Abstract
The mechanistic target of rapamycin–mLST8-raptor complex (mTORC1) functions as a central regulator of cell growth and metabolism in response to changes in nutrient signals such as amino acids. SAMTOR is an S-adenosylmethionine (SAM) sensor, which regulates the mTORC1 activity through its interaction with the GTPase-activating protein activity toward Rags-1 (GATOR1)-KPTN, ITFG2, C12orf66 and SZT2-containing regulator (KICSTOR) complex. In this work, we report the crystal structures of Drosophila melanogaster SAMTOR in apo form and in complex with SAM. SAMTOR comprises an N-terminal helical domain and a C-terminal SAM-dependent methyltransferase (MTase) domain. The MTase domain contains the SAM-binding site and the potential GATOR1-KICSTOR–binding site. The helical domain functions as a molecular switch, which undergoes conformational change upon SAM binding and thereby modulates the interaction of SAMTOR with GATOR1-KICSTOR. The functional roles of the key residues and the helical domain are validated by functional assays. Our structural and functional data together reveal the molecular mechanism of the SAM sensing of SAMTOR and its functional role in mTORC1 signaling.
The structures of SAMTOR reveal the molecular mechanism for how it senses and binds SAM and then regulates mTORC1 signaling.
INTRODUCTION
The mechanistic target of rapamycin (mTOR) is a multifunctional kinase that plays important roles in embryonic development, aging, tumorigenesis, diabetes, and neurodegenerative diseases (1, 2). In mammalian cells, mTOR forms two functionally distinct complexes, the mTOR-mLST8-Raptor complex (mTORC1) and the mTOR-mLST8-Rictor complex (mTORC2) (2, 3). In response to environmental conditions of energy, nutrients, and extracellular growth factors, mTORC1 modulates the anabolic pathway and promotes the initiation and elongation of protein translation through directly phosphorylating specific substrates such as S6 kinase 1 (S6K1) and 4E binding protein 1 (4EBP1) (4, 5). In addition, mTORC1 suppresses the catabolic pathway through inhibition of autophagy and lysosome biogenesis (2, 6).
Two complementary parallel signaling pathways work together to render full activation of mTORC1 at the lysosomal membrane. On one hand, amino acids induce the conversion of small guanosine triphosphatases (GTPases) Ras-related GTP-binding protein A (RagA) to RagD to the active nucleotide-bound state, i.e., the guanosine triphosphate (GTP)–loaded state of RagA/B and the guanosine diphosphate–loaded state of RagC/D (7, 8). After that, a lysosomal multisubunit machinery comprising the vacuolar-type adenosine triphosphatase (v-ATPase), the pentameric Ragulator complex, and the active Rag GTPases recruits mTORC1 to the lysosomal surface (9–11). On the other hand, the growth factor–stimulated kinase Akt phosphorylates and then inhibits the tuberous sclerosis complex, which acts as a GTPase-activating protein (GAP) for the small GTPase Rheb at the lysosomal membrane, where the active GTP-bound Rheb can fully activate mTORC1 (4, 12).
Rag GTPases function as obligate heterodimers such that RagA/B interacts with RagC/D through their C-terminal roadblock domains, and their N-terminal GTPase domains dictate their interactions with the mTORC1 unique component Raptor (10, 11). The GATOR1 complex, comprising three subunits [Nprl2, Nprl3, and DEP domain-containing protein 5 (DEPDC5)], functions upstream of Rag GTPases as a GAP for RagA/B to inactivate mTORC1 when amino acids are deficient (13, 14). The KICSTOR scaffolding complex, consisting of four subunits [Kaptin, integrin-alpha FG-GAP repeat-containing protein 2 (ITFG2), C12orf66, and SZT2], tethers GATOR1 to the lysosomal surface and facilitates the interaction between GATOR1 and Rag GTPases (15, 16). The GATOR2 complex, consisting of five subunits [WD repeat domain 59 (WDR59), WD repeat domain 24 (WDR24), meiosis regulator for oocyte development (MIOS), SEH1 like nucleoporin (SEH1L), and SEC13], functions upstream of GATOR1 as an inhibitor of GATOR1 and thus a positive regulator of mTORC1 (13). The cytoplasmic leucine and arginine activate mTORC1 through regulating the dynamic interplay of GATOR1 and GATOR2. Upon leucine/arginine deprivation, the cytoplasmic leucine sensors Sestrin1/2 and SAR1B or the arginine sensor CASTOR1 (cytosolic arginine sensor for mTORC1 subunit 1) interact with GATOR2 and block the GATOR1-GATOR2 interaction, releasing the GAP activity of GATOR1 for RagA/B, and the binding of leucine to Sestrin1/2 and SAR1B or arginine to CASTOR1 impairs the sensor’s interaction with GATOR2, leading to inactivation of GATOR1 and thus activation of mTORC1 (17–20).
Similar to leucine and arginine, methionine regulates mTORC1 in a Rag GTPase-dependent manner (21). However, the direct cytoplasmic methionine sensor in mTORC1 signaling has not been found so far. Recently, an S-adenosylmethionine (SAM)–binding protein SAMTOR (or C7orf60) was identified as a negative regulator of mTORC1, which functions upstream of Rag GTPases, GATOR1, and KICSTOR: SAMTOR can interact with GATOR1 to prompt the function of GATOR1 and/or KICSTOR in the absence of SAM, and suppresses Rag GTPases and mTORC1; with the supply of SAM, the binding of SAM to SAMTOR disrupts the interaction of SAMTOR with GATOR1-KICSTOR, leading to the inhibition of the GATOR1 GAP activity and, thus, the activation of mTORC1 (22). SAM is synthesized from methionine and adenosine triphosphate (ATP), and the cellular SAM level is directly correlated with methionine (23). The silencing of methionine adenosyltransferase MAT2A, which catalyzes the synthesis of SAM from methionine and ATP, decreases the expression of SAMTOR and the activation of mTORC1 (24). Loss of SAMTOR prevents the inhibition of mTORC1 caused by methionine starvation (22). Thus, SAMTOR serves as a cytoplasmic SAM sensor in the SAM/methionine-mediated mTORC1 signaling.
To illuminate the molecular mechanism of the functional role of SAMTOR in the SAM/methionine-mediated mTORC1 signaling, we determine the crystal structures of Drosophila melanogaster SAMTOR (dSAMTOR) in apo and SAM- and S-adenosyl-l-homocysteine (SAH)–bound forms. Structural analysis shows that SAMTOR comprises an N-terminal helical domain and a C-terminal class I SAM-dependent methyltransferase (MTase) domain. The ligand (SAM/SAH) binds to the MTase domain and makes extensive hydrogen-bonding and hydrophobic interactions with the surrounding residues. The functional roles of the key residues involved in ligand binding are validated by mutagenesis and biochemical assays. In addition, we found that the N-terminal helical domain exhibits a high flexibility and acts as a molecular switch in response to SAM/SAH binding. In the absence of SAM/SAH, the helical domain is positioned away from the ligand-binding site, allowing SAMTOR to interact with GATOR1-KICSTOR. The binding of SAM/SAH appears to induce conformational change of the helical domain to cover the ligand-binding site, blocking the interaction of SAMTOR with GATOR1-KICSTOR. The structural and functional data together provide insight into the molecular mechanism of SAM/SAH sensing by SAMTOR in the SAM/methionine-mediated mTORC1 signaling.
RESULTS
Biochemical characterization of dSAMTOR
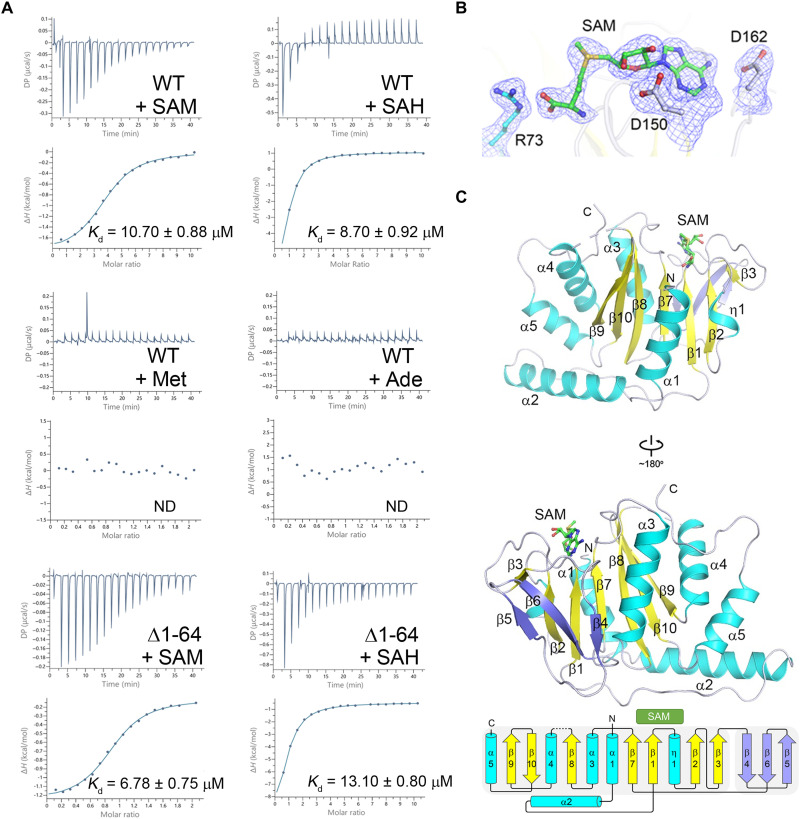
SAMTOR is highly conserved in many metazoans (fig. S1). The Drosophila cells with knockdown of dSAMTOR are resistant to methionine starvation, similar to the human SAMTOR (hSAMTOR)–depleted cells (22), indicating that SAMTOR proteins in different metazoans play a similar regulatory role in mTORC1 signaling. To investigate the structural basis for the functional role of SAMTOR, the full-length dSAMTOR (residues 1 to 302), which shares 47% sequence identity with hSAMTOR, was expressed in Escherichia coli and purified using affinity chromatography and gel filtration chromatography. The recombinant wild-type (WT) dSAMTOR protein exists as a monomer in solution either in the absence or in the presence of SAM as revealed by size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) analyses (fig. S2A). Isothermal titration calorimetry (ITC) measurements show that dSAMTOR can bind both SAM and SAH with a dissociation constant (Kd) of 10.70 ± 0.88 and 8.70 ± 0.92 μM, respectively, which are slightly weaker than those of hSAMTOR (7.0 μM for SAM and 4.6 μM for SAH) (22); however, dSAMTOR has no measurable binding with methionine and adenosine (Fig. 1A and Table 1). These results indicate that like hSAMTOR, dSAMTOR has high binding specificity for SAM and SAH.
Fig. 1. SAM binds exclusively to the MTase domain of dSAMTOR.
(A) ITC measurements for the ligand-binding affinity of the WT and the Δ1-64 mutant dSAMTOR. ND, not detected. The experiments were performed three times for those with measurable binding and two times for those with undetectable binding, and the repeated experiments yielded similar results; for each case, only the result of one representative experiment is shown. (B) Composite simulated annealing Fo − Fc omit map (contoured at 1.5 σ) for the bound SAM and several surrounding residues. (C) Overall structure of the SAM-bound MTase domain of dSAMTOR in two different views. The α helices, major β sheet, and minor β sheet are colored in cyan, yellow, and blue, respectively. The bound SAM is shown with a stick model in green. The topology of the secondary structure elements of the MTase domain is shown below. The linker between β8 and α4 (residues 232 to 238) is disordered and thus is shown with a dashed line.
Table 1. ITC measured thermodynamic parameters.
Kd, dissociation constant; ND, not detected. The experiments were performed three times for those with measurable binding and two times for those with undetectable binding, and the repeated experiments yielded similar results; for each case, only the result of one representative experiment is presented.
| dSAMTOR | Ligand | n value | Kd (μM) | ΔH (kcal mol−1) | TΔS (kcal mol−1) |
| WT | SAM | 0.75 ± 0.08 | 10.70 ± 0.88 | −2.12 ± 0.05 | 4.66 |
| WT | SAH | 0.81 ± 0.09 | 8.70 ± 0.92 | −9.94 ± 1.48 | −2.99 |
| Δ1-64 | SAM | 0.91 ± 0.01 | 6.78 ± 0.75 | −1.16 ± 0.01 | 5.89 |
| Δ1-64 | SAH | 0.67 ± 0.04 | 13.10 ± 0.80 | −14.70 ± 1.05 | −8.08 |
| V66W/E67P | SAM | 0.88 ± 0.02 | 9.47 ± 0.81 | −3.01 ± 0.08 | −3.84 |
| V66W/E67P | SAH | 1.02 ± 0.02 | 11.30 ± 0.37 | −5.09 ± 0.12 | −1.66 |
| R73A | SAH | ND | ND | ND | ND |
| D150A | SAH | ND | ND | ND | ND |
| D162A | SAH | ND | ND | ND | ND |
| L151A | SAH | ND | ND | ND | ND |
| L163A | SAH | ND | ND | ND | ND |
| L197A | SAH | ND | ND | ND | ND |
| Y201A | SAH | 0.74 ± 0.02 | 67.30 ± 6.03 | −2.89 ± 0.17 | 2.80 |
| M202A | SAH | ND | ND | ND | ND |
| F135A | SAM | 0.79 ± 0.01 | 6.46 ± 0.40 | −5.24 ± 0.11 | −1.84 |
| F135A | SAH | 0.80 ± 0.02 | 4.76 ± 0.71 | −8.52 ± 0.51 | −1.26 |
Structures of the SAM- and SAH-bound MTase domain of dSAMTOR
Crystallization of the full-length WT dSAMTOR in the presence of SAM or SAH yielded crystals of the SAM- or SAH-bound dSAMTOR, but no crystals were yielded in the absence of SAM or SAH. The structure of the SAM-bound dSAMTOR was solved by the single-wavelength anomalous dispersion (SAD) method using selenium-labeled protein and was refined to 2.1 Å resolution (Fig. 1, B and C, and Table 2). The structure of the SAH-bound dSAMTOR was solved by the molecular replacement method using the SAM-bound dSAMTOR structure as the search model and was refined to 2.09 Å (fig. S3, A and B, and Table 2). In these structures, the C-terminal region of dSAMTOR and the ligand SAM/SAH are defined with clear electron density (Fig. 1B and fig. S3A); however, the N-terminal region of dSAMTOR (approximately residues 1 to 70 in the SAM-bound structure and residues 1 to 66 in the SAH-bound structure) is invisible in the electron density map. Stability analysis results show that in the absence of SAM, the full-length dSAMTOR is unstable and the N-terminal region is gradually degraded in the storage buffer and the crystallization solution in a time-dependent manner; however, in the presence of SAM, the degradation of the full-length dSAMTOR is substantially alleviated but not completely prevented in both solutions (fig. S4A). These results indicate that SAM binding stabilizes the N-terminal region to some extent but cannot prevent the degradation of the N-terminal domain. SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis shows that the crystals contain mainly the C-terminal fragment corresponding to the N-terminal truncated dSAMTOR (Δ1-64) (fig. S4B). Furthermore, ITC measurements show that compared to the full-length dSAMTOR, the N-terminal truncated dSAMTOR (Δ1-64) exhibits a slightly stronger binding affinity for SAM (6.78 ± 0.75 μM) but a slightly weaker binding affinity for SAH (13.10 ± 0.80 μM), suggesting that the N-terminal region is not required for the binding of SAM/SAH (Fig. 1A and Table 1).
Table 2. Summary of diffraction data and structure refinement statistics.
| SAM-bound | SAH-bound | Apo (V66W/E67P) | |
| Diffraction data | |||
| Wavelength (Å) | 0.9793 | 0.9792 | 0.9792 |
| Space group | P1 | P1 | I23 |
| Cell parameters | |||
| a (Å) | 50.27 | 49.93 | 168.49 |
| b (Å) | 64.10 | 63.99 | 168.49 |
| c (Å) | 79.27 | 80.27 | 168.49 |
| α (°) | 90.10 | 90.26 | 90 |
| β (°) | 93.27 | 93.38 | 90 |
| γ (°) | 96.61 | 96.79 | 90 |
| Resolution (Å) | 43.32–2.10 | 49.49–2.09 | 37.68–3.55 |
| (2.15–2.10)* | (2.14–2.09) | (3.68–3.55) | |
| Observed reflections | 213,131 | 108,874 | 59,066 |
| Unique reflections (I/σ(I) > 0) | 51,008 | 54,989 | 9792 |
| Average redundancy | 4.2 (4.2) | 2.0 (2.0) | 6.0 (6.3) |
| Average I/σ(I) | 9.5 (1.8) | 6.1 (1.7) | 7.9 (1.5) |
| Completeness (%) | 88.5 (59.6) | 94.8 (78.5) | 99.8 (100.0) |
| Rmerge (%)† | 12.7 (84.1) | 10.5 (54.0) | 16.8 (81.3) |
| CC1/2 | 0.995 (0.602) | 0.993 (0.678) | 0.992 (0.686) |
| Refinement and structure model | |||
| Reflections [Fo ≥ 0σ(Fo)] | |||
| Working set | 48,879 | 52,979 | 9236 |
| Test set | 2,018 | 1,995 | 485 |
| Rwork/Rfree (%)‡ | 18.8 / 23.9 | 16.6 / 20.7 | 22.5/27.4 |
| No. of protein atoms | 7,005 | 7,176 | 3843 |
| No. of ligand atoms | 108 | 104 | – |
| No. of water atoms | 427 | 598 | – |
| Average B factor (Å2) | |||
| All atoms | 29.6 | 26.5 | 93.2 |
| Protein | 29.4 | 26.0 | 93.2 |
| Ligand | 22.9 | 16.3 | – |
| Water | 31.1 | 32.1 | – |
| RMSD | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 |
| Bond angles (°) | 1.0 | 0.9 | 1.2 |
| Ramachandran plot (%) | |||
| Favored | 96.8 | 97.6 | 94.3 |
| Allowed | 3.2 | 2.4 | 5.7 |
| Disallowed | 0.0 | 0.0 | 0.0 |
*Numbers in parentheses refer to the highest-resolution shell.
†Rmerge = ∑hkl∑i|Ii(hkl)i − 〈I(hkl)〉|/∑hkl∑iIi(hkl).
‡R factor = ||Fo| − |Fc||/|Fo|.
In the SAM- and SAH-bound dSAMTOR structures, there are four dSAMTOR molecules in the asymmetric unit, which assume almost identical overall structure with a root-mean-square deviation (RMSD) of ~0.6 Å. As molecule A in both structures comprises more defined residues with better electron density, it is used in the structural analysis and discussion hereafter. The defined region of dSAMTOR in the SAM-bound structure (residues 71 to 231 and 239 to 298) assumes a Rossmann fold–like domain comprising a seven-stranded major β sheet (β9↑, β10↓, β8↑, β7↑, β1↑, β2↑, and β3↑) sandwiched by one layer of two α helices (α1 and α2) and a 310 helix (η1) on one side and another layer of three α helices (α3, α4, and α5) on the other side (Fig. 1C), which resembles the typical fold of class I SAM-dependent MTase domains with some variations (25). Most class I MTase domains contain an α helix between strands β2 and β3 (26, 27), whereas the MTase domain of dSAMTOR lacks this α helix, and the corresponding region assumes a loop conformation (Fig. 1C and fig. S5). In addition to the consensus structure elements of class I MTases, the MTase domain of dSAMTOR contains some extra structure elements. Specifically, the α2 helix and the following long loop form a U-shape bracket to buttress the major β sheet from the bottom; the β5, β6, and β4 strands form a minor β sheet to shield the β3, β2, and β1 strands of the major β sheet from the back; the C-terminal α5 helix forms a helical cluster with helices α4 and α3 to flank one side of the major sheet; and the following C-terminal loop protrudes into a cleft between helices α3 and α4 (Fig. 1C and fig. S5). The SAH-bound dSAMTOR structure is almost identical to the SAM-bound structure with an RMSD of 0.6 Å for 204 Cα atoms (fig. S3, B and C).
Structure of the full-length dSAMTOR in apo form
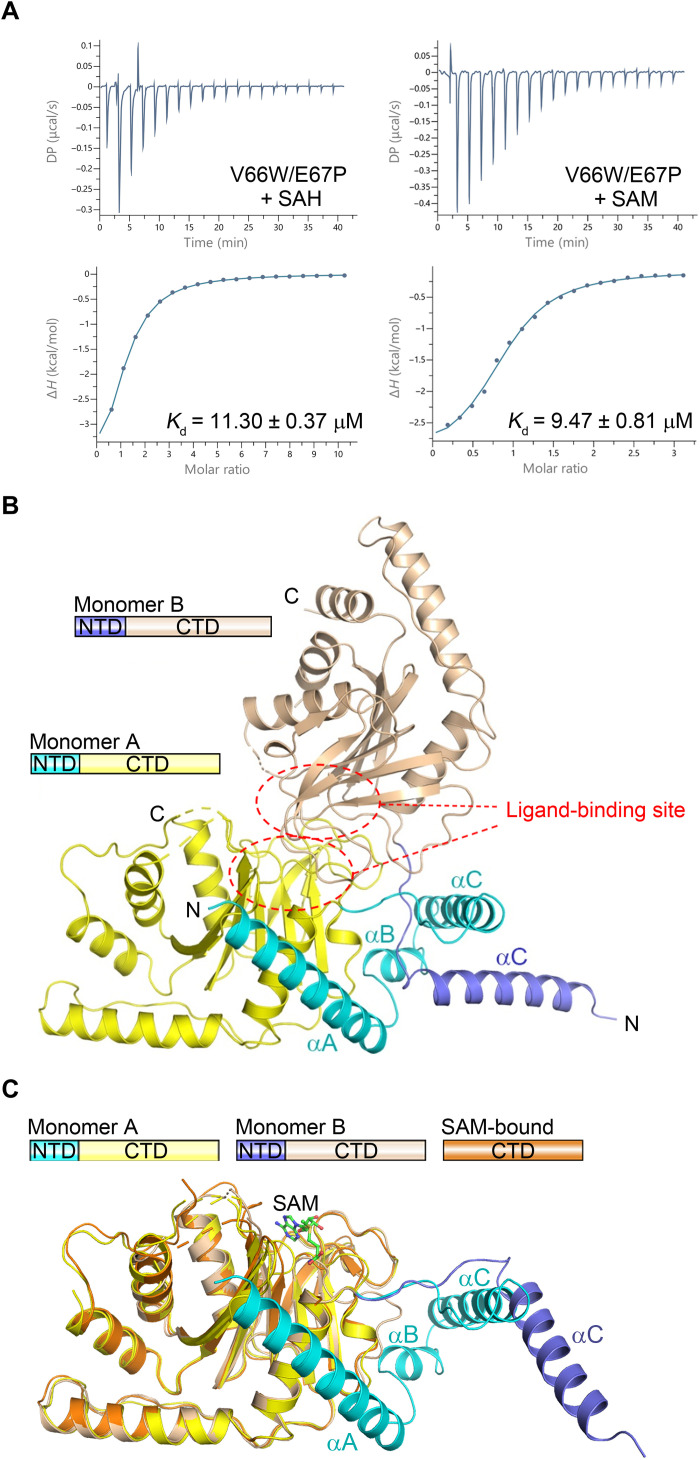
Various attempts at obtaining the structure of the full-length WT dSAMTOR in the absence or presence of SAM/SAH were not successful because of the degradation of the full-length dSAMTOR. As predicted by the PredictProtein server (28), the N-terminal region and the C-terminal MTase domain of dSAMTOR are connected by a linker (residues 61 to 72), and hence, we tried to introduce mutations in the linker to prevent the degradation of the full-length dSAMTOR. After multiple trials, we found that the mutant dSAMTOR containing the V66W/E67P double mutation [mutated to the corresponding residues in nucleomethylin (NML), an MTase, which also consists of an N-terminal helical domain and a C-terminal MTase domain (29, 30)] was stable in the storage buffer and the crystallization solution (fig. S4C). Compared to the WT dSAMTOR, this mutant exhibits a comparable binding affinity for SAM (9.47 ± 0.81 μM) but a slightly weaker binding affinity for SAH (11.30 ± 0.37 μM) (Fig. 2A and Table 1). We successfully determined the structure of the full-length V66W/E67P mutant in apo form (Fig. 2B and Table 2). Unfortunately, crystallization of the full-length V66W/E67P mutant in the presence of SAM or SAH did not yield any crystals.
Fig. 2. The N-terminal domain of the dSAMTOR adopts multiple conformations.
(A) ITC measurements for the ligand-binding affinity of the V66W/E67P mutant dSAMTOR. The experiments were performed three times, which yielded similar results, and for each case, only the result of one representative experiment is shown. (B) Overall structure of the homodimer of the V66W/E67P mutant dSAMTOR in apo form. The two monomers are designated as monomer A and monomer B. The N-terminal helical domain (NTD) and the C-terminal MTase domain (CTD) of monomer A are colored in cyan and yellow, and those of monomer B are in blue and wheat, respectively. The ligand-binding site is indicated by dashed ovals. (C) Superposition of monomer A and monomer B in the V66W/E67P mutant dSAMTOR structure onto the SAM-bound MTase domain of dSAMTOR. The color coding of each structure is shown above.
In this apo V66W/E67P mutant structure, the asymmetric unit contains two dSAMTOR molecules (hereafter designated as monomer A and monomer B), which form an asymmetric homodimer with the ligand-binding site of the MTase domain buried at the dimer interface (Fig. 2B). In monomer A, residues 1 to 200, 205 to 232, and 239 to 297 are defined, and in monomer B, residues 41 to 199, 203 to 231, and 242 to 287 are visible. Although the MTase domains of the two monomers adopt almost identical overall structure (an RMSD of 0.9 Å for 176 Cα atoms), the N-terminal domains assume different conformations (Fig. 2C). In monomer A, the N-terminal domain (residues 1 to 70) comprises three helices named αA to αC, which form a U-shape helical bundle to wrap around one side of the MTase domain. Specifically, helix αA folds on the top of helix α1 of the MTase domain, and helices αB and αC fold along one side of the major sheet (Fig. 2B). In monomer B, only helix αC of the N-terminal domain could be traced, which has no direct interaction with its MTase domain; instead, it extends into and makes interactions with the N-terminal domain of monomer A (Fig. 2B).
Despite the extensive interactions between the two monomers, SEC-MALS analyses show that the WT dSAMTOR always exists as a monomer in solution in both the absence and presence of SAM, and the mutant dSAMTOR exists mainly as a monomer but has a small fraction (about 5%) of dimer in solution in the absence of SAM, whereas it exists solely as a monomer in the presence of SAM (fig. S2B). Structural analysis shows that the αC helices of both monomers participate in crystal packing and make contact with those of the threefold axis-related monomers (fig. S6A). However, the linker regions containing the V66W/E67P double mutation of the two monomers are not involved in crystal packing but are crossed over and make interactions with each other (fig. S6B). These results suggest that the V66W/E67P double mutation facilitates the dimerization in the absence of SAM and the crystal packing appears to favor the homodimer formation, and hence, the conformations of the N-terminal domain are constrained and might not represent the native conformation in solution. Together, the structural and biochemical data suggest that the N-terminal domain of dSAMTOR has a high flexibility in the ligand-free form.
The key residues at the SAM-binding site play a critical role in SAM binding and the interaction with GATOR1-KICSTOR
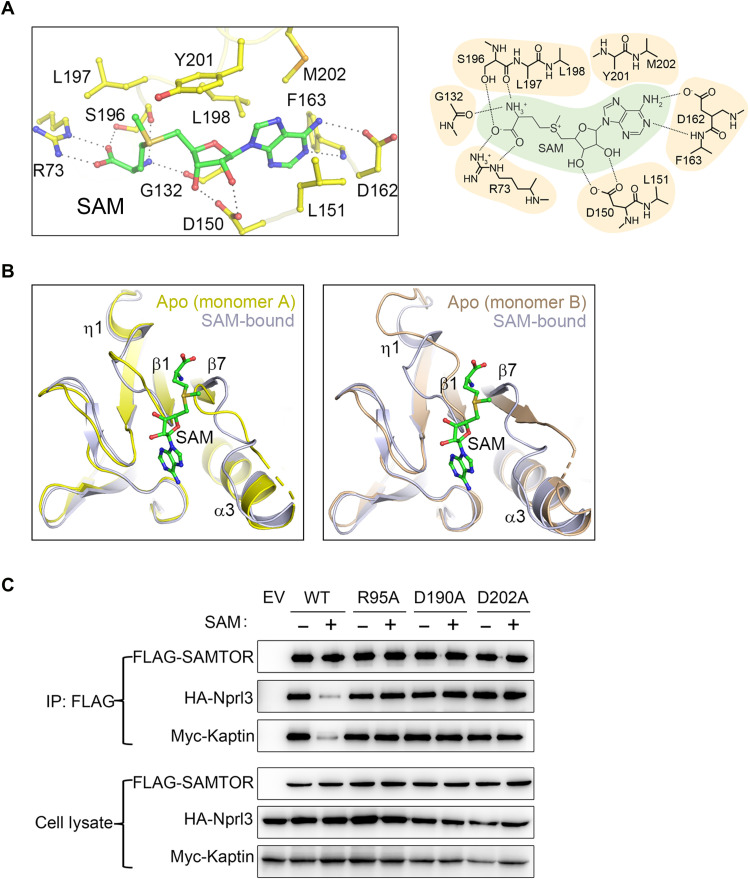
In the structure of the SAM-bound dSAMTOR MTase domain, SAM binds to the C-terminal end of the major sheet (Fig. 1C), which is spatially the common ligand-binding site in class I MTases. The bound SAM assumes an extended conformation, which is stabilized by a network of hydrogen bonds and hydrophobic contacts (Fig. 3A). Specifically, the adenine moiety of SAM is surrounded by several hydrophobic residues including Leu151, Phe163, Leu198, Tyr201, and Met202. The N3 and N6 groups of the adenine moiety form hydrogen bonds with the main chain of Phe163 and the side chain of Asp162, respectively. The 2′- and 3′-hydroxyl groups of the ribose moiety form two hydrogen bonds with the side chain of Asp150. The S-methyl group of the methionine moiety makes hydrophobic contacts with the side chains of Leu197 and Tyr201, the amine group forms hydrogen bonds with the main chains of Gly132 and Ser196, and the carboxyl group forms hydrogen bonds with the side chains of Arg73 and Ser196. Because SAM and SAH differ only by the S-methyl group of SAM, as expected, in the structure of the SAH-bound dSAMTOR MTase domain, SAH binds to the MTase domain with almost identical interactions with the surrounding residues as SAM (fig. S7).
Fig. 3. The key residues at the SAM-binding site play a critical role in SAM binding and the interaction with GATOR1-KICSTOR.
(A) Interactions between SAM and the surrounding residues are shown in a ball-and-stick model (left) and a schematic diagram (right) in the structure of the SAM-bound MTase domain of dSAMTOR. (B) Structural comparison of the ligand-binding site of monomer A (left) and monomer B (right) in the V66W/E67P mutant dSAMTOR and the SAM-bound MTase domain of dSAMTOR. The color coding of each structure is shown above. (C) Co-IP assay to examine the interactions of Flag-hSAMTOR (WT and mutants) with HA-Nprl3 and Myc-Kaptin in the presence and absence of SAM. EV, empty vector. The assay was performed three times, which yielded similar results, and only the result of one representative experiment is shown.
Sequence alignment of the SAMTOR proteins in different species shows that most of the residues involved in the SAM/SAH binding are highly conserved (fig. S1). All class I MTases contain a consensus GxGxG motif in the loop following strand β1, which is the hallmark of the nucleotide-binding site (27, 31, 32). This conserved motif is altered to GSCF/YN in dSAMTOR and other SAMTOR proteins, which is located underneath the bound SAM. The functional implication of this alteration is unclear.
Structural comparison of the MTase domain in the apo V66W/E67P mutant and the SAM-bound MTase domain of dSAMTOR shows that SAM binding does not induce notable conformational changes in the overall structure (an RMSD of 1.0 Å over 195 Cα atoms) (Fig. 2C) except a few small conformational changes at the ligand-binding site, including the loops between β1 and η1 and between β7 and α3 (Fig. 3B). For example, residues 202 to 204 in monomer A and residues 200 to 202 in monomer B in the loop between β1 and η1 are disordered in the apo V66W/E67P mutant structure; the η1 helix in monomer B is unwound into a loop. These results indicate that SAM binding stabilizes the ligand-binding site and induces only some minor local conformational changes.
To validate the functional roles of the key residues involved in ligand binding, we first performed mutagenesis and ITC analyses to examine the binding affinity of dSAMTOR with SAH. The ITC analysis results show that mutations of the residues of dSAMTOR involved in the hydrophilic interactions (R73A, D150A, and D162A) and most of the residues involved in the hydrophobic interactions (L151A, F163A, L197A, and M202A) completely abolish SAH binding (fig. S8 and Table 1). One exception is the Y201A mutant, which retains the ability to bind SAH albeit with a substantially decreased binding affinity (7.7-fold) compared to the WT dSAMTOR, consistent with the structural data showing that the side chain of Tyr201 makes very minor hydrophobic contacts with SAM/SAH.
Then, we performed coimmunoprecipitation (co-IP) assays to examine the interactions of hSAMTOR with the GATOR1 component Nprl3 and the KICSTOR component Kaptin in human embryonic kidney (HEK) 293T cells. The results show that the WT hSAMTOR can interact with the GATOR1 component Nprl3 and the KICSTOR component Kaptin and the supplement of SAM essentially disrupts its interactions with GATOR1-KICSTOR (Fig. 3C), which are consistent with the previous results (22). However, the R95A, D190A, and D202A hSAMTOR mutants, equivalent to the R73A, D150A, and D162A dSAMTOR mutants, respectively, which are deficient in SAM binding, exhibit comparable interactions with GATOR1 and KICSTOR in both the absence and presence of SAM (Fig. 3C), suggesting that these residues of hSAMTOR are critical for SAM sensing and binding. These results are also in agreement with a previous study showing that the D190A hSAMTOR mutant cannot bind SAM and interacts with GATOR1-KICSTOR in a SAM-independent manner and thus inhibits the mTORC1 activity in a dose-dependent manner (22). Together, our structural and functional data indicate that the key residues at the ligand-binding site of SAMTOR play a critical role in SAM sensing and binding and its interaction with GATOR1-KICSTOR in mTORC1 signaling.
The N-terminal domain functions as a molecular switch in mTORC1 signaling
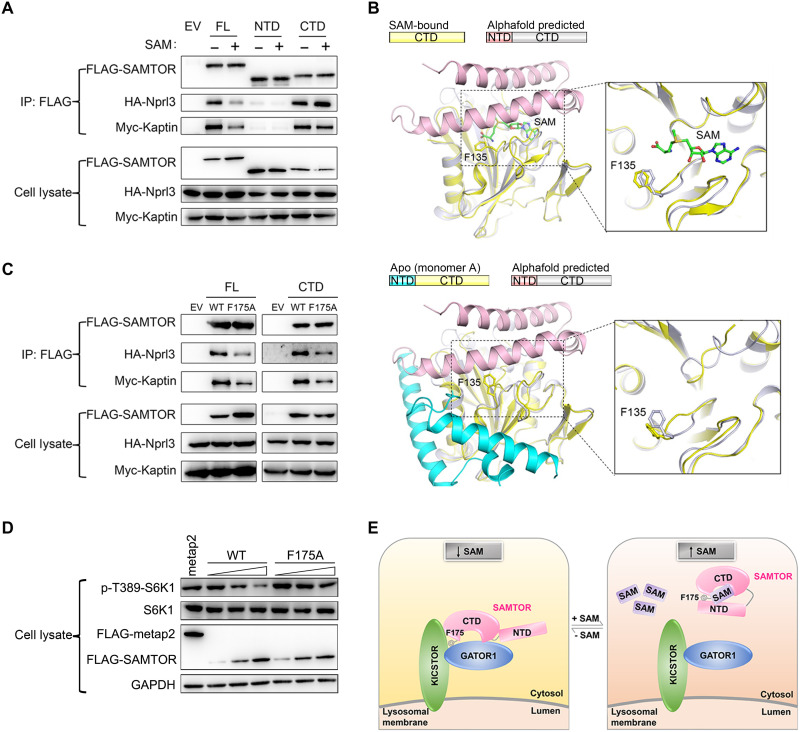
As the N-terminal helical domain of dSAMTOR has a high flexibility in the ligand-free form, we speculated that the flexible helical domain might also play an important role in SAM sensing and mTORC1 signaling. To investigate this possibility, we performed co-IP assays to examine the functional role of the helical domain in the interaction of hSAMTOR with GATOR1-KICSTOR in HEK 293T cells. The results show that the C-terminal MTase domain alone interacts with the GATOR1 component Nprl3 and the KICSTOR component Kaptin with comparable binding abilities to the full-length hSAMTOR in the absence of SAM, but the N-terminal helical domain alone cannot interact with GATOR1-KICSTOR (Fig. 4A). While SAM binding substantially weakens the interaction of the full-length hSAMTOR with GATOR1-KICSTOR, it has no apparent effect on the interaction of the MTase domain with GATOR1-KICSTOR (Fig. 4A). These results indicate that the MTase domain alone can bind to GATOR1-KICSTOR independent of SAM binding, and the helical domain of the full-length hSAMTOR is involved in SAM sensing and the interaction of hSAMTOR with GATOR1-KICSTOR.
Fig. 4. The N-terminal domain functions as a molecular switch in mTORC1 signaling.
(A) Co-IP assay to examine the interactions of the full-length (FL), the N-terminal helical domain (NTD), and the C-terminal MTase domain (CTD) of hSAMTOR with HA-Nprl3 and Myc-Kaptin in the presence and absence of SAM in the HEK 293T cells. The assays in (A), (C), and (D) were performed three times, which yielded similar results, and for each case, only the result of one representative experiment is shown. (B) Structural comparison of the SAM-bound MTase domain of dSAMTOR (left) and the apo V66W/E67P mutant dSAMTOR (monomer A) (right) with the predicted dSAMTOR structure from the AlphaFold Protein Structure Database. The color coding of each structure is shown above. (C) Co-IP assay to examine the interactions of the WT and F175A mutant of the full-length (FL) and the C-terminal MTase domain (CTD) of hSAMTOR with HA-Nprl3 and Myc-Kaptin in the HEK 293T cells. (D) Immunoblotting assay to examine the mTORC1 kinase activity in the HEK 293T cells transiently overexpressing the WT and F175A mutant hSAMTOR at three different levels. The cell lysates were analyzed by immunoblotting of the phosphorylation level (p-T389) of S6K1 and the expression levels of the indicated proteins. (E) A schematic diagram illustrating the proposed molecular mechanism for SAMTOR to sense and bind SAM and then function as a molecular switch through conformational change to regulate its interaction with GATOR1-KICSTOR in mTORC1 signaling.
Because of the lack of a full-length ligand-bound dSAMTOR structure, we retrieved the predicted dSAMTOR structure from the AlphaFold Protein Structure Database (33) and compared it with the dSAMTOR structures solved in this study. The MTase domain of the predicted dSAMTOR structure is essentially identical to that in the SAM-bound dSAMTOR structure (an RMSD of 0.6 Å for 220 Cα atoms) and the apo V66W/E67P mutant structure (an RMSD of 0.9 Å for 220 Cα atoms) (Fig. 4B and fig. S9). The helical domain in the predicted dSAMTOR structure assumes a three-helix bundle and covers the top of the ligand-binding site of the MTase domain. In addition, the search for homologous MTases with similar structures in the Protein Data Bank (PDB) via the DALI (digital addressable lighting interface) server (34) identified two best candidates: the SAH-bound NML (PDB code 2ZFU) with a z-score of 15.0 and 9% sequence identity and the SAH-bound nodulation protein S (NodS) (PDB code 3OFK) with a z-score of 12.0 and 11% sequence identity (29, 30). Similar to dSAMTOR, both NML and NodS comprise two domains: an N-terminal helical domain consisting of several helices (αA to αC in NML and αA and αB in NodS) and a C-terminal MTase domain resembling that of dSAMTOR (an RMSD of 1.4 Å for 127 Cα atoms with NML and 1.8 Å for 124 Cα atoms with NodS) (fig. S9). In the SAH-bound NML and NodS structures, the helical domain also covers the top of the ligand-binding site in a manner similar to that in the predicted dSAMTOR structure. However, in the ligand-free NodS structure (PDB code 3OFJ), the helical domain is disordered (fig. S9) (30). Furthermore, the N-terminal regions in several other MTases also assume a similar helical structure, including the SAM-bound Chim (PDB code 4QDJ) and SAH-bound DhpI (PDB code 3OU2) (35, 36). These results suggest that the N-terminal domains of MTases tend to form helical structures and have high flexibility in the ligand-free form, and the conformation of the helical domain in the predicted dSAMTOR structure might represent the conformation of the full-length dSAMTOR in the ligand-bound form. On the basis of these results, we hypothesized that the N-terminal helical domain of SAMTOR might function as a molecular switch in SAM sensing and mTORC1 signaling through conformational change upon ligand binding, and the potential binding site for GATOR1-KICSTOR might be located in the vicinity of the ligand-binding site of the MTase domain.
Phe175 of hSAMTOR forms part of the GATOR1-KICSTOR–binding site
To identify the potential binding site of hSAMTOR for GATOR1-KICSTOR, we mutated a number of conserved residues of hSAMTOR corresponding to these near the ligand-binding site and on the solvent-exposed surface of dSAMTOR and performed co-IP assay to examine the interactions of these mutants with GATOR1-KICSTOR (fig. S10). The results show that among the tested mutants, only the F175A hSAMTOR mutant exhibits a substantially decreased binding with GATOR1-KICSTOR (fig. S10 and Fig. 4C). In addition, the SAM-insensitive MTase domain bearing the F175A mutation also exhibits a markedly decreased binding with GATOR1-KICSTOR (Fig. 4C). As hSAMTOR functions as a negative regulator of the mTORC1 kinase activity, the phosphorylation of S6K1, a canonical mTORC1 substrate, is suppressed by the WT hSAMTOR in a dose-dependent manner (22). However, the mTORC1 activity is insensitive to the overexpression of the F175A hSAMTOR mutant; i.e., the F175A hSAMTOR mutant cannot inhibit the mTORC1 activity in a dose-dependent manner (Fig. 4D). These results indicate that the F175A hSAMTOR mutant cannot promote the function of GATOR1-KICSTOR and the subsequent inhibition of the mTORC1 activity by GATOR1-KICSTOR, and suggest that Phe175 of hSAMTOR forms part of the potential binding site for GATOR1-KICSTOR.
This suggestion is also supported by the structural data. In the SAM-bound dSAMTOR MTase domain structure, residue Phe135 (equivalent to Phe175 of hSAMTOR) is located in the solvent-exposed loop between β1 and η1 and positioned about 5 Å away from the bound SAM (Fig. 4B). ITC analysis shows that compared to the WT dSAMTOR, the F135A dSAMTOR mutant exhibits slightly stronger binding affinities (<2-fold) for both SAM and SAH (6.46 ± 0.40 and 4.76 ± 0.71 μM, respectively) (fig. S8 and Table 1), indicating that this residue has some indirect effect on ligand binding. In the predicted dSAMTOR structure, which is suggested to represent the ligand-bound form, Phe135 is covered by the N-terminal domain but has no direct interactions (<5 Å) with the N-terminal domain, and thus, the potential GATOR1-KICSTOR-binding site is presumably blocked spatially by the N-terminal domain, prohibiting the binding of dSAMTOR with GATOR1-KICSTOR (Fig. 4B). In the apo V66W/E67P mutant structure, the N-terminal domain packs along one side of the MTase domain and the potential GATOR1-KICSTOR–binding site is exposed on the surface, permitting the binding of dSAMTOR with GATOR1-KICSTOR (Fig. 4B). Together, the structural and functional data indicate that Phe175 of hSAMTOR or Phe135 of dSAMTOR near the ligand-binding site participates in the interaction of SAMTOR with GATOR1-KICSTOR and forms part of the GATOR1-KICSTOR–binding site.
DISCUSSION
The sulfur-containing amino acid methionine presents a key metabolite in many aspects of mammalian physiology, including translation, epigenetics, cell proliferation, and various signaling pathways (37). In metazoans, the mTORC1 signaling pathway senses the cellular level of the methionine metabolite SAM rather than methionine through the SAM sensor SAMTOR to modulate the anabolic and catabolic metabolisms (22). In this work, we determined the crystal structures of the MTase domain of dSAMTOR in SAM- and SAH-bound forms and the full-length V66W/E67P mutant dSAMTOR in apo form. Structural analyses reveal that dSAMTOR comprises an N-terminal helical domain and a C-terminal MTase domain. The helical domain consists of three α helices (αA to αC) and exhibits a high flexibility. The MTase domain adopts a class I MTase fold supplemented with some auxiliary structure elements. In the SAM- and SAH-bound dSAMTOR MTase structures, the SAM and SAH bind to the MTase domain in a similar binding manner and with comparable binding affinities. The MTase domain of dSAMTOR and the full-length dSAMTOR exhibit comparable binding affinities with the ligands, suggesting that the helical domain is not directly involved in ligand binding. Comparison of the SAM-bound MTase domain and the apo V66W/E67P mutant structures shows that ligand binding does not induce notable conformational changes in the overall structure of the MTase domain except a few minor conformational changes at the ligand-binding site. The ligand makes extensive hydrophilic and hydrophobic interactions with several conserved residues, and mutations of these residues impair ligand binding in vitro. Selective mutations with deficient ligand binding also abolish the SAM-sensing ability of hSAMTOR and the mutants could bind to GATOR1-KICSTOR both in the absence and in the presence of SAM in vivo. These results reveal the molecular mechanism of SAM sensing and binding by SAMTOR.
Recently, several direct amino acid sensors upstream of mTORC1 including the arginine sensor CASTOR1 and the leucine sensor Sestrins2 have been identified and the crystal structures of these sensors have been solved. The structural and functional data together have revealed the molecular mechanisms about how CASTOR1 and Sestrins2 sense and bind the ligands. However, as ligand binding does not induce substantial conformational changes of the proteins, the molecular mechanisms about how these sensors function as molecular switches upon ligand binding to interact with the downstream partners and then further regulate mTORC1 signaling remain unclear. On the basis of the structural and functional data of SAMTOR together with the structural comparison with the AlphaFold predicted SAMTOR structure and the structures of several MTases, we can propose a working model for how SAMTOR functions as a molecular switch upon SAM binding to regulate mTORC1 signaling as follows (Fig. 4E). The C-terminal MTase domain of SAMTOR contains both the ligand-binding site and the GATOR1-KICSTOR–binding site. The MTase domain of SAMTOR alone has the ligand binding ability as well as the GATOR1-KICSTOR binding ability in both the presence and absence of the ligand. In other words, it has the ability to sense the SAM level but lacks the switch function to regulate mTORC1 signaling upon SAM binding. On the other hand, while the N-terminal helical domain of SAMTOR is not required for the interactions with the ligand and GATOR1-KICSTOR, it has a high flexibility and functions as a molecular switch to turn on and off the binding ability of the MTase domain with GATOR1-KICSTOR and subsequently mTORC1 signaling. In the absence of SAM, the helical domain is positioned distantly from the ligand-binding site with a flexible conformation, and hence the potential binding site for GATOR1-KICSTOR is exposed and the MTase domain can interact with GATOR1-KICSTOR, leading to the inhibition of mTORC1 signaling. When SAM binds to SAMTOR, the helical domain is positioned on the top of the ligand-binding site and the conformation is stabilized by SAM binding, and hence the potential binding site for GATOR1-KICSTOR is blocked and thus the MTase domain cannot interact with GATOR1-KICSTOR, leading to the activation of mTORC1 signaling (Fig. 4E).
Our biochemical and structural data show that dSAMTOR can bind SAH and SAM with comparable binding affinities (Fig. 1A and Table 1), and SAH binds to the MTase domain with almost identical interactions with the surrounding residues as SAM (Fig. 3A and fig. S7). The previous cell biology data also show that like SAM, the binding of SAH to hSAMTOR can disrupt the interaction of hSAMTOR with GATOR1-KICSTOR and inhibit the GATOR1 GAP activity in mTORC1 signaling (22). As the concentrations of SAM and SAH in cells are estimated to be at similar levels (38–41), these results suggest that SAH might be able to regulate the function of SAMTOR in mTORC1 signaling in the same manner as SAM.
Previous studies show that the arginine sensor CASTOR1 consists of four tandem ACT domains and structurally resembles the C-terminal allosteric domains of aspartate kinases, suggesting that CASTOR1 may have evolved from the ancient aspartate kinase but lost the N-terminal kinase domain during evolution (19, 42–44). The leucine sensor Sentrin2 is a homolog of bacterial reductase Ahpd but lacks a conserved cysteine required for the catalytic activity (18, 45, 46). So far, whether Senstrin2 has a reductase activity remains elusive. SAMTOR contains a conserved class I MTase domain, which is usually known to bind and exert the catalytic activity on DNA or RNA during the regulation of gene expression, repair of mutations, and protection against bacterial restriction enzymes (27). Sequence alignment shows that the SAMTOR proteins have a moderate sequence similarity (~11% identity) to Saccharomyces cerevisiae Bmt2, an MTase responsible for the m1A modification of 25S ribosomal RNA (rRNA) (47). Structural comparison of dSAMTOR with the small RNA MTase Hen1 (PDB code 3HTX) and the long noncoding RNA MTase MePCE (methyl phosphate capping enzyme) (PDB code 6DCB) of class I MTases (48, 49) shows that both Hen1 and MePCE have a positively charged surface cleft around the active site of the MTase domain to bind the RNA substrate (fig. S11). In contrast, the MTase domain of dSAMTOR contains no such kind of positively charged surface cleft; instead, the electrostatic surface around the ligand-binding site is largely negatively charged or hydrophobic, which is apparently unfavorable for the binding of an RNA or DNA substrate, suggesting that SAMTOR is unlikely an RNA or DNA MTase. Nevertheless, whether SAMTOR has an MTase activity on proteins remains elusive and warrants further study.
MATERIALS AND METHODS
Cloning, expression, and purification
The DNA fragment of dSAMTOR was synthesized by Sangon Biotech (Shanghai), China, and cloned into the pET (plasmid expression vector under T7 control)-28a expression vector (Novagen) with an N-terminal His6-SUMO tag. The dSAMTOR mutants were constructed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All constructs were confirmed by DNA sequencing. The His6-SUMO-dSAMTOR plasmid was transformed into E. coli BL21 (DE3) strain (WEIDI) (catalog no. EC1007), and the transformed cells were grown at 37°C in LB medium containing kanamycin (0.05 mg/ml) until OD600 (optical density at 600 nm) reached 0.8 and then induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside at 20°C for 20 hours. The cells were harvested and lysed by sonication in lysis buffer [30 mM tris-HCl (pH 7.5) and 150 mM NaCl]. The dSAMTOR protein was purified by affinity chromatography using a Ni-NTA (nickel-nitrilotriacetic acid) column (Qiagen) with the lysis buffer supplemented with 30 mM imidazole and 200 mM imidazole serving as wash buffer and elution buffer, respectively. The N-terminal His6-SUMO tag of dSAMTOR was removed by a ubiquitin-like protease, and the protein was further purified by gel filtration using a Superdex 200 16/60 column (preparative grade, GE Healthcare) preequilibrated with a storage buffer [10 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM dithiothreitol (DTT)]. The dSAMTOR mutants were expressed and purified with the same methods as the WT dSAMTOR. The purified proteins were of high purity (above 95%) as analyzed by SDS-PAGE (10%). To analyze the stability and degradation of the full-length WT and the V66W/E67P mutant dSAMTOR, the purified protein in the storage buffer or the crystallization solution supplemented without or with SAM (molar ratio of dSAMTOR:SAM = 1:3) was analyzed by SDS-PAGE (10%) at different time intervals in 1 to 3 days. The experiments were performed at least three times to check for reproducibility of the results.
ITC analysis
ITC analysis was performed at 25°C using a PEAQ Micro-calorimeter (MicroCal) to analyze the interaction between dSAMTOR (WT and mutants) and the ligand (SAM and SAH). All of the proteins used in the ITC analysis contain no His6-SUMO tag. An initial injection of 0.8 μl of protein sample was discarded for each dataset to remove the effect of titrant diffusion across the syringe tip during the equilibration process. Each experiment consisted of 20 injections of 2 μl of ligand (1 mM) into the sample cell containing 280 μl of dSAMTOR (0.2 mM). A background titration was performed using identical titrant with the buffer solution. The measured thermal data were analyzed using the MicroCal PEAQ-ITC analysis software v1.1.0.1262 (Malvern). Thermodynamic parameters ΔH° (enthalpy change), n (stoichiometry), and Kd (dissociation constant) were obtained by fitting the data to an equilibrium binding isotherm using a single-site binding model with a nonlinear least-squares method. The free energy of binding (ΔG°) and entropy (ΔS°) were obtained by using the classical thermodynamic formulas: ΔG° = RT × ln Kd and ΔG° = ΔH°–TΔS°, where R is the gas constant and T is the absolute temperature in kelvin. The experiments were performed three times for those with measurable binding and two times for those with undetectable binding to check for reproducibility of the data.
SEC-MALS analysis
The purity, molecular mass, and size distribution of the proteins were analyzed by an analytical light scattering instrument (SEC-MALS) consisting of an Agilent 1260 Infinity Isocratic Liquid Chromatography System, a Wyatt Dawn Heleos II multiangle light scattering detector (Wyatt Technology), and a Wyatt Optilab T-rEX refractive index detector (Wyatt Technology). Analytical SEC was performed at about 25°C using a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with a mobile phase containing 10 mM Hepes (pH 7.5), 200 mM NaCl, and 10 mM DTT. One hundred microliters of protein sample (10 mg/ml) was injected into the column and eluted at a flow rate of 0.5 ml/min. The column effluent was monitored in-line with three detectors that simultaneously monitor ultraviolet absorption, light scattering, and the refractive index. The measurements were analyzed using the ASTRA software (Wyatt Technology) to determine the molecular mass of the protein. The experiments were performed three times to check for reproducibility of the results.
Co-IP assay
About 3,500,000 HEK cells (HEK 293T, SGST.CN, catalog no.SCSP-502) were plated in 10-cm dishes and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone) supplemented with 10% fetal bovine serum (Biochrom). All complementary DNAs (cDNAs) were cloned into the pcDNA3.0 vector. The cells were transfected separately with the plasmids using Lipofectamine 2000 (Invitrogen). For functional analysis of the key residues at the ligand-binding site, the cells were transfected with the following plasmids: 2.5 μg of pcDNA3.0-Flag empty vector, 2.5 μg of Flag-hSAMTOR, 3.5 μg of Flag-hSAMTOR-R95A, 16 μg of Flag-hSAMTOR-D190A, or 20 μg of Flag-hSAMTOR-D202A; 2 μg of hemagglutinin (HA)-Nprl3; and 2 μg of Myc-Kaptin. For functional analysis of the N-terminal helical domain, the cells were transfected with the following plasmids: 2.5 μg of pcDNA3.0-Flag empty vector, 2.5 μg of Flag-hSAMTOR, 5 μg of Flag-GFP-hSAMTORNTD (residues 1 to 90), or 15 μg of Flag-hSAMTORCTD (residues 91 to 405); 2 μg of HA-Nprl3; and 2 μg of Myc-Kaptin.
To identify the residues forming the potential binding site of hSAMTOR for GATOR1-KICSTOR, the cells were transfected with the following plasmids: 2.5 μg of pcDNA3.0-Flag empty vector, 2.5 μg of Flag-hSAMTOR, or the mutant (7.5 μg of Flag-hSAMTOR-F175A, 5 μg of Flag-hSAMTOR-E196A, 5 μg of Flag-hSAMTOR-Y199A, 7.5 μg of Flag-hSAMTOR-Y249A, 5 μg of Flag-hSAMTOR-H282A, 5 μg of Flag-hSAMTOR-K291A, 7.5 μg of Flag-hSAMTOR-D279A/S280A, 7.5 μg of Flag-hSAMTOR-K306A/Y307A, 5 μg of Flag-hSAMTOR-H312A, and 7.5 μg of Flag-hSAMTOR-Q342A/D343A); 2 μg of HA-Nprl3; and 2 μg of Myc-Kaptin. To verify the functional role of F175A in the binding of GATOR1-KICSTOR, the cells were transfected with the following plasmids: 2.5 μg of pcDNA3.0-Flag empty vector, 2.5 μg of Flag-hSAMTOR, 7.5 μg of Flag-hSAMTOR-F175A, 15 μg of Flag-SAMTORCTD, or 15 μg of Flag-SAMTORCTD-F175A; 2 μg of HA-Nprl3; and 2 μg of Myc-Kaptin.
Forty hours after transfection, the cells were collected by centrifugation and lysed with a Triton X-100 lysis buffer [1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 20 mM tris, pH 7.5, 150 mM NaCl, and EDTA-free protease inhibitor (Roche)] for 10 min at 4°C. The cell lysates were centrifuged at 18,000g for 30 min, and then 0.5 mM SAM was added into the cleared lysates. The supernatants were incubated with 30 μl of a 50% slurry of anti-DYKDDDDK G1 Affinity Resin (GenScript) overnight at 4°C. The resin was washed once with a low-salt Triton X-100 wash buffer [1% Triton X-100, 20 mM tris (pH 7.5), and 150 mM NaCl] and three times with a high-salt Triton X-100 wash buffer [1% Triton X-100, 20 mM tris (pH 7.5), and 300 mM NaCl] and then eluted with flag peptide (500 ng/μl). Immunoprecipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting with antibodies specific to Flag (1:2500; Sigma-Aldrich, catalog no. F3165; RRID: AB_259529), HA (1:2500; CST, catalog no. 3724S; RRID: AB_1549585), and Myc (1:2500; Millipore, catalog no. 05-724; RRID: AB_309938). All of the co-IP assays were performed three times to check for reproducibility of the results.
Immunoblotting assay
About 625,000 HEK 293T cells were plated in six-well plates and cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Biochrom). All cDNAs were cloned into the pCAG and pcDNA3.0 vectors. The cells were transfected separately with the plasmids using Lipofectamine 2000 (Invitrogen). To examine whether the WT and mutant hSAMTOR regulate the mTORC1 activity in a dose-dependent manner, the cells were transfected with the following plasmids: 500 ng of pCAG-Flag-metap2 and a varied amount (400, 1000, or 2000 ng) of pcDNA3.0-Flag-hSAMTOR or pcDNA3.0-Flag-hSAMTOR-F175A.
Forty hours after transfection, the cells were collected by centrifugation and lysed with a Triton X-100 lysis buffer [1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 20 mM tris (pH 7.5), 150 mM NaCl, phosphatase inhibitor cocktail 1 (APExBIO), and EDTA-free protease inhibitor (Roche)] for 10 min at 4°C. The cell lysates were centrifuged at 18,000g for 30 min, and the supernatants were analyzed by immunoblotting for the phosphorylation level (p-T389) of S6K1 and the expression levels of the indicated proteins with antibodies specific to Flag (1:2500; Sigma-Aldrich, catalog no. F3165; RRID: AB_259529), S6K1 [1:1000; Cell Signaling Technology (CST), catalog no. 9202S; RRID: AB_331676], p-T389 S6K1 (1:1000; CST, 9234S; RRID: AB_2269803), and glyceraldehyde-3-phosphate dehydrogenase (1:6000; Proteintech, catalog no. 10494-1-AP; RRID: AB_2263076). The immunoblotting assay was performed three times to check for reproducibility of the results.
Crystallization, data collection, and structure determination
Crystallization of the WT and V66W/E67P mutant dSAMTOR was performed using the hanging drop vapor diffusion method by mixing 1.5 μl of protein solution (15 mg/ml) alone or supplemented with 1 mM SAM or SAH and 1.5 μl of reservoir solution at 16°C. Crystals of the SAM- or SAH-bound dSAMTOR were grown from drops consisting of the reservoir solution of 0.1 M sodium citrate (pH 5.0) and 20% (w/v) polyethylene glycol (PEG) 8000. Crystals of the V66W/E67P mutant dSAMTOR were grown from drops consisting of the reservoir solution of 1.8 M ammonium sulfate, 0.1 M bis-tris (pH 6.5), and 2% (v/v) PEG monomethyl ether 550. Diffraction data were collected at −175°C at beamlines 18U1 and 19U1 of the National Facility for Protein Science in Shanghai, China (50), and were processed with XDS (51). The diffraction patterns of the Se-Met–substituted SAM-bound dSAMTOR crystals exhibit strong anisotropic characteristics. The anisotropy of the diffraction data was analyzed by the Diffraction Anisotropy Server (52), which recommends the resolution limits of the data to be truncated to 2.60, 4.40 and 2.10 Å along the reciprocal space directions a*, b*, and c*, respectively. Thus, the original unmerged diffraction data were subjected to anisotropy correction using the STARANISO server (53), and the resultant diffraction data were used for SAD phasing and structure refinement. The diffraction patterns of the SAH-bound dSAMTOR and the V66W/E67P mutant dSAMTOR crystals do not exhibit anisotropic characteristics, and thus, the diffraction data were directly used for structure refinement without anisotropy correction. Statistics of the diffraction data are summarized in Table 2.
The structure of the SAM-bound MTase domain of dSAMTOR was solved using the SAD method implemented in Phenix (54), which yielded an overall figure of merit of 0.30 and identified all 20 Se sites of four SAMTOR molecules in an asymmetric unit. Initial structure model was built by the AutoBuild program in Phenix (54), and the final structure model was built manually using Coot (55). The structures of the SAH-bound MTase domain of dSAMTOR and the V66W/E67P mutant dSAMTOR were solved by the molecular replacement method implemented in Phenix (54). Structure refinement was carried out using Phenix and Refmac5 (54, 56). Structure figures were prepared using PyMOL (57). Statistics of the structure refinement and the quality of structure models are also summarized in Table 2.
Acknowledgments
We thank the staff members at BL18U1 and BL19U1 of the National Facility for Protein Science in Shanghai (NFPSS) and BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF) for technical assistance in diffraction data collection. We are grateful to B. Yang, H. Yang, D. Gao, and other members of our research groups for helpful discussion.
Funding: This study was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB37030305), the National Key Research and Development Program of China (2020YFA0509000 and 2020YFA0803200), and the National Natural Science Foundation of China (31870722).
Author contributions: X.T. carried out the protein expression and purification, crystallization, and ITC experiments. Y.Z. carried out the co-IP and kinase activity experiments. T.Z. carried out the structural studies. G.W. and C.Z. participated in the protein expression and purification and crystallization experiments. F.W. and J.S. participated in the functional studies. J.D. and T.Z. conceived the study, designed the experiments, interpreted the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The crystal structures of the SAM- and SAH-bound MTase domain of dSAMTOR and the apo full-length V66W/E67P mutant dSAMTOR have been deposited in the RCSB Protein Data Bank with accession codes 7VKR, 7VKQ, and 7VKK, respectively.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
References
REFERENCES AND NOTES
- 1.Cornu M., Albert V., Hall M. N., mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 23, 53–62 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H., Rudge D. G., Koos J. D., Vaidialingam B., Yang H. J., Pavletich N. P., mTOR kinase structure, mechanism and regulation. Nature 497, 217–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H., Jiang X., Li B., Yang H. J., Miller M., Yang A., Dhar A., Pavletich N. P., Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wullschleger S., Loewith R., Hall M. N., TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K.-L., Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M., The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-Peled L., Schweitzer L. D., Zoncu R., Sabatini D. M., Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anandapadamanaban M., Masson G. R., Perisic O., Berndt A., Kaufman J., Johnson C. M., Santhanam B., Rogala K. B., Sabatini D. M., Williams R. L., Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogala K. B., Gu X., Kedir J. F., Abu-Remaileh M., Bianchi L. F., Bottino A. M. S., Dueholm R., Niehaus A., Overwijn D., Fils A. P., Zhou S. X., Leary D., Laqtom N. N., Brignole E. J., Sabatini D. M., Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laplante M., Sabatini D. M., mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Peled L., Chantranupong L., Cherniack A. D., Chen W. W., Ottina K. A., Grabiner B. C., Spear E. D., Carter S. L., Meyerson M., Sabatini D. M., A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen K., Huang R. K., Brignole E. J., Condon K. J., Valenstein M. L., Chantranupong L., Bomaliyamu A., Choe A., Hong C., Yu Z., Sabatini D. M., Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 556, 64–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfson R. L., Chantranupong L., Wyant G. A., Gu X., Orozco J. M., Shen K., Condon K. J., Petri S., Kedir J., Scaria S. M., Abu-Remaileh M., Frankel W. N., Sabatini D. M., KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng M., Yin N., Li M. O., SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng M., Yin N., Li M. O., Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 159, 122–133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfson R. L., Chantranupong L., Saxton R. A., Shen K., Scaria S. M., Cantor J. R., Sabatini D. M., Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chantranupong L., Scaria S. M., Saxton R. A., Gygi M. P., Shen K., Wyant G. A., Wang T., Harper J. W., Gygi S. P., Sabatini D. M., The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Ou Y., Luo R., Wang J., Wang D., Guan J., Li Y., Xia P., Chen P. R., Liu Y., SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 596, 281–284 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Meng D., Yang Q., Wang H., Melick C. H., Navlani R., Frank A. R., Jewell J. L., Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 295, 2890–2899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu X., Orozco J. M., Saxton R. A., Condon K. J., Liu G. Y., Krawczyk P. A., Scaria S. M., Harper J. W., Gygi S. P., Sabatini D. M., SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu S. C., Mato J. M., S-Adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 92, 1515–1542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai S., Ye Q., Zeng X., Yang G., Ye C., Chen M., Yu H., Wang Y., Wang G., Huang S., Quan S., Zeng X., Qiao S., CBS and MAT2A improve methionine-mediated DNA synthesis through SAMTOR/mTORC1/S6K1/CAD pathway during embryo implantation. Cell Prolif. 54, e12950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin J. L., McMillan F. M., SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J., Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell 74, 299–307 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Schubert H. L., Blumenthal R. M., Cheng X., Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 28, 329–335 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Honigschmid P., Schafferhans A., Roos M., Bernhofer M., Richter L., Ashkenazy H., Punta M., Schlessinger A., Bromberg Y., Schneider R., Vriend G., Sander C., Ben-Tal N., Rost B., PredictProtein—An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–W343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., Fukamizu A., Kimura K., Shimizu T., Yanagisawa J., Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133, 627–639 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Cakici O., Sikorski M., Stepkowski T., Bujacz G., Jaskolski M., Crystal structures of NodS N-methyltransferase from Bradyrhizobium japonicum in ligand-free form and as SAH complex. J. Mol. Biol. 404, 874–889 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Kozbial P. Z., Mushegian A. R., Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 5, 19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liscombe D. K., Louie G. V., Noel J. P., Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 29, 1238–1250 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Zidek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G. J., Bateman A., Evans R., Pritzel A., Figurnov M., Ronneberger O., Bates R., Kohl S. A. A., Potapenko A., Ballard A. J., Romera-Paredes B., Nikolov S., Jain R., Clancy E., Reiman D., Petersen S., Senior A. W., Kavukcuoglu K., Birney E., Kohli P., Jumper J., Hassabis D., Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Wang X., Feng J., Chen Y., Fang Y., Zhao S., Zhao A., Zhang M., Liu L., Structural insights into the catalytic mechanism of Synechocystis magnesium protoporphyrin IX O-methyltransferase (ChlM). J. Biol. Chem. 289, 25690–25698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. H., Bae B., Kuemin M., Circello B. T., Metcalf W. W., Nair S. K., van der Donk W. A., Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 17557–17562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lauinger L., Kaiser P., Sensing and signaling of methionine metabolism. Metabolites 11, 83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyh-Chang N., Locasale J. W., Lyssiotis C. A., Zheng Y. X., Teo R. Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J. J., Zhu H., Asara J. M., Daley G. Q., Cantley L. C., Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y. J., Yu H., Zhang J., Gao H., Wang S. Y., Li S. X., Wei P., Liang J., Yu G. Z., Wang X. J., Li X. X., Li D. W., Yang W. W., Cul4A-DDB1-mediated monoubiquitination of phosphoglycerate dehydrogenase promotes colorectal cancer metastasis via increased S-adenosylmethionine. J. Clin. Investig. 131, e146187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M., Rho Y., Park R., Jung J. Y., Hwang G. S., Seo Y. K., Seo J. H., Heo Y., Ha T. K., Ha E., Duodenal-jejunal bypass maintains hepatic S-adenosylmethionine/S-homocysteine ratio in diet-induced obese rats. Surg. Obes. Relat. Dis. 17, 1359–1368 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Barber J. R., Morimoto B. H., Brunauer L. S., Clarke S., Metabolism of S-adenosyl-l-methionine in intact human erythrocytes. Biochim. Biophys. Acta 886, 361–372 (1986). [DOI] [PubMed] [Google Scholar]
- 42.Gai Z., Wang Q., Yang C., Wang L., Deng W., Wu G., Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell Discov. 2, 16051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxton R. A., Chantranupong L., Knockenhauer K. E., Schwartz T. U., Sabatini D. M., Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536, 229–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia J., Wang R., Zhang T., Ding J., Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling. Cell Discov. 2, 16035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budanov A. V., Sablina A. A., Feinstein E., Koonin E. V., Chumakov P. M., Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304, 596–600 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Saxton R. A., Knockenhauer K. E., Wolfson R. L., Chantranupong L., Pacold M. E., Wang T., Schwartz T. U., Sabatini D. M., Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma S., Watzinger P., Kotter P., Entian K. D., Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 41, 5428–5443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Y., Ji L., Huang Q., Vassylyev D. G., Chen X., Ma J. B., Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461, 823–827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y., Eichhorn C. D., Wang Y., Cascio D., Feigon J., Structural basis of 7SK RNA 5′-γ-phosphate methylation and retention by MePCE. Nat. Chem. Biol. 15, 132–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W., Tang J., Wang S., Wang Z., Qin W., He J., The protein complex crystallography beamline (BL19U1) at the Shanghai synchrotron radiation facility. Nucl. Sci. Tech. 30, 170 (2019). [Google Scholar]
- 51.Kabsch W., XDS. Acta Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D., Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.I. J. Tickle, C. Flensburg, P. Keller, W. Paciorek, A. Sharff, C. Vonrhein, G. Bricogne, STARANISO (Global Phasing Ltd., 2018); http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi.
- 54.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Murshudov G. N., Skubak P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrödinger, LLC, The PyMOL molecular graphics system, version 2.0 Schrödinger, LLC (2015).
- 58.Gouet P., Courcelle E., Stuart D. I., Metoz F., ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Vidgren J., Svensson L. A., Liljas A., Crystal structure of catechol O-methyltransferase. Nature 368, 354–358 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
References