Abstract
IκBα is a critical regulator of Rel/NF-κB-mediated gene activation. It controls the induction of NF-κB factors by retaining them in the cytoplasm and also functions in the nucleus to terminate the induction process. In this study, we show that IκBα regulates the transcriptional activity of c-Rel in the nuclear compartment. We also demonstrate that discrete functional domains of IκBα are responsible for the cytoplasmic and nuclear regulation of c-Rel. We show that the determinants for the cytoplasmic regulation of c-Rel reside in the N-terminal and central ankyrin regions of IκBα and that the N-terminal domain of IκBα is required to mask the c-Rel nuclear localization signal. Importantly, IκBα sequences necessary to regulate c-Rel in the nucleus map to its central ankyrin domain and to a few negatively charged amino acids that immediately follow in the C-terminal IκBα PEST domain. The mapping of the IκBα determinants that control the cytoplasmic and nuclear activities of c-Rel to specific regions of the molecule suggests that IκBα inhibitors could be designed to antagonize Rel/NF-κB activity in different subcellular compartments or at defined stages of activation.
The Rel/NF-κB family of transcription factors plays a central role in the immune, inflammatory, and acute-phase responses and in the control of cell proliferation (reviewed in references 5–7, 52, 68 and 72). The vertebrate proteins p50/NF-κB1, p52/NF-κB2, RelA, c-Rel, and the viral oncoprotein v-Rel belong to this family (7, 72). These proteins share an N-terminal Rel homology domain that participates in their DNA binding, dimerization, and nuclear localization. The formation of Rel homodimers and heterodimers enables their binding to the major groove of decameric κB DNA sites and the regulation of gene expression (reviewed in references 5–7, 52, 68, and 72).
Rel factors share common pathways of activation that involve their rapid release from inhibitory IκB proteins in response to various stimuli (reviewed in references 7, 32, 52, and 72). IκB factors associated with cytoplasmic Rel dimers mask the Rel nuclear localization signal (NLS), thereby impeding Rel translocation to the nucleus (10, 28, 77). In response to stimuli, IκBα undergoes phosphorylation and rapid degradation via the 26S proteasome (1, 15, 16, 20, 43, 50, 58, 59, 62, 69, 73). These processes allow the nuclear translocation of Rel proteins and the activation of gene expression. In turn, nuclear Rel factors trigger the resynthesis of IκBα, giving rise to an autoregulatory loop that terminates the activation process (17, 21, 46, 64, 66). The transient accumulation of newly synthesized IκBα in the nucleus of stimulated cells correlates with the repression of Rel/NF-κB-dependent gene expression (3, 56). Accumulating evidence supports a model in which the entry of IκBα into the nucleus triggers the relocalization of Rel complexes from the nucleus to the cytoplasm to terminate the activation process (4, 11, 41). The recent identification of a nuclear export signal in IκBα is consistent with this model (4). In light of these findings, it appears that IκBα functions as a dual regulator of Rel proteins by controlling their activation in the cytoplasm and by terminating their activity in the nucleus.
The N-terminal domain of IκBα contains serine and lysine residues that undergo phosphorylation and ubiquitination, respectively, and that are critical for its inducible degradation (1, 15, 16, 20, 24, 43, 50, 58, 59, 62, 69, 73). Mutation or deletion of this domain stabilizes the inhibitor under inducing conditions (2, 15, 43, 65). A central domain of six ankyrin repeats is essential for the interaction of IκBα with Rel factors (34–37). The C-terminal 35 amino acids of IκBα contain a PEST region, rich in negatively charged residues. While this portion of the protein is dispensable for its inducible degradation and for its interaction with Rel, phosphorylation of this portion regulates the constitutive degradation of the protein (2, 8, 48, 51, 63, 65, 71). This portion of IκBα also has been implicated in the inhibition of RelA and c-Rel DNA binding (26, 61). Of the different Rel domains that contact IκBα, the Rel NLS region appears to be the main site for interaction with the inhibitor. Mutation of the NLS decreases the affinity of Rel for IκBα and enables Rel to escape the inhibitory function of IκBα (10, 28, 45, 77).
Among the different Rel/NF-κB dimers, p50-RelA complexes are the most extensively studied. Much less is known, however, about the regulation and function of the transactivating subunit c-Rel. Studies with transgenic mice and knockout animals demonstrated an essential role for c-Rel in lymphocyte proliferation, in immune and inflammatory responses, and in T-cell development (12, 42). As c-rel is the cellular homolog of the viral oncogene v-rel, it is not surprising to find a correlation between the amplification and overexpression of the c-rel gene and oncogenesis (reviewed in references 31 and 49). The activity of c-Rel therefore must be tightly regulated.
In this study, we characterized the functional domains of IκBα involved in the cytoplasmic and nuclear regulation of c-Rel. We show that IκBα can function in vivo to regulate the transcriptional activity of c-Rel in the nuclear compartment. Our studies indicate that the central ankyrin domain of IκBα, together with a few negatively charged C-terminal acidic amino acids, is necessary and sufficient to regulate c-Rel in the nucleus. While the C-terminal domain of IκBα is dispensable for the cytoplasmic retention of c-Rel, we show that its N-terminal domain and ankyrin repeats are essential for this function. We also provide evidence that the N-terminal region of IκBα is required to mask the Rel NLS. Our results indicate that the determinants for the cytoplasmic regulation of c-Rel reside in the N-terminal and central ankyrin domains of IκBα, while those necessary for nuclear Rel protein regulation map to its central ankyrin domain and to a few acidic amino acids that immediately follow in the C-terminal IκBα PEST region.
MATERIALS AND METHODS
Plasmids and mutagenesis.
CCR encodes the wild-type chicken c-Rel protein (18). The CCR.SVNLS mutant was generated by insertion of a double-stranded oligonucleotide (5′-CTAGAGATCACGCCACCAAAGAAGAAAAGGAAAGTGGAGGACCCT-3′) encompassing the simian virus 40 (SV40) large T antigen NLS at the unique HincII site of the chicken c-rel gene. Mutant CCR.KO.SVNLS was derived by site-directed mutagenesis of CCR.SVNLS to inactivate the Rel NLS (KRQR to NWLT). A 1.7-kb EcoRI DNA fragment containing the chicken iκbα gene (23) was cloned into the EcoRI site of the pAlter-1 plasmid (Promega). IκBα deletion mutants were generated by introducing stop codons at defined positions by use of the Altered Sites mutagenesis system (Promega). Mutants Δ267, Δ282, 76-284, Δ285, Δ287, and Δ301, respectively, lack 52, 37, 34, 34, 32, and 18 amino acids from the C terminus of IκBα. Mutants 76-318 and 76-284 were generated by deletion of the first 75 amino acids from the N terminus of full-length IκBα (76-318) or of mutant Δ285 (76-284), followed by the introduction of an ATG codon at position 75 by site-directed mutagenesis. Mutations were confirmed by DNA sequence analysis with Sequenase (U.S. Biochemical).
c-rel and iκbα genes were expressed in vitro from the T7 promoter of the pAlter-1 plasmid. Wild-type and mutant c-Rel and IκBα proteins were expressed in vivo under the control of the cytomegalovirus (CMV) immediate-early promoter of pJDCMV19SV (25) or pcDNA I/Amp (Invitrogen). Plasmid pIL6CAT expresses the chloramphenicol acetyltransferase (CAT) reporter gene from the interleukin-6 promoter and has three copies of the IL6-κB DNA-binding motif (54).
Cell transfection and CAT assays.
Cos-7 SV40-transformed African green monkey kidney cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Cells were maintained at 37°C in an atmosphere of 5% CO2. Transfections were carried out by a modified calcium phosphate procedure (19). Cells (5 × 105) in 60-mm dishes were transfected with 5 μg of CMV rel plasmid DNA in the presence or absence of wild-type or mutant CMV iκbα DNA and 3 μg of pIL6CAT. The total amount of plasmid DNA transfected (13 μg) was kept constant by the addition of the CMV vector. The calcium phosphate precipitate was removed after 16 to 24 h of incubation, and the cells were washed and refed with medium containing 3% fetal bovine serum. Cell extracts were prepared 36 to 48 h after transfection, and the protein concentration was measured by the method of Bradford (14). CAT activities were determined within the linear range of the assay as described previously (29). Assays were performed with 10 μg of total cellular protein for 20 min. Normalized CAT activity from the average of three independent experiments is shown.
Nuclear and cytoplasmic extracts.
Nuclear and cytoplasmic extracts were prepared as described previously (55). Briefly, cells were harvested and resuspended in buffer A (10 mM HEPES [pH 7.8], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% Nonidet P-40). Extracts were incubated for 10 min on ice and fractionated by centrifugation at 10,000 × g. Supernatants containing the cytoplasmic fractions were recovered. The pellets were resuspended in buffer A and centrifuged again at 10,000 × g. The nucleus-containing pellets were lysed in 20 μl of buffer C (20 mM HEPES [pH 7.8], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) and incubated for 20 min on ice. Nuclear extracts were separated from cell debris by centrifugation at 10,000 × g and diluted in 80 μl of buffer D (20 mM HEPES [pH 7.8], 20% glycerol, 50 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF).
Immunofluorescence.
Cells were seeded onto glass coverslips and transfected as described above. Coverslips were harvested at 48 h posttransfection. The cells were fixed in 4% paraformaldehyde and permeabilized in phosphate-buffered saline–0.2% Triton X-100. Coverslips were incubated with rabbit polyclonal antibody Ab1801, specific for the C terminus of c-Rel (45), or with anti-p40/IκBα antibody (39) followed by fluorescein-conjugated goat anti-rabbit antibodies. Coverslips were mounted in the presence of 0.2% para-phenylenediamine (Sigma) and photographed at a magnification of ×600.
Coimmunoprecipitation assays.
Cos-7 cells were harvested at 48 h posttransfection, lysed by sonication in 50 mM Tris HCl (pH 7.5)–150 mM sodium chloride–1% sodium deoxycholate–1% Triton X-100–10 μg of leupeptin per ml–10 mM sodium pyrophosphate–50 mM sodium fluoride–0.5 mM sodium orthovanadate (57), and quantitated for protein concentration by the method of Bradford (14). Equal amounts of whole-cell extracts (400 to 600 μg) were immunoprecipitated with anti–c-Rel antibody Ab1801 (45) or with antibody Ab1507, specific for the c-Rel NLS (26), and protein A-Sepharose (Pharmacia). Samples were washed extensively with ELB buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40) (26) supplemented with Complete protease inhibitor cocktail (Boehringer Mannheim Biochemicals) and resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
Wild-type or mutant c-Rel and IκBα proteins were produced in vitro with a TNT rabbit reticulocyte expression system (Promega) in the presence of 35S-labeled cysteine. Translation reactions were quantitated with a PhosphorImager. c-Rel translation reaction mixtures (3 μl) were incubated in the presence or absence of a threefold molar excess of wild-type or mutant IκBα protein for 15 min at room temperature. Reactions were immunoprecipitated with antibody Ab1801 (45) or Ab1507 (26) and protein A-Sepharose.
DNA-binding assays.
DNA-binding assays of in vivo-synthesized c-Rel proteins were performed with nuclear (3 μg) or whole-cell (20 μg) extracts from transfected cells prepared as described for coimmunoprecipitation assays. Extracts were incubated with 3 × 104 cpm of a 32P-labeled IL6-κB oligonucleotide probe in 12.5 mM HEPES (pH 7.9)–12% glycerol–5 mM MgCl2–60 mM KCl–0.2 mM EDTA–1 mM DTT–1 μg of bovine serum albumin per μl–1 μg of poly(dI-dC) per μl (74) and analyzed on 5% native polyacrylamide gels. In supershift assays, extracts were preincubated with 0.5 to 2 μl of antisera specific for c-Rel (45) or IκBα (39) prior to DNA-binding reactions.
RESULTS
c-Rel mutants that escape cytoplasmic regulation by IκBα.
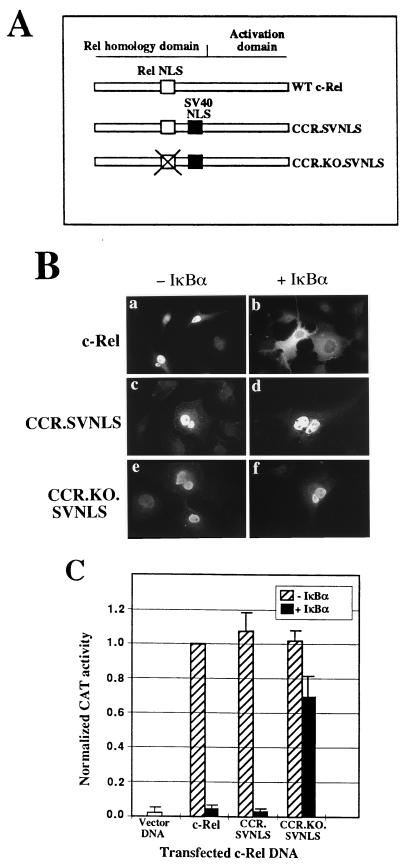
To bypass cytoplasmic regulation by IκBα, c-Rel was constitutively targeted to the nucleus by inserting the NLS of the SV40 large T antigen 3′ to the Rel homology domain of the chicken c-rel gene (38) (CCR.SVNLS; Fig. 1A). As a negative control, the Rel NLS was inactivated by mutagenesis of CCR.SVNLS (CCR.KO.SVNLS; Fig. 1A). The effect of IκBα on the subcellular localization of wild-type and mutant c-Rel proteins was examined by immunofluorescence. As anticipated, c-Rel localized to the nucleus of transiently transfected Cos-7 cells, and the coexpression of the chicken IκBα protein led to the sequestration of c-Rel in the cytoplasm (Fig. 1B, panels a and b) (26, 61). In contrast, CCR.SVNLS and CCR.KO.SVNLS constitutively localized to the nucleus irrespective of the presence or absence of IκBα (Fig. 1B, panels c to f). This result demonstrated the ability of these proteins to escape cytoplasmic regulation by IκBα.
FIG. 1.
c-Rel mutants that bypass cytoplasmic regulation by IκBα. (A) Structures of the CCR.SVNLS and CCR.KO.SVNLS mutants of c-Rel. The Rel NLS and the SV40 NLS are depicted as white and black boxes, respectively. Mutation of the Rel NLS in CCR.KO.SVNLS is indicated by X. WT, wild type. (B) Subcellular localization of c-Rel and c-Rel-derived mutants. Cos-7 cells cotransfected with wild-type or mutant c-rel genes, together with CMV or CMV iκbα expression plasmids, were analyzed by indirect immunofluorescence with an anti–c-Rel antibody. (C) Transcriptional activity of wild-type and mutant c-Rel proteins. Cos-7 cells were transfected with CMV vector DNA or with expression plasmids encoding c-Rel or c-Rel-derived mutants, alone or together with an expression vector for IκBα. The pIL6CAT reporter plasmid, containing three κB DNA sites upstream of the interleukin-6 promoter, was included in the transfection. The average CAT activity from three independent experiments was normalized to that of wild-type c-Rel.
The ability of IκBα to regulate the transcriptional activity of c-Rel and its nuclear mutants was examined in transiently transfected Cos-7 cells. c-Rel, CCR.SVNLS, and CCR.KO.SVNLS transfected alone strongly activated κB DNA site-dependent gene expression in comparison to that obtained with the control pJDCMV19SV vector (Fig. 1C). This result verified that the insertion of the SV40 NLS was not detrimental to the transcriptional activity of c-Rel. Cotransfection with IκBα led to a moderate decrease in the activity of the CCR.KO.SVNLS control, in agreement with the reduced affinity of IκBα for Rel proteins with mutations in the NLS (10, 28; data not shown). In contrast, IκBα abolished transactivation mediated by c-Rel and by the nuclear CCR.SVNLS protein (Fig. 1C). The inhibition of c-Rel-mediated transcription likely resulted from the cytoplasmic retention of c-Rel by IκBα (Fig. 1B). However, the inhibition of CCR.SVNLS activity suggested its regulation by IκBα in the nucleus, consistent with the colocalization of the CCR.SVNLS and IκBα proteins in the nuclear compartment (Fig. 1B; see also Fig. 4). This result indicated that IκBα can regulate the activity of c-Rel in the nucleus and that CCR.SVNLS could be used to map the domains of IκBα necessary for the nuclear regulation of c-Rel.
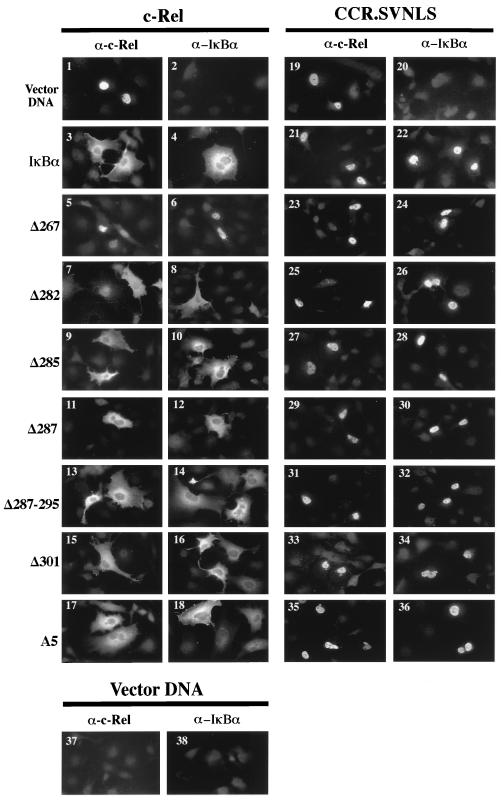
FIG. 4.
Subcellular localization of c-Rel and CCR.SVNLS in the presence of wild-type or mutant IκBα proteins. Cos-7 cells were transfected with expression plasmids encoding c-Rel or CCR.SVNLS, together with control CMV vector DNA or CMV expression plasmids for IκBα or IκBα mutants. Cells were analyzed by immunofluorescence with anti–c-Rel or anti-IκBα antibodies (α). A minimum of 80 positively staining c-Rel-expressing cells were examined. Representative fields are shown.
Mapping of C-terminal IκBα sequences necessary for the cytoplasmic and nuclear regulation of c-Rel.
The C-terminal PEST domain of IκBα was implicated in the inhibition of Rel DNA binding, and the phosphorylation of serine and threonine residues in this region was proposed to be required for this activity (26, 61). The C-terminal sequences of IκBα necessary for the nuclear regulation of c-Rel were mapped by deletion analysis (Fig. 2). The serine and threonine residues in the PEST domain were mutated to alanine to address the role of phosphorylation in the nuclear regulation of c-Rel (A5; Fig. 2). Mutants were assayed for association with c-Rel and CCR.SVNLS, for effects on their subcellular localization, and for inhibition of their DNA-binding and transcriptional activities.
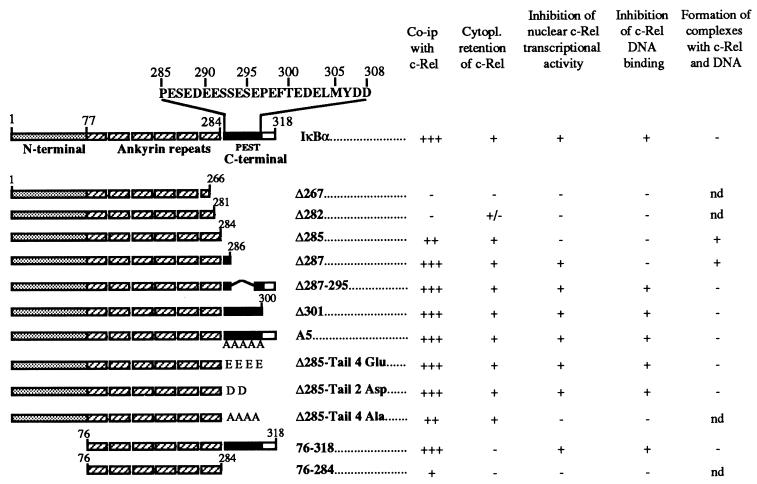
FIG. 2.
Structures and properties of IκBα mutants. The structure of IκBα, comprised of an N-terminal domain (stippled box), ankyrin repeats (hatched boxes), and a C-terminal region (open box) containing a PEST domain (black box), is depicted (not to scale). C-terminal deletion mutants Δ267, Δ282, Δ285, Δ287, Δ301, and 76-284 were generated by introducing stop codons. Translation start codons were introduced at position 75 in mutants 76-318 and 76-284. Mutant A5 contains alanine substitutions for the serines at positions 287, 292, 293, and 295 and the threonine at position 300 of IκBα. Mutant Δ287-295 has an internal deletion of nine amino acids in the C-terminal PEST domain of IκBα. Mutants Δ285-Tail 4 Glu, Δ285-Tail 2 Asp, and Δ285-Tail 4 Ala, respectively, have Glu, Asp, and Ala tails added to the C terminus of mutant Δ285. nd, not determined.
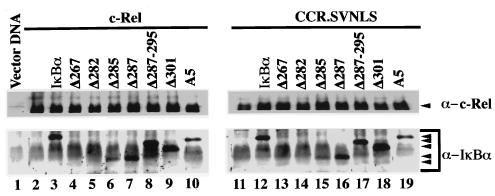
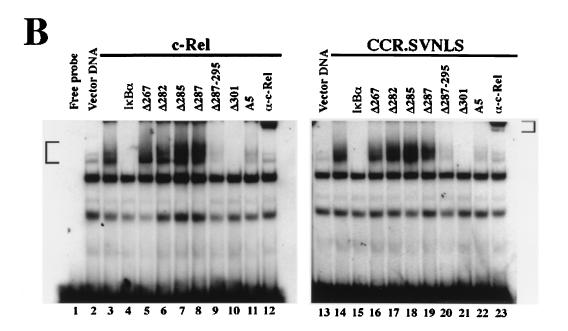
Mutations affecting most of the C terminus of IκBα, including the PEST domain, had no detrimental effect on its association with c-Rel or CCR.SVNLS. As shown in Fig. 3, mutant IκBα proteins affected in this region coprecipitated with c-Rel or CCR.SVNLS as efficiently as wild-type IκBα (Δ287, Δ287-295, Δ301, and A5; compare lanes 3 and 12 to lanes 7 to 10 and lanes 16 to 19, respectively). Whereas c-Rel was nuclear when expressed alone in Cos-7 cells, its coexpression with these mutant IκBα proteins led to their colocalization in the cytoplasm, as seen by immunofluorescence (Fig. 4, panels 1, 3, 4, and 11 to 18). The deletion of all sequences mapping 3′ to the ankyrin domain of IκBα also led to the cytoplasmic retention of c-Rel, despite a slight decrease in Rel protein affinity observed in coimmunoprecipitation assays (Δ285; Fig. 3, lanes 6 and 15, and Fig. 4, panels 9 and 10). In contrast, deletions affecting the ankyrin domain of IκBα compromised its interaction with c-Rel (Δ282 and Δ267). These mutants failed to associate with c-Rel or CCR.SVNLS in coimmunoprecipitation assays (Fig. 3, lanes 4, 5, 13, and 14). Similarly, these mutations respectively decreased or abolished the cytoplasmic retention of c-Rel (Fig. 4, panels 5 to 8). As anticipated, CCR.SVNLS constitutively localized to the nucleus, regardless of the presence or absence of wild-type or mutant IκBα proteins (Fig. 4, panels 19 to 36). Together, these results indicated that the C-terminal domain of IκBα is dispensable for association with c-Rel and for the retention of c-Rel in the cytoplasm.
FIG. 3.
Interaction of c-Rel and CCR.SVNLS with wild-type or mutant IκBα. Extracts from Cos-7 cells transfected with CMV vectors expressing c-Rel (lanes 2 to 10) or CCR.SVNLS (lanes 11 to 19), alone (lanes 2 and 11) or together with expression plasmids for IκBα (lanes 3 and 12) or IκBα mutants (lanes 4 to 10 and lanes 13 to 19), were immunoprecipitated with an anti–c-Rel antibody. Complexes were resolved on 12% polyacrylamide–SDS gels and analyzed by immunoblotting with antibodies specific for c-Rel (top panels) or IκBα (bottom panels). The positions of c-Rel, CCR.SVNLS, and wild-type or mutant IκBα proteins are indicated. Control immunoblots verified that equivalent amounts of wild-type and mutant IκBα proteins were expressed in the cells (data not shown).
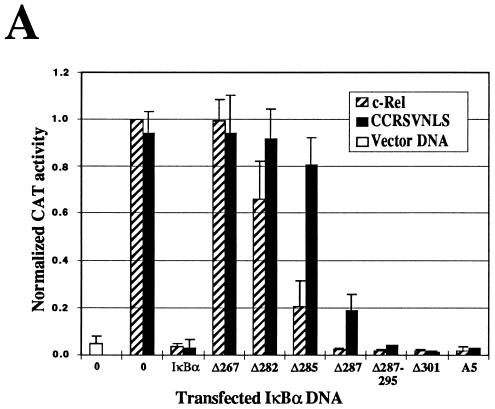
IκBα sequences specifically involved in the regulation of c-Rel in the nucleus were mapped by analyzing the effects of C-terminal IκBα mutations on the transcriptional activity of c-Rel and CCR.SVNLS. As expected, the transactivation potential of c-Rel was significantly inhibited by wild-type and mutant IκBα proteins that led to its efficient retention in the cytoplasm (Δ285, Δ287, Δ287-295, Δ301, and A5; Fig. 5A). Similarly, some of these mutants with deletions or mutations affecting most of the IκBα PEST domain strongly inhibited the activity of the nuclear CCR.SVNLS protein (Δ287, Δ287-295, Δ301, and A5; Fig. 5A). Importantly, however, the elimination of the entire PEST domain in mutant Δ285 dramatically decreased the inhibition of CCR.SVNLS-mediated transactivation but had little effect on the inhibitory activity of IκBα mutant Δ285 toward c-Rel (Fig. 5A). Mutants with deletions affecting the ankyrin domain showed little or no inhibitory activity toward c-Rel or CCR.SVNLS-mediated transcription (Δ267 and Δ282). This result agreed with their impaired association with c-Rel and CCR.SVNLS (Fig. 3, lanes 4, 5, 13, and 14). The fact that mutant Δ285 selectively lost its inhibitory activity toward transcription mediated by the nuclear CCR.SVNLS protein suggested that the first two amino acids of the IκBα PEST domain are essential for regulating c-Rel in the nucleus.
FIG. 5.
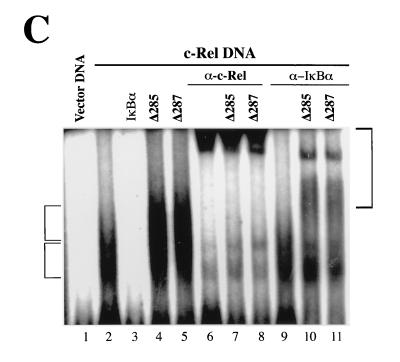
Effects of IκBα mutants on the transcriptional and DNA-binding activities of c-Rel. (A) Differential effects on c-Rel- and CCR.SVNLS-mediated transcription. Cos-7 cells were transfected with CMV vector DNA or CMV expression plasmids encoding c-Rel or CCR.SVNLS, alone (0) or together with expression plasmids for IκBα or IκBα mutants. The pIL6CAT reporter plasmid was included in the transfections. The average CAT activity from three independent experiments was normalized to that of wild-type c-Rel. (B) Inhibition of c-Rel and CCR.SVNLS DNA binding. Whole-cell extracts (20 μg) from Cos-7 cells transfected with CMV expression vectors for c-Rel (lanes 3 to 12) or CCR.SVNLS (lanes 14 to 23), alone (lanes 3 and 14) or together with CMV iκbα (lanes 4 and 15) or iκbα mutants (lanes 5 to 11 and 16 to 22), were analyzed in gel retardation assays. Extracts were incubated with a 32P-labeled IL6-κB DNA probe and resolved in 5% native acrylamide gels. DNA-bound complexes containing c-Rel or CCR.SVNLS were supershifted with an anti–c-Rel antibody (α) (lanes 12 and 23). The bracket on the left indicates the position of DNA-bound c-Rel and CCR.SVNLS complexes. The bracket on the right indicates the position of the supershifted Rel complexes. (C) Formation of complexes containing IκBα C-terminal deletion mutants, c-Rel, and DNA. Whole-cell extracts (20 μg) from Cos-7 cells transfected with vector DNA (lane 1) or CMV c-rel alone (lanes 2, 6, and 9) or together with vectors expressing IκBα (lane 3) or IκBα C-terminal deletion mutant Δ285 (lanes 4, 7, and 10) or Δ287 (lanes 5, 8, and 11) were analyzed for binding to an IL6-κB DNA probe in gel retardation assays. DNA-bound complexes were supershifted with anti–c-Rel (lanes 6 to 8) or anti-IκBα (lanes 9 to 11) antisera (α). Electrophoresis was carried out three times longer than usual to allow the separation of the different complexes. The brackets indicate the positions of DNA-bound Rel proteins (left) and of supershifted complexes (right).
The effects of C-terminal IκBα mutations on the DNA-binding activity of CCR.SVNLS were analyzed in gel retardation assays with whole-cell extracts from transiently transfected Cos-7 cells. As shown in Fig. 5B, the DNA-binding activity of CCR.SVNLS was abolished by wild-type and mutant IκBα proteins that efficiently inhibited its transcriptional activity (Δ287-295, Δ301, and A5; lanes 20 to 22). Interestingly, however, IκBα mutant Δ287, which efficiently blocked transcription by CCR.SVNLS, was unable to inhibit its DNA-binding activity (Fig. 5B, lane 19). Mutants with further deletions were also unable to block CCR.SVNLS DNA binding (Δ267, Δ282, and Δ285; Fig. 5B, lanes 16 to 18). As expected, similar data were obtained in gel retardation assays with nuclear extracts from CCR.SVNLS-expressing cells (data not shown). The analysis of c-Rel-expressing cells was used as a control to verify that the insertion of the SV40 NLS did not interfere with the inhibition of CCR.SVNLS DNA binding by IκBα. These results indicated that the presence of only 2 amino acids C terminal to the ankyrin domain is not sufficient to inhibit c-Rel DNA binding (Δ287). Moreover, our finding that IκBα mutant Δ287 blocked CCR.SVNLS-mediated transcription without interfering with its DNA-binding activity implicates the residues at the very beginning of the IκBα PEST domain in the specific inhibition of transcriptional activation by nuclear c-Rel proteins.
The observation that IκBα mutants Δ285 and Δ287 associated with c-Rel but failed to inhibit its DNA-binding activity led us to investigate whether the interaction of c-Rel with DNA would displace them from the complex. Cells expressing c-Rel alone showed efficient binding to a κB DNA site probe in gel retardation assays (Fig. 5C, lane 2). As expected, IκBα abolished c-Rel DNA binding, whereas mutants Δ285 and Δ287 had no such effect (Fig. 5C, lanes 3 to 5). Importantly, reaction mixtures containing either of these mutants together with c-Rel showed an additional DNA-bound complex of retarded mobility (Fig. 5C, compare lanes 4 and 5 to lane 2). An anti–c-Rel antibody supershifted both bands, indicating the presence of c-Rel in both complexes (Fig. 5C, compare lanes 7 and 8 to lanes 4 and 5, respectively). While an anti-IκBα antibody had little effect on the lower Rel-DNA complex, it supershifted the upper complex in cells expressing c-Rel together with IκBα mutant Δ285 or Δ287 (Fig. 5C, compare lanes 10 and 11 to lanes 4 and 5, respectively). The formation of complexes containing IκBα mutant Δ285 or Δ287 together with c-Rel and DNA agrees with the inability of these mutants to block c-Rel DNA binding.
Negatively charged residues at the beginning of the IκBα PEST domain are essential for regulation of the activity of c-Rel in the nucleus.
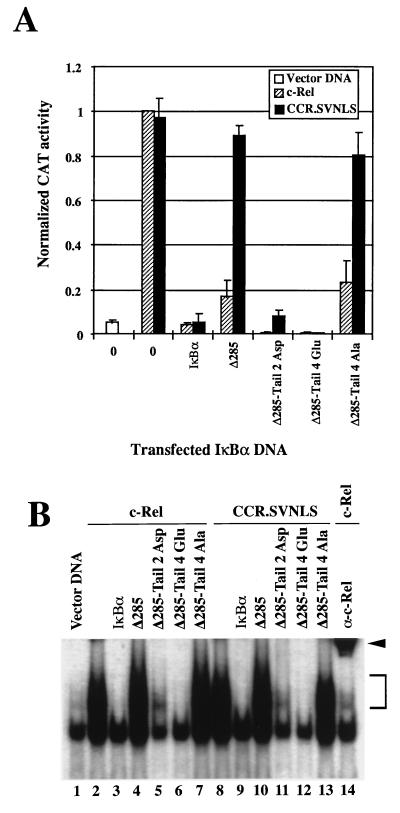
The acidic nature of the IκBα PEST domain and the important role of amino acids immediately C terminal to the ankyrin repeats in the inhibition of CCR.SVNLS-mediated transcription led us to investigate the effect of acidic amino acids C terminal to residue 285 on the nuclear regulation of c-Rel. IκBα mutants with tails of aspartate or glutamate residues added to the C terminus of mutant Δ285 were analyzed for inhibition of c-Rel- or CCR.SVNLS-mediated transcription in transient CAT assays (Δ285-Tail 2 Asp and Δ285-Tail 4 Glu; Fig. 2 and 6A). A neutral tail of alanines was added as a control (Δ285-Tail 4 Ala; Fig. 2). In agreement with the findings described above, mutant Δ285 inhibited c-Rel-mediated transcription via cytoplasmic retention but had no significant inhibitory activity toward the constitutively nuclear CCR.SVNLS protein (Fig. 6A). In sharp contrast, the addition of as little as two aspartate or four glutamate residues to the C terminus of IκBα mutant Δ285 fully restored its ability to inhibit the transcriptional activity of CCR.SVNLS. In contrast, the neutral tail of alanine residues had no inhibitory effect, despite the efficient association of the Δ285-Tail 4 Ala protein with c-Rel and CCR.SVNLS in coimmunoprecipitation assays (Fig. 6A and data not shown).
FIG. 6.
Effects of negatively charged residues in the C terminus of IκBα on the transcriptional and DNA-binding activities of c-Rel. (A) Effects on Rel-mediated transcription. Cos-7 cells were transfected with vector DNA or CMV plasmids encoding c-Rel or CCR.SVNLS, alone (0) or in combination with expression vectors for IκBα or IκBα mutants. Reporter plasmid pIL6CAT was included in the transfections. The normalized CAT activity from the average of three independent experiments is shown. (B) Effects on Rel DNA binding. Shown is a gel retardation analysis of whole-cell extracts (20 μg) from Cos-7 cells transfected with CMV vectors expressing c-Rel (lanes 2 to 7 and 14) or CCR.SVNLS (lanes 8 to 13), alone (lanes 2, 8, and 14) or together with vectors encoding wild-type (lanes 3 and 9) or mutant (lanes 4 to 7 and 10 to 13) IκBα proteins. Proteins were assayed for binding to a 32P-labeled IL6-κB DNA probe. The bracket indicates the position of ectopically expressed Rel-DNA adducts. The arrowhead indicates c-Rel–DNA complexes supershifted with an anti–c-Rel antiserum (α) (lane 14). The lower band present in all lanes corresponds to a DNA-bound complex of endogenous proteins.
The inhibitory activity of the Δ285-Tail mutants toward c-Rel and CCR.SVNLS DNA binding was examined in gel retardation assays. In agreement with the results described above, the addition of two or four acidic residues at the C terminus of Δ285 significantly or fully restored the inhibition of DNA binding by c-Rel and CCR.SVNLS, whereas a tail of alanine residues did not (Fig. 6B, lanes 5 to 7 and 11 to 13). Together, these experiments indicated that negatively charged residues at the C-terminal end of the ankyrin domain of IκBα are necessary to regulate the transcriptional activity of nuclear c-Rel proteins and their binding to DNA. Consistent with our deletion analyses, these assays also demonstrated that only a few acidic residues at this position are sufficient to regulate the activity of c-Rel in the nucleus.
The N terminus of IκBα is required to retain c-Rel in the cytoplasm.
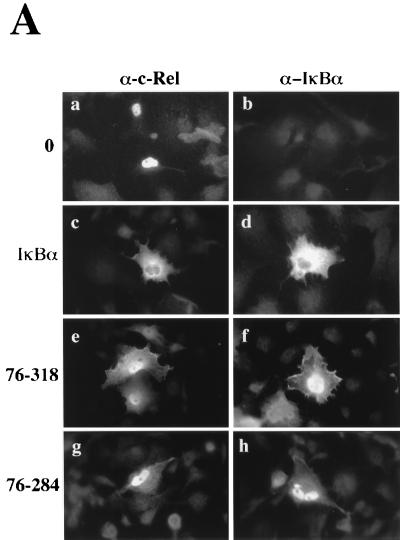
We complemented our study of IκBα by characterizing the activity of mutants lacking its N terminus or both its N- and C-terminal domains (mutants 76-318 and 76-284; Fig. 2). Mutants were analyzed for their effect on the subcellular localization of c-Rel by immunofluorescence. As expected, the coexpression of c-Rel with wild-type IκBα led to its confinement to the cytoplasm (Fig. 7A, compare panels c and a). However, the coexpression of c-Rel with N-terminal IκBα mutant 76-318 or 76-284 resulted in weak cytoplasmic and strong nuclear staining for c-Rel (Fig. 7A, compare panels e and g with panel c). This result suggested that deletion of the N terminus of IκBα impaired its capacity to retain c-Rel in the cytoplasm.
FIG. 7.
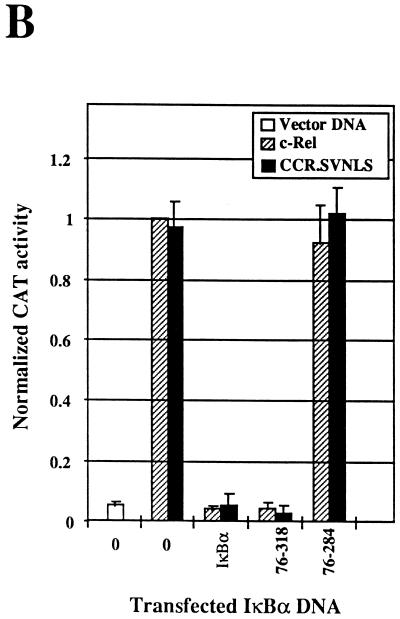
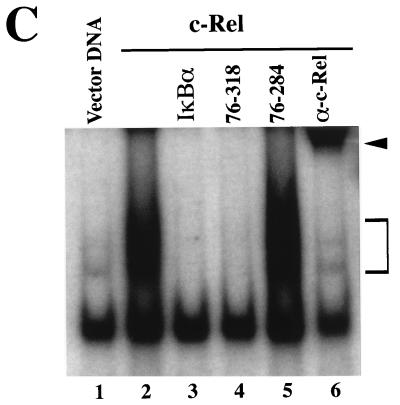
Effect of IκBα N-terminal deletion mutants on the subcellular localization and transcriptional and DNA-binding activities of c-Rel. (A) Effects on subcellular localization. The subcellular localization of c-Rel and IκBα proteins was assessed by immunofluorescence analysis of Cos-7 cells expressing c-Rel alone (0) or together with IκBα or its mutants 76-318 and 76-284, with anti–c-Rel or anti-IκBα antibodies (α). (B) Effects on Rel-mediated transcription. Cos-7 cells were transfected with pIL6CAT together with vector DNA or expression plasmids for c-Rel or CCR.SVNLS, alone (0) or in combination with plasmids encoding wild-type IκBα or mutant 76-318 or 76-284. The average CAT activity from three independent experiments was normalized to that of wild-type c-Rel. (C) Effects on Rel DNA binding. Whole-cell extracts (20 μg) from Cos-7 cells transfected with vector DNA (lane 1) or with CMV c-rel alone (lanes 2 and 6) or together with expression plasmids for IκBα or mutant 76-318 or 76-284 (lanes 3 to 5) were analyzed for binding to a 32P-labeled IL6-κB DNA probe in gel retardation assays. As a control, the DNA-bound c-Rel complex was supershifted with an anti–c-Rel antibody (α). The bracket indicates the position of c-Rel–DNA complexes. The arrowhead indicates supershifted c-Rel complexes. The lower band present in all lanes corresponds to a DNA-bound complex of endogenous proteins.
In spite of its decreased ability to retain c-Rel in the cytoplasm, mutant 76-318 inhibited c-Rel-mediated transcription as efficiently as wild-type IκBα. The inhibition of c-Rel-induced transcription by wild-type IκBα resulted from the retention of c-Rel in the cytoplasm. However, its inhibition by mutant 76-318 occurred primarily in the nucleus, as evidenced by the predominant nuclear localization of both c-Rel and mutant 76-318 (Fig. 7A, panels e and f). Consistent with this model, mutant 76-318 also strongly inhibited the transcriptional activity of the nuclear CCR.SVNLS protein (Fig. 7B). In keeping with these observations, IκBα mutant 76-318 exhibited wild-type inhibitory capacity toward c-Rel DNA binding (Fig. 7C, lane 4). As predicted from our analysis of C-terminal IκBα mutants, the deletion of both the N-terminal and the C-terminal domains of IκBα abolished its inhibitory effect toward c-Rel DNA-binding and transcriptional activities (mutant 76-284; Fig. 7B and C, lane 5). Combined, these experiments indicated that the N terminus of IκBα is important for the cytoplasmic regulation of c-Rel but is dispensable for its regulation in the nucleus.
IκBα mutants with N-terminal deletions fail to mask the c-Rel NLS.
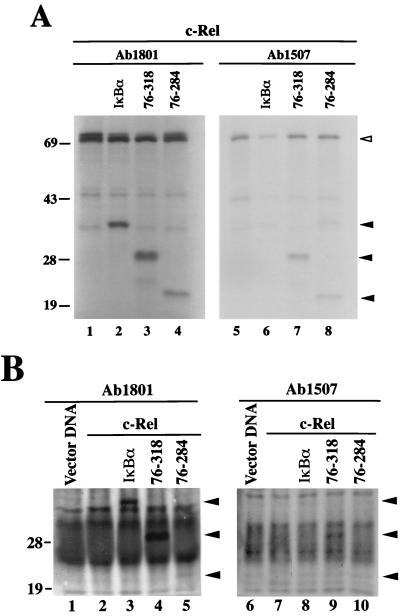
The inefficient retention of c-Rel in the cytoplasm by IκBα mutants with N-terminal deletions suggested that the c-Rel NLS may still be exposed when c-Rel is associated with these mutants. To verify this possibility, complexes of 35S-labeled c-Rel with wild-type IκBα or IκBα mutants with N-terminal deletions were analyzed by immunoprecipitation with an antibody directed against the c-Rel NLS (Ab1507; Fig. 8A). An antibody specific for the C terminus of c-Rel was used as a control (Ab1801). The antibody directed against the C terminus of c-Rel efficiently coprecipitated IκBα and its N-terminal deletion mutants (Fig. 8A, lanes 2 to 4). As anticipated, the anti–c-Rel NLS antibody Ab1507 failed to coprecipitate wild-type IκBα along with c-Rel due to masking of the Rel NLS (Fig. 8A, lane 6). In contrast, IκBα mutants 76-318 and 76-284 complexed with c-Rel were efficiently coprecipitated by this antibody (Fig. 8A, lanes 7 and 8). This result showed that the Rel NLS remained exposed when associated with IκBα N-terminal deletion mutants, whereas it was inaccessible to antibody Ab1507 when complexed with wild-type IκBα. Consistent with this observation, the amount of c-Rel brought down by this antibody in the presence of wild-type IκBα was significantly lower than that brought down in the presence of IκBα mutant 76-318 or 76-284 (Fig. 8A, compare lane 6 with lanes 7 and 8).
FIG. 8.
The c-Rel NLS is unmasked when associated with IκBα N-terminal deletion mutants. (A) Coimmunoprecipitation assay of in vitro-translated proteins. 35S-labeled c-Rel produced by in vitro translation was incubated in the presence of a threefold molar excess of in vitro-translated wild-type IκBα (lanes 2 and 6) or N-terminal deletion mutants of IκBα (lanes 3, 4, 7, and 8). c-Rel was incubated with an equal volume of a mock translation as a control (lanes 1 and 5). Reaction mixtures were immunoprecipitated with antibody Ab1801 against the C terminus of chicken c-Rel (lanes 1 to 4) or anti–c-Rel NLS antibody Ab1507 (lanes 5 to 8). Immunoprecipitated complexes were resolved in a 17% polyacrylamide–SDS gel. The open arrowhead indicates the position of c-Rel. Closed arrowheads indicate the positions of wild-type and mutant IκBα proteins. (B) Coimmunoprecipitation assay of in vivo-labeled proteins. Extracts from [35S]methionine- and [35S]cysteine-labeled Cos-7 cells transfected with CMV vector DNA (lanes 1 and 6) or CMV vectors expressing c-Rel alone (lanes 2 and 7) or together with wild-type or mutant IκBα (lanes 3 to 5 and 8 to 10) were immunoprecipitated with antibody Ab1801 against the C terminus of c-Rel (lanes 1 to 5) or anti–c-Rel NLS antibody Ab1507 (lanes 6 to 10). Reaction mixtures were resolved in a 17% polyacrylamide–SDS gel. Arrowheads indicate the positions of wild-type and mutant IκBα proteins. The sizes of protein molecular mass markers are indicated on the left in kilodaltons.
Similar results were obtained when c-Rel–IκBα complexes were analyzed in transfected cells (Fig. 8B). Since the anti-IκBα antibodies available failed to recognize the IκBα mutants in immunoblots, analysis was performed with proteins labeled in vivo with [35S]methionine and [35S]cysteine. As in our in vitro studies, anti–c-Rel NLS antibody Ab1507 failed to coprecipitate wild-type IκBα (Fig. 8B, compare lanes 3 and 8). However, it coprecipitated IκBα mutant 76-318 along with c-Rel (Fig. 8B, lane 9), indicating exposure of the c-Rel NLS. Our failure to detect the coprecipitation of IκBα mutant 76-284 with antibody Ab1507 presumably resulted from the additive effect of its lower content of methionines and cysteines (which caused its inefficient labeling) and its decreased affinity for c-Rel in vivo, as evidenced by the weak signal that we observed with antibody Ab1801 (Fig. 8B, compare lanes 10 and 5). Nevertheless, the data clearly indicated that the N-terminal domain of IκBα is important for the masking of the Rel NLS and is consequently critical for the cytoplasmic retention of c-Rel.
DISCUSSION
The activity of the Rel/NF-κB factors is tightly regulated. While some physiological conditions require a transient Rel/NF-κB response, others need prolonged activation. Integral parts of the transient activation process are the nuclear translocation of newly synthesized IκBα and the termination of Rel/NF-κB-mediated transcription (3, 11, 41). Here we show that IκBα can function in vivo to inhibit the activity of c-Rel in the nucleus. As previously described for RelA (10), the introduction of an extra NLS into c-Rel helped to circumvent its cytoplasmic regulation by IκBα. By comparing the effects of IκBα on the nuclear regulation of CCR.SVNLS and on the cytoplasmic regulation of c-Rel, we demonstrated that distinct functional domains in IκBα are required for the cytoplasmic and nuclear regulation of c-Rel.
Role of the IκBα C terminus in the cytoplasmic and nuclear regulation of c-Rel.
Our studies showed that the C terminus of IκBα, including the highly acidic PEST region, is dispensable for the cytoplasmic retention of c-Rel. Whereas deletion of this region did not significantly affect the capacity of IκBα to regulate c-Rel in the cytoplasm (Δ285), our deletion studies demonstrated that an intact ankyrin domain is necessary for interaction with c-Rel and its cytoplasmic sequestration. An IκBα C-terminal mutant with a deletion of only three amino acids in the ankyrin domain was compromised in its interaction with c-Rel (Δ282). Although this mutant showed some capacity to retain c-Rel in the cytoplasm, the high transcriptional activity that we observed when c-Rel was coexpressed with Δ282 indicated that some c-Rel dimers escaped cytoplasmic retention. This result emphasized the instability of their interaction. Thus, our failure to detect a physical association between mutant Δ282 and c-Rel in coimmunoprecipitation assays presumably resulted from their reduced affinity for one another and their dissociation under in vitro experimental conditions, as observed by others (26, 60, 61). Further deletion into the ankyrin domain abolished the interaction of IκBα with c-Rel both in vitro and in vivo (Δ267). These findings also agree with those of others (26, 61).
Mutant Δ285, with a deletion of the entire C-terminal domain of IκBα, was highly impaired for the inhibition of CCR.SVNLS-mediated transcription. This result is in contrast to its nearly wild-type capacity to inhibit the transcriptional activity of c-Rel by retaining it in the cytoplasm. The differential effect of this mutant toward c-Rel- and CCR.SVNLS-mediated transcription did not appear to result from differences in their affinity for mutant Δ285, as seen in coimmunoprecipitation assays. Moreover, several observations argue against the possibility that the presence of a foreign NLS altered the conformation and function of CCR.SVNLS: (i) CCR.SVNLS activated transcription as efficiently as wild-type c-Rel, (ii) its affinity for wild-type and mutant IκBα proteins was similar to that of c-Rel, and (iii) its DNA-binding activity was inhibited by IκBα as efficiently as that of c-Rel. Thus, these results indicate that while sequences C terminal to the ankyrin domain of IκBα are dispensable for the cytoplasmic retention of c-Rel, they are essential for inhibition of its transcriptional activity in the nucleus.
The addition of negatively charged tails to the C terminus of mutant Δ285 demonstrated that as few as two acidic amino acids C terminal to the ankyrin domain are sufficient to completely restore the inhibitory activity of IκBα in the nucleus. In agreement with this result, mutant Δ287, which contains a single acidic amino acid C terminal to the ankyrin domain, significantly inhibited the transcriptional activity of the constitutively nuclear CCR.SVNLS protein. Thus, it appears that most of the IκBα PEST domain is dispensable for the inhibition of c-Rel-mediated transcription in the nucleus.
Mechanism for the nuclear regulation of c-Rel.
Nuclear IκBα factors have been shown to promote the relocalization of RelA from the nucleus to the cytoplasm, and a nuclear export sequence was implicated in this process (amino acids 265 to 277 of human IκBα; 4). However, our observation that IκBα inhibited the in vivo activity of the constitutively nuclear CCR.SVNLS protein indicates that IκBα must also be able to actively antagonize nuclear c-Rel function. This antagonism may occur by blocking the transcriptional activity of c-Rel and/or by promoting the dissociation of c-Rel from DNA. In this scenario, the relocalization of Rel dimers to the cytoplasm would likely constitute a second step in the inhibitory process. Reports that IκBα can dissociate Rel-DNA complexes in vitro and block Rel-mediated transcriptional activation agree with our data (44, 70, 76).
The addition of acidic residues to the C terminus of mutant Δ285 simultaneously restored its capacity to inhibit the DNA-binding and transcriptional activities of CCR.SVNLS. This result suggested that IκBα may interfere with the activity of c-Rel in the nucleus by simply inhibiting its binding to κB DNA sites. However, while mutant Δ287 failed to inhibit DNA binding by CCR.SVNLS, it efficiently inhibited the transcriptional activity of this protein. Importantly, this mutant was able to form complexes with c-Rel that bound to DNA. It is thus tempting to speculate that the association of this mutant with DNA-bound c-Rel proteins might interfere with their transcriptional activity by preventing their interaction with components of the transcriptional machinery (40, 75). This model is consistent with a recent report showing that IκBα competes with the basal transcription initiation complex for binding to NF-κB (70). The nuclear regulation of c-Rel by IκBα may thus involve the inhibition of DNA contact and the disruption of c-Rel association with basal transcription factors. IκBα mutant Δ285 also formed complexes with c-Rel and DNA but did not block CCR.SVNLS-mediated transcription. Thus, these results implicate amino acids 285 and 286 at the beginning of the PEST domain in the inhibition of nuclear c-Rel-mediated transcription. Other members of the IκB family, namely, Bcl-3 and IκBβ, have been found in complexes with NF-κB subunits and DNA (13, 27, 67). However, to our knowledge this report is the first indication of an indirect interaction between an IκBα protein and DNA through its association with c-Rel.
Although the mechanism through which IκBα inhibits c-Rel DNA binding remains to be clarified, the absolute requirement for acidic residues at the beginning of the PEST domain suggests that this region may (i) compete with DNA for interaction with the positively charged DNA recognition loop of c-Rel (26, 30, 45, 53); (ii) induce the dissociation of c-Rel from DNA by promoting electrostatic repulsion between c-Rel and the negatively charged DNA backbone, as proposed by others (70); or (iii) induce a conformational change in c-Rel that is incompatible with DNA contact. Future studies will undoubtedly help to address these issues.
Role of phosphorylation of the IκBα PEST domain in the inhibition of c-Rel activity.
The serine and threonine residues found in the IκBα PEST domain are highly phosphorylated by casein kinase II (8, 48, 51, 61, 63). Their mutation in chicken IκBα was reported to dramatically decrease its affinity for c-Rel and to abolish its inhibitory activity toward c-Rel DNA binding (61). These results are in sharp contrast to ours. In our experiments, IκBα mutant A5, with all of the serine and threonine residues in the PEST domain mutated to alanines, exhibited wild-type inhibition of c-Rel DNA binding. This mutant was as effective as wild-type IκBα in interactions with c-Rel, in the cytoplasmic retention of c-Rel, and in the inhibition of c-Rel and CCR.SVNLS DNA binding and transcriptional activation. While the discrepancy between our results and those of Sachdev et al. (61) may derive from differences in experimental conditions, our assays indicate that phosphorylation of the PEST domain is not required for the inhibitory activity of IκBα. Similar findings were recently obtained with human IκBα (9, 22, 70).
The N terminus of IκBα is important for masking of the c-Rel NLS.
An intriguing finding from this study is that the N-terminal domain of IκBα led to masking of the Rel NLS. In contrast to the situation observed with wild-type IκBα, c-Rel associated with IκBα N-terminal deletion mutants was recognized by an NLS-specific antibody. This finding is consistent with the inability of these mutants to retain c-Rel in the cytoplasm. The Rel NLS was previously postulated to be essential for interactions with IκBα, since anti-NLS antibodies abolished the association of Rel with IκBα (77). Likewise, deletion or point mutations in the NLS of various Rel family members dramatically decreased their affinity for IκBα and impaired the inhibition of their DNA binding (10, 28). However, recent evidence suggests that the NLS itself is not required for the interaction of Rel with IκBα. (i) A c-Rel mutant with a deletion of its NLS efficiently interacted with IκBα in a yeast two-hybrid system (60). (ii) Similarly, the NLS of the Drosophila Rel homolog Dorsal is also dispensable for the interaction of Dorsal with the IκB inhibitor Cactus (33). (iii) Here, we showed that the transcriptional activity of a nuclear c-Rel protein with a mutated NLS was slightly inhibited by IκBα but was not abolished (CCR.KO.SVNLS). These results further suggest that NLS mutations only partially interfere with c-Rel protein association with the IκBα inhibitor.
As suggested by others, the ankyrin domain of IκBα may contact sequences proximal to the NLS in the tertiary structure of Rel (47). According to our data, this interaction may position the N-terminal domain of IκBα in a way that masks the Rel NLS and impedes interaction of the NLS with the nuclear translocation machinery. In this context, mutations in the Rel NLS perhaps disrupt the structure of Rel in a way that affects its optimal interaction with the inhibitor. Results similar to ours were recently obtained by others with c-Rel and with p50/NF-κB1 (57a). Interestingly, however, it appears that N-terminal deletions in IκBα may differentially affect the cytoplasmic regulation of c-Rel and RelA (57a, 65). Future studies will help to clarify the basis for these differences.
In conclusion, this study demonstrates that the determinants of IκBα involved in the cytoplasmic and nuclear regulation of c-Rel map to distinct domains of the molecule. The central ankyrin domain of IκBα mediates interactions with c-Rel. The N-terminal domain is essential for the cytoplasmic retention of c-Rel, while a few of the acidic amino acids at the very beginning of the C-terminal PEST domain are necessary to regulate the activity of c-Rel in the nucleus. The identification of these discrete domains may help in the design of inhibitors to antagonize Rel/NF-κB activity in a specific cellular compartment or at a defined stage of activation.
ACKNOWLEDGMENTS
We thank N. Rice (ABL-NCI, Frederick, Md.) for the generous gift of antibody Ab1507 against c-Rel, H. Bose (University of Texas, Austin) for an iκbα cDNA clone and antibodies against the chicken IκBα protein, T. Gilmore (Boston University, Boston, Mass.) for a chicken c-rel cDNA clone, and K. Nakayama (Kanazawa University, Kanazawa, Japan) for the pIL6CAT plasmid. We are grateful to S. Crespo for expert assistance with the construction and characterization of some IκBα mutants. We thank F. Agnès, J. Bash, C. Chen, and W.-X. Zong for fruitful discussions during the course of this work. We also thank J. Bash, C. Chen, I. Martínez-Férez, A. Rabson, and W.-X. Zong for helpful comments on the manuscript and N. Rice for sharing results prior to publication.
This work was supported by a grant from the National Institutes of Health (CA54999) and by the New Jersey Commission on Science and Technology (C.G.). I.L. was supported by postdoctoral fellowships from Ministerio de Educación y Ciencia (Spain)/Fulbright Program and from the International Human Frontier Science Program.
REFERENCES
- 1.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki T, Sano Y, Yamamoto T, Inoue J I. The ankyrin repeats but not the PEST-like sequences are required for signal-dependent degradation of IκBα. Oncogene. 1996;12:1159–1164. [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P A, Baltimore D. A 65-kD subunit of active NF-κB is required for inhibition of NF-κB by IκB. Genes Dev. 1989;3:1689–1698. doi: 10.1101/gad.3.11.1689. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 8.Barroga C F, Stevenson J K, Schwarz E M, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauparlant P, Lin R, Hiscott J. The role of the C-terminal domain of IκBα in protein degradation and stabilization. J Biol Chem. 1996;271:10690–10696. doi: 10.1074/jbc.271.18.10690. [DOI] [PubMed] [Google Scholar]
- 10.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 11.Beg A A, Sha W C, Bronson R T, Baltimore D. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 12.Boothby M R, Mora A L, Scherer D C, Brockman J A, Ballard D W. Perturbation of the T lymphocyte lineage in transgenic mice expressing a constitutive repressor of nuclear factor (NF)-κB. J Exp Med. 1997;185:1897–1907. doi: 10.1084/jem.185.11.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 14.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 17.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-κB and its inhibitor, IκB-α. Proc Natl Acad Sci USA. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capobianco A J, Simmons D L, Gilmore T D. Cloning and expression of a chicken c-rel cDNA: unlike p59v-rel, p68c-rel is a cytoplasmic protein in chicken embryo fibroblasts. Oncogene. 1990;5:257–265. [PubMed] [Google Scholar]
- 19.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 21.Chiao P J, Miyamoto S, Verma I M. Autoregulation of IκBα activity. Proc Natl Acad Sci USA. 1994;91:28–32. doi: 10.1073/pnas.91.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu Z L, McKinsey T A, Liu L, Qi X, Ballard D W. Basal phosphorylation of the PEST domain in IκBβ regulates its functional interaction with the c-rel proto-oncogene product. Mol Cell Biol. 1996;16:5974–5984. doi: 10.1128/mcb.16.11.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis N, Ghosh S, Simmons D L, Tempst P, Liou H C, Baltimore D, Bose H R., Jr Rel-associated pp40: an inhibitor of the rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- 24.Diehl J A, Tong W, Sun G, Hannink M. Tumor necrosis factor-α-dependent activation of a RelA homodimer in astrocytes. Increased phosphorylation of RelA and MAD-3 precedes activation of RelA. J Biol Chem. 1995;270:2703–2707. doi: 10.1074/jbc.270.6.2703. [DOI] [PubMed] [Google Scholar]
- 25.Dougherty J P, Wisniewski R, Yang S L, Rhode B W, Temin H M. New retrovirus helper cells with almost no nucleotide sequence homology to retrovirus vectors. J Virol. 1989;63:3209–3212. doi: 10.1128/jvi.63.7.3209-3212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst M K, Dunn L L, Rice N R. The PEST-like sequence of IκBα is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol Cell Biol. 1995;15:872–882. doi: 10.1128/mcb.15.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita T, Nolan G P, Liou H C, Scott M L, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 28.Ganchi P A, Sun S C, Greene W C, Ballard D W. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gélinas C, Temin H M. The v-rel oncogene encodes a cell-specific transcriptional activator of certain promoters. Oncogene. 1988;3:349–355. [PubMed] [Google Scholar]
- 30.Ghosh G, van Duyne G, Ghosh S, Sigler P B. Structure of NF-κB p50 homodimer bound to a κB site. Nature (London) 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 31.Gilmore T D, Koedood M, Piffat K A, White D W. Rel/NF-κB/IκB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 32.Gilmore T D, Morin P J. The IκB proteins: members of a multifunctional family. Trends Genet. 1993;9:427–433. doi: 10.1016/0168-9525(93)90106-r. [DOI] [PubMed] [Google Scholar]
- 33.Govind S, Drier E, Huang L H, Steward R. Regulated nuclear import of the Drosophila Rel protein Dorsal: structure-function analysis. Mol Cell Biol. 1996;16:1103–1114. doi: 10.1128/mcb.16.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haskill S, Beg A A, Tompkins S M, Morris J S, Yurochko A D, Sampson-Johannes A, Mondal K, Ralph P, Baldwin A S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 35.Hatada E N, Naumann M, Scheidereit C. Common structural constituents confer IκB activity to NF-κB p105 and IκB/MAD-3. EMBO J. 1993;12:2781–2788. doi: 10.1002/j.1460-2075.1993.tb05939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue J, Kerr L D, Rashid D, Davis N, Bose H R, Jr, Verma I M. Direct association of pp40/IκBβ with rel/NF-κB transcription factors: role of ankyrin repeats in the inhibition of DNA binding activity. Proc Natl Acad Sci USA. 1992;89:4333–4337. doi: 10.1073/pnas.89.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaffray E, Wood K M, Hay R T. Domain organization of IκBα and sites of interaction with NF-κB p65. Mol Cell Biol. 1995;15:2166–2172. doi: 10.1128/mcb.15.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 39.Kerr L D, Inoue J, Davis N, Link E, Baeuerle P A, Bose H R, Jr, Verma I M. The Rel-associated pp40 protein prevents DNA binding of Rel and NF-κB: relationship with IκBβ and regulation by phosphorylation. Genes Dev. 1991;5:1464–1476. doi: 10.1101/gad.5.8.1464. [DOI] [PubMed] [Google Scholar]
- 40.Kerr L D, Ransone L J, Wamsley P, Schmitt M J, Boyer T G, Zhou Q, Berk A J, Verma I M. Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-κB. Nature (London) 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- 41.Klement J F, Rice N R, Car B D, Abbondanzo S J, Powers G D, Bhatt P H, Chen C H, Rosen C A, Stewart C L. IκBα deficiency results in a sustained NF-κB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 43.Krappmann D, Wulczyn F G, Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 44.Kretzschmar M, Meisterernst M, Scheidereit C, Li G, Roeder R G. Transcriptional regulation of the HIV-1 promoter by NF-κB in vitro. Genes Dev. 1992;6:761–774. doi: 10.1101/gad.6.5.761. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Gélinas C. IκBα-mediated inhibition of v-Rel DNA binding requires direct interaction with the RXXRXRXXC Rel/κB DNA-binding motif. Proc Natl Acad Sci USA. 1993;90:8962–8966. doi: 10.1073/pnas.90.19.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeBail O, Schmidt-Ullrich R, Israël A. Promoter analysis of the gene encoding the IκB-α/MAD3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehming N, McGuire S, Brickman J M, Ptashne M. Interactions of a Rel protein with its inhibitor. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luque I, Gélinas C. Rel/NF-κB and IκB factors in oncogenesis. Semin Cancer Biol. 1997;8:103–111. doi: 10.1006/scbi.1997.0061. [DOI] [PubMed] [Google Scholar]
- 50.Maggirwar S B, Harhaj E, Sun S C. Activation of NF-κB/Rel by Tax involves degradation of IκBα and is blocked by a proteasome inhibitor. Oncogene. 1995;11:993–998. [PubMed] [Google Scholar]
- 51.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyamoto S, Verma I M. Rel/NF-κB/IκB story. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 53.Muller C W, Rey F A, Sodeoka M, Verdine G L, Harrison S C. Structure of the NF-κB p50 homodimer bound to DNA. Nature (London) 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 54.Nakayama K, Shimizu H, Mitomo K, Watanabe T, Okamoto S, Yamamoto K. A lymphoid cell-specific nuclear factor containing c-Rel-like proteins preferentially interacts with interleukin-6 κB-related motifs whose activities are repressed in lymphoid cells. Mol Cell Biol. 1992;12:1736–1746. doi: 10.1128/mcb.12.4.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osborn L, Kunkel S, Nabel G J. Tumor necrosis factor α and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor κB. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Read M A, Neish A S, Gerritsen M E, Collins T. Postinduction transcriptional repression of E-selectin and vascular cell adhesion molecule-1. J Immunol. 1996;157:3472–3479. [PubMed] [Google Scholar]
- 57.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Rice, N. Personal communication.
- 58.Rodriguez M S, Wright J, Thompson J, Thomas D, Baleux F, Virelizier J L, Hay R T, Arenzana-Seisdedos F. Identification of lysine residues required for signal-induced ubiquitination and degradation of IκB-α in vivo. Oncogene. 1996;12:2425–2435. [PubMed] [Google Scholar]
- 59.Roff M, Thompson J, Rodriguez M S, Jacque J M, Baleux F, Arenzana-Seisdedos F, Hay R T. Role of IκBα ubiquitination in signal-induced activation of NF-κB in vivo. J Biol Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- 60.Rottjakob E M, Sachdev S, Leanna C A, McKinsey T A, Hannink M. PEST-dependent cytoplasmic retention of v-Rel by IκB-α: evidence that IκB-α regulates cellular localization of c-Rel and v-Rel by distinct mechanisms. J Virol. 1996;70:3176–3188. doi: 10.1128/jvi.70.5.3176-3188.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sachdev S, Rottjakob E M, Diehl J A, Hannink M. IκB-α-mediated inhibition of nuclear transport and DNA-binding by Rel proteins are separable functions: phosphorylation of C-terminal serine residues of IκB-α is specifically required for inhibition of DNA-binding. Oncogene. 1995;11:811–823. [PubMed] [Google Scholar]
- 62.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarz E M, Van Antwerp D, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott M L, Fujita T, Liou H C, Nolan G P, Baltimore D. The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 65.Sun S, Elwood J, Greene W C. Both amino- and carboxyl-terminal sequences within IκBα regulate its inducible degradation. Mol Cell Biol. 1996;16:1058–1065. doi: 10.1128/mcb.16.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun S C, Ganchi P A, Ballard D W, Greene W C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 67.Suyang H, Phillips R, Douglas I, Ghosh S. Role of unphosphorylated, newly synthesized IκBβ in persistent activation of NF-κB. Mol Cell Biol. 1996;16:5444–5449. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 69.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tran K, Merika M, Thanos D. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Antwerp D J, Verma I M. Signal-induced degradation of IκBα: association with NF-κB and the PEST sequence in IκBα are not required. Mol Cell Biol. 1996;16:6037–6045. doi: 10.1128/mcb.16.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 73.Whiteside S T, Ernst M K, LeBail O, Laurent-Winter C, Rice N, Israël A. N- and C-terminal sequences control degradation of MAD3/IκBα in response to inducers of NF-κB activity. Mol Cell Biol. 1995;15:5339–5345. doi: 10.1128/mcb.15.10.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X, Gélinas C. A mutant Rel-homology domain promotes transcription by p50/NFκB1. Oncogene. 1997;14:1521–1530. doi: 10.1038/sj.onc.1200985. [DOI] [PubMed] [Google Scholar]
- 75.Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gélinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733–6741. doi: 10.1128/mcb.13.11.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zabel U, Baeuerle P A. Purified human IκB can rapidly dissociate the complex of the NF-κB transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 77.Zabel U, Henkel T, Silva M S, Baeuerle P A. Nuclear uptake control of NF-κB by MAD-3, an IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]