Abstract
Chromosomes in human cancer cells are expected to initiate replication from predictably localized origins, firing reproducibly at discrete times in S phase. Replication products obtained from HeLa cells at different stages of S phase were hybridized to cDNA and genome tiling oligonucleotide microarrays to determine the temporal profile of replication of human chromosomes on a genome-wide scale. About 1,000 genes and chromosomal segments were identified as sites containing efficient origins that fire reproducibly. Early replication was correlated with high gene density. An acute transition of gene density from early to late replicating areas suggests that discrete chromatin states dictate early versus late replication. Surprisingly, at least 60% of the interrogated chromosomal segments replicate equally in all quarters of S phase, suggesting that large stretches of chromosomes are replicated by inefficient, variably located and asynchronous origins and forks, producing a pan-S phase pattern of replication. Thus, at least for aneuploid cancer cells, a typical discrete time of replication in S phase is not seen for large segments of the chromosomes.
Keywords: DNA replication, genome-wide, microarrays, chromatin, human cancer
By general consensus, origins of replication are expected to map at precise locations on chromosomes, fire consistently at the same time in S phase, and produce forks that proceed bidirectionally at predictable and reproducible speeds to eventually replicate entire chromosomes. Based on the average speed of fork movement (1), origins are expected to be distributed at an average spacing of 100 kb, and yet they have been surprisingly difficult to map for single-copy genes in mammalian cells in culture. For the handful of well studied origins, there is one report suggesting use of a single discrete initiation site (2) and a few suggesting use of multiple initiation sites distributed >10-50 kb, with different sites used in different cells with variable efficiency (3-10). In addition, origins are expected to fire at discrete times in S phase (11). For example, Giemsa R bands on metaphase chromosomes mostly replicate earlier in S phase compared with the Giemsa G bands (12). Heterochromatin regions near the centromeres and telomeres are usually replicated late in S phase (13, 14). Examination of a few genes suggest that housekeeping genes like actin and nuclear lamin replicate early in S phase, whereas differentiation-specific genes like immunoglobulin or β-globin replicate late in cells where they are not expressed and early in cells where they are expressed (7, 15, 16). It has been postulated that the open nucleosome structure associated with euchromatin and with expressed genes is conducive to early replication in S phase.
With the advent of complete genome sequences, we can now ask what fraction of replication origins behaves in the ideal fashion described above and whether the rules regarding time of origin firing apply on a genome-wide scale. In Saccharomyces cerevisiae, the time of replication of all 6,000 genes was determined, and the results were modeled to estimate the distribution of origins of replication along the yeast chromosomes (17). Although such studies revealed the locations and time of firing of origins, they produced the surprising result that expression of a yeast gene did not correlate with time of replication in S phase. A similar study on cultured cells in Drosophila, however, demonstrated that there was a correlation between the expression of a gene and its likelihood of being replicated early (18).
We have now carried out a study in human cancer cells in culture to estimate the time of replication of hundreds of genes. Thymidine-aphidicolin block was used to arrest cells at the G1/S boundary. After confirming that cells released from the block progress synchronously through S phase, we harvested timed replication pools of DNA and used these as probes first on cDNA arrays to validate this method for establishing time of replication. The pools were subsequently used on newly available high-density genome-tiling arrays covering all unique sequences on chromosomes 21 and 22. The high definition obtained by use of finer timed replication pools and high density genomic arrays shows that, whereas gene density and chromatin states predict time of replication of many segments, many other segments are replicated in a pan-S phase pattern suggestive of significant flexibility in origin usage, origin timing, and fork movement.
Methods
High-Density Genome-Tiling Array. The arrays comprise 1,020,643 probe pairs (i.e., Perfect Match, Mis-Match) interrogating the repeat masked sequence of chromosomes 21 and 22 (≈35 Mb) (Affymetrix, Santa Clara, CA). Thus, on average, there is a probe pair every 35 bp in the non-repeat regions of the two chromosomes. There is no bias regarding exonic/coding regions versus noncoding regions. Heavy/light (H/L) DNA from each time point was prepared as mentioned in Fig. 1, fragmented with DNaseI to ≈50 bp, and then end-labeled with biotin-ddATP by using terminal transferase. The labeled DNA was hybridized to the genome-tiling arrays. The array results were normalized and analyzed by using g-trans software (Affymetrix). The graphs from these processed data were generated by using integrated genome browser software (Affymetrix).
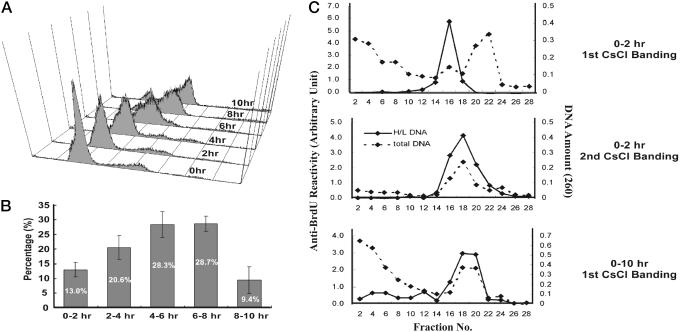
Fig. 1.
Synchronous progression of HeLa cell through S phase and harvesting of timed replication pools. (A) HeLa cells released from a G1-S block followed by FACS for DNA content. (B) Cells synchronously released in the same way as in A were pulsed with [3H]thymidine (20 μCi/ml) (1 Ci = 37 GBq) at indicated time intervals, and thymidine incorporation was measured by liquid scintillation counting. The sum total of cpm values was regarded as 100% of replication. Results were obtained in triplicate from two independent experiments. (C) Isolation of newly replicated DNA on CsCl density gradient. (Top) Genomic DNA labeled during 0-2 hr of S phase. Solid line is the anti-BrdUrd reactivity in an ELISA, and dashed line is the absorbance at 260 nm, which reflect distribution of H/L DNA and total DNA, respectively. (Middle) Second purification of H/L fractions obtained from Top. Complete overlap of two peaks indicates reliable purity of H/L DNA. (Bottom) First round of CsCl centrifugation of genomic DNA from cells labeled with BrdUrd for 0-10 hr of S phase. All genomic DNA was labeled into H/L DNA within 10 hr.
Measurement of Enrichment of Replication in a 2-hr Interval Relative to the Entirety of S Phase. Because the distribution of array intensities should be the same across all replicate arrays, all replicate arrays were quantile-normalized (19). For each genomic position to which a probe pair (PM, MM) mapped, i, a data set, Si, was generated consisting of all (PM, MM) pairs mapping within a window of ±5 kb. We test for the statistically significant enrichment of treatment (signal over the 2-hr labeling period) over control (signal over the 8-hr labeling period) by applying a Wilcoxon rank sum test (20) to these estimates of abundance. Genomic positions belonging to regions that were specifically enriched over a given 2-hr interval of labeling were defined by applying a stringent P value cutoff of 10-5. Resultant positive positions that were <5 kb apart (less than the half-width of the testing window) were merged to form a predicted region that was replicated selectively in a given 2-hr interval. Greater details are provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Probes for Interphase FISH. For interphase FISH, the pan-S phase pattern probes were RP1-5O6 and RP11-247I13 (representing chromosome 22 region 36,675,000-36,775,000 and 30,301,231-30,313,718 as per NCBI30 build). RP4-671O14 and RP11-91E22 (chromosome 22 region 42,550,000-42,725,000, and chromosome 11 gene 23998:T78107:Hs.167185) were from segments with monophasic replication, early and late replicating, respectively.
Results and Discussion
Synchronization of HeLa Cells and Harvesting of Timed Replication Products. Thymidine-aphidicolin block was used to arrest human cervical carcinoma cells (HeLa) at the G1/S boundary. Release of the cells from the block followed by FACS for DNA content showed that DNA content began to increase by 2 hr and eventually doubled after 8-10 hr (Fig. 1 A). The bell-shaped distribution of DNA synthesis during S phase was consistent with normal progression through S phase (Fig. 1B). Cells progressing synchronously through S phase were labeled with bromodeoxyuridine for 2-hr intervals and BrdUrd-incorporated H/L double-stranded DNA separated from unlabeled light-light (L/L) DNA by CsCl density gradient centrifugation (Fig. 1C). Two cycles of centrifugation were sufficient to purify the H/L DNA, and labeling of cells for the entire duration of S phase converted all of the DNA in the population to H/L DNA.
Hybridization to cDNA Arrays. DNA replicated in each time interval was purified as in Fig. 1C and labeled with Cy5 whereas total genomic DNA (H/L DNA obtained by labeling for 0-10 hr) was labeled with Cy3 as a negative control. The two probes were mixed in equal molar ratios and hybridized to a human cDNA array containing 1,589 genes spotted in duplicate on glass slides. Only 625 of the 1,589 spotted genes (39%) showed reproducible patterns of replication that allowed them to be clustered according to their time of replication in S phase (Fig. 2A). Examples of genes replicated preferentially during a given time interval are shown in the boxes on the side.
Fig. 2.
Analysis of time of replication with a human cDNA array 1.7k7 (University Health Network, Toronto). (A) The array results were clustered by using cluster software (Stanford University) and presented by treeview (Stanford University). Red indicates enrichment in H/L fraction from indicated interval relative to total genomic DNA, and green indicates depletion of gene in H/L fraction of that interval relative to total genomic DNA. Information about each gene is given in three ways (Image clone ID:NCBI accession ID:Unigene ID). (B) Confirmation of microarray analysis by dot blot hybridization. The graph represents mean values of two independent experiments with the highest signal from a dot in the series (cpm) normalized to 1. The 305120 (Image clone ID) is thus confirmed to be early (0-2 hr) replicating. (C) The 501492 is confirmed by dot blot hybridization to be late (6-8 hr) replicating. Details are the same as in B.
Validation of Time of Replication. Because these methods have not been applied for these analyses before, we confirmed the classification of a subset of the genes by two approaches. First, the timed replication products were immobilized on membranes, and selected genes were used as radioactive probes. The bar graphs of the hybridization signals confirmed the classification of early replicating (Fig. 2B) and late-replicating (Fig. 2C) genes. Nine (five early and four late candidates) of 10 selected cDNAs were confirmed by dot blotting to be correctly classified (data not shown). The second approach, interphase FISH, will be discussed later.
Hybridization to Genome-Tiling Arrays Reveals Segments That Replicate in a Defined Part of S Phase and Other Segments That Replicate in a Pan-S Phase Pattern. Despite the success in classifying time of replication for 40% of the genes, the failure to classify the time of replication for 60% of the genes was surprising and could be because we were interrogating genomic events with spliced cDNAs. We therefore turned to a high-density genome-tiling Affymetrix array comprising all unique oligonucleotides in chromosomes 21 and 22 representing ≈35 Mb of DNA (21, 22). These experiments were confined to DNA replicated during the first 8 hr after release from aphidicolin-thymidine block. To minimize the noise in the signal obtained from individual oligonucleotides, we developed a statistical method that gives a confidence value to the signal from a given set of oligonucleotides (see Methods). Briefly, we assess whether contiguous signals are coenriched in a particular part of S phase relative to the entirety of S phase. We mapped high confidence (P < 10-5) regions of replication for each quarter of S phase applying the Wilcoxon rank sum test in a sliding window of 10 kb. We found 40, 39, 17, and 77 regions on chromosome 21, and 70, 87, 28, and 45 regions on chromosome 22 that replicated in a single quarter of S phase in the entire population of cells. These regions cover 3.504 Mb (<10% of the region interrogated). Even with a lower P value of <10-2, 35.1% of the total amount of DNA detected by the genome tiling arrays was replicated selectively in one quarter of S phase, a percentage consistent with the cDNA array data. Areas with significant enrichment of the replication signal early or late in S phase are shown (Fig. 3 A and B). Genomic regions and genes clearly replicated in a temporally monophasic pattern in one part of S phase (another example in Fig. 6, box B, which is published as supporting information on the PNAS web site) are indicative of replication from well defined efficient origins and uniformly migrating forks that are used in most of the cells in the culture at a similar time in S phase. In contrast to the 35.1% of the sequences that replicate in a temporally monophasic pattern, many regions gave a robust replication signal over the entire 0-8 hr interval but were not selectively replicated in one particular quarter of S phase (Figs. 3C and 6 and Supporting Methods; see also Fig. 7, which is published as supporting information on the PNAS web site). We suggest that this pan-S phase pattern of replication is due to variably located replication initiation sites that fire asynchronously and due to replication forks that migrate at variable speeds over the same genomic segments in different chromosomes and cells.
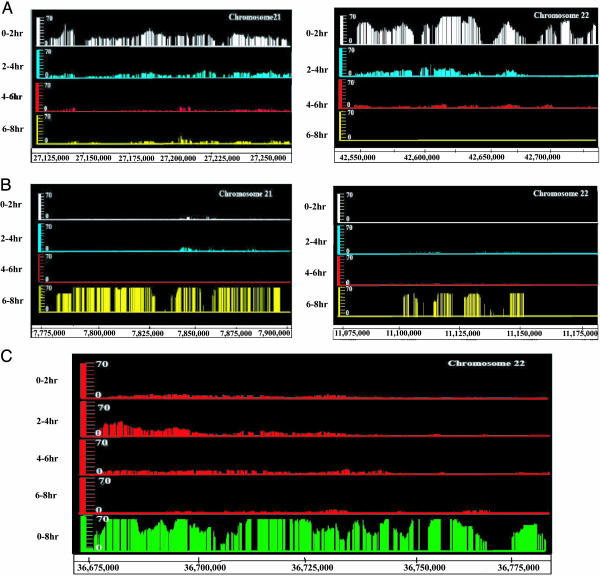
Fig. 3.
Genomic replication profile. (A) Comparative replication profiles of two early replicating segments on chromosomes 21 and 22. Each line shows the significant enrichment of replication signal in the indicated 2-hr interval relative to the 0-8 hr interval. (B) Profile of two late replicating segments at the telomeric ends of the p arms of the two chromosomes as in A.(C) Examples of segments that replicate in all quarters of S phase without significant enrichment of signal in any one quarter. The top four lines are the same as in A. The green bars in the bottom line indicate P values for replication with the 0-8 hr labeled probe indicating that replication of the entire segment was detected easily. The probability that significant replication was detected in all time intervals over this stretch is shown in Fig. 7. Another example of pan-S phase replication is shown in Fig. 6, box A.
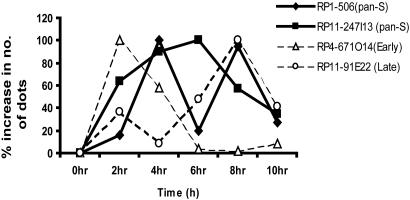
FISH Analyses to Confirm the Monophasic and Pan-S Pattern of Replication. We confirmed the time of replication of regions selected from the cDNA and genome-tiling arrays by interphase FISH (23). Synchronously progressing HeLa cells were harvested at indicated time points during S phase, and genomic clones were labeled and hybridized to the denatured interphase chromosomes. A single hybridization signal indicates that the targeted DNA region is unreplicated, and a doublet hybridization signal indicates that the targeted DNA region is replicated. The replication of each probe during a time interval is estimated by the increase in the number of dots during that time interval. For all of the regions shown in Fig. 4, cells at the G1/S boundary (0 h) contained three single dots due to the aneuploidy of HeLa cells. After entry into S phase, however, cells showed hybridization signals ranging from three single dots to three doublets (six dots) (Fig. 8, which is published as supporting information on the PNAS web site). The appearance of intermediate numbers of dots/cell (four, five, and six) indicate that all three copies of the gene do not replicate at exactly the same time in a cell. The early replicating pattern of RP4-671O14 is clearly evident with a robust increase in the number of dots only during the first 4 hr of S phase. In contrast, the late replicating region RP11-91E22 showed maximum increase in number of dots in the last 4 hr of S phase. For the probes representing pan-S pattern of replication, RP1-5O6 showed a biphasic pattern of replication as the increment in dots showed two peaks separated by one interval with only a modest amount of replication. RP11-247I13 showed a pan-S phase pattern of replication with a considerable amount of replication distributed over multiple time intervals of S phase.
Fig. 4.
Confirmation of pan-S and temporally monophasic pattern of replication with interphase FISH. Synchronously progressing cells through S phase were hybridized with fluorescence-labeled probes of PAC/BAC clone DNA RP1-5O6 and RP11-247I13 (for pan-S pattern of replication) and RP4-671O14 and RP11-91E22 (for early and late replicating areas, respectively). Increase in number of dots from ≥80 cells was calculated at each interval of S phase. To facilitate comparison of replication profiles, the maximum increase in dots in any interval for a given probe (100%) was used for normalizing the increases in the other intervals.
Early Replication Correlates with High Gene Density. We next compared the time of replication of a chromosomal segment with the exon density in that segment (Supporting Methods). For each mapped ideally replicating region, the exon density was estimated by computing the median of the exon density values that fall within the region. The exon densities are high (≈0.033) for regions replicated during the first 4 hr of S phase compared with the areas replicated in the last 4 hr (0.009 for 4-6 hr; 0 for 6-8 hr replication periods) (Fig. 5A). The drop in exon density from either of the two early replicating regions to the later replicating regions is significant (P < 10-15).
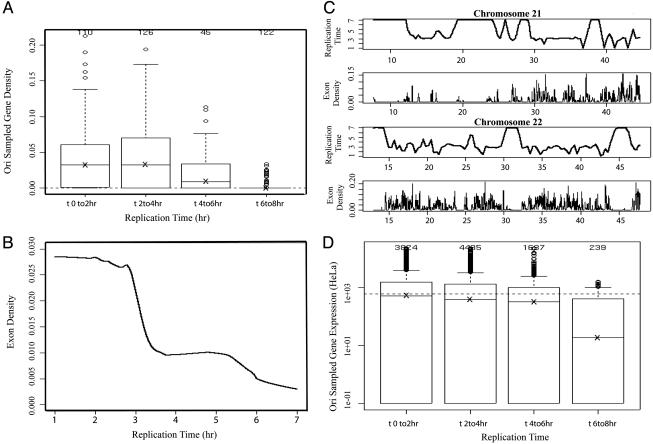
Fig. 5.
Correlation between gene density and replication time. (A) Box plots showing the distribution of exon densities in the segments replicated in each quartile of S phase. (B) Smoothed TR50 profile (using lowess algorithm with f = 0.01 fraction of the data included in the running local fit) of chromosomes 21 and 22 (with the position of each chromosome shown on the x axis in Mb) compared with the exon density profile. The lowest point in each valley indicates a site that is replicated before its adjoining segments and so likely to contain origins of replication. (C) Lowess smoothed profile of exon density (y axis) versus TR50 (x axis) scatter plot (with f = 0.3) indicates an acute decrease in gene density for segments replicated after ≈3.1 hr of S phase. (D) Box plots showing the distribution of cytoplasmic RNA levels (Methods) on a log scale in the segments replicated in each quartile of S phase. The dashed line corresponds to a cutoff above which exons are classified as expressed.
Time of 50% Cumulative Replication (TR50) Profile of Chromosomal Segments. The replication time at each interrogated position on the array was estimated more precisely by calculating the in crease in the signal for each of the four time periods and then calculating the time that corresponds to 50% cumulative replication, TR50 (Supporting Methods). Plotting the TR50 against the distance along the chromosome produces a temporal profile of replication that identifies sites that are replicated earlier than their adjoining segments (Fig. 5B). These sites will be useful places to search for origins of replication. Plotting of the exon density and TR50 shows a striking anticorrelation between exon density and replication timing on a chomosomal scale (Fig. 5B). The Spearman correlation coefficient between the two profiles was -0.34 and -0.58 for chromosomes 21 and 22, respectively (Supporting Methods). The profile also suggests that centromeric regions replicate late in S phase (Fig. 9, which is published as supporting information on the PNAS web site), consistent with earlier cytogenetic studies and with the paucity of genes in these areas. Although a clear transition from low to high A + T content is evident for regions that replicate early versus late in S phase (Fig. 10, which is published as supporting information on the PNAS web site), this correlation could be explained by the association of high exon density with early replicating regions and the fact that exon-dense regions tend to have low A + T content compared with the genome average.
At Least Two Discrete Chromatin Environments Could Account for the Replication Times. High exon density could correlate with a more open chromatin structure. Assuming there are a finite number of chromatin structures on a genomic scale (e.g., euchromatin and heterochromatin), if chromatin structure drives replication time, the exon density versus replication time profile might be expected to assume a staircase profile where relatively open chromatin with high exon density is replicated first followed by sharp decreases (or steps) in exon density for every subsequent state of chromatin. Indeed, we find a relatively strong staircase-like profile in the running average of exon density versus replication time scatter plot (Fig. 5C). For regions replicated in 0-2.8 hr, the average exon density is flat around 0.028. For regions replicated at 2.8-3.5 hr, the exon density drops 64% to 0.01 where it remains flat until we reach regions replicated about the fifth hour of replication. This transition in exon density occurs across regions that differ in replication time by ≈40 min and is very sharp given cell synchonization errors that are on the order of tens of minutes. The inflexion point (i.e., where the second derivative in the averaged exon density versus replication timing profile is zero) in the transition occurs at ≈3.1 hr at an averaged exon density of 0.0174. This time likely corresponds to a first wave of replication over the most accessible states of chromatin and not necessarily all of euchromatin. Regions of much more compact chromatin are likely beginning to be replicated after 3.1 hr.
Early Replication Corresponds to Higher Gene Expression. Given that we expect gene expression to be suppressed in heterochromatin relative to euchromatin, we further tested the hypothesis that replication timing is determined by chromatin structure by comparing the gene expression levels against replication times for the efficient origins (Supporting Methods). The trend is clearly toward decreasing exon expression level with increasing replication time (Fig. 5D). The median exon expression level (in arbitrary intensity units) for the regions replicated in the first quarter of S phase is 515, which is very close to the cutoff that defines expressed exons. The median expression levels in loci replicating in the remaining three quarters of S phase are 388, 315, and 18. The drop in expression levels from loci replicated in the first two quarters of S phase to the last is significant (P < 10-15).
Conclusions
DNA microarrays are being developed into a powerful tool for genome-wide assay of patterns of DNA replication in yeasts and Drosophila (17, 18). Experiments on cosmid-based genomic arrays to assay DNA abundance of human cells sorted into G1 and S by flow cytometry also find that high gene density is correlated with early replication (24). Here, we used cDNA arrays to demonstrate that the finer timed replication pools from synchronized cells give valid results regarding time of replication of genes. The pools were then applied to very high-density genome-tiling DNA arrays. Forty percent of the genomic sequences in chromosomes 21 and 22, as well as the genes distributed throughout the genome, replicate reproducibly in a precise part of S phase. Even with a higher stringency cutoff (P < 10-5), the results identify at least 400 segments where we can fruitfully search for ideal origins of replication. In addition, we identify hundreds of early and late replicating segments that can be studied molecularly to elucidate the mechanisms that control time of origin firing in human cells. The time of replication in S phase of these segments is correlated with gene density in a manner that suggests that two to three discrete chromatin environments contribute to the time of DNA replication.
A surprise from the study is the discovery that 60% of the interrogated genomic segments with a good replication signal replicate through the four quarters of S phase without any enrichment in one particular time period. Replication at several origins has been reported to initiate from delocalized initiation zones controlled by redundant replicator elements (25-28), and the initiator protein, human origin recognition complex (ORC), does not have detectable sequence specificity (29). In view of this finding, the simplest explanation for the pan-S phase pattern of replication is that large stretches of chromosomes in HeLa cells are replicated by distributed and inefficient origins (or replication forks) that fire (or move) variably in space and time during S phase. Although some of this pan-S phase replication might be due to aneuploidy, malignant transformation, mono-allelic gene expression, and imprinting, our results suggest significant flexibility in origin selection and firing and replication fork movement in mammalian cancer cells. It will be worthwhile to examine whether the pan-S phase pattern of replication is as widespread in diploid cells synchronized by other methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 CA60499 and UO1 HG003157 (to A.D.) and an award from the Brain Korea program of the Korean Ministry of Education (to Y.J. to work with A.D.). D.S.H. and Y.J. were partly supported by Systems Biology, Ministry of Science and Technology, Republic of Korea. This project was also funded in part by Federal funds from the National Cancer Institute, National Institutes of Health, under Contract N01-CO-12400, and by Affymetrix, Inc. (to T.R.G.).
Author contributions: Y.J., N.K., T.R.G., and A.D. designed research; Y.J., N.K., P.K., C.L., and D.S.H. performed research; Y.J., S.B., N.K., S.G., D.M., and A.D. analyzed data; S.B., S.G., and T.R.G. contributed new reagents/analytic tools; and S.B., N.K., and A.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: H/L, heavy/light; TR50, time of 50% cumulative replication.
References
- 1.Huberman, J. A. & Riggs, A. D. (1968) J. Mol. Biol. 32, 327-341. [DOI] [PubMed] [Google Scholar]
- 2.Abdurashidova, G., Deganuto, M., Klima, R., Riva, S., Biamonti, G., Giacca, M. & Falaschi, A. (2000) Science 287, 2023-2026. [DOI] [PubMed] [Google Scholar]
- 3.Dijkwel, P. A. & Hamlin, J. L. (1995) Mol. Cell. Biol. 15, 3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dijkwel, P. A., Mesner, L. D., Levenson, V. V., d'Anna, J. & Hamlin, J. L. (2000) Exp. Cell Res. 256, 150-157. [DOI] [PubMed] [Google Scholar]
- 5.Dijkwel, P. A., Wang, S. & Hamlin, J. L. (2002) Mol. Cell. Biol. 22, 3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waltz, S. E., Trivedi, A. A. & Leffak, M. (1996) Nucleic Acids Res. 24, 1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, J., Ermakova, O. V., Riblet, R., Birshtein, B. K. & Schildkraut, C. L. (2002) Mol. Cell. Biol. 22, 4876-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePamphilis, M. L. (1999) BioEssays 21, 5-16. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis, M. L. (2003) Cell 114, 274-275. [DOI] [PubMed] [Google Scholar]
- 10.Anglana, M., Apiou, F., Bensimon, A. & Debatisse, M. (2003) Cell 114, 385-394. [DOI] [PubMed] [Google Scholar]
- 11.Goren, A. & Cedar, H. (2003) Nat. Rev. Mol. Cell Biol. 4, 25-32. [DOI] [PubMed] [Google Scholar]
- 12.Hand, R. (1978) Cell 15, 317-325. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll, R. M. & Fangman, W. L. (1988) Cell 54, 505-513. [DOI] [PubMed] [Google Scholar]
- 14.Ofir, R., Wong, A. C., McDermid, H. E., Skorecki, K. L. & Selig, S. (1999) Proc. Natl. Acad. Sci. USA 96, 11434-11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, D. M. (2002) Curr. Opin. Cell Biol. 14, 377-383. [DOI] [PubMed] [Google Scholar]
- 16.Simon, I., Tenzen, T., Mostoslavsky, R., Fibach, E., Lande, L., Milot, E., Gribnau, J., Grosveld, F., Fraser, P. & Cedar, H. (2001) EMBO J. 20, 6150-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- 18.Schubeler, D., Scalzo, D., Kooperberg, C., van Steensel, B., Delrow, J. & Groudine, M. (2002) Nat. Genet. 32, 438-442. [DOI] [PubMed] [Google Scholar]
- 19.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185-193. [DOI] [PubMed] [Google Scholar]
- 20.Hollander, M. & Wolfe, D. (1999) Nonparametric Statistical Methods (Wiley, New York).
- 21.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916-919. [DOI] [PubMed] [Google Scholar]
- 22.Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A. J., et al. (2004) Cell 116, 499-509. [DOI] [PubMed] [Google Scholar]
- 23.Simon, I., Tenzen, T., Reubinoff, B. E., Hillman, D., McCarrey, J. R. & Cedar, H. (1999) Nature 401, 929-932. [DOI] [PubMed] [Google Scholar]
- 24.Woodfine, K., Fiegler, H., Beare, D. M., Collins, J. E., McCann, O. T., Young, B. D., Debernardi, S., Mott, R., Dunham, I. & Carter, N. P. (2004) Hum. Mol. Genet. 13, 191-202. [DOI] [PubMed] [Google Scholar]
- 25.Mesner, L. D., Li, X., Dijkwel, P. A. & Hamlin, J. L. (2003) Mol. Cell. Biol. 23, 804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalejta, R. F., Li, X., Mesner, L. D., Dijkwel, P. A., Lin, H. B. & Hamlin, J. L. (1998) Mol. Cell 2, 797-806. [DOI] [PubMed] [Google Scholar]
- 27.Altman, A. L. & Fanning, E. (2004) Mol. Cell. Biol. 24, 4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman, A. L. & Fanning, E. (2001) Mol. Cell. Biol. 21, 1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vashee, S., Cvetic, C., Lu, W., Simancek, P., Kelly, T. J. & Walter, J. C. (2003) Genes Dev. 17, 1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.