Abstract
Cytochrome c (CYC) oxidase (COX), a multisubunit enzyme that functions in mitochondrial aerobic energy production, catalyzes the transfer of electrons from CYC to oxygen and participates in creating the electrochemical gradient used for ATP synthesis. Modeling three-dimensional structural data on COX and CYC reveals that 57 of the >1,500 COX residues can be implicated in binding CYC. Because of the functional importance of the transfer of electrons to oxygen, it might be expected that natural selection would drastically constrain amino acid replacement rates of CYC and COX. Instead, in anthropoid primates, although not in other mammals, CYC and COX show markedly accelerated amino acid replacement rates, with the COX acceleration being much greater at the positions that bind CYC than at those that do not. Specifically, in the anthropoid lineage descending from the last common ancestor of haplorhines (tarsiers and anthropoids) to that of anthropoids (New World monkeys and catarrhines) and that of catarrhines (Old World monkeys and apes, including humans), a minimum of 27 of the 57 COX amino acid residues that bind CYC were replaced, most frequently from electrostatically charged to noncharged residues. Of the COX charge-bearing residues involved in binding CYC, half (11 of 22) have been replaced with uncharged residues. CYC residues that interact with COX residues also frequently changed, but only two of the CYC changes altered charge. We suggest that reducing the electrostatic interaction between COX and CYC was part of the adaptive evolution underlying the emergence of anthropoid primates.
Keywords: mitochondria, electron transport chain, molecular coevolution
Cytochrome c (CYC) and the subunits of CYC oxidase (COX) are mitochondrial-functioning proteins that play a central role in aerobic energy production. By catalyzing the transfer of electrons from CYC to oxygen, COX greatly increases an electrochemical gradient used for ATP synthesis. As a consequence of their critical function, mutations altering the structures of either CYC or COX must face the close scrutiny of natural selection. Among vertebrates, relatively few amino acid replacements have occurred in the structures of CYC and COX. However, against this background of slow rates of CYC and COX evolution, upsurges in COX and CYC amino acid replacement rates occurred during the emergence and evolution of the larger brained primates (members of Anthropoidea) (1-12). Here, by using atomic resolution structural data on bovine COX (13) and horse CYC (14) and an experimentally verified model of the interaction between COX and CYC (15, 16), we identify, among the >1,500 amino acid residues of COX, 57 residues that are likely to bind CYC during delivery of electrons to oxygen. We then demonstrate that the amino acid replacement rate acceleration in anthropoid lineages is especially pronounced in the subset of COX residues that can bind CYC. We further demonstrate that, among these CYC-binding COX residues, many amino acid replacements were from charge-bearing [electrostatically significant (ES)] residues to non-charge-bearing residues. This finding, when added to the findings of previous studies, allows us to suggest that the reduction in electrostatic interaction between COX and CYC altered the kinetics of electron transfer as an adaptation to the needs of the emerging anthropoid primates.
The previous studies established that the accelerated amino acid replacement rates in anthropoid lineages encompassed nuclear DNA (nDNA)-encoded proteins and mtDNA-encoded proteins. Mammalian COX has 13 different protein subunits, three that are mtDNA-encoded and 10 that are nDNA-encoded. Accelerated rates in anthropoids were detected for two mtDNA-encoded subunits, COX1 and COX2; for seven nDNA-encoded COX subunits, COX4, COX5, COX6B, COX6C, COX7A, COX7C, and COX8; and for nDNA-encoded CYC (1-12, 17). Other studies pointed to the importance of electrostatic interactions between CYC and COX (16, 18-22) and indicated that the amino acid replacements in anthropoids altered the nature of the binding of CYC to COX (23).
Earlier findings established that CYC is a dipole (24, 25) with a lysine-rich, positively charged surface complementary to negatively charged domains of COX. These COX domains are shaped by the mtDNA-encoded subunits. The nDNA-encoded subunits provide a positively charged surface that probably interacts with the negatively charged part of the CYC dipole. More recently, extensive mutational analysis (16, 21, 26) and simulated binding of horse CYC to bovine COX (15) further demonstrated the importance of electrostatic interactions between CYC and COX. The electrostatic interactions participate in long-range protein-protein association and short-range orientation, such as through salt bridge formation (15). Osheroff et al. (23) examined by enzyme kinetic measurements the functional interaction between CYC and COX from different mammalian species. The results (23) depicted slow loris (a nonanthropoid primate) to be much more similar to horse (a nonprimate mammal) than to rhesus monkey (an anthropoid primate in the infraorder Catarrhini), even though phylogenetically slow loris is more closely related to rhesus monkey than to horse. When CYC and COX were from catarrhine sources, the enzyme kinetics were comparable with those measured when CYC and COX were from nonanthropoid mammals. The descriptions of the bovine COX (27) and horse CYC (14) crystal structures, investigations into the CYC-binding site of COX (15), and availability of a range of comparative COX and CYC sequence data from anthropoids and other vertebrate taxa now allow us to further examine evolutionary changes in electrostatic interactions between CYC and COX and to test whether CYC and COX coadaptively evolved in anthropoids.
Materials and Methods
DNA Sequences. RNA was prepared from either heart, kidney, lung, or liver tissues, and isolated with either a phenol-chloroform method (28) or with the Qiagen Midi kit. Some RNAs were obtained from the Center for Reproduction of Endangered Species, San Diego. RACE PCR (29) was performed on these RNAs, and the products were either sequenced directly or cloned into the pGEM-T easy vector (Promega) and sequenced. CYC was amplified for sequencing by PCR from genomic DNA. Additional sequences are from previous studies (1, 4, 6, 8, 10-12, 17) and from GenBank (see Tables 3 and 4, which are published as supporting information on the PNAS web site). Alignments of inferred amino acid sequences were performed with clustalx, version 1.81 (30). Insertions relative to the bovine amino acid sequence were removed from the data set to avoid speculation as to the three-dimensional position of amino acids not included in the known crystal structures. COX8L was substituted for COX8H because COX8H is absent in catarrhine primates (11).
Binding-Site Residues. COX residues that form the CYC binding site were extracted from these alignments by two criteria: that they lie on the surface of COX and that an atom of a surface COX residue side chain lies within 10 Å of any CYC atom when CYC is bound to COX (15). Surface residues were determined with the program getarea 1.1 (31) by using a radius for water of 1.4 Å. Polar interactions of up to 8.5 Å have been hypothesized in the docking model of CYC on COX (15). We have added 1.5 Å to allow for the possibility of longer range indirect interactions as well as minor conformational changes. Using a more restricted distance between amino acid side chains does not affect the conclusions presented here, although fewer residues would be included.
ES Residue Positions and ES Changes. If a positively charged (Arg or Lys) or negatively charged (Asp or Glu) residue in the binding site residue positions occurred among the taxa examined, that position was treated as an ES position. Among ES positions, ES changes were amino acid replacements that changed electrostatic state, there being three states: negatively charged, noncharged, and positively charged.
Evolutionary Analyses. Relative rate tests comparing tarsier (a nonanthropoid primate) COX to human or other anthropoid primate COX were performed by using Tajima's (32) method in mega 2.1 (33). Results obtained by using cow, pig, rat, or mouse as the outgroup did not substantially change the results. This analysis excluded COX5B, COX6A, and COX7B, for which the tarsier sequence was unavailable. Placement of amino acid replacements in the evolutionary history of primates was accomplished by using generally accepted phylogenetic relationships among primates (34) and among eutherian mammals (35, 36). By using this phylogeny, the number and location of changes along lineages were estimated in two ways: the most parsimonious solutions and Bayesian solutions as implemented in mrbayes 2.01 (37) using the Equalin amino acid replacement option. This analysis excluded the eight binding site residues of COX6A. For human, these residues differ from the other mammals in the data set by only one non-ES residue. Dates of divergence for calculating amino acid replacements over time were taken from generally accepted sources (34-36). CYC coding sequences were aligned and ancestral nodes were reconstructed by maximum likelihood using TrN+г model parameters as chosen by both hierarchical likelihood ratio test and Akaike Information Criterion by modeltest (38). Inferred ancestral sequences were then translated to protein.
Complete mitochondrial genomes were used to obtain the mtDNA docking site residues in a range of tetrapods. One representative species was sampled for each available tetrapod order. African lungfish (Protopterus dolloi) and coelacanth (Latimeria chalumnae) were used as outgroups. Accession numbers and taxon names are given in Table 4. A phylogenetic tree representing the ordinal relationships among the species was used to infer amino acid changes during descent of the tetrapods. Phylogenetic relationships were taken from previously published studies (35, 36, 39-42). Inferred amino acid changes were reconstructed with deltran in a maximum parsimony framework by using paup* 4.0b10 (43). This same procedure was used to infer amino acid changes in available complete mtDNA genomes for primates.
Results
Definition and Evolution of the Binding Site. We have identified 57 residues of COX that are likely to interact with CYC (Fig. 1). These binding site residues, i.e., the COX residues on which CYC docks, include 33 mtDNA-encoded residues (10 COX1, 20 COX2, and 3 COX3 residues) and 24 nDNA-encoded residues (eight COX6A, nine COX6B, five COX7A, one COX7C, and one COX8 residue). Results of relative rate tests for these residues indicate a statistically significant anthropoid rate acceleration relative to the most closely related nonanthropoid, tarsier (44, 45), for the entire COX monomer, for the 57 residues that are the binding site, and for residues that are not part of the binding site (Table 1). However, binding site residues have sustained the most pronounced rate acceleration, undergoing over four times the number of changes per amino acid than have amino acids that are not part of the binding site (0.43 vs. 0.10 for human vs. tarsier, respectively).
Fig. 1.
COX residues at the binding site of CYC. (A) The position of the docked CYC (shown in red). (B) Residues that form the binding site for CYC on COX. Green residues are those that have been neutralized in anthropoid primate evolution, yellow residues are those that have changed from neutral to positive, and gray residues are the remainder of residues that form the binding site. (C) Alignment of residues at the binding site. The point of electron transfer (COX2W104) is shown highlighted in orange. Residues 119 and 158, involved in salt bridge formation (15), are shown in boldface.
Table 1. Relative rates of change to the residues of COX near CYC when docked.
| Comparison
|
Replacements
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Region | A | B | Outgroup | Residues | A | B | Outgroup | P | Replacements/Residues |
| Full monomer | Human | Tarsier | Cow | 1,541 | 164 | 28 | 39 | 0.00 | 0.11 |
| Nondocking residues | Human | Tarsier | Cow | 1,492 | 143 | 27 | 39 | 0.00 | 0.10 |
| Docking residues | Human | Tarsier | Cow | 49 | 21 | 1 | 0 | 0.00 | 0.43 |
| OWM | Tarsier | Cow | 49 | 23 | 1 | 0 | 0.00 | 0.47 | |
| NWM | Tarsier | Cow | 49 | 14 | 0 | 0 | 0.00 | 0.29 | |
| Tarsier | Cow | Mouse | 49 | 0 | 2 | 5 | 0.16 | 0.00 | |
| Nondocking, ES | Human | Tarsier | Cow | 252 | 24 | 5 | 8 | 0.00 | 0.10 |
| Docking, ES | Human | Tarsier | Cow | 20 | 13 | 0 | 0 | 0.00 | 0.65 |
| Docking, non-ES | Human | Tarsier | Cow | 29 | 8 | 1 | 0 | 0.02 | 0.28 |
A and B refer to lineages being compared relative to the indicated outgroup. OWM, Old World monkeys; NWM, New World monkeys.
Electrostatic Changes. Because binding CYC to COX predominately involves an electrostatic interaction (15), the expectation is that ES residues are functionally important and, thus, under the scrutiny of natural selection but not always the purifying form. We observed that the rate acceleration in anthropoids is most pronounced at ES residue positions. For example, as may be inferred by parsimony from an interspecies alignment of residues at the binding site (Fig. 1C), there are a minimum of 27 changes (31 changes by Bayesian inference) from the earlier eutherians (cetartiodactyl-rodent-primate ancestor) to humans, of which 13 are at 22 ES residue positions (59%), compared with 14 changes at 35 non-ES residue positions (31%).
Of these 13 ES changes, 11 were from charged to neutral, thus reducing the number of charged residues at the CYC binding site. Only two ES changes were from neutral to charged (Table 2). This level of ES change to COX residues near the CYC binding site is unique in vertebrates examined to date. For example, the amino acid sequences from cow, pig, rat, mouse, and tarsier do not have ES differences. The ES changes in anthropoids appear to reduce electrostatic interaction between CYC and COX: The seven changes from negatively charged to neutral residues are located on the mitochondrially encoded subunits that face the positively charged residues of docked CYC, whereas the four changes from positively charged residues to neutral are to nuclear-encoded residues near the negatively charged pole of docked CYC. Overall, ES changes are over six times more common at the binding site than in other regions of COX (0.65 vs. 0.10 on the human lineage) (Table 1), showing that the high rate of ES change at the CYC binding site does not arise from an overall high rate of ES change to COX.
Table 2. Evolution of charged residues at the CYC docking site of COX in primates.
| Subunit | COX position | COX residue | COX change | CYC position | CYC residue | CYC change | COX-CYC, Å |
|---|---|---|---|---|---|---|---|
| COX1 | 50 | D→N | Catarrhine | 28 | T | — | 6.6 |
| COX1 | 119 | E | — | 49 | T | — | 6.6 |
| COX1 | 221 | D | — | 73 | K | — | 5.7 |
| COX2 | 114 | E→G | Catarrhine | 87 | K | — | 8.7 |
| COX2 | 115 | D→G | Catarrhine | 87 | K | — | 5.5 |
| COX2 | 119 | D→N | Catarrhine | 13 | K | — | 1.6 |
| COX2 | 127 | D→Y | Anthropoid | 11 | V→I | Anthropoid | 7.0 |
| COX2 | 127 | Y→F | Catarrhine | 11 | V→I | Anthropoid | 7.0 |
| COX2 | 139 | D | — | 12 | Q→M | Catarrhine | 3.9 |
| COX2 | 157 | E→Q | Anthropoid | 86 | K | — | 2.8 |
| COX2 | 158 | D | — | 83 | A→V | Anthropoid | 2.5 |
| COX3 | 111 | E→Q | Anthropoid | 73 | K | — | 8.6 |
| COX6A | 58 | K | — | 76 | P | — | 4.2 |
| COX6A | 64 | D | — | 54 | N | — | 9.9 |
| COX6B | 61 | K→Q | Catarrhine | 87 | K | — | 7.8 |
| COX6B | 71 | A→D | Anthropoid | 69 | E | — | 3.5 |
| COX6B | 73 | D | — | 88 | K | — | 8.8 |
| COX6B | 74 | D→E | Anthropoid | 69 | E | — | 5.2 |
| COX6B | 78 | E | — | 66 | E | — | 7.8 |
| COX7A | 57 | H→R | Anthropoid | 50 | D→E | Platyrrhine | 5.8 |
| COX7A | 57 | H→R | Anthropoid | 50 | D→A | Catarrhine | 5.8 |
| COX7A | 58 | K→N | Anthropoid | 47 | S→T | Platyrrhine | 0.8 |
| COX7C | 47 | K→Q | OWM | 27 | K | — | 4.4 |
| COX7C | 47 | K→T | Human | 27 | K | — | 4.4 |
| COX8 | 43 | R→P | Catarrhine | 26 | H | — | 9.3 |
Complete lists of docking site residues in CYC and COX are shown in Tables 5 and 6. COX change refers to the lineage on which the amino acid replacement occurred. Catarrhine refers to the stem catarrhine lineage; anthropoid refers to the stem anthropoid lineage; platyrrhine refers to the lineage from the platyrrhine-catarrhine divergence node to the squirrel monkey, a representative extant New World monkey; OWM (Old World monkey) refers to the lion-tailed macaque. CYC position shows the nearest amino acid residue position of CYC that is within 10 Å of the listed residue of COX.
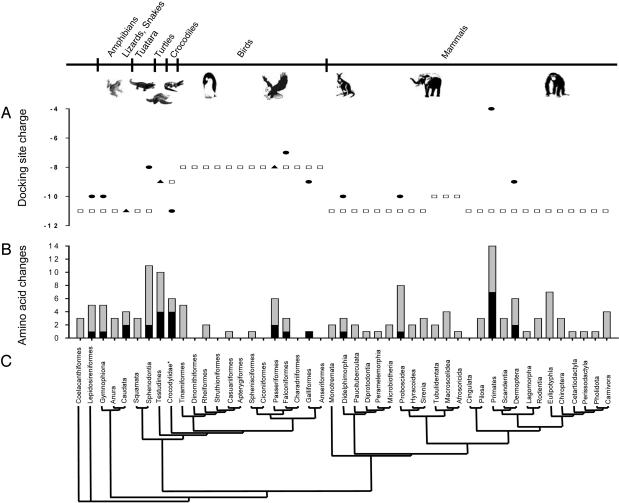
The public availability of a large comparative body of mitochondrial genome sequence data (Table 4) allows us to further demonstrate that ES changes rarely occur among mammals and other tetrapods (Fig. 2). Note that the negatively charged surface of the CYC-binding site of COX in most orders of amphibians and mammals has remained at the net electrostatic charge of -11. However, in the catarrhine primates (Fig. 3), this net charge was reduced to -4. Except for birds, which show a limited charge reduction in the mitochondrially coded COX residues that bind CYC, no other clade of tetrapods as represented by the 45 species examined in 45 orders (Fig. 2) show such a dramatic reduction of net charge. A similar analysis for nuclear subunits was also conducted and is shown in Figs. 5 and 6, which are published as supporting information on the PNAS web site.
Fig. 2.
Evolution of the 33 mitochondrially encoded COX residues that putatively bind CYC in tetrapods. (C) Phylogenetic tree depicting the presumed interordinal relationships among tetrapods (35, 36, 39-42). *, The familial name Crocodylidae is given because no commonly agreed on named ordinal rank exists for this clade. (B) Number of amino acid replacements in the mtDNA-encoded portion of COX that binds CYC. Bars indicate the total number of deltran-inferred amino acid changes on each terminal lineage from the ancestral node for each order to the terminal node; the number of inferred ES changes in the total number of changes are indicated by black fill in the bar graph. (A) Inferred ordinal ancestral docking site total charge (see Materials and Methods) is shown by open squares; filled circles correspond to the current charge in each ordinal representative species (Table 4). Orders in which only a square is shown have no inferred ES changes in their terminal lineage. Triangles indicate cases in which ES changes were compensatory such that the terminal and ancestral nodes for the order maintain the same total charge at the docking interface.
Fig. 3.
Evolution of the 33 mitochondrially encoded COX residues that putatively bind CYC in primates. (Left) Phylogenetic relationships among different species of primates (33). (Center) Number of amino acid replacements in the mtDNA-encoded portion of COX that binds CYC. Bars indicate the total number of deltran-inferred amino acid changes on each terminal lineage from the ancestral crown primate to the terminal species; the number of inferred ES changes in the total number of changes are indicated by black fill in the bar graph. (Right) Docking site total charge. The vertical line represents the inferred total charge of the crown primate ancestor; filled ovals are the inferred total charge of the indicated primate species.
Phylogenetic analysis of the CYC data set revealed a total of 18 amino acid replacements, of which 14 occurred within Anthropoidea, a majority in the stem anthropoid and stem catarrhine lineages. Moreover, 12 of the replacements were at positions that are within 10 Å of a COX ES position when CYC is docked (Table 5, which is published as supporting information on the PNAS web site). Strikingly, unlike the COX changes, in most instances no charge change is found in CYC. The exceptions are a D to A replacement at CYC-50 on the catarrhine lineage, nearest to a gain of a positive charge (H to R) in COX (Table 2), and a G to E replacement at CYC-89 on the catarrhine lineage, near an S to T change in COX (Table 5; see also Table 6, which is published as supporting information on the PNAS web site).
Evolutionary Analysis. Amino acid replacements in the CYC-binding site of COX have occurred predominantly on three lineages (Fig. 4): the stem lineage of anthropoids (platyrrhines and catarrhines), the stem lineage of catarrhines (Old World monkeys and apes, including humans), and in platyrrhines (New World monkeys). However, the ES changes occurred only in the stem anthropoids and stem catarrhines. In the lineage encompassing the stem anthropoids and the stem catarrhines there were, according to the Bayesian analysis, 29 changes from the most recent common ancestor of tarsier and anthropoids (≈58 million years ago) to the most recent common ancestor of Old World monkeys and apes (≈25 million years ago), whereas zero and four changes have occurred to the ape and Old World monkey lineages, respectively. The observation that the amino acid replacement rate of COX residues at the CYC binding site is increased in stem anthropoid and stem catarrhine lineages but has been slow in more recent lineages (Fig. 4) is consistent with the hypothesis that the changes in the ancestral lineages were advantageous and positively selected and, in descendent lineages, have been maintained by purifying selection. This pattern of accelerated rates followed by decelerated rates is inconsistent with the alternate hypothesis that the amino acid replacement rate increase is due to a relaxation of functional constraints.
Fig. 4.
Rates of amino acid replacement by lineage, showing the increase in the amino acid replacement rate in ancestral (i.e., stem) anthropoid primates, followed by a decrease in the amino acid replacement rate in descendent anthropoid primate lineages. (A) Vertical bars depict the number of amino acid replacements per amino acid residue per billion years (BY). Gray bars depict the rates for the 57 binding site residues, and the rates for the 1,578 nonbinding site residues are shown in white. (B) Changes to COX at the CYC-binding site as determined by Bayesian analysis. The numbers of inferred changes to the binding site residues are shown above the branches (ES changes are shown in parentheses), and the numbers of nonbinding site changes are shown below the branches.
Discussion
In cross-species experiments, Osheroff et al. (23) found overly tight binding of human CYC on bovine COX and greatly reduced steady-state reaction kinetics. They found less tight binding and much higher steady-state reaction kinetics when either horse CYC or slow loris CYC docked on bovine COX or, alternatively, when human CYC docked on rhesus monkey COX. Our results on the dual upsurges of amino acid replacements in COX and CYC within the Anthropoidea provide insights on the findings of Osheroff et al. (23). In view of the fact that there are very few amino acid differences between tarsier (a nonanthropoid primate) and cow at the 57 COX residue positions implicated in binding CYC (Fig. 1), the cause of the overly tight binding of human CYC on bovine COX may reside in the stem anthropoid and stem catarrhine CYC amino acid replacements. These replacements would not appreciably alter salt bridge formation between human CYC and bovine COX, as can be deduced from the data in Table 2. However, at least some of the replacements might have strengthened the nonelectrostatic bonding between the two proteins. In turn, the extensive charge neutralizing replacements in stem anthropoid and stem catarrhine COX would have appreciably weakened salt bridge formation between human CYC and rhesus monkey COX and, thus, resulted in less tight binding than in the case of human CYC and bovine COX. To recapitulate, our results show that the CYC binding site of nonanthropoid primate (tarsier, lemur, and slow loris) COX shows greater overall similarity to bovine or other nonprimate mammalian COX than to catarrhine COX and that the nonanthropoids lack the extensive ES changes found in catarrhines. Nevertheless, changes in reaction kinetics do not appear to result exclusively from ES changes to the CYC-binding site of COX. The activity of horse CYC with rhesus monkey COX is only about half the activity of human CYC with rhesus monkey COX (23), yet the electrostatic interactions in both must be similar. These data further suggest that nonelectrostatic changes to anthropoid CYC also affect the reaction kinetics between CYC and COX.
As noted, for the extensive charge neutralizations found at the COX/CYC interaction site, the reduction in electrostatic attraction is carried out almost wholly by COX residues. Chemically significant changes to CYC residues, such as charge alterations, may be constrained because CYC has the known additional roles of interacting with the mitochondrial bc1 complex and with apoptotic protease activating factor 1. Despite these constraints, apparently coevolving changes to CYC are seen and appear to compensate for COX charge changes in a more subtle way. Examination of the COX/CYC interacting surfaces suggests that at least some of the paired substitutions increase hydrophobic contacts near the site of electron transfer (for example V→I in CYC-11 paired with D→Y,F in COX2-127; Table 2). Although the effects of altering the hydrophobic surface will need to be addressed experimentally, they are likely to affect CYC's on rate and off rate and the rate of electron transfer.
Are changes to the CYC-binding site of COX the only accelerated change in the ancestry of anthropoid primates? Results of the relative rate tests (Table 1) indicate that there is a statistically significant rate acceleration in the nonbinding site residues of COX during anthropoid evolution as well. Additional changes to other regions of COX are not surprising, because the primary function of the nuclear-encoded subunits is thought to be regulatory in nature (46, 47). If so, changes in the regulatory scheme would be called for to accommodate a significant shift in enzyme kinetics.
We suggest that the accelerated amino acid replacement rates of COX and CYC in the stem anthropoid and stem catarrhine lineages were coadaptive and part of the organismal evolution out of which emerged a constellation of new phenotypic features. Two correlated features linked to the mitochondrial electron transport chain are extended life spans and greatly enlarged brains. Considerable evidence (48, 49) has aging and diseases of old age associated with damage from mitochondrial-generated radical species, i.e., delaying the onset of excessive damage should extend life spans. A possibility worth exploring is that an effect of the marked changes in the electron transport chain proteins of anthropoids was to reduce the radical flux which accompanies oxidative energy production. Brain tissues have potentially the greatest stake in changes to energetically important proteins. Although the human brain accounts for only ≈2% of the adult human body weight, it utilizes ≈16% of the total oxygen consumed (50) and as much as 65% during fetal development (51). Most of the energy demand in a neuron is oxidative (52). Moreover, many deleterious mutations in mitochondrial genes are associated with neuropathological conditions (53). Thus, we suggest that there is a nonfortuitous association between molecular changes in aerobic energy metabolism proteins, such as COX and CYC, and organismal changes, such as brain enlargements in anthropoids. In fact, the same charge reduction tendency is possible to see in birds (Fig. 2), a group with recognized higher metabolic demands than mammals. We realize that this hypothesis is not amenable to direct experimental testing. However, indirect phylogenetic tests are possible because evolution of enlarged brains occurred separately in a number of clades but not in their sister clades. Thus, it is possible to explore in each such pair of closely related clades whether the proteins important for brain function evolved faster in the clade showing the most encephalization.
Supplementary Material
Acknowledgments
We thank Victoria A. Roberts and Shelagh Ferguson-Miller for comments and discussion and Jeffrey Doan, Allon Goldberg, and Maik Hüttemann for assistance and discussion. We thank the Duke University Primate Center (Durham, NC) for tissue samples and the Center for Reproduction of Endangered Species (San Diego) for RNA. This research was supported by National Institutes of Health Grant GM65580 and National Science Foundation Grants BCS-9910679, MCB-9816923, and BCS-0318375.
Author contributions: T.R.S., D.E.W., M.U., J.C.O., M.G., and L.I.G. designed research; T.R.S., D.E.W., M.U., and J.C.O. performed research; T.R.S., D.E.W., J.C.O., M.G., and L.I.G. analyzed data; and T.R.S., D.E.W., J.C.O., M.G., and L.I.G. wrote the paper.
Abbreviations: CYC, cytochrome c; COX, CYC oxidase; ES, electrostatically significant; nDNA, nuclear DNA.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY236506, AY585857, AY585861, AY585862, AY585864, and AY918493-AY918495).
References
- 1.Baba, M. L., Darga, L. L., Goodman, M. & Czelusniak, J. (1981) J. Mol. Evol. 17, 197-213. [DOI] [PubMed] [Google Scholar]
- 2.Goodman, M., Romero-Herrera, A. E., Dean, H., Czelusniak, J. & Tashian, R. E. (1982) in Macromolecular Sequences in Systematic and Evolutionary Biology, ed. Goodman, M. (Plenum, New York), pp. 115-191.
- 3.Adkins, R. M. & Honeycutt, R. L. (1994) J. Mol. Evol. 38, 215-231. [DOI] [PubMed] [Google Scholar]
- 4.Wu, W., Goodman, M., Lomax, M. I. & Grossman, L. I. (1997) J. Mol. Evol. 44, 477-491. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, T. D., Jermiin, L. S. & Easteal, S. (1998) J. Mol. Evol. 47, 249-257. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt, T. R., Goodman, M. & Grossman, L. I. (1999) Mol. Biol. Evol. 16, 619-626. [DOI] [PubMed] [Google Scholar]
- 7.Andrews, T. D. & Easteal, S. (2000) J. Mol. Evol. 50, 562-568. [DOI] [PubMed] [Google Scholar]
- 8.Wu, W., Schmidt, T. R., Goodman, M. & Grossman, L. I. (2000) Mol. Phylogenet. Evol. 17, 294-304. [DOI] [PubMed] [Google Scholar]
- 9.Grossman, L. I., Schmidt, T. R., Wildman, D. E. & Goodman, M. (2001) Mol. Phylogenet. Evol. 18, 26-36. [DOI] [PubMed] [Google Scholar]
- 10.Wildman, D. E., Wu, W., Goodman, M. & Grossman, L. I. (2002) Mol. Biol. Evol. 19, 1812-1815. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg, A., Wildman, D. E., Schmidt, T. R., Huttemann, M., Goodman, M., Weiss, M. L. & Grossman, L. I. (2003) Proc. Natl. Acad. Sci. USA 100, 5873-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doan, J. W. (2004), Ph.D. thesis (Wayne State University, Detroit, MI).
- 13.Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1996) Science 272, 1136-1144. [DOI] [PubMed] [Google Scholar]
- 14.Bushnell, G. W., Louie, G. V. & Brayer, G. D. (1990) J. Mol. Biol. 214, 585-595. [DOI] [PubMed] [Google Scholar]
- 15.Roberts, V. A. & Pique, M. E. (1999) J. Biol. Chem. 274, 38051-38060. [DOI] [PubMed] [Google Scholar]
- 16.Zhen, Y., Hoganson, C. W., Babcock, G. T. & Ferguson-Miller, S. (1999) J. Biol. Chem. 274, 38032-38041. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, T. R., Goodman, M. & Grossman, L. I. (2002) Gene 286, 13-19. [DOI] [PubMed] [Google Scholar]
- 18.Brautigan, D. L., Ferguson-Miller, S. & Margoliash, E. (1978) J. Biol. Chem. 253, 130-139. [PubMed] [Google Scholar]
- 19.Brautigan, D. L., Ferguson-Miller, S., Tarr, G. E. & Margoliash, E. (1978) J. Biol. Chem. 253, 140-148. [PubMed] [Google Scholar]
- 20.Osheroff, N., Borden, D., Koppenol, W. H. & Margoliash, E. (1980) J. Biol. Chem. 255, 1689-1697. [PubMed] [Google Scholar]
- 21.Witt, H., Wittershagen, A., Bill, E., Kolbesen, B. O. & Ludwig, B. (1997) FEBS Lett. 409, 128-130. [DOI] [PubMed] [Google Scholar]
- 22.Witt, H., Malatesta, F., Nicoletti, F., Brunori, M. & Ludwig, B. (1998) Eur. J. Biochem. 251, 367-373. [DOI] [PubMed] [Google Scholar]
- 23.Osheroff, N., Speck, S. H., Margoliash, E., Veerman, E. C., Wilms, J., Konig, B. W. & Muijsers, A. O. (1983) J. Biol. Chem. 258, 5731-5738. [PubMed] [Google Scholar]
- 24.Koppenol, W. H., Vroonland, C. A. & Braams, R. (1978) Biochim. Biophys. Acta 503, 499-508. [DOI] [PubMed] [Google Scholar]
- 25.Koppenol, W. H. & Margoliash, E. (1982) J. Biol. Chem. 257, 4426-4437. [PubMed] [Google Scholar]
- 26.Witt, H., Malatesta, F., Nicoletti, F., Brunori, M. & Ludwig, B. (1998) J. Biol. Chem. 273, 5132-5136. [DOI] [PubMed] [Google Scholar]
- 27.Tsukihara, T., Itoh-Shinzawa, K. & Yoshikawa, S. (1996) Tanpakushitsu Kakusan Koso 41, 1353-1362. [PubMed] [Google Scholar]
- 28.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 29.Hüttemann, M. (2002) BioTechniques 32, 730-736. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraczkiewicz, R. & Braun, W. (1998) J. Comp. Chem. 19, 319-333. [Google Scholar]
- 32.Tajima, F. (1993) Genetics 135, 599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 34.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9, 585-598. [DOI] [PubMed] [Google Scholar]
- 35.Madsen, O., Scally, M., Douady, C. J., Kao, D. J., DeBry, R. W., Adkins, R., Amrine, H. M., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Nature 409, 610-614. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, W. J., Eizirik, E., O'Brien, S. J., Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Science 294, 2348-2351. [DOI] [PubMed] [Google Scholar]
- 37.Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. (2001) Science 294, 2310-2314. [DOI] [PubMed] [Google Scholar]
- 38.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson, M. A., Gullberg, A., Spotorno, A. E., Arnason, U. & Janke, A. (2003) J. Mol. Evol. 57, S3-S12. [DOI] [PubMed] [Google Scholar]
- 40.Paton, T., Haddrath, O. & Baker, A. J. (2002) Proc. R. Soc. London Ser. B 269, 839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rest, J. S., Ast, J. C., Austin, C. C., Waddell, P. J., Tibbetts, E. A., Hay, J. M. & Mindell, D. P. (2003) Mol. Phylogenet. Evol. 29, 289-297. [DOI] [PubMed] [Google Scholar]
- 42.Cotton, J. A. & Page, R. D. (2002) Proc. R. Soc. London B 269, 1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swofford, D. L. (2000) paup* (Sinauer, Sunderland, MA), Version 4.0.
- 44.Meireles, C. M., Czeluzniak, J., Page, S. L., Wildman, D. E. & Goodman, M. (2003) in Tarsiers: Past, Present, and Future, eds. Wright, P. C., Simons, E. L. & Gursky, S. (Rutgers Univ. Press, New Brunswick, NJ), pp. 145-160.
- 45.Schmitz, J., Ohme, M. & Zischler, H. (2001) Genetics 157, 777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadenbach, B., Kuhn-Nentwig, L. & Buge, U. (1987) Curr. Top. Bioenerg. 15, 114-162. [Google Scholar]
- 47.Arnold, S., Goglia, F. & Kadenbach, B. (1998) Eur. J. Biochem. 252, 325-330. [DOI] [PubMed] [Google Scholar]
- 48.Mandavilli, B. S., Santos, J. H. & Van Houten, B. (2002) Mutat. Res. 509, 127-151. [DOI] [PubMed] [Google Scholar]
- 49.Harper, M. E., Bevilacqua, L., Hagopian, K., Weindruch, R. & Ramsey, J. J. (2004) Acta Physiol. Scand. 182, 321-331. [DOI] [PubMed] [Google Scholar]
- 50.Aiello, L. C. & Wheeler, P. (1995) Curr. Anthropol. 36, 199-221. [Google Scholar]
- 51.Holliday, M. (1986) in Human Growth: A Comprehensive Treatise, eds. Flakner, F. & Tanner, J. M. (Plenum, New York), pp. 117-139.
- 52.Kasischke, K. A., Vishwasrao, H. D., Fisher, P. J., Zipfel, W. R. & Webb, W. W. (2004) Science 305, 99-103. [DOI] [PubMed] [Google Scholar]
- 53.Betts, J., Lightowlers, R. N. & Turnbull, D. M. (2004) Neurochem. Res. 29, 505-511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.