment plots depicting fold change of cell type enrichment per TC, and TC prevalence. (E) Individual heatmap of the TC composition (percentage of each cell phenotype per TC) after SOM clustering. Individual TCs are denoted by the color bar at the top of the graph. (F) Individual cytokine profiles resulting from multiplexed proximity extension assays, displayed as fold change in relation to untreated controls. Values are sorted from low to high based on the combinatorial treatment condition (nivolumab + anti-CD47).
The higher frequency of TC5 in tumor center samples suggests a higher activity of adaptive immunity there. Vascular TCs (TC4 and TC7) were more abundant in the tumor periphery, which could portend neovascularization at the tumor invasive front (27). When attempting to stratify the composition of the individual TCs according to individual treatments and location of the biopsies, no clear trends of composition changes were noticed (fig. S7).
Cell-cell correlations within TCs have distinct patterns depending on biopsy location
Next, we computed cell-cell correlations for each TC across all explants, comparing tumor center and periphery. These relationships were quantified using the Pearson correlation coefficients of the numbers of cells per TC (Fig. 3E). These cell-cell spatial correlation analyses indicate cell populations preferentially colocalizing in the same TC. Notable differences of cell-cell correlation were observed in certain TCs. For example, adaptive immune cells correlated more strongly with vasculature in TC4 (glomeruloid vascular proliferations) in samples from the tumor center compared to those from the periphery (Fig. 3E, TC4, center). This indicates a higher lymphoid cell infiltration of vascular proliferates in the tumor center. Furthermore, CD4+ T cells negatively correlated with tumor cells, vasculature, and M2 macrophages in TC5 (adaptive immune compartment) in samples from the periphery, suggesting a blunted CD4+ T cell response at the tumor invasive front (Fig. 3E, TC5, periphery). In TC6 (microglia-enriched compartment), microglia and M2 macrophages correlated more strongly with tumor cells in samples from the tumor center, as did M2 microglia and M1 macrophages (Fig. 3E, TC6, center). These correlations point toward a stronger GAM-mediated immune response within the tumor center.
In summary, our spatial and cell-cell correlation analyses comparing explants from tumor center and periphery suggest that innate and adaptive antitumoral immune responses are enriched within the tumor center, whereas neovascularization and vasculature-associated processes are more prominent at the tumor periphery. This could have implications for designing locally targeted immunotherapies and antiangiogenic therapies, especially for tumor recurrences.
Patient-personalized immunotherapy assessment by integrating explant-specific TC immune cell enrichment and secreted protein profiles
To pinpoint the effects of immunotherapies targeting innate and adaptive immune cell types in our patient-personalized model, we integrated the multidimensional information from both spatial imaging data and soluble factor profiles after 7 days of bioreactor treatment. Despite carefully navigated biopsies, explants from patient 580’s tumor periphery contained areas with higher cell density, whereas patient 583’s center explants partly showed a lower cell density. This indicates a greater regional heterogeneity in these tumors, which was not fully captured by 5-ALA neuronavigation (fig. S4C). Moreover, tumor heterogeneity was prominent across patients, was higher in tumor center than periphery, but was relatively low within each patient’s center and periphery explants under the different treatment conditions (fig. S6). For example, in patient 587, we observed a relatively homogeneous distribution of cell densities in all tumor center explants (Fig. 4A), whereas some variation was seen in the explants from other patients (fig. S4). Treatment-specific cell type and TC compositions were analyzed, and results are displayed as cell type pie charts (Fig. 4B and fig. S6), TC overlay plots (Fig. 4C and fig. S6), TC-specific cell type enrichment heatmaps (Fig. 4D and fig. S6), and TC-specific cell type composition heatmaps (Fig. 4E and fig. S6). In this exemplary tumor, immunotherapy-treated tumor center explants showed increased enrichment of CD8+ and especially CD4+ T cells in TC5 (adaptive immune compartment) compared to the untreated control (Fig. 4D, TC5 orange, immunotherapies versus control). Moreover, we observed an increased enrichment of M1 macrophages, granulocytes, and lymphocytes in TC2 (myeloid compartment) in immunotherapy-treated explants (Fig. 4D, TC2 light blue, immunotherapies versus control).
Interferon-γ levels distinguish immunotherapy responder and nonresponder tumor explants
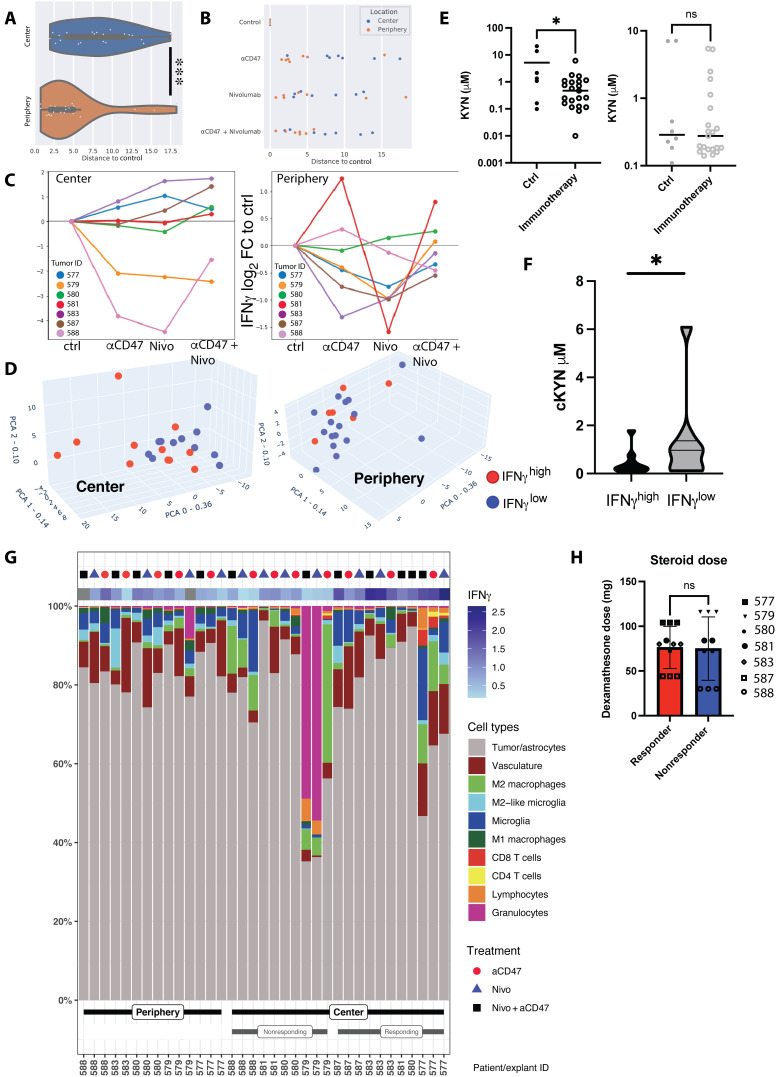
To establish additional criteria for the differentiation between explants responding or not to immunotherapy, we measured 92 soluble proteins including cytokines and chemokines in the media of each bioreactor. For example, in patient 587, we observed increases in several cytokines including interferon-γ (IFNγ) after immunotherapy (Fig. 4F and fig. S5). Explants from the tumor center displayed higher global soluble protein levels than those from the periphery when integrating all 92 analytes per sample (Fig. 5A). This was irrespective of the type of immunotherapy applied; however, when comparing soluble protein levels of center versus periphery in each individual treatment group, no significant differences were identified (Fig. 5B). Nevertheless, many soluble proteins were present at significantly higher levels in immunotherapy-treated tumor center versus periphery explants, including IFNγ, interleukin-7 (IL-7), IL-15, and CXCL-13, among others (fig. S8).
Fig. 5. Stratification based on IFNγ levels in culture media reveals a subset of tumor center explants responding to immunotherapy.
(A) Global soluble protein analysis of bioreactor media in relation to their reference (control samples). Distances to control correspond to Euclidean distances between sample and the corresponding control on the first two components of the PCA of global cytokine response—36% and 14% of the total variance, respectively. (B) Relative distance to control of global soluble protein expression values among immunotherapy-treated center and periphery explants. (C) Log2 fold change of IFNγ secretion versus control in center and periphery, per individual patient, condition, and biopsy location. Samples with a treated/control ratio of >1 were counted as IFNγhigh, independent of the applied treatment regimen. (D) 3D PCA of global cytokine response in the center (left) and periphery (right) samples, overlaid with information on IFNγ secretion. Red dots, IFNγhigh; blue dots, IFNγlow. For individual PC weights, see fig. S9. (E) Measurement of kynurenine (KYN) levels in bioreactor supernatants after local immunomodulatory treatments depending on biopsy location. The left graph represents center biopsies, and the right graph represents periphery biopsies. For individual measurements per treatment modality, refer to fig. S11. (F) Pooled KYN values among immunotherapy-treated IFNγhigh responder and IFNγlow nonresponder explants (Student’s t test). (G) Cellular composition (as percentage of total cell count per explant) of individual treated explants stratified according to periphery, IFNγlow center, and IFNγhigh center. IFNγ values represent log2 fold change to untreated control explant. (H) Cumulative pre- and intrasurgical steroid doses (dexamethasone) in responding and nonresponding center explants. Statistics: (A) Mann-Whitney U test, (E) two-tailed Welch’s test, (F and H) Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.005.
To define immunotherapy “responder” and “nonresponder” explants, we used IFNγ levels as a surrogate marker of an antitumoral immune response (20). Compared to untreated controls, IFNγ levels were higher in media from a subset of tumor center explants in all immunotherapy conditions (Fig. 5C, left). In contrast, only few explants from the tumor periphery showed enhanced IFNγ levels upon immunotherapy (Fig. 5C, right). In tumor center explants, combination immunotherapy increased IFNγ levels more than did single treatments, and blocking PD-1 generally induced higher IFNγ levels than did blocking CD47 (Fig. 5C, left). Combination immunotherapy of explants from the tumor periphery only increased IFNγ levels in patient 580, whose peripheral samples had a high cell density similar to tumor center biopsies (fig. S4), and in patient 581 (tissue from tumor periphery not available for analysis).
In addition, to stratify explants and investigate immune response–related protein signatures in an unbiased manner, we performed PCA analysis and displayed explants with high IFNγ levels (IFNγhigh) as blue symbols and those with low IFNγ levels (IFNγlow) as red symbols (Fig. 5D). In the periphery, no clear segregation pattern was observed, and IFNγ levels were generally uniformly distributed (Fig. 5D, right). In contrast, in the tumor center, IFNγhigh responder explants were associated with higher values in the two most highly weighted PCs (Fig. 5D, left). PC0 with the highest weight (36%) was characterized by CASP-8, LAP-TGFβ, CSF1, CD40, TNF, MMP7, GAL9, and TNFRSF12A, while PC1 (weight: 14%) was determined by IL-8, followed by myeloid chemokines MCP1, MCP3, CCL4, MCP2, IL-6, and CCL20. In PC2 (weight: 9%), TIE, TRAIL, and CD244 were the most important determinants (fig. S9).
Furthermore, in association with IFNγ, area under the curve (AUC) receiver-operating characteristic (ROC) analysis identified other mediators that were potentially associated with an immunotherapy-induced immune response: VEGFA, CXCL10, CXCL9, CCL23, GZMH, IL10, TNFRSF12A, and CD4 reached an AUC of >0.78 (fig. S10A). Heatmap clustering of IFNγhigh and IFNγlow explants from the tumor center and the immune-blunted periphery samples visualized the distinct, patient-specific cytokine profiles per treatment (fig. S10B).
Reduced kynurenine levels in media from immunotherapy-treated tumor center explants
As another surrogate marker for immunotherapy response, we measured the metabolite kynurenine (KYN) by liquid chromatography–mass spectrometry (LC-MS) in bioreactor media after 7 days of culture. KYN is produced from the amino acid l-tryptophan by the enzymes IDO and tryptophan 2,3-dioxygenase (TDO), which have been linked to immunosuppressive iTME states (28). In untreated control explants, KYN concentrations tended to be higher in the media of tumor center compared to periphery (mean, 5.5 versus 2.1 μM; P = 0.08; fig. S11). Immunotherapy-treated center explants had significantly lower KYN levels than untreated explants (mean, 0.8 versus 5.1 μM; P = 0.01); this effect was not observed in explants from the periphery (mean, 1.0 versus 1.6 μM; P = 0.23; Fig. 5E). KYN levels in bioreactor media inversely correlated with IFNγ levels when stratifying the immunotherapy-treated explants from tumor centers into IFNγhigh responders and IFNγlow nonresponders (Fig. 5F). Individualized assessment showed changes per treatment and location, and in a subset of explants, lower KYN levels after both anti-CD47, anti–PD-1, or combination immunotherapy were observed, even in explants from the periphery (fig. S11). These findings support the notion of an overall stronger activation of antitumoral immunity upon immunotherapy in explants from the tumor center.
Neoadjuvant GBM therapy and high numbers of intratumoral myeloid cells might portend tumor explant immunotherapy nonresponse
We compared the cellular composition of IFNγhigh responder and IFNγlow nonresponder explants in our cohort (Fig. 5G) and attempted to correlate the explant response to clinical behavior. While no statistically significant differences in cell type composition were seen between responding and nonresponding explants, the iTME composition of individual explants could potentially bear important information on the later clinical disease course. The nonresponder explants from patient 579 [mesenchymal subtype, methylguanine DNA methyltransferase (MGMT) unmethylated, overall survival 505 days] had very high numbers of granulocytes and M2 macrophages, which might be responsible for an intense immunosuppression in this patient’s explants. The other nonresponding explants were from patient 588 (mesenchymal subtype, MGMT methylated), who was initially biopsied and underwent neoadjuvant chemotherapy with temozolomide and radiation therapy before surgical resection. This patient’s tumor center explants contained high numbers of immunosuppressive M2 macrophages, which could explain nonresponse. Moreover, the overall survival of patient 588 was only 204 days. In patients 580 and 581 (both classical subtype, MGMT methylated), we observed an IFNγhigh response in tumor center explants only upon combination immunotherapy. Treated explants from the tumor periphery also displayed IFNγhigh responses in those patients, despite their rather low immune cell contents. Those two patients had better clinical outcomes, with an overall survival of 651 days (patient 580) and >700 days (patient 581), with patient 581 being alive at the time of concluding this study.
IFNγhigh responder explants show distinct immune cell compositions in their TCs
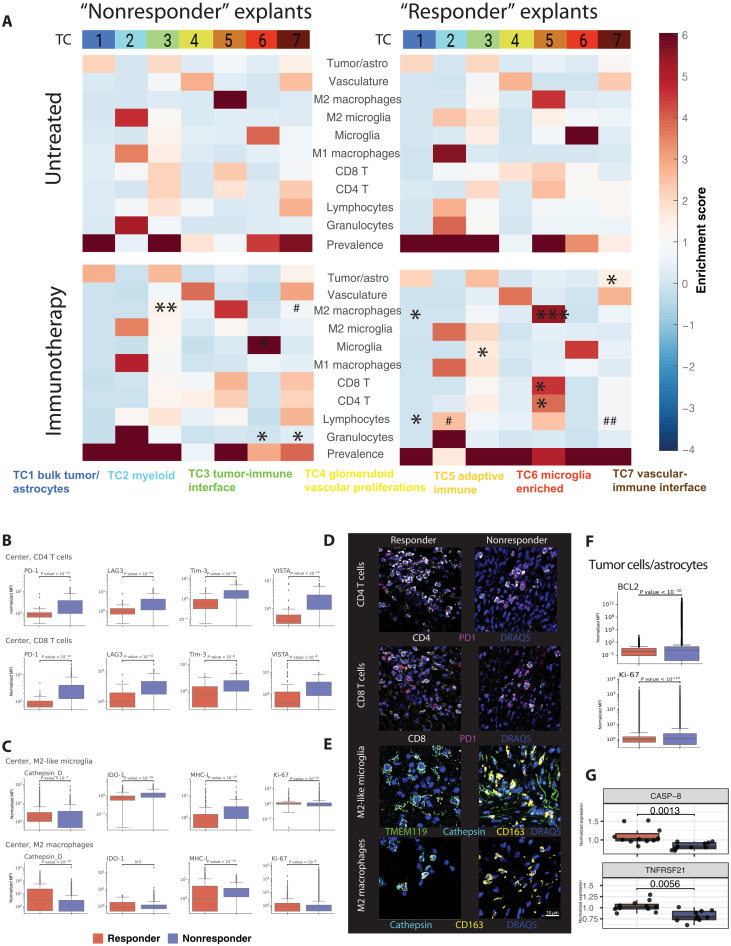
We next used the information derived from the soluble protein analysis to identify potential immunotherapy-induced changes in iTME architecture. Because an overall treatment response was mainly detectable in explants from the tumor center, we focused our analysis on this group and differentiated between immunotherapy-treated responder and nonresponder explants. Differences between these groups were assessed in TC enrichment diagrams to depict TC prevalence and cell type composition per TC (fold change; Fig. 6A). In nonresponders, no significant changes within the TCs and their prevalence were detected after treatment. In responders, immunotherapy led to a significant lymphocyte diminution in TC2 (myeloid compartment) and TC7 (vascular-immune interface; Fig. 6A, bottom right, # in TC2, ## in TC7).
Fig. 6. Immunotherapy induces distinct cellular composition shifts within TCs and changes cellular expression levels of functional markers in responding explants.
(A) Enrichment score (ES) of cellular composition in center samples, stratified by IFNγ secretion. Horizontal analysis between treated samples, IFNγlow versus IFNγhigh (asterisks in condition with higher FC display significant results): TC5: CD8+ T cells, mean ES from 1.8 to 4.24; CD4+ T cells, mean ES from 1.9 to 4.17; M2 macrophages, mean ES from 4.07 to 5.69; TC6 microglia, mean ES from 5.03 to 4.38. Vertical analysis between untreated and treated samples (# in conditions with higher ES display significant results). (B) Normalized inhibitory checkpoint molecule expression (mean fluorescent intensity within a specific cell type) on CD4+ and CD8+ T cells in IFNγhigh (responder) versus IFNγlow (nonresponder) center explants. (C) Normalized expression of functional markers and surrogates of immunosuppression in M2-like microglia and M2 macrophages in IFNγhigh versus IFNγlow center explants. For other cell types and markers, refer to fig. S12. (D) Immunofluorescent PD-1 expression on CD4+ and CD8+ T cells from CODEX-stained explants in a responding explant (patient 577, combinatorial condition) and a nonresponding explant (patient 579, aCD47 condition). Cells were identified on the basis of cell passports in fig. S3. (E) Immunofluorescent cathepsin D expression on M2-like microglia and M2 macrophages from a responding explant (patient 583, combinatorial condition) and a nonresponding explant (patient 588, combinatorial condition). Scale bar, 10 μm. (F) Normalized expression of anti-apoptotic protein BCL-2 and proliferation marker Ki-67 in IFNγhigh versus IFNγlow center explants. (G) Normalized protein expression from explant culture supernatant of CASP-8 and TNFRSF21 in IFNγhigh versus IFNγlow center explants. Statistics: (A) *P < 0.05 or #P < 0.05, **P < 0.01 or ##P < 0.01, ***P < 0.005 or ###P < 0.005, two-tailed Welch t tests, (B and C) Kruskal-Wallis H test with correction for multiple comparisons for median marker expression values.
Moreover, when comparing treatment outcomes between nonresponders and responders, the most prominent differences affected TC5 (adaptive immune compartment), where a significantly higher enrichment of CD4+ and CD8+ T cells, as well as M2 macrophages was observed in immunotherapy-treated responders (Fig. 6A, * in bottom panels). Conversely, in immunotherapy-treated nonresponder explants, M2 macrophages were more enriched in TC3 (tumor-immune interface). Among treated samples, region prevalences of TC4 (responders, 5.8%; nonresponders, 1%; P = 0.001) and TC7 (responders, 7.5%; nonresponders, 3.7%; P = 0.01) were increased in the responding samples (fig. S7C). This confirmed not only intra-TC composition shifts but also overall TC prevalence changes upon immunomodulatory treatments.
Cell type–specific functional marker expression profiles indicate innate and adaptive immune activation in explants responding to immunotherapy
We then investigated whether individual cell types within explants changed their phenotypic and functional properties upon immunotherapy. In line with our previous observations, diverging activation patterns among cell types per tumor region were noted: CD4+ T cells in center explants from responders displayed lower surface levels of the immune checkpoints PD-1, LAG-3, TIM-3, and VISTA. In contrast, CD4+ T cells from the periphery showed no significant changes in the expression of those immune checkpoints between immunotherapy and control conditions (Fig. 6, B and D, and fig. S12).
Similar observations were made for CD8+ T cells (Fig. 6, B and D). Furthermore, CD8+ T cells expressed lower levels of IDO-1, which is consistent with our findings of lower KYN levels in this group (figs. S10 and S12). The “lymphocyte” cluster showed an alternate pattern in terms of inhibitory immune checkpoint expression with partial up-regulation of PD-1, LAG-3, and TIM-3 in both center and periphery. This subset was driven to proliferate in the responder center explants as assessed by enhanced Ki67 staining intensity (fig. S12, lymphocytes).
M1 macrophages displayed a double-sided response among responders and nonresponders. Overall, a stronger proliferation (Ki67) was observed in the responding center explants, albeit accompanied by higher levels of IDO-1. The immune checkpoints PD-1, LAG-3, TIM-3, and VISTA were up-regulated in responders; conversely, the lysosomal enzyme cathepsin D, suggestive for a higher phagocytic activity, was also elevated (29). We did not observe marked differences between center and periphery (fig. S12, M1 macrophages).
Changes in M2 macrophages, which were the most abundant myeloid immune phenotype in the cohort and are an important cell population to address in GBM immunotherapy, were more pronounced in responders: Ki67 and cathepsin D were significantly enhanced in IFNγhigh samples. Major histocompatibility complex I (MHC-I), a potential “don’t eat me signal” (30), was down-regulated in responding center explants. Also, IDO-1 was down-regulated in the responder periphery upon treatment. M2 macrophages might be the most important source of IDO-1 activity in the tumors under investigation, and the suppression of IDO-1 in this cell type could be a surrogate for an effective intratumoral immune response. In terms of other inhibitory checkpoints, a down-regulation of PD-1, TIM-3, and LAG-3 and, in contrast, an up-regulation of the macrophage checkpoint VISTA were observed (Fig. 6, C and E, and fig. S12, M2 macrophages), which might equally have implications on M2 macrophage tumoricidal activity (31).
Last, we assessed marker emergence in microglia and M2-like microglia. In line with findings in M2 macrophages, the phagocytic marker cathepsin D was elevated in responding samples, whereas expression of IDO was lower in this group, pointing toward reduced immunosuppressive features. Moreover, responsive microglia tended to proliferate more in both center and periphery, independent of their polarization state. In all three examined treatments, we found a profound down-regulation of MHC-I. Concerning classical immune checkpoint expression, microglia and M2-like microglia tended to down-regulate PD-1, but the potential compensatory immune checkpoint VISTA was increased in responding samples (Fig. 6, C and E, and fig. S12, microglia and M2-like microglia).
Although we were focusing on the iTME in this study, we also noted specific marker intensity changes in tumor cells/astrocytes as a consequence of iTME remodeling. Surrogates of an antitumoral response included the down-regulation of CD47 and PD-L1 in both center and periphery responder explants, lower levels of tumor cell–expressed IDO-1, and a less pronounced cellular proliferation as detected by lower Ki-67 staining intensities (Fig. 6F and fig. S12). Furthermore, the anti-apoptotic protein BCL-2 was down-regulated in responder explants (32), thus indicating a potential propensity of these tumor cells to undergo apoptosis (Fig. 6F). Confirmative, the levels of soluble proapoptotic caspase-8 (CASP-8) (33) and death receptor 6 (TNFRSF21) (34) were higher in the media of responder explants, corroborating the antitumoral effect of the applied immunotherapies (Fig. 6G). Together, this study provides an in-depth proof-of-concept analysis of a potential immunotherapy response in patient-derived GBM explants with divergence between tumor center and periphery.
DISCUSSION
The iTME in GBM is a complex, highly immunosuppressive ecosystem. To date, immunotherapies against GBM have mostly failed. However, data in systemic, neoadjuvant application of T cell checkpoint inhibitors point toward a tumor-intrinsic immune response (24).
Here, we modeled the iTME of IDH wild-type GBM and its potential amenability to local immunomodulatory treatments in a patient-individualized way. For this, we combined multiple technologies including neuronavigated intraoperative tissue acquisition, live, intact 3D tissue cultures, GBM-relevant immunotherapy, FFPE-CODEX, soluble protein analysis, and metabolomics.
Our integrative framework enabled us to describe components of the GBM iTME in depth and to monitor immune-mediated responses on a combined spatial and biomarker-based basis. We identified cell composition differences between center and periphery, described seven conserved TCs of the GBM iTME across explants, and probed their organization, functional states, and communication.
In a subset of tumor center explants, we identified solid surrogates of an intratumoral immune response by TC shifts, intra- and inter-TC composition changes, distinct functional and phenotypic changes of the annotated cell types under question, and a favorable reversion of immunosuppressive features. To our surprise, a strong adaptive immune response was almost exclusively ensuing in the center explants, whereas the peripheral invasion zone acted as an “immune desert” zone and displayed much lower responses in both cellular rearrangements and soluble factor levels. This observation might be of translational clinical relevance and might support the concept of local neoadjuvant immunomodulatory treatments before bulk tumor resection or after reemergence of contrast-enhancing tumor areas.
However, the periphery was not completely inert to immunotherapies. At least in responding tumors, the dominating M2-like microglia and M2 macrophages substantially changed their marker profile toward antitumoral action. Although the study was iTME centered, we also observed perturbations of the tumor cells/astrocytes in responding explants. Responding explant tumor cells/astrocytes had lower levels of CD47, PD-L1, IDO-1, and CD73. These expression level differences on the tumor cells might impede a successful antitumoral immune response but could also have been induced by a successful reversion of the iTME by the treatments under investigation. Clearly, responding explants displayed lower tumor cell proliferation, which serves as a direct surrogate of antitumor efficacy. The detection of higher levels of cell death–associated molecules such as CASP-8 or TNFRSF21 in the media of responding explants suggested the presence of antitumoral activity.
Before surgery and intraoperatively, patients were exposed to a short course of relatively high doses of corticosteroids. In studies of immune checkpoint inhibitors for recurrent GBM, long-term steroid use was negatively associated with a successful immune response (35). In our cohort, there was no difference in the steroid dose the patients received for responding and nonresponding explants, and the varying patient-specific doses were unlikely to be responsible for the observed differences in the immunological and antitumoral phenomena.
Recent discoveries in ex vivo culture systems have equally unraveled the potential of tumor-resident T cells to contribute to an intratumoral immune response upon T cell checkpoint inhibition, albeit in extracerebral tumors (20). However, patient-derived data on innate immune modulators in a prototypic, myeloid-dominated, immunologically “cold” tumor such as GBM are scarce.
Efforts to model GBM ex vivo in a patient-centered fashion are needed to find potential treatment combinations that are directly translatable to patient care. Few reports on other 3D GBM ex vivo culture systems have been described: For example, GBM organoids serve as a valid tool to study personalized therapies and can even be cocultured with chimeric antigen receptor T cells (36). However, organoids lack the integration of microenvironment constituents and are therefore more suited to study direct tumor targeting drugs. Furthermore, 3D bioprinted and vascularized models have been established, which artificially add components of the GBM iTME such as microglia to the bioprinted system (37). Other authors used spheroids, organotypic slice cultures, or tumoroids to study the tumor complexity, as reviewed by Soubéran and Tchoghandjian (38). However, most of the current models lack an integrative GBM iTME, which has important implications for immunotherapy response assessment. The perfused bioreactor approach partly overcomes this limitation with the caveat that the contribution influx of immune cells from the peripheral circulation was not assessed in our proof-of-concept study. Supplementing the bioreactor system with, e.g., patient-matched peripheral blood mononuclear cells, expanded tumor-infiltrating lymphocytes, natural killer (NK) cells, or tumor-specific chimeric antigen receptor T cells could overcome this limitation. This would significantly expand the scope of the bioreactor system and enable the analysis of interactions of added cells with the iTME, and their dynamics could be studied more in detail to elucidate their contribution to successful antitumoral immunity.
Our proof-of-concept study has several limitations. Although we were analyzing more than 800,000 single cells in depth, the patient sample size of the study was small, limiting the interpretation of systematic correlations with clinical outcomes, survival, or transcriptomic GBM subtype or methylation data. However, we identified two nonresponding patients who either were pretreated by standard of care therapy (patient 588) or had a massive immunosuppressive granulocyte and M2 macrophage infiltration (patient 579). These explants were not able to raise a sufficient IFNγ-associated response and had no spatial rearrangement of adaptive immune cells within their iTME despite their high cellularity and presence of adaptive immune cells. Furthermore, two patients (patents 580 and 581) displayed an intermediate or “partial” response based on the cytokine exploration, and combinatorial immunotherapies with both anti-CD47 and anti–PD-1 antibodies raised an IFNγ response, which hinted toward a synergistic effect of targeting both innate and adaptive immune compartments. These two patients had the longest overall survival among this cohort, but this might also be related to other more favorable prognostic factors including the patients’ relatively young age and positive MGMT status.
Furthermore, intratumoral heterogeneity even within patient-matched experimental series posed a problem to the interpretation of the results, necessitating more biopsies per treatment condition. The small sample size forced us to pool different immunotherapy modalities (e.g., T cell checkpoint inhibition versus myeloid checkpoint inhibition) in the TC composition analysis and precluded us from identifying cellular reactions to either modality alone.
The granularity of our clustering strategy, especially concerning T cell subsets, was lower than expected in our study. We were not able to identify NK T cells, Tregs, or B cells with certainty, and summarized these cells, if they were present, into a lymphocyte cluster that precluded us from a more detailed T or B cell analysis. Moreover, deeper subclustering of macrophage and microglia populations and their states could be achieved by methods such as single-cell RNA sequencing (scRNA-seq) (6, 39, 40) or targeted mass cytometry panels. In that regard, our study was limited in the overall tissue availability and parallelization of bioreactor experiments to perform scRNA-seq of a matched series of explants alongside CODEX. In addition, as opposed to CODEX, spatial cell-cell interactions can only be partially inferred from scRNA-seq data (41). However, we are confident that the normalized functional marker expression analysis in the T cells within explants strictly speaks in favor of phenotype switches in the cell types under question. Hence, GBM is dominated by myeloid cells, and we provide a solid phenotypic characterization of microglia and macrophage subsets within GBM, despite the absence of transcriptional activation state information. After treatment, compensatory checkpoints were up-regulated, e.g., in the case of the checkpoint protein VISTA (V-type immunoglobulin domain-containing suppressor of T cell activation) in both center and periphery M2 macrophages and microglia responding explants. Whether this observation represents an important immune escape or resistance mechanism is subject to further investigation but underscores the plasticity of the innate iTME upon therapeutic interventions. While disruption of the CD47-Sirpa axis (17–19) and PD-1 blockade (31) lead to tumor cell phagocytosis, we were unable to directly visualize this primordial mode of action in our explant system because of several reasons: (i) A universal GBM-specific tumor cell marker or a fluorescent label for tumor cells, used, e.g., in 2D phagocytosis assays, is missing in our system; (ii) these markers might be disintegrated after tumor cell phagocytosis by macrophages; (iii) the very dense composition of tumor cells and their overlay with intermingled macrophages/microglia in histoarchitectonically intact explants makes it almost impossible to tell apart multinucleated macrophages from macrophages with ingested tumor cells. On the other hand, we are confident that the assessed surrogate markers for macrophage activation, such as lysosomal cathepsin D induction, as well as the cytokine profiles in responding explants are suggestive of an active macrophage and microglia response after anti-CD47 (and anti–PD-1) treatment.
Our study focused on immune cells and included only few tumor cell markers that impeded us from discriminating tumor cells from reactive astrocytes and from assigning different oncogenic driver mutations to tumor cell subclones. Nevertheless, we were able to assess potential predictive markers such as CD47 or PD-L1 quantitatively on tumor cells/astrocytes with implications on immunotherapy response.
Overall, the proposed strategy of combining complex 3D tissue culturing with multiplexed imaging and targeted secretome analysis is not yet standardized and far from reaching implications for clinical practice. Here, demonstrated the feasibility of this concept, and future developments may implicate to concentrate patient stratification efforts on miniaturization of bioreactor systems, more center biopsies per patient, fewer informative functional iTME markers, and IFNγ-associated cytokines with predictive capacity. This approach would enable us to test a variety of immunotherapy combinations and tumor-targeting drugs on a larger patient cohort and lead to key data that could be correlated with clinical outcomes.
MATERIALS AND METHODS
GBM tissue collection and patient characteristics
Biopsy samples from contrast medium–enhancing tumor center and nonenhancing tumor periphery were collected during surgery via intraoperative navigation. From each patient, 5-ALA–positive, vital tumor center biopsies as well as biopsies from the 5-ALA–low/negative tumor periphery/infiltration zone were obtained. Tissue acquisition was documented by intraoperative imaging (taking into consideration 5-ALA positivity and neuronavigation; fig. S1). In total, seven tumor samples from isocitrate dehydrogenase 1/2 wild-type primary GBM were included in this study. One patient (ID 588) was pretreated with temozolomide and radiation therapy before resection. Patient characteristics, survival data, molecular and histopathological data, as well as pre-, intra-, and postoperative steroid exposure are summarized in Fig. 1B and table S1. Magnetic resonance images from the localizations of patients’ tumor biopsies are shown in fig. S1. Written informed consent was obtained from all patients, and the study was approved by and conducted according to the guidelines of the local ethics committee (Ethikkommission Nordwestschweiz, #42/10).
Ex vivo perfusion bioreactor cultures
Bioreactor cultures under perfusion were performed according to the manufacturer’s instructions (Cellec Biotek AG, Basel, Switzerland). Briefly, fresh, intact GBM tissue explants were placed into ice-cold Neurobasal-A medium (Life Technologies, #21103049) and immediately taken to the laboratory (less than 30 min). Tissues from the center and periphery of the tumor were cut into ~20- to 30-mm3 fragments. This size was selected on the basis of preliminary experiments, which showed improved survival with limited cutting-related damage. Tissue fragments were placed between two 10-mm-diameter discs of microfibrillar collagen hemostat sheets (Ultrafoam, Avitene) prewet with phosphate-buffered saline (PBS) for 1 hour at 37°C, 5% CO2. Silicone adaptors and ethylene-tetrafluoroethylene copolymer mesh grids were arranged on the top and bottom of the collagen scaffold, and the tissues were placed into U-CUP perfusion chambers (Cellec Biotek AG). The perfusion media consisted of a 50:50 mix of Neurobasal-A medium (Life Technologies, #21103049) and Dulbecco’s modified Eagle’s medium/F12 medium (Gibco, #21331020) supplemented with nonessential amino acids (1×; Sigma-Aldrich, #M7145), 1 mM sodium pyruvate (Sigma-Aldrich, #S8636), 44 mM sodium bicarbonate (Gibco, #5080094), 25 mM Hepes (Gibco, #156301), 4 mM l-alanyl-l-glutamine (Corning, #25-015-CI), antibiotic-antimycotic (1×; Gibco, #LS15240062), human recombinant epidermal growth factor (20 ng/ml; Gibco, #PHG0314), human recombinant fibroblast growth factor (20 ng/ml; Gibco, #AA10-155), heparin sulfate (10 ng/ml; STEMCELL Technologies, #07980), and 5% human serum (Sigma-Aldrich, #H4522-100ML). The perfusion flow rate was set at 0.47 ml/min, resulting in a superficial flow velocity of 100 μm/s. Starting from day 0, bioreactor media were either left untreated or supplemented with the anti–PD-1 antibody nivolumab (Opdivo, Bristol Myers Squibb; 5 μg/ml) or anti-CD47 antibody (Bio X Cell, #BE0019-1; 5 μg/ml), alone or in combination. On day 7, culture media were frozen at −80°C and tissues were fixed in 10% neutral buffered formalin (Sigma-Aldrich, #HT501128) for 24 hours and processed according to standard histopathology procedures for paraffin embedding (42).
Generation of CODEX DNA-conjugated antibodies
Purified, carrier-free monoclonal and polyclonal antibodies (data S1) were conjugated to short DNA oligonucleotides (Trilink Biotechnologies) as described before (25, 43, 44). Briefly, antibodies were concentrated on 50-kDa molecular weight cutoff centrifugal filter columns (Amicon Ultra, EMD Millipore, #UFC505096), and sulfhydryl groups were activated using a mixture of 2.5 mM TCEP (Sigma-Aldrich, #C4706-10G) and 2.5 mM EDTA (Sigma-Aldrich, #93302) in PBS (pH 7.0) for 30 min at room temperature. Toluene-deprotected, lyophilized, maleimide-modified DNA oligonucleotides were then conjugated to the antibodies at a 2:1 weight/weight ratio for 2 hours, with at least 100 μg of antibody per reaction. Conjugated antibodies were washed, eluted in PBS-based antibody stabilizer (Thermo Fisher Scientific, #NC0436689) containing 5 mM EDTA and 0.1% NaN3 (Sigma-Aldrich, #S8032), and stored at 4°C.
CODEX antibody validation, titration, and staining
Validation and titration of the CODEX antibodies, as well as the stainings of FFPE GBM bioreactor tissues were performed as described before (25, 43). Briefly, DNA-conjugated antibodies were first validated on tonsil tissues or a multitumor tissue microarray under the supervision of a board-certified surgical pathologist (C.M.S.), and staining patterns were confirmed with online databases [The Human Protein Atlas, www.proteinatlas.org (45); Pathology Outlines, www.pathologyoutlines.com] and the published literature. GBM bioreactor tissues were punched out of their respective paraffin blocks using 4-mm-diameter punches, assembled into arrays, sectioned at 4 μm, and placed on 22 × 22 mm glass coverslips (Electron Microscopy Sciences, #72204-01) pretreated with Vectabond (Vector Labs, #SP-1800) according to the manufacturer’s instructions. Coverslips were baked at 70°C for 1 hour and deparaffinized, and heat-induced epitope retrieval was performed in a LabVision PT module (Thermo Fisher Scientific) using Dako target retrieval solution (pH 9) (Agilent, #S236784-2) at 97°C for 10 min. After blocking for 1 hour at room temperature, 100 μl of antibody cocktail was added to the coverslip and staining was performed in a sealed humidity chamber overnight at 4°C on a shaker, followed by washing and fixation in 1.6% paraformaldehyde (10 min, room temperature), 100% methanol (5 min, 4°C), and BS3 (Thermo Fisher Scientific, #21580; 20 min, room temperature).
CODEX multicycle reaction and image acquisition
CODEX multicycle reactions and image acquisition were performed as described before (25, 43). Briefly, stained coverslips were mounted onto custom-made acrylic plates (Bayview Plastic Solutions) using coverslip mounting gaskets (Qintay, #TMG-22), and the tissue was stained with Hoechst 33342 (Thermo Fisher Scientific, #62249; 1:1000). Acrylic plates were mounted onto a custom-designed plate holder and secured onto the stage of a BZ-X710 inverted fluorescence microscope (Keyence). Fluorescent oligonucleotides (concentration: 400 nM) were aliquoted in black 96-well plates (Thermo Fisher Scientific, #07-200-762) in 250 μl of H2 buffer containing Hoechst nuclear stain (1:600) and sheared salmon sperm DNA (0.5 mg/ml; Thermo Fisher Scientific, #AM9680). DRAQ5 nuclear stain (Cell Signaling Technologies, #4084 L) was added to the last well at a dilution of 1:100. For details on the order of fluorescent oligonucleotides and microscope light exposure times, see data S1. Automated image acquisition and fluidics exchange were performed using a CODEX instrument and driver software (Akoya Biosciences) according to the manufacturer’s instructions, with slight modifications. Tissue overview images were acquired manually using a CFI Plan Apo λ 2×/0.10 objective (Nikon), and automated imaging was performed using a CFI Plan Apo λ 20×/0.75 objective (Nikon). After each multicycle reaction, H&E stainings were performed according to standard pathology procedures, and tissues were reimaged in bright-field mode. Staining quality, marker expression, and distribution were verified on each individual section according to (25, 43).
Computational image processing
Raw TIFF image files were processed, deconvolved, and background-subtracted using the CODEX Toolkit uploader and Microvolution software (Microvolution) as described before (25, 43, 46). Antibody stainings were visually assessed for each channel and cycle in each spot using ImageJ software (Fiji, version 2.0.0). Final figures of cell passports and composites for main cell clusters were generated using OMERO.web app (www.openmicroscopy.org/omero/figure/).
Cell segmentation
TIFF hyperstacks were segmented based on DRAQ5 nuclear stain, pixel intensities were quantified, and spatial fluorescence compensation was performed using the CODEX Toolkit segmenter as described previously (25), using the following settings: radius, 7; maximum cutoff, 1.0; minimum cutoff, 0.07; relative cutoff, 0.2; cell size cutoff factor, 0.4; nuclear stain channel, 4; nuclear stain cycle, 23; membrane stain channel, 1; membrane stain cycle, −1 (i.e., not used); use membrane, false; inner ring size, 1.0; Delaunay graph, false; anisotropic region growth, false. This generated comma-separated value (CSV) and flow cytometry standard (FCS) files for further downstream analysis.
Cleanup gating, unsupervised hierarchical clustering, and cluster validation
All background-subtracted FCS files were imported into CellEngine (https://cellengine.com). Gates were tailored for each file individually in a blinded manner by two experts in flow and mass cytometry (T.S. and C.M.S.). Nucleated cells were positively identified, and artifacts were removed by gating on Hoechst1/DRAQ5 double-positive cells, followed by gating on focused cells in the Z plane. After cleanup gating, FCS files were reexported and subsequently imported into VorteX clustering software, where they were subjected to unsupervised hierarchical X-shift clustering using an angular distance algorithm (47). The following data import settings were applied: numerical transformation, none; noise threshold, no; feature rescaling, none; normalization, none; merge all files into one dataset, yes. Clustering was based on the following antibody markers: CD8, Iba1, MPO, HLA-DR, CD45RA, GFAP, collagen IV, ɑ-SMA, CD15, CD206, TMEM119, CD11c, CD11b, S100A9, CD4, CD3, CD45, CD73, CD34, CD45RO, CD31, Ki-67, NGFR, CD2, CD163, CD68, and EGFR. The following settings were used for clustering: distance measure, angular distance; clustering algorithm, X-shift (gradient assignment); density estimate, N nearest neighbors (fast); number of neighbors for density estimate (K), from 150 to 5, steps 30; number of neighbors, determine automatically. The optimal cluster number was determined using the elbow point validation tool and was at K = 40, resulting in 173 clusters. Clusters and corresponding data were exported as a CSV file and were manually verified and assigned to cell types by overlaying the single cells from each individual cluster onto the stitched bioreactor images in ImageJ, based on the unique cluster identifiers and cellular X/Y position, using custom-made ImageJ scripts (available at https://github.com/bmyury/CODEX-fiji-scripts). Clusters with similar morphological appearance in the tissue and similar marker expression profiles were merged, and artifacts were removed. Unclear clusters were reclustered on the basis of the markers highlighted in Fig. 1D, rechecked individually on the stitched tissue slides, and merged back to the already annotated cell types, resulting in 10 final clusters.
CODEX marker expression analysis
Data normalization was achieved by log-transformed median normalization of each marker in each sample to their respective baseline control sample to focus on expression changes and allow comparisons between samples. Differences in median expression were assessed using the nonparametric Kruskal-Wallis H test (48), using Benjamini-Hochberg procedure to control the false discovery rate.
CytoMAP spatial analysis to identify TCs
CytoMAP (26) was written using MATLAB version 2018b (MathWorks). A detailed description of the workflow and functions built into CytoMAP is available in the online user manual (https://gitlab.com/gernerlab/cytomap/-/wikis/home). Below is a brief discussion of the analysis used for the datasets described in this manuscript. The annotated cell types for each dataset were loaded into CytoMAP by importing the corresponding CSV files. These collectively contain the cell type annotations derived from the final clustering, mean fluorescent pixel intensity values per cell for each marker, and spatial positions (centroids) for each cell object. Once imported, the following functions were used in CytoMAP to analyze the data.
Raster scan neighborhoods
This function identifies the local composition of cell types within a circular area in the tissue, which we here refer to as TC. This function calculates the number of cells and the mean fluorescent intensity of each channel summed over all cells in each TC. The positions of the TCs are evenly distributed throughout the tissue in a grid pattern with a distance between TC centers of half of the user-defined radius (r = 75 μm). The TC information was used for further analysis (e.g., local cellular densities and cell-cell associations).
Cell-cell correlation analysis
The local cell density within individual TCs was used to correlate the location of different cell types, revealing which cell populations preferentially associate with one another, or conversely avoid one another. This function calculates the Pearson correlation coefficient of the number of cell or object types within the scanned TCs and graphs these on a heatmap plot. This correlation analysis can be performed across multiple samples and can be done either over entire tissues or within specified tissue regions. This is important, as cells may have distinct associations with one another in different TCs. For analysis of TC frequencies and distributions among samples, TC prevalence and cell type composition per sample were exported and analyzed per treatment condition or location.
Multiplexed secreted protein analysis in explant culture media
Bioreactor media were centrifuged to remove cell debris, and 92 secreted proteins, including cytokines, chemokines, and soluble cell membrane proteins, were measured externally by proximity extension technology using the Olink immuno-oncology panel (www.olink.com/products/immuno-oncology). Data are presented as normalized protein expression values, Olink Proteomics’ arbitrary unit on a log2 scale. Missing data were associated with a lower median expression. They were imputed either as half the molecule detection threshold or such that the sum of all imputed values for a molecule is 0.1 of the sum of the molecule’s expressions (whichever is the smallest). The PCA was performed on centered and scaled data.
Determination of KYN in explant culture media by LC-MS
Stock solutions of KYN (Sigma-Aldrich) and D6-kynurenine (Cambridge Isotope Laboratories Inc.) were prepared at 5 and 10 mM in water. A series of seven standard solutions for the calibration curve were prepared at concentrations from 5000 to 39 nM by serial dilution. D6-kynurenine was diluted at 600 nM in LC-MS–grade methanol (Sigma-Aldrich). Supernatants and calibration solutions (5 μl) were quenched with ice-cold methanol containing the internal standard (D6-kynurenine at 600 nM). Samples were mixed and centrifuged at 3700g for 15 min. Twenty microliters of ultrapure water (supplied by a MilliQ Advantage A10 purification system, Merck Millipore) was added before injection, and the total dilution was ×10. High-performance LC (HPLC) was set up to inject directly from the supernatant 8 mm above the pellet. For instrumentation and chromatographic conditions, measurements were performed on HPLC Nexera X2 HPLC (Shimadzu) coupled to API 5500 (AB Sciex) with positive ion electrospray ionization and operated in multiple reaction monitoring (MRM) mode. Data were collected and processed by Analyst 1.6.2, and the chromatographic separation was carried out on Acquity HSS T3 1.8 mm, 2.1 × 50 mm (Waters Corp.) at 40°C. The separation method consisted of a gradient elution of the mobile phase (0.1% formic acid in A: water and B: acetonitrile) as follows: 0% B from 0 to 0.25 min, 10% B at 0.3 min, 15% B at 1.1 min, 90% B at 1.2 min, and held for 0.3 min before reequilibration. Total run time was 2 min, and all samples were analyzed with an injection of 2 ml. The source parameters were curtain gas (CUR), nitrogen at 20 pounds-force per square inch (PSIG), collision gas at 6 PSIG, ion source gas-1 at 20 PSIG; ion source gas-2 at 20 PSIG, ion spray voltage (IS) at 4500 V, turbo heater temperature (TEM) at 500°C, and entrance potential (EP) at 10 V. The electrospray ionization was operated in positive MRM mode after optimization according to standard procedure. Compound-specific values (mass transitions, declustering potential, and collision energy) of MS parameters were as follows for the two analytes: KYN: Q1 [mass/charge ratio (m/z)] 209.05; Q3 (m/z) 192.05; DP (V) 43.0; CE (V) 12.5; D6-kynurenine: Q1 (m/z) 215.05; Q3 (m/z) 198.05; DP (V) 40.0; CE (V) 14.0.
Statistical analysis
Data are reported as means ± SD or means ± SEM, and statistical significance was determined using the Mann-Whitney U test, two-tailed Wilcoxon test, Student’s t test, or Kruskal-Wallis test using Prism v9.0e (GraphPad), as specified in the respective figure legends. Differences were considered statistically significant if P < 0.05. Only significant P values are displayed. Correlations were evaluated using Pearson correlation. Computational analyses were performed in R v.4.0.2, CytoMAP, Prism v9.0e, or Python 3.8.8 (using packages: scipy 1.7.1, numpy 1.21.1, statsmodels 0.12.2, pandas 1.3.1, scikit-learn 0.24.1, matplotlib 3.4.2, seaborn 0.11.1). Experiments were performed without duplicates because of material restrictions.
The immunotherapy response based on IFNγ cytokine measurements was assessed by first calculating the ROC curves based on the delta values (immunotherapy-treated condition minus untreated condition) for each parameter measured. Parameters that were strongly associated with IFNγ were selected on the basis of the AUC-ROC curve (fig. S10A).
Acknowledgments
We thank A. Zippelius (Department of Oncology, University Hospital Basel, Switzerland), D. Merkler (Department of Pathology, University Hospital Geneva, Switzerland), J. Schittenhelm (Department of Pathology and Neuropathology, University Hospital Tübingen, Germany), C. Le Magnen (University of Basel), and S. Singh (Neurosurgery Department, University of Hamilton, Ontario, Canada) for critically reading the manuscript. The anti–PD-1 and anti-CD47 antibodies for bioreactor treatment were a gift from H. Läubli (Department of Oncology, University Hospital Basel, Switzerland). We are grateful to S. Frank and J. Hench (Department of Pathology, University Hospital Basel, Switzerland) for histopathological and molecular workup of the GBM specimens.
Funding: This work was supported by a Swiss National Science Foundation Professorial Fellowship (PP00P3_176974); the ProPatient Forschungsstiftung, University Hospital Basel (Annemarie Karrasch Award 2019); and the Department of Surgery, University Hospital Basel, to G.H. and by The Brain Tumour Charity Foundation, London, UK (GN-000562) to G.H. and C.M.S. Research in the G.P.N. laboratory was supported by the U.S. NIH (5U54CA20997103, IDIQ17X149), the U.S. DOD (W81XWH-14-1-0180), the U.S. FDA (DSTL/AGR/00980/01), Cancer Research UK (C27165/A29073), the Bill and Melinda Gates Foundation (OPP1113682), the Parker Institute for Cancer Immunotherapy, the Beckman Center for Molecular and Genetic Medicine, and the Rachford & Carlotta A. Harris Endowed Chair. C.M.S. was supported by an Advanced Postdoc Mobility Fellowship from the Swiss National Science Foundation (P300PB_171189, P400PM_183915) and an International Award for Research in Leukemia from the Lady Tata Memorial Trust, London, UK. T.S. was supported by an UICC Technical Fellowship (TF/18/625070). D.P. was supported by an NIH T32 Fellowship (AR007422), an NIH F32 Fellowship (CA233203), a Stanford Dean’s Postdoctoral Fellowship, and Stanford’s Dermatology Department. S.S.B. was supported by a Stanford Bio-X Interdisciplinary Graduate Fellowship and Stanford’s Bioengineering Department. G.L.B. was supported by an NIH T32 Fellowship (5T32AI007290-34).
Author contributions: Conceptualization: G.H. and C.M.S. Methodology: T.S., C.P.Z., E.M.B., W.D., A.T.W., T.A.M., S.Z., F.L.G., M.G.M., M.-F.R., C.M.S., and G.H. Clinical data and patient integration: P.S. and G.H. Software: C.P.Z., E.M.B., W.D., S.S.B., and G.L.B. Validation: T.S., C.P.Z., E.M.B., W.D., J.F., C.P., D.P., C.M.S., and G.H. Formal analysis: T.S., C.P.Z., E.M.B., W.D., A.T.W., M.M.E., C.M.S., and G.H. Investigation: T.S., C.P.Z., E.M.B., and G.H. Resources: T.S., M.G.M., C.M.S., and G.H. Writing—original draft: G.H. and C.M.S. Writing—review and editing: all authors. Supervision: G.H. and C.M.S. Project administration: G.H. and C.M.S. Funding acquisition: T.S., G.P.N., C.M.S., and G.H.
Competing interests: G.P.N. has equity in and is a scientific board member of Akoya Biosciences Inc. C.M.S. has received research funds from and is a scientific advisor to Enable Medicine Inc., both outside the current work. M.G.M. is a scientific advisor to Cellec Biotek AG and is inventor on a patent related to this work filed by University Hospital Basel and Cellec Biotek AG (nos. WO2015181185A1, EP3149154B1; US10473646B2; JP6833678B2; CA2950353A1, filed on 26 May 2015, published on 3 December 2015). G.H. has equity in and is a cofounder of Incephalo Inc. All other authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The single-cell data table of clustered, annotated cell types with metadata can be downloaded from Mendeley (https://data.mendeley.com/datasets/f9hfcfyt93/1).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J. B., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O., Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Allavena P., Mantovani A., Immunology in the clinic review series; focus on cancer: Tumour-associated macrophages: Undisputed stars of the inflammatory tumour microenvironment. Clin. Exp. Immunol. 167, 195–205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman R. L., Klemm F., Akkari L., Pyonteck S. M., Sevenich L., Quail D. F., Dhara S., Simpson K., Gardner E. E., Iacobuzio-Donahue C. A., Brennan C. W., Tabar V., Gutin P. H., Joyce J. A., Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 17, 2445–2459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyonteck S. M., Akkari L., Schuhmacher A. J., Bowman R. L., Sevenich L., Quail D. F., Olson O. C., Quick M. L., Huse J. T., Teijeiro V., Setty M., Leslie C. S., Oei Y., Pedraza A., Zhang J., Brennan C. W., Sutton J. C., Holland E. C., Daniel D., Joyce J. A., CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darmanis S., Sloan S. A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., Li G., Chang S. D., Connolly I. D., Li Y., Barres B. A., Gephart M. H., Quake S. R., Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klemm F., Maas R. R., Bowman R. L., Kornete M., Soukup K., Nassiri S., Brouland J.-P., Iacobuzio-Donahue C. A., Brennan C., Tabar V., Gutin P. H., Daniel R. T., Hegi M. E., Joyce J. A., Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebel E., Kapolou K., Unger S., Núñez N. G., Utz S., Rushing E. J., Regli L., Weller M., Greter M., Tugues S., Neidert M. C., Becher B., Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Reardon D. A., Omuro A., Brandes A. A., Rieger J., Wick A., Sepulveda J., Phuphanich S., de Souza P., Ahluwalia M. S., Lim M., Vlahovic G., Sampson J., OS10.3 randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: CheckMate 143. Neuro Oncol. 19, iii21 (2017). [Google Scholar]
- 9.Cloughesy T. F., Mochizuki A. Y., Orpilla J. R., Hugo W., Lee A. H., Davidson T. B., Wang A. C., Ellingson B. M., Rytlewski J. A., Sanders C. M., Kawaguchi E. S., Du L., Li G., Yong W. H., Gaffey S. C., Cohen A. L., Mellinghoff I. K., Lee E. Q., Reardon D. A., O’Brien B. J., Butowski N. A., Nghiemphu P. L., Clarke J. L., Arrillaga-Romany I. C., Colman H., Kaley T. J., de Groot J. F., Liau L. M., Wen P. Y., Prins R. M., Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 25, 477–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Rourke D. M., Nasrallah M. P., Desai A., Melenhorst J. J., Mansfield K., Morrissette J. J. D., Martinez-Lage M., Brem S., Maloney E., Shen A., Isaacs R., Mohan S., Plesa G., Lacey S. F., Navenot J. M., Zheng Z., Levine B. L., Okada H., June C. H., Brogdon J. L., Maus M. V., A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seliger B., Combinatorial approaches with checkpoint inhibitors to enhance anti-tumor immunity. Front. Immunol. 10, 999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuvolone M., Kana V., Hutter G., Sakata D., Mortin-Toth S. M., Russo G., Danska J. S., Aguzzi A., SIRPα polymorphisms, but not the prion protein, control phagocytosis of apoptotic cells. J. Exp. Med. 210, 2539–2552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takenaka K., Prasolava T. K., Wang J. C., Mortin-Toth S. M., Khalouei S., Gan O. I., Dick J. E., Danska J. S., Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat. Immunol. 8, 1313–1323 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Willingham S. B., Volkmer J. P., Gentles A. J., Sahoo D., Dalerba P., Mitra S. S., Wang J., Contreras-Trujillo H., Martin R., Cohen J. D., Lovelace P., Scheeren F. A., Chao M. P., Weiskopf K., Tang C., Volkmer A. K., Naik T. J., Storm T. A., Mosley A. R., Edris B., Schmid S. M., Sun C. K., Chua M. S., Murillo O., Rajendran P., Cha A. C., Chin R. K., Kim D., Adorno M., Raveh T., Tseng D., Jaiswal S., Enger P. O., Steinberg G. K., Li G., So S. K., Majeti R., Harsh G. R., van de Rijn M., Teng N. N., Sunwoo J. B., Alizadeh A. A., Clarke M. F., Weissman I. L., The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. U.S.A. 109, 6662–6667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schürch C. M., Forster S., Brühl F., Yang S. H., Felley-Bosco E., Hewer E., The “don’t eat me” signal CD47 is a novel diagnostic biomarker and potential therapeutic target for diffuse malignant mesothelioma. Oncoimmunology 7, e1373235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schürch C. M., Roelli M. A., Forster S., Wasmer M.-H., Brühl F., Maire R. S., Di Pancrazio S., Ruepp M.-D., Giger R., Perren A., Schmitt A. M., Krebs P., Charles R.-P., Dettmer M. S., Targeting CD47 in anaplastic thyroid carcinoma enhances tumor phagocytosis by macrophages and is a promising therapeutic strategy. Thyroid 29, 979–992 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gholamin S., Mitra S. S., Feroze A. H., Liu J., Kahn S. A., Zhang M., Esparza R., Richard C., Ramaswamy V., Remke M., Volkmer A. K., Willingham S., Ponnuswami A., McCarty A., Lovelace P., Storm T. A., Schubert S., Hutter G., Narayanan C., Chu P., Raabe E. H., Harsh G., Taylor M. D., Monje M., Cho Y. J., Majeti R., Volkmer J. P., Fisher P. G., Grant G., Steinberg G. K., Vogel H., Edwards M., Weissman I. L., Cheshier S. H., Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl. Med. 9, eaaf2968 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhang M., Hutter G., Kahn S. A., Azad T. D., Gholamin S., Xu C. Y., Liu J., Achrol A. S., Richard C., Sommerkamp P., Schoen M. K., McCracken M. N., Majeti R., Weissman I., Mitra S. S., Cheshier S. H., Anti-CD47 treatment stimulates phagocytosis of glioblastoma by M1 and M2 polarized macrophages and promotes M1 polarized macrophages in vivo. PLOS ONE 11, e0153550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutter G., Theruvath J., Graef C. M., Zhang M., Schoen M. K., Manz E. M., Bennett M. L., Olson A., Azad T. D., Sinha R., Chan C., Assad Kahn S., Gholamin S., Wilson C., Grant G., He J., Weissman I. L., Mitra S. S., Cheshier S. H., Microglia are effector cells of CD47-SIRPα antiphagocytic axis disruption against glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 116, 997–1006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voabil P., de Bruijn M., Roelofsen L. M., Hendriks S. H., Brokamp S., van den Braber M., Broeks A., Sanders J., Herzig P., Zippelius A., Blank C. U., Hartemink K. J., Monkhorst K., Haanen J. B. A. G., Schumacher T. N., Thommen D. S., An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat. Med. 27, 1250–1261 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Capper D., Jones D. T. W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D. E., Kratz A., Wefers A. K., Huang K., Pajtler K. W., Schweizer L., Stichel D., Olar A., Engel N. W., Lindenberg K., Harter P. N., Braczynski A. K., Plate K. H., Dohmen H., Garvalov B. K., Coras R., Hölsken A., Hewer E., Bewerunge-Hudler M., Schick M., Fischer R., Beschorner R., Schittenhelm J., Staszewski O., Wani K., Varlet P., Pages M., Temming P., Lohmann D., Selt F., Witt H., Milde T., Witt O., Aronica E., Giangaspero F., Rushing E., Scheurlen W., Geisenberger C., Rodriguez F. J., Becker A., Preusser M., Haberler C., Bjerkvig R., Cryan J., Farrell M., Deckert M., Hench J., Frank S., Serrano J., Kannan K., Tsirigos A., Brück W., Hofer S., Brehmer S., Seiz-Rosenhagen M., Hänggi D., Hans V., Rozsnoki S., Hansford J. R., Kohlhof P., Kristensen B. W., Lechner M., Lopes B., Mawrin C., Ketter R., Kulozik A., Khatib Z., Heppner F., Koch A., Jouvet A., Keohane C., Mühleisen H., Mueller W., Pohl U., Prinz M., Benner A., Zapatka M., Gottardo N. G., Driever P. H., Kramm C. M., Müller H. L., Rutkowski S., von Hoff K., Frühwald M. C., Gnekow A., Fleischhack G., Tippelt S., Calaminus G., Monoranu C. M., Perry A., Jones C., Jacques T. S., Radlwimmer B., Gessi M., Pietsch T., Schramm J., Schackert G., Westphal M., Reifenberger G., Wesseling P., Weller M., Collins V. P., Blümcke I., Bendszus M., Debus J., Huang A., Jabado N., Northcott P. A., Paulus W., Gajjar A., Robinson G. W., Taylor M. D., Jaunmuktane Z., Ryzhova M., Platten M., Unterberg A., Wick W., Karajannis M. A., Mittelbronn M., Acker T., Hartmann C., Aldape K., Schüller U., Buslei R., Lichter P., Kool M., Herold-Mende C., Ellison D. W., Hasselblatt M., Snuderl M., Brandner S., Korshunov A., von Deimling A., Pfister S. M., DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt C., Papadimitropoulos A., Mele V., Muraro M. G., Mengus C., Iezzi G., Terracciano L., Martin I., Spagnoli G. C., “In vitro” 3D models of tumor-immune system interaction. Adv. Drug Deliv. Rev. 79-80, 145–154 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Duerinck J., Schwarze J. K., Awada G., Tijtgat J., Vaeyens F., Bertels C., Geens W., Klein S., Seynaeve L., Cras L., D’Haene N., Michotte A., Caljon B., Salmon I., Bruneau M., Kockx M., Dooren S. V., Vanbinst A.-M., Everaert H., Forsyth R., Neyns B., Intracerebral administration of CTLA-4 and PD-1 immune checkpoint blocking monoclonal antibodies in patients with recurrent glioblastoma: A phase I clinical trial. J. Immunother. Cancer 9, e002296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schalper K. A., Rodriguez-Ruiz M. E., Diez-Valle R., López-Janeiro A., Porciuncula A., Idoate M. A., Inogés S., de Andrea C., López-Diaz de Cerio A., Tejada S., Berraondo P., Villarroel-Espindola F., Choi J., Gúrpide A., Giraldez M., Goicoechea I., Gallego Perez-Larraya J., Sanmamed M. F., Perez-Gracia J. L., Melero I., Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 25, 470–476 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Schürch C. M., Bhate S. S., Barlow G. L., Phillips D. J., Noti L., Zlobec I., Chu P., Black S., Demeter J., McIlwain D. R., Kinoshita S., Samusik N., Goltsev Y., Nolan G. P., Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoltzfus C. R., Filipek J., Gern B. H., Olin B. E., Leal J. M., Wu Y., Lyons-Cohen M. R., Huang J. Y., Paz-Stoltzfus C. L., Plumlee C. R., Poschinger T., Urdahl K. B., Perro M., Gerner M. Y., CytoMAP: A spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 31, 107523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cribaro G. P., Saavedra-López E., Romarate L., Mitxitorena I., Díaz L. R., Casanova P. V., Roig-Martínez M., Gallego J. M., Perez-Vallés A., Barcia C., Three-dimensional vascular microenvironment landscape in human glioblastoma. Acta Neuropathol. Commun. 9, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai L., Ladomersky E., Lenzen A., Nguyen B., Patel R., Lauing K. L., Wu M., Wainwright D. A., IDO1 in cancer: A Gemini of immune checkpoints. Cell. Mol. Immunol. 15, 447–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bewley M. A., Pham T. K., Marriott H. M., Noirel J., Chu H.-P., Ow S. Y., Ryazanov A. G., Read R. C., Whyte M. K. B., Chain B., Wright P. C., Dockrell D. H., Proteomic evaluation and validation of cathepsin D regulated proteins in macrophages exposed to Streptococcus pneumoniae. Mol. Cell. Proteomics 10, M111.008193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkal A. A., Weiskopf K., Kao K. S., Gordon S. R., Rosental B., Yiu Y. Y., George B. M., Markovic M., Ring N. G., Tsai J. M., McKenna K. M., Ho P. Y., Cheng R. Z., Chen J. Y., Barkal L. J., Ring A. M., Weissman I. L., Maute R. L., Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 19, 76–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon S. R., Maute R. L., Dulken B. W., Hutter G., George B. M., McCracken M. N., Gupta R., Tsai J. M., Sinha R., Corey D., Ring A. M., Connolly A. J., Weissman I. L., PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Maji, S. Panda, S. K. Samal, O. Shriwas, R. Rath, M. Pellecchia, L. Emdad, S. K. Das, P. B. Fisher, R. Dash, in Advances in Cancer Research, K. D. Tew, P. B. Fisher, Eds. (Academic Press, 2018), vol. 137, pp. 37–75. [DOI] [PubMed] [Google Scholar]
- 33.Fritsch M., Günther S. D., Schwarzer R., Albert M.-C., Schorn F., Werthenbach J. P., Schiffmann L. M., Stair N., Stocks H., Seeger J. M., Lamkanfi M., Krönke M., Pasparakis M., Kashkar H., Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575, 683–687 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Zeng L., Li T., Xu D. C., Liu J., Mao G., Cui M.-Z., Fu X., Xu X., Death receptor 6 induces apoptosis not through type I or type II pathways, but via a unique mitochondria-dependent pathway by interacting with Bax protein. J. Biol. Chem. 287, 29125–29133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorgulescu J. B., Gokhale P. C., Speranza M. C., Eschle B. K., Poitras M. J., Wilkens M. K., Soroko K. M., Chhoeu C., Knott A., Gao Y., Lim-Fat M. J., Baker G. J., Bonal D. M., Nguyen Q.-D., Grant G. R. L., Ligon K. L., Sorger P. K., Chiocca E. A., Anderson A. C., Kirschmeier P. T., Sharpe A. H., Freeman G. J., Reardon D. A., Concurrent dexamethasone limits the clinical benefit of immune checkpoint blockade in glioblastoma. Clin. Cancer Res. 27, 276–287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob F., Salinas R. D., Zhang D. Y., Nguyen P. T. T., Schnoll J. G., Wong S. Z. H., Thokala R., Sheikh S., Saxena D., Prokop S., Liu D., Qian X., Petrov D., Lucas T., Chen H. I., Dorsey J. F., Christian K. M., Binder Z. A., Nasrallah M., Brem S., O’Rourke D. M., Ming G., Song H., A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neufeld L., Yeini E., Reisman N., Shtilerman Y., Ben-Shushan D., Pozzi S., Madi A., Tiram G., Eldar-Boock A., Ferber S., Grossman R., Ram Z., Satchi-Fainaro R., Microengineered perfusable 3D-bioprinted glioblastoma model for in vivo mimicry of tumor microenvironment. Sci. Adv. 7, eabi9119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soubéran A., Tchoghandjian A., Practical review on preclinical human 3D glioblastoma models: Advances and challenges for clinical translation. Cancers 12, 2347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sankowski R., Böttcher C., Masuda T., Geirsdottir L., Sagar, Sindram E., Seredenina T., Muhs A., Scheiwe C., Shah M. J., Heiland D. H., Schnell O., Grün D., Priller J., Prinz M., Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 22, 2098–2110 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Masuda T., Sankowski R., Staszewski O., Böttcher C., Amann L., Sagar, Scheiwe C., Nessler S., Kunz P., van Loo G., Coenen V. A., Reinacher P. C., Michel A., Sure U., Gold R., Grün D., Priller J., Stadelmann C., Prinz M., Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Phillips D., Schürch C. M., Khodadoust M. S., Kim Y. H., Nolan G. P., Jiang S., Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging. Front. Immunol. 12, 687673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. R. Goldblum, L. W. Lamps, J. K. McKenney, J. L. Myers, Rosai and Ackerman’s Surgical Pathology E-Book (Elsevier Health Sciences, 2017). [Google Scholar]
- 43.Black S., Phillips D., Hickey J. W., Kennedy-Darling J., Venkataraaman V. G., Samusik N., Goltsev Y., Schürch C. M., Nolan G. P., CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy-Darling J., Bhate S. S., Hickey J. W., Black S., Barlow G. L., Vazquez G., Venkataraaman V. G., Samusik N., Goltsev Y., Schürch C. M., Nolan G. P., Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur. J. Immunol. 51, 1262–1277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F., Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G. P., Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samusik N., Good Z., Spitzer M. H., Davis K. L., Nolan G. P., Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruskal W. H., Wallis W. A., Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621 (1952). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S1
Data S1