Abstract
The “QuantitatEVs: multiscale analyses, from bulk to single vesicle” workshop aimed to discuss quantitative strategies and harmonized wet and computational approaches toward the comprehensive analysis of extracellular vesicles (EVs) from bulk to single vesicle analyses with a special focus on emerging technologies. The workshop covered the key issues in the quantitative analysis of different EV‐associated molecular components and EV biophysical features, which are considered the core of EV‐associated biomarker discovery and validation for their clinical translation. The in‐person‐only workshop was held in Trento, Italy, from January 31st to February 2nd, 2023, and continued in Milan on February 3rd with “Next Generation EVs,” a satellite event dedicated to early career researchers (ECR). This report summarizes the main topics and outcomes of the workshop.
Keywords: multiomics analysis, multiscale analysis, quantitative biology, single vesicle
1. THE QUANTITATEVS ISEV WORKSHOP OVERVIEW
The “QuantitatEVs: multiscale analyses, from bulk to single vesicle” workshop was dedicated to the challenges in analyzing EVs when studied in bulk or as single entities. The event was organized by the International Society for Extracellular Vesicles (ISEV) with the support of two nonprofit organizations, the Pezcoller Foundation (https://www.pezcoller.it/en/about‐us/) and the Italian Society for Extracellular Vesicles (EVIta) (https://www.evitasociety.org). Additionally, the workshop received financial support from private companies, namely Abbelight, Alfatest, Bio‐Techne, NanoFCM, Quanterix, Schaefer, DBA Italia, and Diatech Lab Line. Discussions were encouraged at any time during the workshop. Roundtables, chaired by experts and ISEV board members, were the major focus of the event, as expected in all ISEV workshops. This report briefly summarizes the goals of the workshop and highlights the viewpoints and the future perspectives that emerged during the roundtable discussions that engaged early career researchers (ECR) and senior participants from academia, healthcare organizations, and industry. The overarching aim of the “QuantitatEVs” workshop was to discuss existing methodologies, identify major analytical gaps, and propose novel strategies to analyze omics data in the EV field with the final aim to identify and characterize specific proteins or genomic material at the single EV level. The subject of discussion ranged from the pros and cons of common EV purification approaches to the existing computational pipelines of omics data analyses, the newest technologies for single EV studies, and how to combine bulk EV data analysis with single EV analysis to nominate new disease‐associated biomarkers. While protocols in the analysis of EVs from biofluids have progressed tremendously, largely thanks to the dedicated EV task forces within ISEV, there is a lack of consensus on the best pipelines for the computational analysis to apply to the EV data (Witwer et al., 2021). At the same time, the analysis of single EVs appeals to many researchers from academia and industry. Given the challenges posed by the size resolution, continuous improvement and technological advancement are needed, and recognition of technical limits is required to differentiate between reliable results and artefacts (Bordanaba‐Florit et al., 2021; Nolan & Duggan, 2018). Based on this premise, the QuantitatEVs International Organizing Committee (IOC) identified five specific topics to be discussed during the sessions and the relevant roundtables to foster fruitful discussions and to reach a consensus on how the community intends to move forward.
1.1. Workshop participants
The ISEV Workshop “QuantitatEVs: multiscale analyses, from bulk to single vesicle” was attended by applicants selected by the IOC based on their interest statement and/or their submitted abstract. The workshop was attended by 59 selected delegates, 11 sponsor representatives from industries, and 8 members of the IOC (Figure 1a). The ISEV chapters were represented as follows, Europe–Africa with 78%, America with 16.9%, and Asia with 5.1% (Figure 1b). Participants travelled from more than twenty countries around the globe (Figure 1c). The workshop's participants were well balanced between junior and senior investigators (52.5% and 47.5%, respectively) (Figure 2a), working either in academia (67.8%), healthcare institutions (18.6%), or industry (13.6%) (Figure 2b). When asked to express their preference for one of the topics, the attendees distributed their choices equally (Figure 2c). Among the attendees, there were eight invited speakers, while 17 and 18 were selected among the abstract applicants for the Trento (main workshop) and the Milan (ECR workshop) sessions, respectively. The workshop in Milan was open to ECRs registered for the main workshop and to additional investigators from the local area.
FIGURE 1.
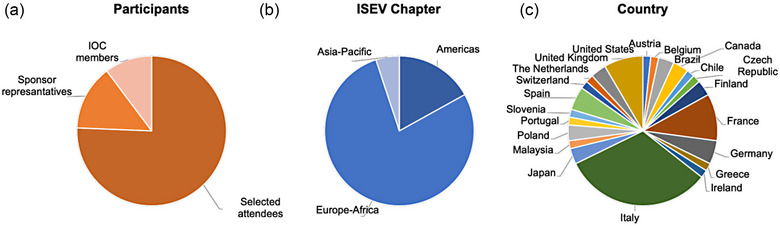
ISEV QuantitatEVs workshop demographics. Graphs of participant subdivision according to (a) selected attendees (75.6%), sponsor representatives (14.1%), and IOC members (10.3%); (b) the ISEV geographical chapters’ representation (Europe‐Africa‐78%; Americas‐16.9%; Asia‐Pacific‐5.1%), and (c) the Country of origin (Italy‐32%; France‐10.2%; United States of America‐8.5%; Germany‐5.1%; Spain‐5.1%; Brasil‐3.4%; Canada‐3.4%; Poland‐3.4%; United Kingdom‐3.4%; Finland‐3.4%; Japan‐3.4%; Austria‐1.7%; Belgium‐1.7%; Chile‐1.7%; Czech Republic‐1.7%; Ireland‐1.7%; Portugal‐1.7%; Slovenia‐1.7%; Greece‐1.7%; Malaysia‐1.7%; Switzerland‐1.7%; The Netherlands‐1.7%).
FIGURE 2.
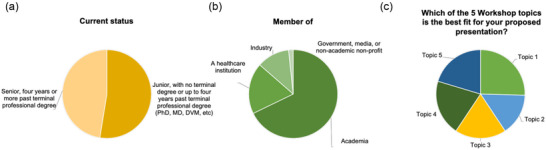
A balanced international workshop. Graphs of participant subdivision according to (a) stage of career classified as junior (Medical doctor Degree ‐MD, Philosophy doctor degree, PhD, Doctor of Veterinary Medicine or equivalent degree not yet obtained or obtained within the previous 4 years‐52.5%) and senior (more than 4 years from MD, Ph.D. or equivalent degree‐47.5%); (b) affiliation to academics (67.8%), industries (11.9%), healthcare institutions (18.6%), or government, media, or nonacademic nonprofit (1.7%); (c) Intention to present in one of the five proposed topics during the workshop (Topic 1 ‘Preanalytics’−25.4%; Topic 2 ‘Use of existing computational tools for ‐omics EV data analysis’−15.3%; Topic 3 ‘New computational tools for ‐omics EV data analysis’−18.6%; Topic 4 ‘Single EV analysis’−20.3%; Topic 5 ‘From bulk to single analysis for biomarkers identification and validation’−20.3%).
1.2. Workshop organization
The workshop was organized in five thematic sessions with related roundtables. Each session was introduced by one invited speaker per topic. The sessions focused on:
The impact of preanalytical steps on downstream data generation.
The quantitative approaches that can be adopted from the cell data analysis field into the EV field in terms of computational biology and bioinformatics.
The EV omics data analyses that currently require the development of novel EV‐tailored approaches/pipelines.
The state‐of‐the‐art technologies to perform single EV characterization and relatively challenges.
The contribution of bulk versus single EV analysis for biomarker validation.
Each session lasted on average 1 h and a half and they all ran in the mornings. Two parallel round tables were planned in the afternoon of day 1 and day 2 to discuss the topics presented in Sessions 1–2 and 3–4, respectively. Parallel round tables were followed by a joint discussion of about 1 h—attended by all participants. The round tables lasted about 2 h. Each participant attended the round tables chosen at the time of registration. Session 5 round table—on the morning of day 3 ‐ was attended by all participants.
A few weeks before the workshop, the participants were asked to answer 24 questions in a pre‐workshop survey (78% participation). The questions covered all five sessions, and the results were used as a starting point for discussion during the roundtables. To track the post‐workshop attendees' mindsets, the same scientific survey was proposed after the workshop (37% participation). Aggregated comparative assessment of the responses highlighted partial agreement among the attendees on a few highly debated points, evidencing some resolution and the definition of new goals to achieve. Of relevance, 77% of the responders declared that the workshop had a great impact on their knowledge base; for 63% of the responders, the discussions were perceived as helpful in improving their performance, and for almost 70%, what learned at the workshop was considered relevant to improve their research outcomes.
2. QUANTITATEVS WORKSHOP POINTS OF DISCUSSION AND RELATED RAISED ISSUES
Several steps in the EV purification and processing steps were proven to affect the downstream analysis, particularly for biofluids (Coumans et al., 2017; Erdbrugger et al., 2021; Geeurickx & Hendrix, 2020; Holcar et al., 2021). As part of ISEV, EV task forces are dedicated to improving EV research reproducibility by enhancing rigour and standardization and thus helping the EV community attain robustness, reliability, credibility, and sustainability (https://www.isev.org/taskforces). Guidelines on standardized reporting were established by the EV task forces on specific topics of investigation, and MISEV 2018 guidelines (Théry et al., 2018) are in place. The QuantitatEVs workshop was intended to bring together experts with different backgrounds to jointly consider all biological and methodological variables and diverse analysis perspectives for their potential impact on the downstream results. This section summarizes the main topics that were discussed, the questions raised, and the possible solutions proposed during the workshop.
2.1. Session 1. Impact of pre‐analytical steps on downstream data generation. Importance of knowing the boundaries and the constraints
During the first session, the impact of preanalytical steps on downstream data generation was discussed, along with the importance of knowing the boundaries and the constraints. Dr. Guido Jenster (Erasmus MC, Netherlands) provided examples of how the EV community recently developed biofluid collection standard operating procedures (SOPs) to minimize the effect on the downstream EV characterization (https://brd.nci.nih.gov/brd/sop‐compendium/show/1583; Van Royen et al., 2023). He also mentioned the need to standardize as many variables as possible, to test the effect of the variables on the EV characterization, to follow the MISEV 2018 and the upcoming MISEV 2023 guidelines, and to report detailed protocols while also making them open access through repositories, such as Vesiclepedia, EVpedia, ExoCarta, exRNA, miRandola, Zenodo, and the Biospecimen Research Database (https://brd.nci.nih.gov/brd/). His recommendation to deal with known and unknown variables was to (i) make an inventory and order them on their expected impact, (ii) test the effects of most relevant variables and publish the results, (iii) standardize the variables using SOPs and make them open access, (iv) record deviations from the SOPs, (v) reinterpret the data, learn continuously and adapt the protocols if needed, (vi) accept that it is not possible to control all known variables—and many new unknown ones are encountered ‐, and (vii) enjoy this challenging journey into the unknown. The opening lecture was followed by the talks of Mari Palviainen (University of Helsinki, Finland) and Ioannis Sotiropoulos (National Centre for Scientific Research [NCSR] Demokritos, Greece). The talk by Dr. Palviainen underscored the need for collaboration and harmonization of preanalytical workflows in EV research to improve the potential of interstudy comparative assessments. Dr. Sotiropoulos provided examples of how protocols need to be adapted to the specific intended downstream analysis and further presented a novel isolation method for spontaneously released brain exosomes (Gomes et al., 2023). Dr. Holcar (University of Ljubljana, Slovenia) presented on the topic of plasma‐EV in healthy adults, discussing the need to monitor the quality of plasma preparations and EV enrichment steps to account for potential EV‐ and non‐EV contaminants. She also delved into the investigation of various biospecimen variables, including demographics, lifestyle, and clinical characteristics of study subjects, which may need to be carefully matched between patient and healthy control groups in EV biomarker studies. Importantly, while specific study subjects’ characteristics, such as smoking, were found to exhibit correlations with differences in EV subtypes, none of them had a significant impact on the overall composition of plasma EVs. Moreover, the analyzed study subjects’ features did not account for the entirety of the observed variability within the analyzed plasma‐EV subsets in healthy adults.
The pre‐analytical steps upstream of proper bio‐banking procedures were discussed during the roundtable. The participants and panelists agreed that any EV isolation step likely introduces biases and variations in downstream analyses; thereby methods allowing analysis without pre‐isolation steps are highly desirable. The participants contributed to the discussion by bringing their own experiences and agreed on the need to be (i) knowledgeable/aware of the SOPs before starting any EV bulk analysis, (ii) aware of the variables that need to be considered for any scientific question to address, and (iii) open to revisiting the data in case new tools or technologies become available. It was highlighted that all these aspects are especially relevant when the downstream analysis includes proteomics, transcriptomics, genomics, or lipidomics, which can detect up to picograms of molecules and are particularly affected by small differences among the analyzed samples.
2.2. Session 2. EV omics data analysis: Which quantitative approaches can we adopt from the canonical cell data analysis field into the EV field in terms of computational biology and bioinformatics?
The second session was dedicated to EV‐omics data analysis, focusing on which quantitative approaches can be adopted from the canonical cell data analysis field into the EV field regarding computational biology and bioinformatics. Dr. Levi Waldron (CUNY Graduate School of Public Health and Health Policy) opened the session by describing challenges and solutions explored in other fields (e.g., metagenomics) and suggested that the EV community would likely benefit from curated repositories with harmonized, labeled, and curated data. The selected talk by Sylwia Bobis–Wozowicz (Jagiellonian University, Poland), Vito D'Agostino (University of Trento, Italy), and Elena Casarotto (University of Milan, Italy) provided examples of EV bulk analyses that were instrumental in defining novel pathological pathways in different disease conditions and in determining which EV‐enriched bioactive molecules may play a role in regeneration. Dr. Bobis–Wozowicz demonstrated the utility of the multiomics approach to delineate the molecular cargo of stem cell‐derived EVs, which is responsible for their pro‐regenerative capacity in the context of heart regeneration. Comprehensive data from proteomics, transcriptomics, and miRN‐ome analyses revealed a distinct molecular footprint of EVs derived from different oxygen concentrations, which shaped their biological activity. Based on this approach, she identified the most abundant EV‐associated bioactive molecules, which allowed her to establish a network of signaling pathways regulating cell survival. Dr. D'Agostino showed that a DNA base‐excision repair protein, that is, APE1, modulates the secretion of EV‐RNA (Mangiapane et al., 2021), providing evidence that a functional base‐excision repair protein is delivered through exosomes in response to genotoxic stresses. Dr. Elena Casarotto used microRNA analysis to investigate whether and how the protein quality control system (PQC) can affect not only the secretion of proteins into large and small EVs (Casarotto et al., 2022), but also the EV microRNA content, especially in the context of neurodegenerative diseases, where the PQC is usually impaired.
During the discussion, it emerged that the lack of annotated, updated, harmonized databases dedicated to the EV field should be overcome to interpret better the results of bulk analysis on different material sources. The roundtable offered the opportunity to discuss the need to share relevant information regarding the computational analysis and to deposit the data to allow for cross‐study comparison.
2.3. Session 3. EV omics data analysis: Which topics require the development of novel EV‐tailored approaches?
The third session questioned the need to develop novel EV‐tailored analytical and computational approaches instead of inheriting those from cell‐based analysis. Dr. Aleksandar Milosaviljevic (Baylor College of Medicine) shared his experience from the exRNA Consortium and questioned whether it is advisable to focus only on EVs or to broaden the scope of studies to include other particles, like lipoproteins and ribonucleoproteins, as these Non‐Vesicular Extracellular Particles (NVEPs) share protein and exRNA cargo and may not always be separated experimentally. One example is provided by the extracellular RNA‐Binding Proteins (exRBPs) and associated exRNA, which may be found in both EVs and NVEPs across human biofluids, as evidenced by the recent intersection analysis of eCLIP and exRNA Atlas data (LaPlante et al., 2023). Yari Ciani (University of Trento, Italy), Shiro Suetsugu (Nara Institute of Science and Technology, Japan), and Alice Gualerzi (IRCCS Fondazione Don Carlo Gnocchi, Italy) described the application of three different omics approaches (transcriptomics, proteomics, and lipidomics) to address their biological questions with biomolecular, spectrometric and spectroscopic methods, respectively. Dr. Ciani highlighted the potential relevance of plasma‐derived EV‐RNAs as disease progression markers in a cohort of highly annotated castration‐resistant prostate cancer patients’ liquid biopsies. Specifically, he showed that the deconvolution of the transcriptomic signal revealed differential contributions from multiple prostate and immune cell types when comparing groups of patients stratified based on differential responses to androgen receptor signaling inhibitor treatment. Furthermore, he showed quantitative data supporting the differential integrity of RNA molecules in EV based on the RNA biotype and the putative tissue of origin. Next, Dr. Suetsugu talked about EVs that can be derived from cellular protrusions, the majority of which are dependent on the membrane‐sculpting I‐BAR domain proteins (D'Angelo et al., 2023). He compared proteomes of EVs from various cells with altered filopodia formations by the modulation of the I‐BAR domain proteins (Hu et al., 2020; Nishimura et al., 2021) by using the isotope labeling of the various EV samples. The EVs were found to deliver cellular signaling proteins, including small GTPases (Nishimura et al., 2021). Dr. Gualerzi presented the results from the application of Raman spectroscopy to the study of microglia‐derived EVs (MEV). Raman spectroscopy is a sensitive optical technique that can provide information on the chemical content of EVs using limited volumes of EV preparations, highlighting the main molecular constituents of an EV population (Gualerzi et al., 2019, 2017). The spectra of MEV from multiple activated microglia states showed major variations in the spectral intervals that can be attributed to the lipid components (Lombardi et al., 2019). The spectra from single lipid components, including endocannabinoids, were compared to the MEV spectra, thereby providing the relative quantitation of lipid components, both on intact and broken EVs. The Raman investigation provided hints for the subsequent standard lipidomic characterization approach, highlighting the role of specific lipid subfamilies in intercellular communication within the brain and their involvement in MEV‐mediated responses to inflammation and injury.
During the roundtable, the discussion focused on how omics analysis may support EV research toward clinical application and biomarker discovery. A consensus was reached on the need to create reference sets and baseline data, together with metadata for each specific omic, to accurately detect relevant individuals’ perturbations. The discussion ended on the need for single EV profiling and the related methodological challenges. It was highlighted that to properly empower single EV analysis, EV subpopulations need to be fully characterized in terms of associated biological markers. In turn, it would be crucial to define the biological relevance of each subpopulation and the possibility of properly isolating and examining it in a reproducible manner.
2.4. Session 4. Single EV characterization
The workshop next focused on the single EV characterization for the fourth session. The introductory talk by Dr. Juan M. Falcon–Perez (CIC bioGUNE‐BRTA, Bilbao, Spain) highlighted the need for understanding the content of a single vesicle to unravel the different subpopulations of EV secreted by the cells. The current technologies for single vesicle characterization were presented with special mention for the Raman Tweezers Microspectroscopy, able to inform of the global composition of single vesicles in terms of the proportion of proteins, nucleic acids, and lipids. Afterward, Dr. Marco Monopoli (RCSI, Dublin, Ireland), an expert of nanoparticles and their biomolecular corona, described the factors that affect the corona composition, such as the effect of specific dyes on the composition of the corona and the glycans and the differences between hard and soft corona. The selected talks by Lorena Martin Jaular (Institut Curie‐Inserm U932, France), John Nolan (Scintillon Institute), and Daniela Boselli (San Raffaele Scientific Institute, Milano, Italy) reported the challenges and the need for careful controls in the analysis of single vesicles. Dr. Martin Jaular presented the optimization steps for single EV detection by imaging flow cytometry. For the detection, EVs were either labeled using a lipidic probe (MG‐488), carboxyfluorescein succinimidyl ester (CFSE), or obtained from cells expressing MyrPalm‐sfGFP and then stained with fluorescent antibodies to target the tetraspanins CD9, CD63, and CD81. As negative controls, EV from Hela cells knocked out of the different tetraspanins were used. The work identified several critical technical points to obtain reliable EV detection by imaging flow cytometry, including a possible interaction of the lipidic probe with antibodies. Dr. Nolan described the calibration of flow cytometers and validation of assays to produce single EV measurements (and their limits of detection, LODs) reported in quantitative terms that can be compared between laboratories. Dr. Boselli underlined the importance of controls in the analysis of EVs with flow cytometry and showed how it is possible to carry out a multiparametric analysis of EVs thanks to the use of spectra flow cytometry. The spectral approach minimized spreading error and spectral overlay, thus allowing the simultaneous identification of several EV subpopulations, such as the most common EV subsets (erythrocyte‐derived EV, platelet‐derived EV, leukocyte‐derived EV, and endothelial‐derived EV) and novel EV subsets, characterized by activation and exhaustion markers. Overall, the data demonstrated the high heterogeneity of the circulating plasma‐derived EVs, thus opening the door for the single EV analysis as well as for their use as potential diagnostic/prognostic biomarkers.
The open discussion during the roundtable started by the attendees’ enumeration of the techniques considered the most useful and/or widespread for single EV analysis, such as microscopy (e.g., high sensitivity, spectral, and super‐resolution, and atomic force microscopy), flow cytometry (e.g., imaging flow cytometry) and single EV microarrays. Several questions were next raised on possible technique‐specific biases introduced by pre‐enrichment steps and whether a single EV should be analyzed after labeling or label‐free. Attendants agreed on a possible source of unintended error by using antibodies directed against tetraspanins as a proxy of the general labeling/capturing method due to the extremely high heterogeneity of their expression on the EV surface. The consensus was that a bias is always introduced either in the selection of EV populations by the labeling or by the technology in the label‐free and that the use of positive and negative controls is of utmost importance in any label‐based experiment.
2.5. Session 5. What is the contribution of bulk versus single EV analysis for biomarker validation?
During the last session, the topic of discussion was the contribution of bulk versus single EV analysis for biomarker validation. The invited speaker, Dr. Dolores Di Vizio (Cedars‐Sinai, USA), presented her work as an example of how, starting from the analysis of large EV (large oncosomes) purified from a disease model, it is possible to identify a disease biomarker to validate it in biofluids and to characterize its abundance at the single EV level. She underlined how this process presents numerous technical challenges and requires cross‐validation with different approaches, thereby highlighting the need for well‐trained computational biologists who can either apply the pipelines for bulk EV analysis to single EV or develop new ones. The selected talks by Anna Kashkanova (Max Planck Institute for the Science of Light, Germany), Pietro Parisse (IOM‐CNR; Italy), and Lien Lippens (Ghent University, Belgium) showed new technological approaches that could improve the EV quantification in a label‐free context and the understanding of EV fusogenic properties at the single EV level. In brief, the selected talk by Dr. Kashkanova introduced interferometric nanoparticle tracking analysis (iNTA) (Kashkanova et al., 2022) with significantly higher detection sensitivity for smaller particles, compared to traditional NTA, as well as the ability to report on the scattering cross‐sections and refractive index of nanoparticles. It was shown that iNTA could differentiate EVs from large lipoproteins based on their size and refractive index (Kashkanova et al., 2023). Dr. Parisse focused on the fusogenicity properties of EVs through Atomic Force Microscopy imaging of EV in interaction with supported lipid membranes of different compositions. The technique uniquely allows following the kinetics of the docking, fusion, and diffusion of EV down to a single vesicle level, shedding new light on the role of different membrane phases and different EV origins on the vesicle internalization routes (Paba et al., 2023; Perissinotto et al., 2021). Dr. Lippens demonstrated the potential of the asymmetric flow field‐flow fractionation multiangle light scattering (AF4‐MALS) technique for the separation and characterization of EVs.
During the final discussion, the participants agreed on the need to perform bulk analysis to shortlist potential new biomarkers; nevertheless, integrating bulk and single EVs would be ideal for accelerating biomarker discovery. Finally, the take‐home message was that while performing a single EV analysis, it is essential to use, share, and produce reference material that can be used across laboratories and platforms and to support interlaboratory efforts in this direction.
2.6. Next generation EVs satellite event—ECR symposium
After the main workshop, a satellite event was held in Milano to forecast the future of the EV field through the eyes of the new generation of scientists. On this dedicated day, they had additional opportunities to present their work either via regular or flash talks. Dr. Martin Blessing, from the Max Planck Institute for the Science of Light, Germany, received the best ECR symposium talk prize consisting of a complimentary registration to ISEV2023 in Seattle. The symposium was opened by two keynote lectures, one from academia and one from industry. The first lecture was given by Dr. Paolo Bergese (University of Brescia, Italy) on neglected and unnoticed problems associated with EV characterization, spanning molecular and nanoscale stoichiometry, times, and energies. The seven “out‐of‐the‐box” EV problems that were touched on can be summarized as follows: (1) Finding the link between molecular profiles and mesoscale signatures of EV, which is related to (2) the need to develop multiscale algorithms and models that provide structure and dynamics of EV in complex environments, including those within organisms. After being secreted by cells, most EV moves inside fluids, (3) which may contain other biotic and abiotic nanoparticles (e.g., lipoproteins and nanoplastics). These fluids are nanostructured, crowded, and sticky (Busatto et al., 2020, 2022; Zendrini et al., 2023) prompting questions on (4) the meaning and physical achievability of purity (Zendrini et al., 2020) in EV formulations, as well as issues related to (5) handling, (6) the role in EV surface engineering (Musicò et al., 2023) and (7) the potential for diagnostics (Radeghieri et al., 2022; Radeghieri & Bergese, 2023) of the EV biomolecular corona. The talk demonstrated how physical chemistry and nanotechnology are important aids to advance intuition and insight in EV research, also sharpening the interplay between theory and experiment.
The second presentation by Dr. Davide Zocco (Lonza), provided the industry perspective on EV trends and future opportunities. Briefly, the EV therapy market is an emerging field, with rapid growth and a Compounded Annual Growth Rate of 62% and a total of 59 allogeneic EV products in the pipeline as of 2022. Of these products, 86% are at a preclinical stage, 7% are in Phase I, 5% in Phase II, and 2% in Phase III. No commercial EV therapies are currently available. Dr. Zocco also presented the situation in terms of EV production by cell lines. A total of 40 EV Total Experience (Tx) companies have been identified, 16 of which are exploiting mesenchymal stem cells (MSCs), eight are using HEK 293, and the rest a plethora of different cell types including iPSCs and primary fibroblasts. In parallel with the growth of EV market, an increasing number of scientific discoveries and intellectual property (IP) around EV have been published in recent years, with seminal publications on EV cargo loading (Ferreira et al., 2022), biodistribution in nonhuman primates (Driedonks et al., 2022) and cancer tissue tropism (Garofalo et al., 2021). However, while EV science is rapidly advancing, the industrialization of EV therapies is still in its infancy, and there is an urgent need for innovative technologies that would bring down the cost of manufacturing while driving up quality at scale. Next, the event was complemented by a story of success from the start‐up world with EVerZom, a spin‐off company developed in 2019 with the academic patent on massive EV production in bioreactor stimulated by turbulence (Patents WO 2019/002608 & WO 2020/136362). Dr. Thibaut Fourniols presented the story from the lab discovery to the company establishment and highlighted the need for initial key elements such as a useful innovation for the community protected by intellectual property, and a team of dedicated founders supported by academic representatives, specific infrastructure for start‐up companies and seed funding. Some details about the EV downstream process, characterization and quality control by potency assays have also been presented. After that, Dr. Marcella Chiari (SCITEC‐CNR) highlited that within the European Innovation Council (EIC) concerted efforts are driving multidisciplinary advancements in EV research. In her lecture on the innovation pathway in EV, she exemplified the need for multidisciplinarity through the integration of diverse scientific expertise: biologists, engineers, materials scientists, and entrepreneurs to develop new detection methodologies (Daaboul et al., 2016) to advance the Technology Readiness Level (TRL) until the market is reached.
The symposium was open to the local community and was highly participated with more than 90 attendees.
3. THE POST‐WORKSHOP SURVEY
The participants were asked to express their opinion on the effect of the sample preparation (from the preprocessing to the EV purification) on the downstream ‐omics analysis. By the end of the workshop, most attendees agreed that the EV purification steps affect the downstream results and that the choice of the technique depends on the specific scientific question to be addressed. Similarly, the participants discussed the possibility of exploiting computational tools already used for the omics analysis of cell components. In this respect, the consensus was that there is a need to precisely report the analytical steps used during the computational analysis to allow harmonization across all the generated data. Taking into account that bulk analysis provides a list of candidates that need to be validated by single vesicle analysis, a part of the workshop was dedicated to the approaches now in use for the study of single EVs. The importance of using standards and reference materials (Geeurickx & Hendrix, 2020; Welsh et al., 2023) during the single EV analysis, the need for a step for EV enrichment (Bordanaba‐Florit et al., 2021), and the role of EV corona (Tóth et al., 2021) were proposed as alternatives in the surveys. Interestingly, in the post‐survey, most of the attendees conveyed the need for reference material and the possibility of analyzing the “sample” as such without a purification step to avoid downstream bias in the analysis. Finally, the QuantitatEVs IOC asked if the participants envisioned a single EV analysis methodology for translation into the clinics and, eventually, which technology. In the pre‐survey, most of the responders were positive about this avenue. They considered flow cytometry and high‐resolution microscopy the prominent technologies to be brought to the clinics. In the postmeeting survey, a consensus was reached on the need to improve the existing technologies further. The questions that resulted in a change in the post‐workshop survey attendees’ opinions are summarized in Figure 3. Finally, numerous participants recognized the need to cross‐fertilize the EV field with the computational area to design analysis pipelines and SOPs for the bulk data generated in the EV field. During the discussion, it emerged that, despite the presence of various ATLASes dedicated to the EV field (EV‐TRACK Consortium et al., 2017; Griffiths‐Jones, 2006; Kalra et al., 2012; Keerthikumar et al., 2016; Kim et al., 2015; S. Li et al., 2018; J. R. Li et al., 2019; Liu et al., 2019; Lötvall et al., 2012; Murillo et al., 2019; Pisitkun et al., 2004; Russo et al., 2018; Wang et al., 2021; Hulstaert et al., 2020; also summarized and discussed in Mugoni et al., 2022. Additional ATLASes are presented in Huang et al., 2023; Lai et al., 2022; Xie et al., 2021; Bockorny et al., 2023; Cheung et al., 2016; Subramanian et al., 2015; Xu et al., 2020; Russo et al., 2012; Chandy et al., 2020) the lack of detailed annotation, updating, and standardization in such databases should be overcome to better interpret the results of bulk analyses on different material sources. This can be facilitated by detailed metadata standardization and annotation. One of the long‐term goals of QuantitatEVs is to harmonize the data and keep the existing EV ATLASes constantly updated (Figure 4).
FIGURE 3.
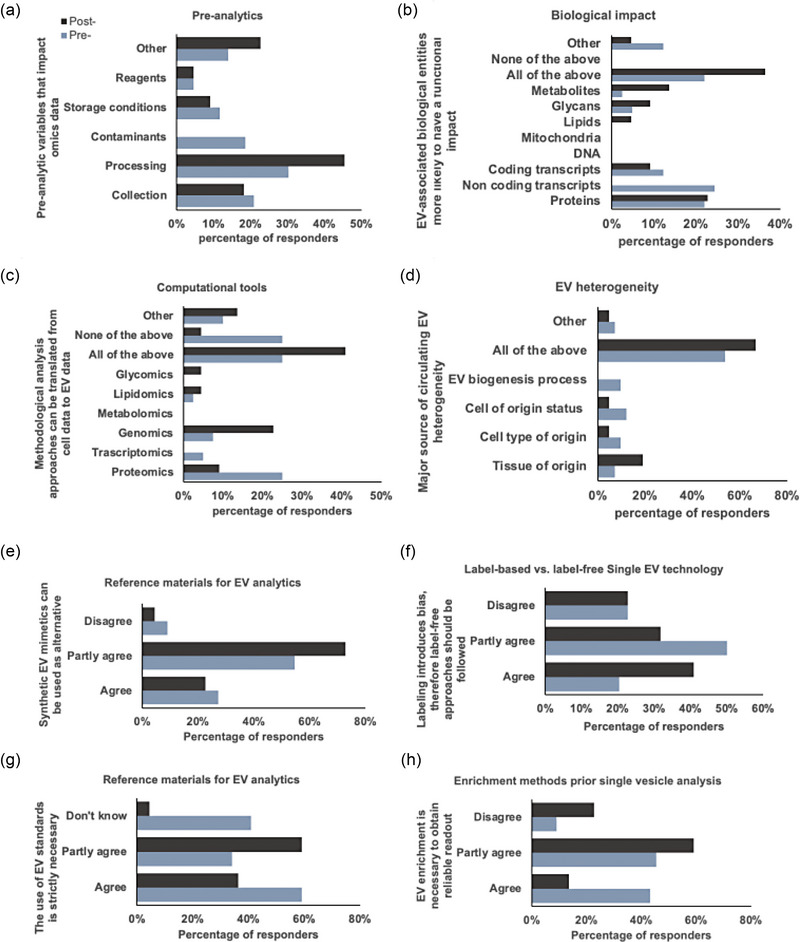
Results of the pre‐and postworkshop survey. Only questions in which the consensus changed after the workshop were reported. The participants expressed their opinion on (a) which preanalytic variables affect most the omics data; (b) which component of EVs has a functional impact on receiving cells; (c) what computational tools can be shared between cell data and EV data; (d) major source of EV heterogeneity; (e) need to use synthetic reference material for EV analytics; (f) impact of labelling on EV analysis; (g) The need to use EV standards to compare the analysis; (h) need to purify EV before analyzing them at the single EV level.
FIGURE 4.
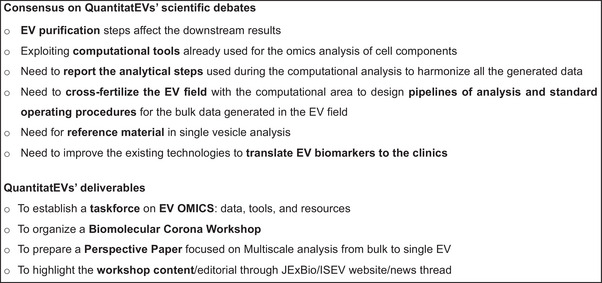
Summary of consensus reached after the round tables (top), and the workshop deliverables (bottom).
4. WORKSHOP'S MAJOR OUTCOMES
The workshop ended with a summary of the main outcomes, such as the intention to initiate an ISEV task force on EV OMICS to organize data, tools, and resources and the proposal of a future ISEV workshop focused on the EV biomolecular corona. Last, it was decided to write this short report to share a snapshot of the workshop and the postworkshop activities with the community and to provide a summary of the main outcomes (Figure 4). The current report was prepared by the QuantitatEVs IOC, the local organizing committee, with the contribution of the invited speakers. A comprehensive perspective paper on the topics addressed during the “QuantitatEVs” is to be considered.
AUTHOR CONTRIBUTIONS
Manuela Basso: Conceptualization; data curation and analysis; writing—original draft; writing—review and editing. Alessandro Gori: Writing—review and editing. Caterina Nardella: Conceptualization; data curation; writing—review and editing. Mari Palviainen: Writing—review and editing. Marija Holcar: Writing—review and editing. Ioannis Sotiropoulos: Writing—review and editing. Sylwia Bobis‐Wozowicz: Writing—review and editing. Vito G. D'Agostino: Writing—review and editing. Elena Casarotto: Writing—review and editing. Yari Ciani: Writing—review and editing. Shiro Suetsugu: Writing—review and editing. Alice Gualerzi: Writing—review and editing. Lorena Martin‐Jaular: Writing—review and editing. Daniela Boselli: Writing—review and editing. Anna Kashkanova: Writing—review and editing. Pietro Parisse: Writing—review and editing. Lien Lippens: Writing—review and editing. Martina Pagliuca: Writing—review and editing. Martin Blessing: Writing—review and editing. Roberto Frigerio: Writing—review and editing. Thibaut Fourniols: Writing—review and editing. Ana Meliciano: Writing—review and editing. Anna Fietta: Writing—review and editing. Paolo Vincenzo Fioretti: Writing—review and editing. Karolina Soroczyńska: Writing—review and editing. Silvia Picciolini: Writing—review and editing. Amanda Salviano‐Silva: Writing—review and editing. Paolo Bergese: Writing—review and editing. Davide Zocco: Writing—review and editing. Marcella Chiari: Writing—review and editing. Guido Jenster: Writing—review and editing. Levi Waldron: Writing—review and editing. Aleksandar Milosavljevic: Writing—review and editing. John Nolan: Writing—review and editing. Marco P Monopoli: Writing—review and editing. Kenneth W. Witwer: Writing—review and editing. Benedetta Bussolati: Writing—review and editing. Dolores Di Vizio: Writing—review and editing. Juan Falcon Perez: Writing—review and editing. Metka Lenassi: Writing—review and editing. Marina Cretich: Writing—review and editing. Francesca Demichelis: Conceptualization; data curation and analysis; writing—original draft; writing—review and editing.
CONFLICTS OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
The IOC likes to acknowledge the financial support by nonprofit organizations (the Pezcoller Foundation and EVita) and industrial sponsors (Abbelight, Alfatest, Bio‐Techne, NanoFCM, Quanterix, Schaefer, DBA Italia, and Diatech Lab line); to thank the personnel from the Events, Branding, and Graphic Design Division at the University of Trento, the Centro Interdipartimentale di Scienze Mediche (CISMed) at the University of Trento for providing the workshop's location, members of the Basso's and Demichelis’ laboratories for contributing as notetakers (Lorenzo Messa, Luisa Donini, Vera Mugoni, and Federico Vannuccini), and all attendees for their active participation. We would also like to thank Juçara Gastaldi Cominal, Shin Yong Lee, Alexis Germán Murillo Carrasco, Gaia Gavioli, Modeline Longjohn, Ritesh Khanna, Norhayati Binti Liaqat Ali Khan, and Federica Busi for presenting their data during the flash talk presentations. This work was supported by Cancer Research UK and Fondazione AIRC per la Ricerca sul Cancro ETS: Accelerator Award 20218, Grant n.: A26822/A22792; NCI Prostate Cancer SPORE, Grant n.: P50CA211024‐01A..
Basso, M. , Gori, A. , Nardella, C. , Palviainen, M. , Holcar, M. , Sotiropoulos, I. , Bobis‐Wozowicz, S. , D'Agostino, V. G. , Casarotto, E. , Ciani, Y. , Suetsugu, S. , Gualerzi, A. , Martin‐Jaular, L. , Boselli, D. , Kashkanova, A. , Parisse, P. , Lippens, L. , Pagliuca, M. , Blessing, M. , … Demichelis, F. (2024). International society for extracellular vesicles workshop. QuantitatEVs: multiscale analyses, from bulk to single extracellular vesicle. Journal of Extracellular Biology, 3, e137. 10.1002/jex2.137
DATA AVAILABILITY STATEMENT
A summary of the survey results is presented in Figure 3. The full material regarding anonymized survey results is available upon request to the corresponding author.
REFERENCES
- Bockorny, B. , Muthuswamy, L. , Huang, L. , Hadisurya, M. , Lim, C. M. , Tsai, L. L. , Gill, R. R. , Wei, J. L. , Bullock, A. J. , Grossman, J. E. , Besaw, R. J. , Narasimhan, S. , Tao, W. A. , Perea, S. , Sawhney, M. S. , Freedman, S. D. , Hidalgo, M. , Iliuk, A. , & Muthuswamy, S. K. (2023). A large‐scale proteomics resource of circulating extracellular vesicles for biomarker discovery in pancreatic cancer (preprint). Oncology, 10.1101/2023.03.13.23287216 medRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordanaba‐Florit, G. , Royo, F. , Kruglik, S. G. , & Falcón‐Pérez, J. M. (2021). Using single‐vesicle technologies to unravel the heterogeneity of extracellular vesicles. Nature Protocols, 16, 3163–3185. 10.1038/s41596-021-00551-z [DOI] [PubMed] [Google Scholar]
- Busatto, S. , Yang, Y. , Iannotta, D. , Davidovich, I. , Talmon, Y. , & Wolfram, J. (2022). Considerations for extracellular vesicle and lipoprotein interactions in cell culture assays. Journal of Extracellular Vesicles, 11, e12202. 10.1002/jev2.12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto, S. , Zendrini, A. , Radeghieri, A. , Paolini, L. , Romano, M. , Presta, M. , & Bergese, P. (2020). The nanostructured secretome. Biomaterials Science, 8, 39–63. 10.1039/C9BM01007F [DOI] [PubMed] [Google Scholar]
- Casarotto, E. , Sproviero, D. , Corridori, E. , Gagliani, M. C. , Cozzi, M. , Chierichetti, M. , Cristofani, R. , Ferrari, V. , Galbiati, M. , Mina, F. , Piccolella, M. , Rusmini, P. , Tedesco, B. , Gagliardi, S. , Cortese, K. , Cereda, C. , Poletti, A. , & Crippa, V. (2022). Neurodegenerative disease‐associated TDP‐43 fragments are extracellularly secreted with CASA complex proteins. Cells, 11, 516. 10.3390/cells11030516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandy, M. , Rhee, J.‐W. , Ozen, M. O. , Williams, D. R. , Pepic, L. , Liu, C. , Zhang, H. , Malisa, J. , Lau, E. , Demirci, U. , & Wu, J. C. (2020). Atlas of exosomal microRNAs secreted from human iPSC‐derived cardiac cell types. Circulation, 142, 1794–1796. 10.1161/CIRCULATIONAHA.120.048364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, K.‐H. , Keerthikumar, S. , Roncaglia, P. , Subramanian, S. L. , Roth, M. E. , Samuel, M. , Anand, S. , Gangoda, L. , Gould, S. , Alexander, R. , Galas, D. , Gerstein, M. B. , Hill, A. F. , Kitchen, R. R. , Lötvall, J. , Patel, T. , Procaccini, D. C. , Quesenberry, P. , Rozowsky, J. , … Laurent, L. C. (2016). Extending gene ontology in the context of extracellular RNA and vesicle communication. Journal of Biomedical Semantics, 7, 19. 10.1186/s13326-016-0061-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumans, F. A. W. , Brisson, A. R. , Buzas, E. I. , Dignat‐George, F. , Drees, E. E. E. , El‐Andaloussi, S. , Emanueli, C. , Gasecka, A. , Hendrix, A. , Hill, A. F. , Lacroix, R. , Lee, Y. , van Leeuwen, T. G. , Mackman, N. , Mäger, I. , Nolan, J. P. , van der Pol, E. , Pegtel, D. M. , Sahoo, S. , … Nieuwland, R. (2017). Methodological Guidelines to Study Extracellular Vesicles. Circ Res, 120, 1632–1648. 10.1161/CIRCRESAHA.117.309417 [DOI] [PubMed] [Google Scholar]
- Daaboul, G. G. , Gagni, P. , Benussi, L. , Bettotti, P. , Ciani, M. , Cretich, M. , Freedman, D. S. , Ghidoni, R. , Ozkumur, A. Y. , Piotto, C. , Prosperi, D. , Santini, B. , Ünlü, M. S. , & Chiari, M. (2016). Digital detection of exosomes by interferometric imaging. Scientific Reports, 6, 37246. 10.1038/srep37246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, G. , Raposo, G. , Nishimura, T. , & Suetsugu, S. (2023). Protrusion‐derived vesicles: New subtype of EVs? Nature Reviews Molecular Cell Biology, 24, 81–82. 10.1038/s41580-022-00555-x [DOI] [PubMed] [Google Scholar]
- Driedonks, T. , Jiang, L. , Carlson, B. , Han, Z. , Liu, G. , Queen, S. E. , Shirk, E. N. , Gololobova, O. , Liao, Z. , Nyberg, L. H. , Lima, G. , Paniushkina, L. , Garcia‐Contreras, M. , Schonvisky, K. , Castell, N. , Stover, M. , Guerrero‐Martin, S. , Richardson, R. , Smith, B. , … Witwer, K. W. (2022). Pharmacokinetics and biodistribution of extracellular vesicles administered intravenously and intranasally to Macaca nemestrina . Journal of Extracellular Biology, 1, e59. 10.1002/jex2.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A. , … Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J of Extracellular Vesicle, 10, e12093. 10.1002/jev2.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, J. V. , Da Rosa Soares, A. , Ramalho, J. , Máximo Carvalho, C. , Cardoso, M. H. , Pintado, P. , Carvalho, A. S. , Beck, H. C. , Matthiesen, R. , Zuzarte, M. , Girão, H. , Van Niel, G. , & Pereira, P. (2022). LAMP2A regulates the loading of proteins into exosomes. Science Advances, 8, eabm1140. 10.1126/sciadv.abm1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M. , Villa, A. , Brunialti, E. , Crescenti, D. , Dell'Omo, G. , Kuryk, L. , Vingiani, A. , Mazzaferro, V. , & Ciana, P. (2021). Cancer‐derived EVs show tropism for tissues at early stage of neoplastic transformation. Nanotheranostics, 5, 1–7. 10.7150/ntno.47226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeurickx, E. , & Hendrix, A. (2020). Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Molecular Aspects of Medicine, 72, 100828. 10.1016/j.mam.2019.10.005 [DOI] [PubMed] [Google Scholar]
- Gomes, P. A. , Bodo, C. , Nogueras‐Ortiz, C. , Samiotaki, M. , Chen, M. , Soares‐Cunha, C. , Silva, J. M. , Coimbra, B. , Stamatakis, G. , Santos, L. , Panayotou, G. , Tzouanou, F. , Waites, C. L. , Gatsogiannis, C. , Sousa, N. , Kapogiannis, D. , Costa‐Silva, B. , & Sotiropoulos, I. (2023). A novel isolation method for spontaneously released extracellular vesicles from brain tissue and its implications for stress‐driven brain pathology. Cell Communication and Signaling, 21, 35. 10.1186/s12964-023-01045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths‐Jones, S. (2006). miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Research, 34, D140–D144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, A. , Kooijmans, S. A. A. , Niada, S. , Picciolini, S. , Brini, A. T. , Camussi, G. , & Bedoni, M. (2019). Raman spectroscopy as a quick tool to assess purity of extracellular vesicle preparations and predict their functionality. Journal of Extracellular Vesicles, 8, 1568780. 10.1080/20013078.2019.1568780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, A. , Niada, S. , Giannasi, C. , Picciolini, S. , Morasso, C. , Vanna, R. , Rossella, V. , Masserini, M. , Bedoni, M. , Ciceri, F. , Bernardo, M. E. , Brini, A. T. , & Gramatica, F. (2017). Raman spectroscopy uncovers biochemical tissue‐related features of extracellular vesicles from mesenchymal stromal cells. Scientific Reports, 7(9820), 10.1038/s41598-017-10448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcar, M. , Kandušer, M. , & Lenassi, M. (2021). Blood Nanoparticles ‐ Influence on Extracellular Vesicle Isolation and Characterization. Front. Pharmacol., 12, 773844. 10.3389/fphar.2021.773844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. T. , Sasakura, N. , Matsubara, D. , Furusawa, N. , Mukai, M. , Kitamura, N. , Obayashi, T. , Nishimura, T. , Oono‐Yakura, K. , Funato, Y. , Okamura, Y. , Tarao, K. , Nakano, Y. , Murakami, Y. , Kinoshita, K. , Takahashi, C. , Miki, H. , Gonda, K. , Scita, G. , … Suetsugu, S. (2020). Involvement of I‐BAR protein IRSp53 in tumor cell growth via extracellular microvesicle secretion. bioRxiv 2020.04.20.050492. 10.1101/2020.04.20.050492 [DOI]
- Huang, Y. , Arab, T. , Russell, A. E. , Mallick, E. R. , Nagaraj, R. , Gizzie, E. , Redding‐Ochoa, J. , Troncoso, J. C. , Pletnikova, O. , Turchinovich, A. , Routenberg, D. A. , & Witwer, K. W. (2023). Towards a human brain EV atlas: Characteristics of EVs from different brain regions, including small RNA and protein profiles (preprint). Neuroscience, 10.1101/2023.05.06.539665 bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert, E. , Morlion, A. , Avila Cobos, F. , Verniers, K. , Nuytens, J. , Vanden Eynde, E. , Yigit, N. , Anckaert, J. , Geerts, A. , Hindryckx, P. , Jacques, P. , Brusselle, G. , Bracke, K. R. , Maes, T. , Malfait, T. , Derveaux, T. , Ninclaus, V. , Van Cauwenbergh, C. , Roelens, K. , … Mestdagh, P. (2020). Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Reports, 33, 108552. 10.1016/j.celrep.2020.108552 [DOI] [PubMed] [Google Scholar]
- Kalra, H. , Simpson, R. J. , Ji, H. , Aikawa, E. , Altevogt, P. , Askenase, P. , Bond, V. C. , Borràs, F. E. , Breakefield, X. , Budnik, V. , Buzas, E. , Camussi, G. , Clayton, A. , Cocucci, E. , Falcon‐Perez, J. M. , Gabrielsson, S. , Gho, Y. S. , Gupta, D. , Harsha, H. C. , … Mathivanan, S. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biology, 10, e1001450. 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkanova, A. D. , Blessing, M. , Gemeinhardt, A. , Soulat, D. , & Sandoghdar, V. (2022). Precision size and refractive index analysis of weakly scattering nanoparticles in polydispersions. Nature Methods, 19, 586–593. 10.1038/s41592-022-01460-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkanova, A. D. , Blessing, M. , Reischke, M. , Baur, J. , Baur, A. S. , Sandoghdar, V. , & Van Deun, J. (2023). Label‐free discrimination of extracellular vesicles from large lipoproteins. Journal of Extracellular Vesicles, 12, 12348. 10.1002/jev2.12348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar, S. , Chisanga, D. , Ariyaratne, D. , Al Saffar, H. , Anand, S. , Zhao, K. , Samuel, M. , Pathan, M. , Jois, M. , Chilamkurti, N. , Gangoda, L. , & Mathivanan, S. (2016). ExoCarta: A web‐based compendium of exosomal cargo. Journal of Molecular Biology, 428, 688–692. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.‐K. , Lee, J. , Simpson, R. J. , Lötvall, J. , & Gho, Y. S. (2015). EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Seminars in Cell & Developmental Biology, 40, 4–7. 10.1016/j.semcdb.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Lai, H. , Li, Y. , Zhang, H. , Hu, J. , Liao, J. , Su, Y. , Li, Q. , Chen, B. , Li, C. , Wang, Z. , Li, Y. , Wang, J. , Meng, Z. , Huang, Z. , & Huang, S. (2022). exoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Research, 50, D118–D128. 10.1093/nar/gkab1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante, E. L. , Stürchler, A. , Fullem, R. , Chen, D. , Starner, A. C. , Esquivel, E. , Alsop, E. , Jackson, A. R. , Ghiran, I. , Pereira, G. , Rozowsky, J. , Chang, J. , Gerstein, M. B. , Alexander, R. P. , Roth, M. E. , Franklin, J. L. , Coffey, R. J. , Raffai, R. L. , Mansuy, I. M. , … Milosavljevic, A. (2023). exRNA‐eCLIP intersection analysis reveals a map of extracellular RNA binding proteins and associated RNAs across major human biofluids and carriers. Cell Genomics, 3, 100303. 10.1016/j.xgen.2023.100303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.‐R. , Tong, C.‐Y. , Sung, T.‐J. , Kang, T.‐Y. , Zhou, X. J. , & Liu, C.‐C. (2019). CMEP: A database for circulating microRNA expression profiling. Bioinformatics, 35, 3127–3132. 10.1093/bioinformatics/btz042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Li, Y. , Chen, B. , Zhao, J. , Yu, S. , Tang, Y. , Zheng, Q. , Li, Y. , Wang, P. , He, X. , & Huang, S. (2018). exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Research, 46, D106–D112. 10.1093/nar/gkx891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Zhang, Q. , Zhang, J. , Li, C. , Miao, Y.‐R. , Lei, Q. , Li, Q. , & Guo, A.‐Y. (2019). EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Research, 47, D89–D93. 10.1093/nar/gky985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, M. , Parolisi, R. , Scaroni, F. , Bonfanti, E. , Gualerzi, A. , Gabrielli, M. , Kerlero De Rosbo, N. , Uccelli, A. , Giussani, P. , Viani, P. , Garlanda, C. , Abbracchio, M. P. , Chaabane, L. , Buffo, A. , Fumagalli, M. , & Verderio, C. (2019). Detrimental and protective action of microglial extracellular vesicles on myelin lesions: Astrocyte involvement in remyelination failure. Acta Neuropathologica, 138, 987–1012. 10.1007/s00401-019-02049-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall, J. , Rajendran, L. , Gho, Y. , Thery, C. , Wauben, M. , Raposo, G. , Sjöstrand, M. , Taylor, D. , Telemo, E. , & Breakefield, X. O. (2012). The launch of Journal of Extracellular Vesicles (JEV), the official journal of the International Society for Extracellular Vesicles—about microvesicles, exosomes, ectosomes and other extracellular vesicles. Journal of Extracellular Vesicles, 1, 18514. 10.3402/jev.v1i0.18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiapane, G. , Parolini, I. , Conte, K. , Malfatti, M. C. , Corsi, J. , Sanchez, M. , Pietrantoni, A. , D'Agostino, V. G. , & Tell, G. (2021). Enzymatically active apurinic/apyrimidinic endodeoxyribonuclease 1 is released by mammalian cells through exosomes. Journal of Biological Chemistry, 296, 100569. 10.1016/j.jbc.2021.100569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugoni, V. , Ciani, Y. , Nardella, C. , & Demichelis, F. (2022). Circulating RNAs in prostate cancer patients. Cancer Letters, 524, 57–69. 10.1016/j.canlet.2021.10.011 [DOI] [PubMed] [Google Scholar]
- Murillo, O. D. , Thistlethwaite, W. , Rozowsky, J. , Subramanian, S. L. , Lucero, R. , Shah, N. , Jackson, A. R. , Srinivasan, S. , Chung, A. , Laurent, C. D. , Kitchen, R. R. , Galeev, T. , Warrell, J. , Diao, J. A. , Welsh, J. A. , Hanspers, K. , Riutta, A. , Burgstaller‐Muehlbacher, S. , Shah, R. V. , … Milosavljevic, A. (2019). exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell, 177, 463–477. e15. 10.1016/j.cell.2019.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicò, A. , Zenatelli, R. , Romano, M. , Zendrini, A. , Alacqua, S. , Tassoni, S. , Paolini, L. , Urbinati, C. , Rusnati, M. , Bergese, P. , Pomarico, G. , & Radeghieri, A. (2023). Surface functionalization of extracellular vesicle nanoparticles with antibodies: A first study on the protein corona “variable”. Nanoscale Advances, 5, 4703–4717. 10.1039/D3NA00280B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T. , Oyama, T. , Hu, H. T. , Fujioka, T. , Hanawa‐Suetsugu, K. , Ikeda, K. , Yamada, S. , Kawana, H. , Saigusa, D. , Ikeda, H. , Kurata, R. , Oono‐Yakura, K. , Kitamata, M. , Kida, K. , Hikita, T. , Mizutani, K. , Yasuhara, K. , Mimori‐Kiyosue, Y. , Oneyama, C. , … Suetsugu, S. (2021). Filopodium‐derived vesicles produced by MIM enhance the migration of recipient cells. Developmental Cell, 56, 842–859. e8. 10.1016/j.devcel.2021.02.029 [DOI] [PubMed] [Google Scholar]
- Nolan, J. P. , & Duggan, E. (2018). Analysis of individual extracellular vesicles by flow cytometry, In: Hawley, T. S. , Hawley, R. G. (Eds.), Flow cytometry protocols, methods in molecular biology. Springer, pp. 79–92. 10.1007/978-1-4939-7346-0_5 [DOI] [PubMed] [Google Scholar]
- Paba, C. , Dorigo, V. , Senigagliesi, B. , Tormena, N. , Parisse, P. , Voitchovsky, K. , & Casalis, L. (2023). Lipid bilayer fluidity and degree of order regulates small EVs adsorption on model cell membrane. Journal of Colloid and Interface Science, 652, 1937–1943. 10.1016/j.jcis.2023.08.117 [DOI] [PubMed] [Google Scholar]
- Perissinotto, F. , Rondelli, V. , Senigagliesi, B. , Brocca, P. , Almásy, L. , Bottyán, L. , Merkel, D. G. , Amenitsch, H. , Sartori, B. , Pachler, K. , Mayr, M. , Gimona, M. , Rohde, E. , Casalis, L. , & Parisse, P. (2021). Structural insights into fusion mechanisms of small extracellular vesicles with model plasma membranes. Nanoscale, 13, 5224–5233. 10.1039/D0NR09075A [DOI] [PubMed] [Google Scholar]
- Pisitkun, T. , Shen, R.‐F. , & Knepper, M. A. (2004). Identification and proteomic profiling of exosomes in human urine. Proceedings of the National Academy of Sciences of the United States of America, 101, 13368–13373. 10.1073/pnas.0403453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeghieri, A. , Alacqua, S. , Zendrini, A. , Previcini, V. , Todaro, F. , Martini, G. , Ricotta, D. , & Bergese, P. (2022). Active antithrombin glycoforms are selectively physiosorbed on plasma extracellular vesicles. Journal of Extracellular Biology, 1, e57. 10.1002/jex2.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeghieri, A. , & Bergese, P. (2023). The biomolecular corona of extracellular nanoparticles holds new promises for advancing clinical molecular diagnostics. Expert Review of Molecular Diagnostics, 23, 471–474. 10.1080/14737159.2023.2215927 [DOI] [PubMed] [Google Scholar]
- Russo, F. , Di Bella, S. , Nigita, G. , Macca, V. , Laganà, A. , Giugno, R. , Pulvirenti, A. , & Ferro, A. (2012). miRandola: Extracellular circulating MicroRNAs database. PLoS ONE, 7, e47786. 10.1371/journal.pone.0047786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, F. , Di Bella, S. , Vannini, F. , Berti, G. , Scoyni, F. , Cook, H. V. , Santos, A. , Nigita, G. , Bonnici, V. , Laganà, A. , Geraci, F. , Pulvirenti, A. , Giugno, R. , De Masi, F. , Belling, K. , Jensen, L. J. , Brunak, S. , Pellegrini, M. , & Ferro, A. (2018). miRandola 2017: A curated knowledge base of non‐invasive biomarkers. Nucleic Acids Research, 46, D354–D359. 10.1093/nar/gkx854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. L. , Kitchen, R. R. , Alexander, R. , Carter, B. S. , Cheung, K. , Laurent, L. C. , Pico, A. , Roberts, L. R. , Roth, M. E. , Rozowsky, J. S. , Su, A. I. , Gerstein, M. B. , & Milosavljevic, A. (2015). Integration of extracellular RNA profiling data using metadata, biomedical ontologies and Linked Data technologies. Journal of Extracellular Vesicles, 4, 27497. 10.3402/jev.v4.27497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth, E. Á. , Turiák, L. , Visnovitz, T. , Cserép, C. , Mázló, A. , Sódar, B. W. , Försönits, A. I. , Petővári, G. , Sebestyén, A. , Komlósi, Z. , Drahos, L. , Kittel, Á. , Nagy, G. , Bácsi, A. , Dénes, Á. , Gho, Y. S. , Szabó‐Taylor, K. É. , & Buzás, E. I. (2021). Formation of a protein corona on the surface of extracellular vesicles in blood plasma. Journal of Extracellular Vesicles, 10, e12140. 10.1002/jev2.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B. , Casert, C. , Cheng, L. , Cmoch, A. , … Hendrix, A. , EV‐TRACK Consortium . (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14, 228–232. 10.1038/nmeth.4185 [DOI] [PubMed] [Google Scholar]
- Van Royen, M. E. , Soekmadji, C. , Grange, C. , Webber, J. P. , Tertel, T. , Droste, M. , Buescher, A. , Giebel, B. , Jenster, G. W. , Llorente, A. , Blijdorp, C. J. , Burger, D. , Erdbrügger, U. , & Martens‐Uzunova, E. S. (2023). The quick reference card “Storage of urinary EVs”—A practical guideline tool for research and clinical laboratories. Journal of Extracellular Vesicles, 12, 12286. 10.1002/jev2.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Chai, Z. , Pan, G. , Hao, Y. , Li, B. , Ye, T. , Li, Y. , Long, F. , Xia, L. , & Liu, M. (2021). ExoBCD: A comprehensive database for exosomal biomarker discovery in breast cancer. Briefings in Bioinformatics, 22, bbaa088. 10.1093/bib/bbaa088 [DOI] [PubMed] [Google Scholar]
- Welsh, J. A. , Arkesteijn, G. J. A. , Bremer, M. , Cimorelli, M. , Dignat‐George, F. , Giebel, B. , Görgens, A. , Hendrix, A. , Kuiper, M. , Lacroix, R. , Lannigan, J. , Van Leeuwen, T. G. , Lozano‐Andrés, E. , Rao, S. , Robert, S. , De Rond, L. , Tang, V. A. , Tertel, T. , Yan, X. , … Van Der Pol, E. (2023). A compendium of single extracellular vesicle flow cytometry. Journal of Extracellular Vesicles, 12, e12299. 10.1002/jev2.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Goberdhan, D. C. , O'Driscoll, L. , Théry, C. , Welsh, J. A. , Blenkiron, C. , Buzás, E. I. , Di Vizio, D. , Erdbrügger, U. , Falcón‐Pérez, J. M. , Fu, Q. , Hill, A. F. , Lenassi, M. , Lötvall, J. , Nieuwland, R. , Ochiya, T. , Rome, S. , Sahoo, S. , & Zheng, L. (2021). Updating MISEV: Evolving the minimal requirements for studies of extracellular vesicles. Journal of Extracellular Vesicles, 10, e12182. 10.1002/jev2.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, F. , Liu, S. , Wang, J. , Xuan, J. , Zhang, X. , Qu, L. , Zheng, L. , & Yang, J. (2021). deepBase v3.0: Expression atlas and interactive analysis of ncRNAs from thousands of deep‐sequencing data. Nucleic Acids Research, 49, D877–D883. 10.1093/nar/gkaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhang, L. , Wang, T. , Wu, Y. , Pu, X. , Li, M. , & Guo, Y. (2020). ExoceRNA atlas: A database of cancer ceRNAs in human blood exosomes. Life Sciences, 257, 118092. 10.1016/j.lfs.2020.118092 [DOI] [PubMed] [Google Scholar]
- Zendrini, A. , Cardellini, J. , Frigerio, R. , Bertoni, M. , Berti, D. , & Bergese, P. (2023). On the interaction and nanoplasmonics of gold nanoparticles and lipoproteins. JCIS Open, 11, 100088. 10.1016/j.jciso.2023.100088 [DOI] [Google Scholar]
- Zendrini, A. , Paolini, L. , Busatto, S. , Radeghieri, A. , Romano, M. , Wauben, M. H. M. , Van Herwijnen, M. J. C. , Nejsum, P. , Borup, A. , Ridolfi, A. , Montis, C. , & Bergese, P. (2020). Augmented COlorimetric NANoplasmonic (CONAN) method for grading purity and determine concentration of EV microliter volume solutions. Frontiers in Bioengineering and Biotechnology, 7, 452. 10.3389/fbioe.2019.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A summary of the survey results is presented in Figure 3. The full material regarding anonymized survey results is available upon request to the corresponding author.


