Abstract
In earlier studies, we identified short (6- to 22-nt) sequences that functioned as internal ribosome entry sites (IRESes) and enhanced translation. The size of these IRES elements suggested that they might be prevalent within the messenger population and that individual elements might affect the translation of different groups of mRNAs. To begin to assess the number of different IRES elements in mammalian cells, we have developed a powerful method that uses a positive feedback mechanism to amplify the activities of individual IRES elements. This method uses a vector that encodes a dicistronic mRNA with a reporter gene (Renilla luciferase or the EGFP) as the first cistron and the yeast Gal4/viral protein 16 (VP16) transcription factor as the second cistron. Transcription of this mRNA is driven by a minimal promoter containing four copies of the Gal4 upstream activation sequence. In this method, the presence of an IRES in the intercistronic region facilitates the translation of Gal4/VP16, which binds to the upstream activation sequences and triggers a positive feedback loop that escalates the production of dicistronic mRNA and Gal4/VP16. A corresponding increase in the translation of the first cistron (luciferase or EGFP) is monitored either by measuring luciferase activity or by using FACS. The latter enables IRES-positive cells to be isolated. We present tests of the feedback mechanism by using an IRES module from Gtx homeodomain mRNA and an IRES from hepatitis C virus and demonstrate the utility of this vector system for the screening, identification, and analysis of IRES elements.
Keywords: internal ribosome entry site, selection
Eukaryotic mRNAs can initiate translation by either cap-dependent or cap-independent mechanisms. Presently, the relative contributions of these mechanisms to the proteome are unknown; however, some studies suggest that cap-independent mechanisms may account for the translation of many mRNAs (e.g., ref. 1). For some mRNAs, cap-independent translation is facilitated by sequence elements termed internal ribosome entry sites (IRESes). IRESes were first discovered in uncapped picornavirus RNAs (2, 3) and were subsequently identified in other viral and cellular mRNAs from mammals, insects, and yeast (4, 5). For some mRNAs, IRESes facilitate translation when cap-dependent initiation is less efficient or blocked (e.g., refs. 6-9). Internal initiation also facilitates the translation of particular mRNAs with 5′ leaders that are encumbered by numerous upstream AUGs or RNA secondary structures (10, 11).
A variety of evidence suggests that different IRESes vary in length, sequence composition, and in their requirements for initiation factors or other trans-acting factors, suggesting that internal initiation of translation occurs by a number of different mechanisms (5, 12). In earlier studies, we and others showed that some IRESes are modular in composition (10, 13-17). We identified an IRES module from the 5′ leader of the Gtx homeodomain mRNA and showed that maximal activity was obtained with sequences of 7 nucleotides. Various lines of evidence suggested that the mechanism underlying the activity of this sequence element involves base pairing to a complementary segment of 18S rRNA (18). In another study, we identified a 22-nt IRES module in the 5′ leader of the Rbm3 mRNA (14). In addition, it has been reported by others that the 5′ leader of c-myc mRNA contains two short IRES elements (19).
The short size of some IRES modules suggests the hypothesis that they may be prevalent within mRNA populations; if so, the identification and analysis of IRES elements is critical to understanding how they affect translation initiation. Moreover, the identification of IRES elements with particular properties is of practical significance by providing means for enhancing protein production. In earlier studies, we generated synthetic IRESes containing multiple individual IRES elements and showed that this multimerization led to higher, and in some cases exponential, increases in IRES activity. To facilitate the discovery process, we and others have developed a number of methods to screen for IRES elements in mammalian cells (20, 21) and in yeast (22). In all of these studies, dicistronic mRNAs containing a library of random nucleotide sequences in the intercistronic sequence (ICS) were expressed in cells, and those cells containing IRES elements were identified on the basis of the expression of the second cistron. The mammalian methods used a fluorescent reporter protein as the second cistron, and positive cells were identified with FACS. However, a limitation of these methods was that the activities of individual IRES elements were relatively low, leading to the identification of large numbers of false positive cells.
To circumvent this signal-to-noise problem, we have developed a positive feedback vector based on a dicistronic mRNA that encodes a reporter protein as the first cistron and the Gal4/viral protein 16 (VP16) transcription factor as the second cistron. An IRES in the ICS of this mRNA facilitates translation of Gal4/VP16 and triggers a positive feedback loop in which Gal4/Vp16 binds to UAS sequences in the upstream promoter of the dicistronic mRNA and increases the transcription of this mRNA. More dicistronic mRNA results in more Gal4/VP16, leading to ever-increasing amounts of both the dicistronic mRNA and the encoded proteins. We show here evidence for this amplification by monitoring the activity of the first cistron and we analyze the gains and mechanisms entailed by use of this method.
Methods
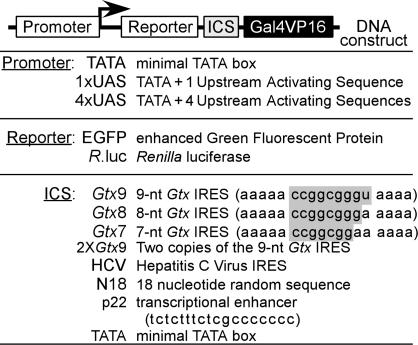
Construction of Dicistronic Vectors. The constructs used in this study express dicistronic mRNAs that encode a reporter protein as the first cistron and a transcription factor as the second cistron. As shown in Fig. 1, the promoters used to drive transcription of the dicistronic mRNAs consist of a minimal promoter (TATA box), either alone or in combination with one or four copies of the GAL4 upstream activating sequence (UAS). The first cistron of the dicistronic mRNAs encodes a reporter protein, either the EGFP or Renilla luciferase, and the second cistron encodes the Gal4/VP16 fusion protein. The ICS contains one or another of various sequence elements, including the IRES from the hepatitis C virus (HCV) (23, 24), 7- to 9-nt segments of the Gtx IRES module, two copies of the 9-nt Gtx IRES module (18), and a library of 18 random nucleotides (N18).
Fig. 1.
Positive feedback vectors. A schematic representation of the positive feedback vector is shown along with the various promoter sequences, reporter cistrons, and ICSes used in these studies. Promoter elements used are a minimal TATA box with one or four copies of a UAS. The reporters used in this study are EGFP and Renilla luciferase. The ICSes are 7-, 8-, and 9-nt versions of an IRES module from the Gtx mRNA (the Gtx nucleotides are shaded), two copies of the 9-nt Gtx IRES module, the HCV IRES, and the N18 sequences.
The vector backbone is based on the plasmid pHRG-B (Promega), and the different promoters were cloned by using HindIII and NcoI restriction sites. The first cistron was cloned by using the NcoI and MluI restriction sites, and Gal4/VP16 was cloned by using the XbaI restriction site. The original BamHI site in pHRG-B was mutated so that both EcoRI and BamHI sites in the ICS were unique. Most of the IRESes and random N18 fragments were cloned into this amplification vector by using the EcoRI and BamHI restriction sites.
Cell Culture and Transfection Analysis. Reporter constructs (0.5 μg) were transfected into Chinese hamster ovary (CHO) cells (2 × 104) by using FuGENE 6 (Roche). Transfection efficiencies were normalized by cotransfection with 0.2 μg of a LacZ reporter gene construct (pCMVβ, Clontech). Cells were harvested 2 days after transfection and assayed for luciferase activity. For time course experiments, cells were harvested at the time points indicated. For cells transfected with constructs expressing the luciferase protein, luciferase activities were determined as described in ref. 13. Cells expressing EGFP were sorted by FACS on a FACSVantage SE (Becton Dickinson) (20). FACS analysis was performed 2 days after transfection. β-galactosidase assays were performed as described in ref. 13. The integrity and size of mRNAs were determined by Northern blot analyses by using a Renilla luciferase probe (13, 25).
Double-stranded oligonucleotides containing N18 sequences were cloned into the ICS of the positive feedback vector by using EcoRI and BamHI restriction sites. Overnight ligations used T4 DNA ligase at 16°C. The resulting ligation mix was transfected into CHO cells, and FACS analyses were performed 3 days later. For each FACS analysis, the first 100,000 cells were analyzed and a sorting window was drawn to select the cells with highest EGFP expression. DNA was extracted from cells recovered by FACS and PCR reactions were carried out by using primers to sequences that flank the EcoRI and BamHI restriction sites. After digestion with both EcoRI and BamHI restriction enzymes, the resulting fragments were recloned in the same amplification vector and retested.
For determining the number of plasmids per transfected cell, equal amounts of two plasmids were mixed, CMV-EGFP and CMV-enhanced cyan fluorescent protein (CMV-ECFP; Clontech). The cloning vector pBluescript-KS II (Stratagene) was used as filler for cotransfection. CHO cells were transfected with these different mixtures and FACS analysis was performed 2 days later to assess the expression of both EGFP and ECFP.
Results
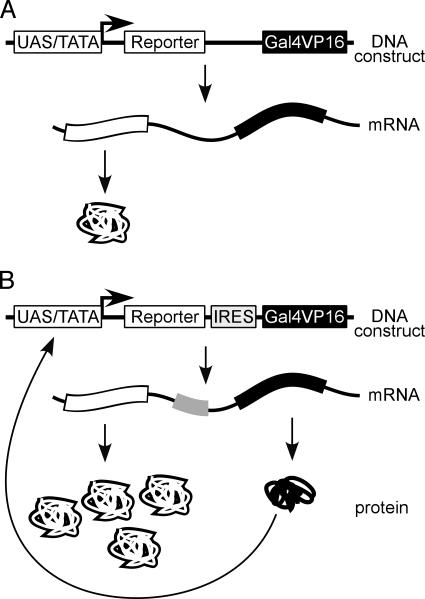
Positive Feedback Dicistronic Reporter Vector. Gal4/VP16 is a highly active transcription factor generated by fusing the DNA-binding domain of the yeast Gal4 transcription factor with the acidic activating region from the herpes simplex virus protein VP16 transcription factor (26). Gal4/VP16 binds to a DNA sequence termed the UAS. This sequence is not a target of mammalian transcription factors, making it possible to specifically monitor the expression of Gal4/VP16 in mammalian cells by monitoring the activity of a reporter gene under the transcriptional control of UAS sequences. These properties of Gal4/VP16 enabled us to generate a mammalian positive feedback vector for the identification and analysis of IRES elements (see Fig. 2).
Fig. 2.
Schematic of positive feedback vector. (A) In the absence of an IRES in the ICS of the dicistronic vector, the expression of the reporter cistron should be proportional to the strength of the upstream promoter. The UAS/TATA promoter will produce a low level of expression of the dicistronic mRNA and of the reporter protein. In the absence of an IRES, Gal4/VP16 should not be expressed from this dicistronic mRNA and should not affect its transcription. (B) The presence of an IRES in the ICS will greatly enhance the expression of the reporter protein by a positive feedback mechanism. The IRES will facilitate the translation of the Gal4/VP16 transcription factor, which will bind to UAS sequences in the promoter and activate transcription of the dicistronic mRNA, enhancing the expression of both cistrons.
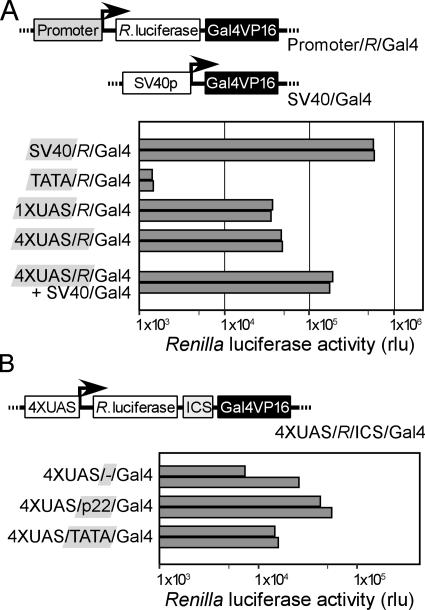
The parent positive feedback vector encodes a dicistronic mRNA with Renilla luciferase as the first cistron and Gal4/VP16 as the second cistron. The transcription of this dicistronic mRNA occurs by way of a minimal TATA box promoter but can be enhanced in cells expressing the Gal4/VP16 second cistron by means of the four UAS sequences in the promoter. Control constructs containing the minimal TATA box promoter alone or containing this minimal promoter along with one or four copies of the UAS were tested in transiently transfected CHO cells. Cells were harvested after 48 h and luciferase activities were measured. The results showed that all three vectors expressed a low level of Renilla luciferase (Fig. 3A), reflecting a low level of mRNA production by means of the minimal promoter. The construct with the minimal TATA box promoter was ≈0.2% as active as the simian virus 40 (SV40) promoter, whereas the 1XUAS and 4XUAS promoters were ≈6% and 7.5% as active as the SV40 promoter, respectively.
Fig. 3.
Transfection analysis and initial characterization of positive feedback vectors. (A) Reporter assays with different configurations of promoter elements controlling the expression of the dicistronic mRNA. Four promoter elements were tested in the promoter/R/Gal4 vector: the SV40 promoter, a minimal TATA box, a minimal TATA box with one UAS, and a minimal TATA box with four UASes. The 4XUAS/R/Gal4 vector was also tested in a cotransfection experiment along with the SV40/Gal4 expression vector. (B) Reporter assays with different control ICSes. Sequences tested in this study were a no-insert control (-), a transcriptional enhancer (p22), and a minimal TATA box. CHO cells were transfected with the indicated constructs, and the cells were harvested 2 days later. Reporter activities were determined. For each case, the results of two independent experiments, each presented as a shaded bar, are shown in the histogram.
To assess the ability of Gal4/VP16 to enhance Renilla luciferase activity, CHO cells were cotransfected with the p4XUAS/R/Gal4 construct along with a second plasmid (pSV40/Gal4) that expresses Gal4/VP16 by means of the SV40 promoter. The presence of the second vector led to an increase in Renilla luciferase activity of ≈5-fold, reflecting the increased transcription of the dicistronic mRNA by means of the UAS promoter sequences (Fig. 3A). This effect on luciferase activity was shown to depend on the presence of the four UAS sequences in the promoter of the dicistronic mRNA (data not shown).
Short Transcriptional Promoter or Enhancer Sequences in the ICS Do Not Trigger Positive Feedback. A limitation of the use of dicistronic mRNAs to assess IRESes is that non-IRES sequences might enhance the expression of the second cistron by mechanisms that generate monocistronic mRNAs, e.g., by transcription of a monocistronic mRNA corresponding to the second cistron. The positive feedback mRNA in this study was designed to minimize possible false-positive events because maximal expression of the reporter protein encoded by the first cistron will occur only when the second cistron (Gal4/VP16) is translated from the amplified dicistronic mRNA. In an earlier study performed in yeast (22), we screened libraries of dicistronic mRNAs containing N18 in the ICS and found that spacing the N18 sequences <40 nt upstream of the initiation codon of the second cistron dramatically reduced the number of false-positive selection events because of the presence of N18 sequences with promoter activity. We therefore maintained this spacing in these new vectors. To further minimize false-positive selection events resulting from N18 sequences that function as transcriptional enhancers and lead to the generation of monocistronic transcripts through cryptic transcriptional promoters located 5′ of the second cistron, we introduced the nucleotide sequence AUG upstream of the N18 sequence as a decoy initiation codon. Depending on the N18 sequence, the ORF resulting from this upstream AUG will either overlap the Gal4/VP16 cistron in a different reading frame or will terminate within the N18 sequence. These aspects of the vector design were tested by using two different N18 sequences in the ICS: a TATA box promoter sequence (TATAAA) and p22, an 18-nt transcriptional enhancer identified in an earlier study (27); p22 was able to enhance the transcription of a minimal promoter by ≈100-fold in CHO cells (data not shown). The results (Fig. 3B) showed that neither the TATA box promoter nor the p22 element enhanced Renilla luciferase activity.
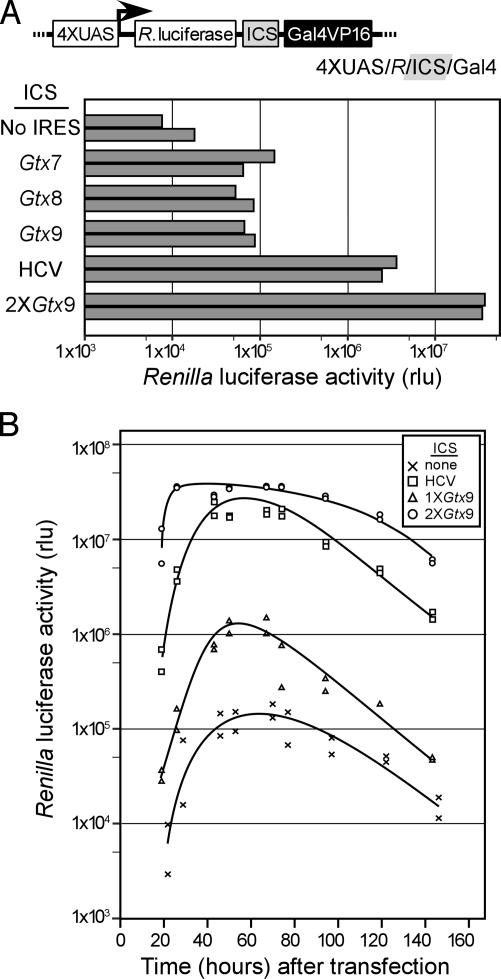
Tests of the Positive Feedback Mechanism. Known IRES sequences were tested in the ICS of the p4xUAS/R/GVP16 vector. The IRESes tested were from the HCV, and also 7- to 9-nt of an IRES element from the 5′ leader of the mouse Gtx homeodomain mRNA; in addition, we tested a construct that contained two linked copies of the 9-nt Gtx IRES element. In an earlier study, the 9-nt IRES element enhanced translation of a second cistron by ≈2.5-fold over background in mouse neuroblastoma N2a cells, whereas synthetic constructs containing two copies of this IRES element were ≈2.8 times as active as a single element (13). In CHO cells, the Gtx IRES element was less active, and two linked copies had a level of activity close to background (data not shown).
Through use of the positive feedback vector, all of the IRESes tested were found to increase translation dramatically (Fig. 4A). Maximal activity in this study was obtained with the two Gtx IRES elements. A time course analysis over 6 days indicated that the activity obtained with this Gtx-IRES element was maximal 2 days after transfection, whereas the activities obtained with the other IRESes were maximal after 3 days (Fig. 4B).
Fig. 4.
Analysis of IRESes in the positive feedback vector. (A) Reporter activity assay with different IRESes in the ICS of the 4XUAS/R/ICS/Gal4 positive feedback vector. IRESes tested in the ICS were 7-, 8-, and 9-nt variations of the Gtx IRES module, which were tested as single copies in the ICS. Two copies of the 9-nt Gtx IRES module and the HCV IRESes were also tested. CHO cells were transfected with the indicated constructs, cells were harvested 2 days later, and reporter activities were determined. The results of two independent experiments are shown in the histogram. (B) Time course of expression for individual IRESes in the 4XUAS/R/ICS/Gal4 vector. The IRESes in this study included one copy of the Gtx9 IRES, two copies of the Gtx9 IRES, and the HCV IRES. CHO cells were transfected with the indicated constructs, and the cells were harvested at the indicated time points. The results of two independent experiments are shown.
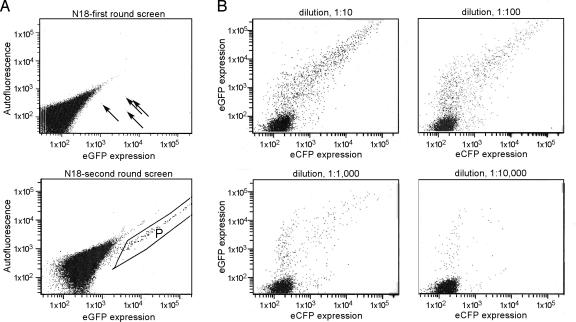
For use as a screening tool, we replaced the Renilla luciferase cistron from the parent vector (p4xUAS/R/GVP16) with a cistron encoding EGFP. A library of constructs containing random 18-nt sequences was then used to transfect CHO cells (Fig. 5A). In contrast to our earlier selection study in which positive cells were close to background (20), positive cells in this study were clearly above background because of the high levels of EGFP produced by means of the positive feedback mechanism. In this example, 100,000 cells were transfected and sorted by FACS 3 days later. The four highest expressing cells were isolated, and ICSes were recovered by genomic PCR, recloned into the dicistronic vector, and reselected. Fig. 5B shows the enrichment obtained in such a procedure.
Fig. 5.
FACS analysis of transfected CHO cells. (A) First- and second-round enrichment of plasmids containing random N18 sequences in the ICS of the positive feedback vector (4XUAS/R/N18/Gal4). A library of constructs was transfected into CHO cells and first-round FACS was performed 3 days after transfection. DNA was extracted from recovered cells and amplified by PCR. The PCR products were digested and cloned into the same vector for a second round of cell sorting. Both sorting graphs show a total of 100,000 cells from the transfected CHO cells pool. (Lower) The area labeled “P” shows an enrichment of positive cells. The ordinate axis indicates the expression of EGFP. The abscissa is a measure of autofluorescence. In both cases, the units are arbitrary. Four positive cells are indicated with arrows. (B) Determining the approximate number of plasmids per transiently transfected cell. Two reporter constructs encoding EGFP and ECFP were mixed (1:1) and transfected into CHO cells with a neutral plasmid (pBluescript-KSII). The reporter constructs were diluted at 1:10, 1:100, 1:1,000, and 1:10,000 concentrations with the pBluescript plasmid. FACS profiles were taken 2 days after transfection. The ordinate axis indicates the expression of ECFP, and the abscissa indicates the expression of EGFP.
A consideration in these studies is that transfection results in the introduction of multiple plasmids per cell. Therefore, a positive cell, i.e., a cell containing a construct with an IRES, will also contain many other plasmids that do not possess IRESes. As a result, the identification of active sequences may require multiple rounds of selection, dilution of the plasmids with a neutral filler plasmid, or a combination of these approaches to obtain individual sequence elements. To determine approximately how many different plasmids were contained per transfected cell, CHO cells were transfected with two different plasmids at a 1:1 molar ratio: one that expresses EGFP and the other that expresses the ECFP. These plasmids were diluted with up to a 10,000-fold molar excess of a neutral filler plasmid that does not express EGFP (pBluescript-KSII). The transfected cells were analyzed by FACS. At the lower dilutions (Fig. 5B), most cells expressed both fluorescent proteins, indicating the presence of both plasmids in these cells. At a 1:1,000 dilution, many cells expressed only EGFP or ECFP, and at a 1:10,000-fold dilution most cells expressed only one fluorescent protein. These results indicate that each transfected cell contained between ≈500 and 5,000 plasmids.
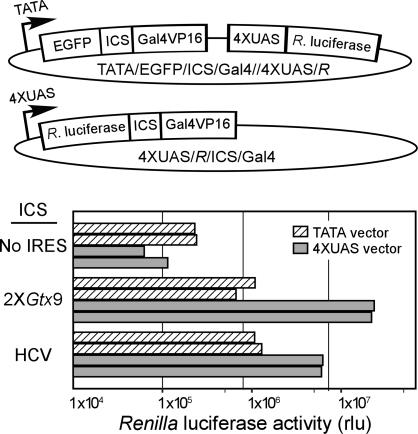
Use of Positive Feedback to Distinguish Between IRES and Promoter Activities. To differentiate further between sequences with IRES activity and sequences with transcriptional promoter or enhancer activities, we separated the Renilla luciferase and Gal4/VP16 cistrons from the 4XUAS/R/ICS/Gal4 vector (TATA/EGFP/ICS/Gal4/4XUAS/R) (Fig. 6). In this configuration, Gal4/VP16 is still the second cistron of a dicistronic mRNA but with a different first cistron, in this case, EGFP. This dicistronic mRNA is transcribed by a minimal TATA box promoter and cannot be induced because it lacks UAS sequences. In contrast to this transcription unit, the Renilla luciferase gene is under the transcriptional control of a minimal TATA box promoter with 4XUAS sequences. In this vector, expression of Renilla luciferase requires Gal4/Vp16. The presence of an ICS that facilitates the expression of Gal4/Vp16 monocistronic mRNAs, e.g., promoter or enhancer elements, will lead to similar levels of Renilla luciferase activity from both vectors, i.e., in the positive feedback vector and in the vector in which the Renilla luciferase and Gal4/Vp16 cistrons are separate transcription units. In contrast, an ICS that functions in the context of the dicistronic mRNA, e.g., an IRES element, will only enhance Renilla luciferase expression in the positive feedback vector.
Fig. 6.
Analysis of IRESes in vector systems with or without positive feedback to assess whether the activity of an ICS requires the production of the dicistronic mRNA or is independent of it. (Top) The vector lacking positive feedback. (Middle) The vector with positive feedback. (Bottom) The IRESes tested were two copies of the Gtx9 IRES module and the HCV IRES, which were cloned into the ICS of both vectors. Constructs were transfected into CHO cells and harvested 2 days later, and the reporter activities were determined. The results of two independent experiments for each example are shown.
The 2XGtx9 and HCV IRESes were tested in these two vectors in transfected CHO cells (Fig. 6), and both showed a high level of Renilla luciferase activity in the positive feedback vector and a low level of expression in the TATA/EGFP/ICS/Gal4/4XUAS/R vector. Constructs containing two copies of the 9-nt Gtx IRES element or containing the HCV IRES showed ≈96% and 84% less activity when the promoter of the dicistronic mRNA lacked the UAS sequences, indicating that the activities of the Gtx and HCV sequences depended on the production of the dicistronic mRNA, a finding that is consistent with their functioning as IRESes.
Discussion
In these studies, we developed a positive feedback vector system designed to facilitate IRES identification and analysis. This vector system amplifies the activities of relatively weak individual IRES elements and facilitates their identification and analysis. In this vector system, for example, two copies of the 9-nt Gtx IRES module enhanced the expression of a reporter protein >300-fold over background in CHO cells, despite the fact that this IRES module, when tested in the dicistronic Renilla Photinus hairpin mRNA (13), was not very active in these cells. Indeed, without amplification, the activity of two copies of this IRES module was near background. Two copies of the Gtx IRES module in the positive feedback vector were more active than the stronger HCV IRES in this same vector. This finding suggests that the shorter Gtx IRES module was more efficient in amplifying the dicistronic mRNA than the longer, more highly structured HCV IRES. The fact that a minimal TATA box promoter and a transcription enhancer did not function in this feedback system indicates that the use of this vector is not compromised by short transcriptional elements. This property should minimize false-positive selection events.
Earlier studies by our laboratory and others have indicated that some IRESes are modular in composition (e.g., refs. 13, 14, and 19). In addition, we showed that other short cis-acting sequences can modulate IRES activity without functioning as IRESes themselves (see ref. 14). The activities of most of the individual IRES elements that have been identified thus far are relatively weak, although we have shown that multimerization of individual short elements can lead to exponential increases in activity (13, 20). The positive amplification method overcomes the limitation of the weak activity of small IRES modules. In the context of native mRNAs, we expect that the activity of an IRES will depend on the activities of the individual IRES modules as well as on their interactions, which may be additive, synergistic, or interfering.
The present development of a feedback method to identify individual IRES elements is an extension of our earlier studies (20, 22) and is motivated by the possibility that short sequences, such as the 7-nt Gtx IRES module are widespread within the mRNA population and that combinations of these mRNA sequences may have important global effects on the proteome. The development of an efficient selection method to identify IRES elements will enable their sequences to be characterized on the basis of sequence motifs and expression properties; it will also enable an assessment of the frequency of these elements in the messenger population.
The investigation of selected IRES elements will also facilitate analyses of translational mechanisms. The small size of these elements reduces the complexity inherent in the analysis of naturally occurring cellular IRESes, which tend to be much larger in size. Thus far, we have obtained evidence that two IRES elements, from the Rbm3 and Gtx mRNAs (14, 18), bind directly to ribosomes. However, indirect mechanisms involving binding to initiation factors or other RNA-binding proteins are also likely, in some cases, to facilitate translation initiation. In the ribosome filter hypothesis (28), we discussed the possibility that IRES elements may increase the local concentration of the translation machinery by a variety of means and that this increase in local concentration will lead to additional translation initiation events.
In addition to these fundamental issues related to the search for IRES elements, there are practical issues related to the enhancement of protein production in cell lines. Individual IRES modules can also serve as valuable building blocks for the generation of synthetic IRESes and translational enhancers having specific expression properties. In addition, this selection methodology facilitates the identification of IRES elements with specific properties such as cell-type specificity. Synthetic IRESes with such properties may be useful, for example, in gene therapy applications.
Acknowledgments
We thank Dr. Robyn Meech for critical reading of the manuscript and Luke Burman for excellent technical assistance. This work was supported by National Institutes of Health Grant GM61725 (to V.P.M.), the G. Harold and Leila Y. Mathers Charitable Foundation (V.P.M.), U.S. Public Health Service Grant NS39837 (to G.M.E.), and The Skaggs Institute for Chemical Biology (W.Z.).
Abbreviations: IRES, internal ribosome entry site; ICS, intercistronic sequence; UAS, upstream activated sequence; HCV, hepatitis C virus; N18, library of 18 random nucleotides; CHO, Chinese hamster ovary; SV40, simian virus 40; ECFP, enhanced cyan fluorescent protein; VP16, viral protein 16.
References
- 1.Keiper, B. D. & Rhoads, R. E. (1997) Nucleic Acids Res. 25, 395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier, J. & Sonenberg, N. (1988) Nature 334, 320-325. [DOI] [PubMed] [Google Scholar]
- 3.Jang, S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593-1612. [DOI] [PubMed] [Google Scholar]
- 5.Vagner, S., Galy, B. & Pyronnet, S. (2001) EMBO Rep. 2, 893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannes, G. & Sarnow, P. (1998) RNA 4, 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyronnet, S., Pradayrol, L. & Sonenberg, N. (2000) Mol. Cell 5, 607-616. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, S., Bruynooghe, Y., Denecker, G., Van Huffel, S., Tinton, S. & Beyaert, R. (2000) Mol. Cell 5, 597-605. [DOI] [PubMed] [Google Scholar]
- 9.Pinkstaff, J. K., Chappell, S. A., Mauro, V. P., Edelman, G. M. & Krushel, L. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2770-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappell, S. A., Owens, G. C. & Mauro, V. P. (2001) J. Biol. Chem. 276, 36917-36922. [DOI] [PubMed] [Google Scholar]
- 11.Le Quesne, J. P., Stoneley, M., Fraser, G. A. & Willis, A. E. (2001) J. Mol. Biol. 310, 111-126. [DOI] [PubMed] [Google Scholar]
- 12.Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I. & Hellen, C. U. (2001) Proc. Natl. Acad. Sci. USA 98, 7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappell, S. A., Edelman, G. M. & Mauro, V. P. (2000) Proc. Natl. Acad. Sci. USA 97, 1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappell, S. A. & Mauro, V. P. (2003) J. Biol. Chem. 278, 33793-33800. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein, J., Sella, O., Le, S.-Y. & Elroy-Stein, O. (1997) J. Biol. Chem. 272, 9356-9362. [DOI] [PubMed] [Google Scholar]
- 16.Huez, I., Creancier, L., Audigier, S., Gensac, M.-C., Prats, A.-C. & Prats, H. (1998) Mol. Cell. Biol. 18, 6178-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoneley, M., Paulin, F. E. M., Le Quesne, J. P. C., Chappell, S. A. & Willis, A. E. (1998) Oncogene 16, 423-428. [DOI] [PubMed] [Google Scholar]
- 18.Chappell, S. A., Edelman, G. M. & Mauro, V. P. (2004) Proc. Natl. Acad. Sci. USA 101, 9590-9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cencig, S., Nanbru, C., Le, S. Y., Gueydan, C., Huez, G. & Kruys, V. (2004) Oncogene 23, 267-277. [DOI] [PubMed] [Google Scholar]
- 20.Owens, G. C., Chappell, S. A., Mauro, V. P. & Edelman, G. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesan, A. & Dasgupta, A. (2001) Mol. Cell. Biol. 21, 2826-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou, W., Edelman, G. M. & Mauro, V. P. (2003) Proc. Natl. Acad. Sci. USA 100, 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. & Nomoto, A. (1992) J. Virol. 66, 1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellen, C. U. & Pestova, T. V. (1999) J. Viral. Hepat. 6, 79-87. [DOI] [PubMed] [Google Scholar]
- 25.Mauro, V. P. & Edelman, G. M. (1997) Proc. Natl. Acad. Sci. USA 94, 422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. (1988) Nature 335, 563-564. [DOI] [PubMed] [Google Scholar]
- 27.Edelman, G. M., Meech, R., Owens, G. C. & Jones, F. S. (2000) Proc. Natl. Acad. Sci. USA 97, 3038-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauro, V. P. & Edelman, G. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12031-12036. [DOI] [PMC free article] [PubMed] [Google Scholar]