Abstract
UDP-glucuronosyltransferase (UGT) isozymes catalyze detoxification of numerous chemical toxins present in our daily diet and environment by conjugation to glucuronic acid. The special properties and enzymatic mechanism(s) that enable endoplasmic reticulum-bound UGT isozymes to convert innumerable structurally diverse lipophiles to excretable glucuronides are unknown. Inhibition of cellular UGT1A7 and UGT1A10 activities and of [33P]orthophosphate incorporation into immunoprecipitable proteins after exposure to curcumin or calphostin-C indicated that the isozymes are phosphorylated. Furthermore, inhibition of UGT phosphorylation and activity by treatment with PKCε-specific inhibitor peptide supported PKC involvement. Coimmunoprecipitation, colocalization by means of immunofluorescence, and cross-linking studies of PKCε and UGT1A7His revealed that the proteins reside within 11.4 Å of each other. Moreover, mutation of three PKC sites in each UGT isozyme demonstrated that T73A/G and T202A/G caused null activity, whereas S432G-UGT1A7 caused a major shift of its pH-8.5 optimum to 6.4 with new substrate selections, including 17β-estradiol. S432G-UGT1A10 exhibited a minor pH shift without substrate alterations. PKCε involvement was confirmed by the demonstration that PKCε overexpression enhanced activity of UGT1A7 but not of its S432 mutant and the conversion of 17β-[14C]estradiol by S432G-UGT1A7 but not by UGT1A7. Consistent with these observations, treatment of UGT1A7-transfected cells with PKCε-specific inhibitor peptide or general PKC inhibitors increased 17β-estradiol catalysis between 5- and 11-fold, with parallel decreases in phosphoserine-432. Here, we report a mechanism involving PKC-mediated phosphorylation of UGT such that phosphoserine/threonine regulates substrate specificity in response to chemical exposures, which possibly confers survival benefit.
Keywords: PKC, PKC inhibitors, detoxifying enzymes, UDP-glucuronosyltransferase mutants, immunofluorescence
Mammalian UDP-glucuronosyltransferase (UGT) isozymes detoxify numerous lipid-soluble chemicals, including metabolites, such as neurotoxic bilirubin, as well as dietary and environmental agents/toxins and medicines (1). UGT facilitates detoxification of chemicals by attaching glucuronic acid molecules to form water-soluble, readily excretable glucuronides. Failure to attach glucuronic acid (glucuronidation) to toxins allows high levels to accumulate in tissues, causing deleterious effects, including mutations of genes and the development of cancer (2). Mutations in UGT1 genes lead to lethal hyperbilirubinemic Crigler-Najjar disease (3) and to toxic tissue levels of the commonly used analgesic acetaminophen (Tylenol) in rats (4). The fact that a limited number of UGT isozymes facilitate excretion of a vast number of structurally diverse chemicals suggests a mechanism evolved to confer flexibility on an isozyme to metabolize multiple toxins. Because UGTs are bound to membranes of the endoplasmic reticulum (ER) that cause difficulties associated with purification and structural characterization, the molecular mechanism controlling detoxification has, until now, remained unresolved. After the rapid down-regulation of UGT activity in human colon cells treated with the common condiment curcumin and calphostin-C (5, 6), known as a general kinase (7) and PKC inhibitor (8), respectively, we sought to determine whether: (i) PKC-mediated phosphorylation of UGT is demonstrable in these cells and (ii) this posttranslational modification affects the catalytic activities of these isozymes. Here, we report a mechanism involving PKC-mediated phosphorylation of UGT such that phosphoserine/threonine regulates substrate selection in response to chemical exposures.
Methods
Cell Cultures. LS180 colon and COS-1 (American Type Culture Collection) cells were grown in DMEM with 10% and 4% FCS, respectively. LS180 cells have significant constitutive levels of UGT substrate activities, and COS-1 cells have significant ER but are essentially UGT-free. Because of sequence similarities but differences in biochemical properties (5), we selected UGT1A7 and UGT1A10 for comparisons. Transfection and transduction efficiencies are given in Supporting Text, which is published as supporting information on the PNAS web site.
In Vitro Glucuronidation Using Different Acceptor Substrates. The glucuronidation assay, described in refs. 6 and 9, used 100 μg of cellular protein. Buffers for the pH curves and product processing and quantitation are described in refs. 9 and 10. Reaction mixtures were incubated for 2-4 h at 37°C (5).
Western Blot Analysis of UGT-Transfected COS-1 Cells. All transfections used pSVL-based UGTcDNAs in COS-1 cells, which were incubated for 72 h; cells were incubated for 60 h for UGT radiolabeling and the PKCε-overexpression study. Cells were untreated or treated with curcumin or calphostin-C; equal cellular protein was resolved in SDS-10%/PAGE and Western-blotted with anti-UGT1 (6). All experiments showing PKCε protein, except the cross-linking experiment, were immunocomplexed with anti-UGT (common end) and trapped with protein-A-Sepharose (coimmunoprecipitation) before Western blotting with different antibodies.
To determine the phosphorylation status of position 432 in UGT1A7, UGT1A10, and their mutants, microsomes were prepared from transfected COS-1, solubilized, and immunocomplexed with anti-UGT (6). Duplicate samples were resolved by SDS/12% PAGE and Western-blotted with anti-UGT1 and anti-phosphoserine (4A9, Calbiochem). Membranes were blocked with 5% BSA/0.1% Tween 20 in 25 mM Tris/137 mM NaCl, pH 7.5 (TBS), washed in TBS-Tween 20, and exposed to antibody-horseradish peroxidase conjugate for visualization (6).
After treatment of UGT1A7-transfected cells with PKCε-specific inhibitor peptide or general PKC inhibitors, cells were cross-linked with 11.4-Å spacer-arm disuccinimidyl suberate (DSS) (Pierce), according to the manufacturer's protocol. Cells were solubilized and resolved in 4-15% gradient SDS/PAGE before Western blotting as described above.
In Cellulo Labeling of UGTs with [33P]Orthophosphate. Sixty hours after transfection with UGT1A7, UGT1A10, or their triple PKC-sites mutants, cells were conditioned (6) before exposure to 5.0 mCi (1 Ci = 37 GBq) of [33P]orthophosphate per ml of medium for 8 h with or without calphostin-C in the last hour. Equal solubilized cellular protein (11, 12) was immunocomplexed with anti-UGT to make duplicate gels, processed, and resolved in SDS/PAGE. One gel was Western-blotted with anti-UGT (12); the other was fixed and exposed to x-ray film (6). Parallel unlabeled cultures were analyzed for glucuronidation.
Inhibition of UGT in LS180 and UGT-Transfected COS-1 Cells Treated with PKCε-Specific Translocation-Inhibitor Peptide. Confluent LS180 or UGT1A7-transfected cells were treated with Antennapedia-conjugated PKCε-specific peptide (KAI Pharmaceuticals, South San Francisco, CA) derived from the unique region V1 of PKCε, its scrambled control, Antennapedia carrier (13), or select PKC inhibitors. Before treatment, cells were serum- and phosphate-starved for 16 h. Glucuronidation was determined with eugenol and capsaicin (data not shown) or 17β-estradiol.
Colocalization of UGT1A7 and PKCε. To determine whether UGT1A7 and PKCε colocalize, COS-1 cells grown on Lab-Tek slides (Nalge) were transfected (14) with pUGT1A7cDNA fused at the 3′ end with His-Tag derived from pcDNA3.1 vector (Invitrogen), generating pUGT1A7His. Cells were processed for immunofluorescence (15) and exposed to primary antibody: mouse anti-His-Tag (Amersham Pharmacia) and either rabbit anti-PKCε or rabbit anti-phospho(Ser-729)PKCε (Upstate Biotechnology, Lake Placid, NY). Controls were not treated with primary antibody. UGT1A7His and PKCε were visualized with donkey anti-mouse-FITC conjugate or donkey anti-rabbit-rhodamine conjugate (The Jackson Laboratory), respectively, by using a photometric Zeiss microscope; images were assembled by computer. All cells were exposed to DAPI fluorescein to identify nuclei.
Mutagenesis of PKC Sites in UGT1A7 and UGT1A10. Site-directed mutagenesis at amino acid 73, 202, or 432 in UGT1A7 and UGT1A10 was carried out as described in refs. 6 and 9. Oligonucleotides to create S432G were identical to those for S435G in UGT1A1 at common residue serine-432/435 (6). Mutations at serine-432 in UGT1A7 and UGT1A10 replaced acc in the antisense (463C) primer and ggt in the sense (435C) primer.
For T202A in UGT1A7, the primers were as follows: sense, 202A7S (5-gacgccatggctttcaaggagagagtacgg-3); antisense, 202A7S (complement). For T73A mutation in UGT1A7, primers were as follows: sense, 73A7S (5-tgcgcagtgaagacttactcaacctca-3); antisense, 73A7S (complement). Primers for the T202A and T73A mutations in UGT1A10 are reported in ref. 12.
In Cellulo Glucuronidation of [14C]17β-Estradiol by UGT1A7-Transfected COS-1 Cells. Seventy-two hours after transfection with UGT1A7, UGT1A10, or a variant (T202A or S432G), cells were conditioned in DMEM with 4% charcoal-stripped FBS for 24 h before adding 40 μM [14C]17β-estradiol (2.5 μCi/40 μM). Triplicate 500-μl samples of medium were collected at 3 h and diluted 1:2 with 100% ethanol. Supernatants were recovered after centrifugation, vacuum-dried, and solubilized in PBS (pH 6.8); one-half were treated with 50 units of β-glucuronidase for 2 h at 37°C or with buffer and processed for quantitation (10, 19). Duplicate unlabeled cultures were Western-blotted as described above.
Results and Discussion
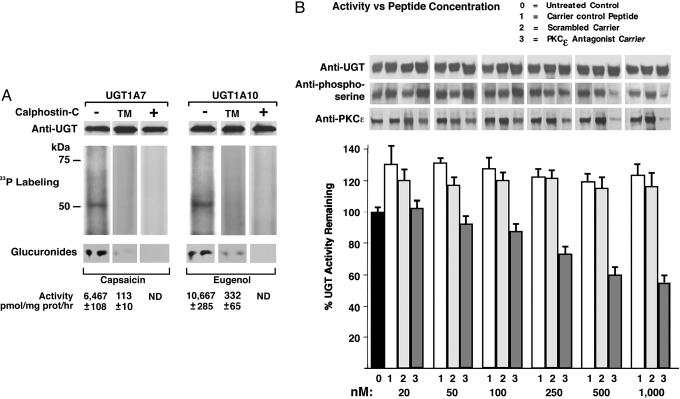
UGT Inhibition by Curcumin and Calphostin-C and [33P]Orthophosphate Incorporation. While studying the effects of different UGT substrates on glucuronidating activity in LS180 or HT29 colon cells, we uncovered the fact that exposure to curcumin transiently inhibited activity measured in vitro (6, 12). Moreover, preliminary data revealed that 10 recombinant UGTs were also inhibited by curcumin and/or the PKC inhibitor calphostin-C. Hence, we studied the inhibition in detail using UGT1A7 and UGT1A10. Calphostin-C treatment of cells transfected with wild-type UGT or its triple PKC-sites mutant showed both inhibition of incorporation of [33P]orthophosphate into immunoprecipitable UGT1 (12) resolved in SDS/PAGE and UGT activity (Fig. 1A). This result suggests that the active state of UGT depends on PKC-mediated phosphorylation of the protein.
Fig. 1.
Radiolabeling and inhibition of UGTs. (A)[33P]Orthophosphate labeling of UGT1A7 and UGT1A10 in COS-1 cells and their triple-PKC sites mutants (TM) and after calphostin-C treatment. Eight-hour labeling of UGT-transfected cells was with or without 1.0 μM calphostin-C in the final hour. ND, not detectable. (B) Effect of PKCε-specific inhibitor peptide on UGT in LS180 colon cells. After 3-h exposure to PKCε-specific inhibitor peptide, scrambled peptide, or Antennapedia carrier control, cell homogenates were assayed for eugenol glucuronidation (pH 8.0). Western blot analysis is described in Methods.
Effect of PKCε-Specific Translocation-Inhibitor Peptide on UGT in LS180 cells. To establish whether UGT modification is PKCε-specific, we determined UGT and PKCε content trapped by UGT immunocomplexes, UGT phosphorylation status, and glucuronidating activity after treatment of LS180 cells with PKCε-specific translocation-inhibitor peptide. Translocation is mediated by a receptor for activated C-kinaseε (β-COP) by means of attachment to the unique segment-V1 region of the PKC isozyme (13, 16); V1 region-specific peptide prevents the targeted PKC from anchoring near its substrates. Despite 20-30% activation by Antennapedia carrier or scrambled amino acid peptide control, the PKCε-specific peptide caused concentration-dependent inhibition of glucuronidation (pH 8.0) that culminated at 50% (Fig. 1B). Furthermore, UGT phosphorylation and PKCε content decreased in parallel with treatment without affecting UGT content (Fig. 1B). Also, PKCα- and PKCβ-specific peptides inhibited glucuronidation, but higher concentrations were required than used for the PKCε peptide (data not shown). In separate in vitro experiments, we demonstrated that T73, T202, and S432 in UGT1A7 and UGT1A10 are PKC sites through phosphorylation of the corresponding peptides by PKCα, PKCβII, and PKCε, but not their respective alanine mutants (Fig. 6A, which is published as supporting information on the PNAS web site). Taken together, these results strongly indicate that PKC isozymes phosphorylate UGTs.
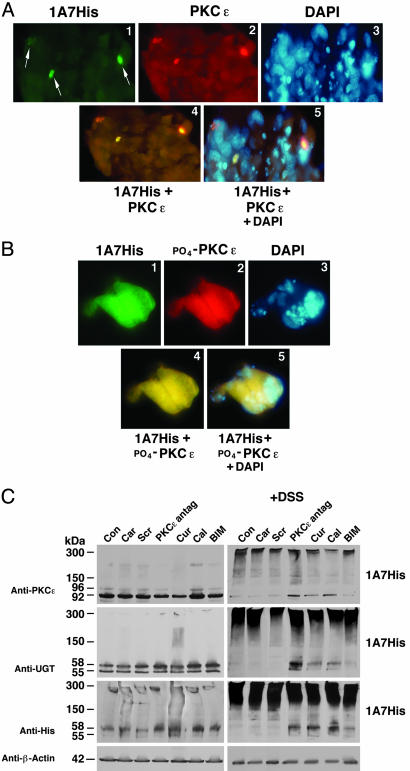
Colocalization of UGT1A7 and PKCε. Although we have strong evidence that PKCs are involved in supporting UGT activity, the kinases are not commonly known to associate with the ER where UGTs reside. Therefore, we carried out PKCε and UGT colocalization studies in UGT1A7His-transfected cells by using immunofluorescence from anti-His-UGT1A7-FITC-conjugate and anti-PKCε-rhodamine-conjugate (Fig. 2A) that generated a yellow fused image (Fig. 2A, images 1, 2, and 4), indicating that the two proteins colocalize. Furthermore, yellow fused immunofluorescence assembled from anti-His-UGT1A7-FITC-conjugate and anti-phospho-(serine-729)-PKCε-rhodamine-conjugate confirmed that activated PKCε (17) colocalizes with UGT (Fig. 2B, images 1, 2, and 4) outside the nucleus, according to DAPI fluorescence (Fig. 2 A and B, images 3 and 5). PKCα and its phospho version also show colocalization with UGT1A7 (data not shown). Inasmuch as UGTs are ER-bound, the results indicate that activated PKC also attaches to this organelle, which is consistent with their translocation to cellular structures (16).
Fig. 2.
Immunofluorescence and cross-linking of PKCε and UGT1A7. (A) Colocalization of UGT1A7His and PKCε in COS-1 cells. Colocalization was processed 72 h after transfection. (B) Colocalization of UGT1A7His and phospho(Ser-729)PKCε in COS-1 cells. Colocalization was processed as described in Methods. Omission of primary antibody prevented secondary antibody-conjugate binding (not shown). Images are magnified ×40. (C) Cross-linking UGT1A7His and PKCε. UGT1A7His-transfected COS-1 cells were untreated or treated with curcumin (200 μM for 1 h), calphostin-C (500 nM for 1 h), BIM (5 μM for 1 h), or Antennapedia-conjugated PKCε-specific antagonist peptide (500 nM for 3 h). Finally, cells were treated for 30 min with a cross-linker, 1.25 mM DSS. “Car” and “Scr” denote carrier and scrambled peptide controls, respectively. Western blot analysis is described in Methods.
Cross-Linking of UGT1A7 and PKCε. To assess whether UGT1A7 and PKCε reside on the same side of the ER membrane and whether PKCε-specific peptide prevents PKCε location proximal to UGT, we attempted to cross-link the proteins with the 11.4-Å linker arm, DSS, in UGT1A7-transfected cells in the absence and presence of inhibitors. Whereas proteins exhibited typical migration in SDS/PAGE when cells lacked DSS exposure before Western blotting with anti-UGT and anti-PKCε (Fig. 2C Left), complexes were formed that ranged from 150 to 300 kDa after DSS exposure (Fig. 2C Right). Moreover, pretreatment with PKCε-specific translocation-inhibitor peptide (13), the general kinase inhibitor curcumin (7), PKC-specific inhibitor calphostin-C (8), or general PKC inhibitor bisindolylmaleimide (BIM) (18) prevented total complexing of the proteins (Fig. 2C Right). β-Actin was not affected by treatment. Carrier and scrambled peptide represent controls. Given DSS linker arm length and competition by PKCε-specific inhibitor, the results indicate that PKCε and UGT are in close proximity, most likely on the luminal side of ER.
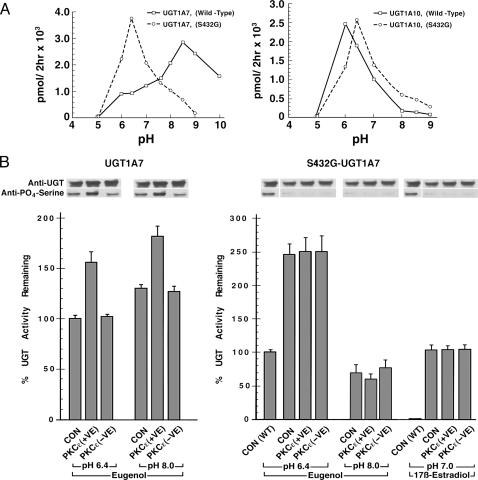
Effects of T73A, T202A, and S432G Mutations on UGT1A7 and UGT1A10 Activity. Consistent with our recent demonstration of substrate-dependent pH optima for UGT isozymes that range from 5.5 to 8.5 (5), we inquired whether this flexibility can be correlated with the phosphorylation pattern of an isozyme. This low in vitro pH range for certain UGTs is consistent with that for unrelated but ER-bound enzymes (19, 20). Comparisons of catalytic activity at multiple pH values for wild type and T73, T202, and S432 mutants of UGT1A7 and UGT1A10 proved to be highly informative. Whereas T73A/G and T202A/G mutants of UGT1A7 and UGT1A10 showed no activity (Fig. 6B), the S432G mutant at the single serine PKC site in UGT1A7 and UGT1A10 retained full activity. pH studies showed a minor shift for S432G-UGT1A10 from 6.0 to 6.4 (Fig. 3A Right), whereas there was a dramatic shift for S432G-UGT1A7 from pH 8.5 to 6.4 (Fig. 3A Left) to resemble UGT1A10. (See Fig. 7, which is published as supporting information on the PNAS web site, for clarity of mutation sites.) To our knowledge, this finding provides the first evidence that phospho-group(s) in UGT operationally control pH of catalysis and that phosphoserine-432 in UGT1A7 controls its alkaline pH activity. Mutations at PKA phosphorylation sites in mouse CYP2E1 have also shown changes in substrate preference (21).
Fig. 3.
pH studies and effects of PKCε overexpression on UGTs. (A) Effect of mutation of the serine-432 PKC site in UGT1A7 and UGT1A10. UGT isozymes were assayed with eugenol. SEs for three determinations are ±1-5%. (B) Effect of overexpression of PKCε on UGT1A7 or S432G-UGT1A7-transfected COS-1 cells. Transfected cells were exposed to positive PKCε (+ve)-GFP-cDNA-Semliki Forest viral transductant (22) or dominant-negative PKCε (-ve)-GFP (23) transductant for 12 h. Two μM phorbol-12-myristate-13-acetate was added 15 min before harvest; activity was measured with eugenol and 17β-estradiol. UGT1A7 activity at pH 6.4, 7.0, and 8.0 for 17β-estradiol was ±2% of S432G-UGT1A7 activity at pH 7.0. Western blot analysis is described in Methods.
Effect of PKCε Overexpression on UGT. Because UGT inhibition by translocation-inhibitor peptide (Fig. 1B) and colocalization and cross-linking studies (Fig. 2) indicated that PKCε supports UGT activity, we compared PKCε overexpression on UGT1A7 and S432G-UGT1A7 levels, their phosphorylation at position 432, and corresponding activities in transfected COS-1 after infection with PKCε (+ve)- or PKCε (-ve)-GFPcDNA viral construct (22). PKCε (+ve) caused a 30-50% increase in UGT1A7 activity at different pH values (Fig. 3B Left), whereas it had no effect on S432G-UGT1A7 (Fig. 3B Right). Dominant-negative PKCε (-ve) with a defective ATP binding site (23) had no effect on either version of UGT1A7. Results indicate that PKCε phosphorylates only serine-432 in UGT1A7 and that this phosphoserine supports eugenol conversion primarily at alkaline pH (Fig. 3A Left). Moreover, specific PKC isozyme(s) appear to phosphorylate select PKC sites in UGT.
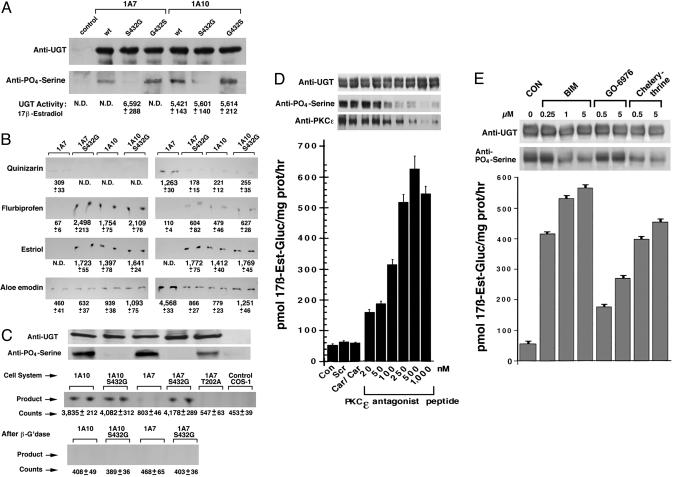
Phosphorylation Status of the Serine-Specific PKC Site and Associated Substrate Activities of UGT1A7 and UGT1A10. To delineate further how the single phosphoserine, position 432, affects activity, we compared the phosphorylation status of S432G in both UGT1A7 and UGT1A10 expressed in COS-1 cells with their substrate selections. Aided by a Western blot with anti-UGT1 that showed equal UGT proteins (Fig. 4A Upper), its duplicate blot with anti-phosphoserine showed that both wild types and revertants (lanes 2, 4, 5, and 7) were phosphorylated, whereas there was no detectable phosphate in either S432G mutant (Fig. 4A, lanes 3 and 6). Unexpectedly, S432G-UGT1A7, but not UGT1A7, metabolized new substrates, 17β-estradiol (pH 7.0) (Fig. 4A, row 3) (12), flurbiprofen, and estriol (pH 6.4 and 7.6) (Fig. 4B); in addition, S432G-UGT1A7 failed to convert quinizarin and expressed reduced activity toward aloe-emodin, similar to results for UGT1A10 and S432G-UGT1A10 mutant (Fig. 4B) (5, 12). The fact that serine-432 in UGT1A7 and UGT1A10 (Fig. 4A) was phosphorylated, and S432G-UGT1A7 was dephosphorylated but acquired new substrates at multiple pH values, supports the conclusion that the phospho-pattern operationally controls pH of catalysis and substrate selection.
Fig. 4.
Altered substrates of UGT1A7 after dephosphorylation. (A) Phosphorylation status of serine-432 in UGT1A7, UGT1A10, and variants versus substrate selections. Transfected COS-1 cell microsomes were processed for Western blot by using anti-UGT or anti-phosphoserine. 17β-Estradiol-glucuronides are shown. (B) Autoradiograms of TLC-resolved [14C]glucuronides of other substrates. Glucuronides of 17β-estradiol and estriol represent pmol/mg of protein per hr; other glucuronides represent pmol/mg of protein per 4 h. ND, not detectable. (C) In cellulo glucuronidation of 17β-estradiol. After 3-hr exposure to 17β-[14C]estradiol, medium from transfected or control cells was analyzed for 17β-[14C]estradiol-glucuronide before and after β-glucuronidase (β-G'dase) treatment; unlabeled cultures were used for Western blot as described in Methods after resolving in a 12% gel. (D and E) Elicitation of 17β-estradiol glucuronidation by treatment of UGT1A7-transfected COS-1 cells with PKCε-specific inhibitor peptide (D) and general PKC inhibitors (E). Cells were exposed to PKCε-specific inhibitor for 3 h and general PKC inhibitors for 45 min. Solubilized samples were immunocomplexed with anti-UGT before Western blotting as described in Methods. Con, control; Scr, scrambled; Car, carrier.
Conversion of 17β-Estradiol in Cellulo. To determine whether S432G-UGT1A7 also converts 17β-estradiol in cellulo, we added 17β-[14C]estradiol to COS-1 control and to those cells expressing UGT1A7, UGT1A10 wild type, or a variant and compared 17β-[14C]estradiol-glucuronide production in the medium. Aided by the demonstration of equal specific protein in parallel cultures (Fig. 4C Top) and appropriately dephosphorylated S432 mutants, we found that S432G-UGT1A7, UGT1A10, and S432G-UGT1A10 generated >3,400 net counts of putative 17β-[14C]estradiol-glucuronide compared with 350 or fewer counts by UGT1A7 and T202A-UGT1A7 (Fig. 4C Middle). Sensitivity of the radiolabeled product to β-glucuronidase treatment confirmed that it was a glucuronide (Fig. 4C Bottom) and established that S432G-UGT1A7, but not UGT1A7, converts 17β-estradiol under in vitro and in cellulo conditions, similar to UGT1A10 and its mutant.
Conversion of 17β-Estradiol by UGT1A7 After Treatment of Transfected Cells with PKCε-Specific or General PKC Inhibitors. Although we confirmed that PKCε overexpression positively affected only phosphoserine-432 in UGT1A7 and that S432G-UGT1A7 acquired new activity toward 17β-estradiol and other chemicals, a remaining question is whether treatment of UGT1A7-expressing cells with PKCε-specific inhibitor to prevent serine-432 phosphorylation would support 17β-estradiol glucuronidation. To that end, treatment of UGT1A7-transfected cells with PKCε-specific inhibitor peptide did not affect UGT level (Fig. 4D Upper, row 1), but it did cause a concentration-dependent decrease in its phosphoserine-432 and in PKCε content (Fig. 4D Upper, rows 2 and 3) in parallel with an 11-fold increase in 17β-estradiol-glucuronide production. Earlier, we showed that increasing levels of this same inhibitor peptide caused a progressive inhibition of eugenol conversion at pH 8.0 (Fig. 1B) after treatment of LS180 cells; now we find that treatment of UGT1A7-transfected cells increases 17β-estradiol conversion at pH 7.0 (Fig. 4D), an event associated with the pH shift (Fig. 3A Left). Also, treatment of UGT1A7-transfected cells with increasing concentrations of general PKC inhibitors BIM or chelerythrine caused between 11- and 9-fold increases in 17β-estradiol-glucuronide production, respectively (Fig. 4E Lower), in parallel with decreases in phosphoserine-432 without affecting UGT levels (Fig. 4E Upper). Despite modest decreases in phosphoserine-432 after treatment with classical PKC inhibitor Gö-6976 (Fig. 4E), there was a remarkable 3- to 5-fold increase in 17β-estradiol-glucuronide. Consistent with changes in substrate selections after treatment of UGT1A7 with BIM, chelerythrine, and Gö-6976 (Fig. 4E), eugenol metabolism at pH 8.0 versus 6.4 showed parallel decreases and increases, respectively (Fig. 8, which is published as supporting information on the PNAS web site). This prominent increase in 17β-estradiol glucuronidation that progressed in parallel with serine-432 dephosphorylation, which is analogous to S432G-UGT1A7 and S432A-UGT1A7 (Fig. 3A) mutants, demonstrates that inhibition of select PKC isozymes can alter UGT substrate specificity and that alternative phosphorylation and dephosphorylation of serine-432 expands substrate selections as depicted in Fig. 5. Moreover, the capacity to trap PKCε in immunocomplexes by anti-UGT, combined with colocalization and cross-linking studies, signifies that the two proteins are in close proximity or in a complex (Figs. 1B, 2, and 4D). This finding is also consistent with a demonstration that PKC isoforms phosphorylate essential components of the transport machinery of rough ER (24).
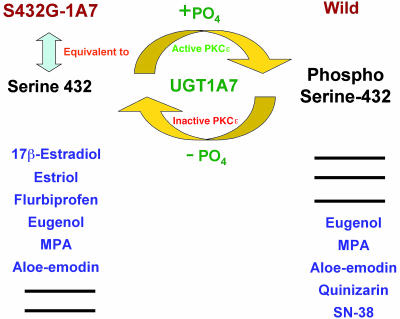
Fig. 5.
Substrate variation associated with phosphorylation and dephosphorylation of serine-432 in UGT1A7. Phosphorylation of serine-432 in wild-type UGT1A7 was shown to be mediated by constitutive (active) PKCε, whereas dephosphorylation of serine-432 was associated with S432G/A or treatment of UGT1A7-transfected cells with PKCε-specific translocation-inhibitor peptide or general PKC inhibitors. For UGT1A7, Sn-38 metabolism mimics quinizarin (27), and mycophenolic acid (MPA) mimics eugenol (11). Hence, phosphoserine-432-UGT1A7 and serine-432-UGT1A7 catalyzed the substrate sets as shown.
We have demonstrated that UGT1A7 and UGT1A10 require phosphorylation, which controls aspects of catalysis. Moreover, the sensitivity of 10 UGTs to curcumin and/or calphostin-C suggests that phosphorylation is likely a universal requirement for UGTs. Whereas null activity for T73A/G and T202A/G mutants in UGT1A7 and UGT1A10 indicates that those phospho-modifications are strictly required for unknown reasons, the emergence of new activity at multiple pH values for S432G-UGT1A7 indicates that phosphoserine in UGT1A7 controls catalysis and substrate selection. Moreover, in cellulo glucuronidation of 17β-[14C]estradiol by S432G-UGT1A7, but not UGT1A7, indicates that the mutant is “competent” to function under physiological conditions.
The question arises as to the prevalence of altered substrate selections associated with mutants of serine-434 ± 2 in different UGT1 isozymes. Whereas S432G-UGT1A7 exhibited a dramatic pH shift to resemble UGT1A10 and, thus, new substrate selections that resemble UGT1A10, we have preliminary evidence that S434G/A-UGT1A6, S432G-UGT1A7, and S432G/A-UGT1A10, but not their wild types, catalyze bilirubin between 25% and 40% of the level of bilirubin-metabolizing UGT1A1. Similarly, S435G-UGT1A1 has reduced bilirubin activity (6). Hence, other examples of variable/new substrate profiles are associated with dephosphorylation/phosphorylation patterns (Fig. 5) of the different isozymes, which are under investigation.
Although understanding the role that phospho-group(s) play in UGT catalysis requires structural characterization of the protein, it is possible that the phospho group, despite attachment to threonine/serine in UGT, generates different ionizing state(s) (i.e., multiple pK values), which operationally control catalytic pH and substrate selections. Another possibility is that UGT catalysis, which requires partially negatively charged (nucleophilic) acceptor substrates (e.g., eugenol and 17β-estradiol) (9), utilizes a phosphothreonine/serine operationally to enhance proton abstraction (25, 26) from hydroxyl or carboxyl group(s) in the numerous substrates with wide structural variations. Such an effect could enhance the interaction of an acceptor substrate with partially positively charged carbon-1 of the common donor substrate, UDP-glucuronic acid. A recently modified but structurally characterized enzyme system demonstrated that newly added phosphoserine inhibited activity by attracting the catalytic proton (26, 27). In summary, this report describes a major breakthrough that shows that PKC-mediated phosphorylation of serine/threonine in UGT controls catalytic pH and substrate specificity, which can be altered by chemical exposures that disrupt PKC signaling (N.K.B. and I.S.O., unpublished data). Such regulated substrate selection may provide an efficient chemical defense system of major survival benefit to mammalians.
Supplementary Material
Acknowledgments
We express our appreciation to Dr. Anil B. Mukherjee for critical discussions concerning the manuscript.
Author contributions: N.K.B., T.S., J.R., and I.S.O. designed research; N.K.B., M.K., A.G., S.K., P.S.M., R.B., and I.S.O. performed research; N.K.B., T.S., J.R., and I.S.O. contributed new reagents/analytic tools; N.K.B., M.K., A.G., S.K., J.R., and I.S.O. analyzed data; I.S.O. wrote the paper; N.K.B. critiqued the manuscript and helped write the paper; A.G., S.K., P.S.M., and R.B. were involved in experiments; and I.S.O. critiqued the manuscript and results.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: UGT, UDP-glucuronosyltransferase; ER, endoplasmic reticulum; DSS, disuccinimidyl suberate; BIM, bisindolylmaleimide.
References
- 1.Dutton, G. J. (1980) in Glucuronidation of Drugs and Other Compounds, ed. Dutton, G. J. (CRC, Boca Raton, FL), pp. 69-78.
- 2.Vinneau, D. S., DeBoni, U. & Wells, P. G. (1995) Cancer Res. 55, 1045-1051. [PubMed] [Google Scholar]
- 3.Ritter, J. K., Yeatman, M. T., Ferreira, P. & Owens, I. S. (1992) J. Clin. Invest. 90, 150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Morais, S. M. F. & Wells, P.G. (1989) Hepatology 10, 163-167. [DOI] [PubMed] [Google Scholar]
- 5.Basu, N. K., Ciotti, M., Hwang, M. S., Kole, L., Mitra, P. S., Cho, J. W. & Owens, I. S. (2004) J. Biol. Chem. 279, 1429-1441. [DOI] [PubMed] [Google Scholar]
- 6.Basu, N. K., Kole, L. & Owens, I. S. (2003) Biochem. Biophys. Res. Commun. 303, 98-104. [DOI] [PubMed] [Google Scholar]
- 7.Liu, J.-Y., Lin, S.-J. & Lin, J.-K. (1993) Carcinogenesis 14, 857-861. [DOI] [PubMed] [Google Scholar]
- 8.Tillman, D. M., Izeradjene, K., Szucs, K. S., Douglas, L. & Houghton, J. A. (2003) Cancer Res. 63, 5118-5125. [PubMed] [Google Scholar]
- 9.Ciotti, M. & Owens, I. S. (1996) Biochemistry 31, 10119-10124. [DOI] [PubMed] [Google Scholar]
- 10.Ciotti, M., Yeatman, M. T., Sokol, R. J. & Owens, I. S. (1995) J. Biol. Chem. 270, 3284-3291. [DOI] [PubMed] [Google Scholar]
- 11.Basu, N. K., Kole, L., Kubota, S. & Owens, I. S. (2004) Drug. Metab. Dispos. 32, 768-773. [DOI] [PubMed] [Google Scholar]
- 12.Basu, N. K., Kubota, S., Meselhy, M. R., Ciotti, M., Chowdhury, B., Hartori, M. & Owens, I. S. (2004) J. Biol. Chem. 279, 28320-28329. [DOI] [PubMed] [Google Scholar]
- 13.Yedovitzky, M., Mochly-Rosen, D., Johnson, J. A., Gray, M.O., Ron, D., Abramovitch, E., Cerasi, E. M. & Nesher, R. (1997) J. Biol. Chem. 272, 1417-1420. [DOI] [PubMed] [Google Scholar]
- 14.Ritter, J. K., Crawford, J. M. & Owens, I. S. (1992) J. Biol. Chem. 266, 1043-1047. [PubMed] [Google Scholar]
- 15.Tinhofer, I., Bernhard, D., Senfter, M., Anether, G., Loeffler, M., Kroemer, G., Kofler, R., Csordas, A. & Greil, R. (2001) FASEB J. 15, 1613-1615. [DOI] [PubMed] [Google Scholar]
- 16.Mochly-Rosen, D. (1995) Science 268, 247-251. [DOI] [PubMed] [Google Scholar]
- 17.Cenni, V., Doppler, H., Sonnenburg, E. D., Maraldi, N., Newton, A. C. & Toker, A. (2002) Biochem. J. 363, 537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu, G., Wang, D., Lin, Y.-H., McMahon, T., Koo, E. H. & Messing, R. O. (2001) Biochem. Biophys. Res. Commun. 285, 997-1006. [DOI] [PubMed] [Google Scholar]
- 19.Arion, W. J., Wallin, B. K., Carlson, P. W. & Lange, A. J. (1972) J. Biol. Chem. 247, 2558-2565. [PubMed] [Google Scholar]
- 20.Thigpen, A. E., Davis, D. L., Milatovich, A., Mendonca, B. B., Imperato-McGinley, J., Griffin, J. E., Francke, U., Wilson, J. D. & Russell, D. W. (1992) J. Clin. Invest. 90, 799-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oesch-Bartlomowicz, B., Padma, P. R., Becker, R., Richter, B. Hengstler, J. G., Freeman, J. E., Wolf, C. R. & Oesch, F. (1998) Exp. Cell Res. 242, 294-302. [DOI] [PubMed] [Google Scholar]
- 22.Chang, E.-Y. Szallasi, Z., Acs, P., Raizada, V., Wolfe, P. C., Fewtrell, C., Blumberg, P. M. & Rivera, J. (1997) J. Immunol. 159, 2624-2632. [PubMed] [Google Scholar]
- 23.Brodie, C., Bogi, K., Acs, P., Lazarovici, P., Petrovics, G., Anderson, W. B. & Blumberg, P. M. (1999) Cell Growth Differ. 10, 183-191. [PubMed] [Google Scholar]
- 24.Gruss, O. J., Feick, P., Frank, R. & Dobberstein, B. (1999) Eur. J. Biochem. 260, 785-793. [DOI] [PubMed] [Google Scholar]
- 25.Kim, D.-Y., Stauffacher, C. V. & Rodwell, V. W. (2000) Biochemistry 39, 2269-2275. [DOI] [PubMed] [Google Scholar]
- 26.Omkumar, R. V. & Rodwell, V. W. (1994) J. Biol. Chem. 269, 16862-16866. [PubMed] [Google Scholar]
- 27.Ciotti, M., Basu, N., Brangi, M. & Owens, I. S. (1999) Biochem. Biophys. Res. Commun. 260, 199-202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.