Abstract
Tcf transcription factors interact with β-catenin and Armadillo to mediate Wnt/Wingless signaling. We now report the characterization of genes encoding two murine members of the Tcf family, mTcf-3 and mTcf-4. mTcf-3 mRNA is ubiquitously present in embryonic day 6.5 (E6.5) mouse embryos but gradually disappears over the next 3 to 4 days. mTcf-4 expression occurs first at E10.5 and is restricted to di- and mesencephalon and the intestinal epithelium during embryogenesis. The mTcf-3 and mTcf-4 proteins bind a canonical Tcf DNA motif and can complex with the transcriptional coactivator β-catenin. Overexpression of Wnt-1 in a mammary epithelial cell line leads to the formation of a nuclear complex between β-catenin and Tcf proteins and to Tcf reporter gene transcription. These data demonstrate a direct link between Wnt stimulation and β-catenin/Tcf transcriptional activation and imply a role for mTcf-3 and -4 in early Wnt-driven developmental decisions in the mouse embryo.
Tcf-1 and Lef-1 are the two founding members of a small subfamily of vertebrate high-mobility-group (HMG) box transcription factor genes (5, 24, 36, 37, 42). Tcf-1 and Lef-1 were originally defined as lymphoid-specific transcription factors based on their affinity for the enhancers of the CD3ɛ and the T-cell receptor α genes, respectively. Tcf-1 and Lef-1 were later found to be expressed in largely overlapping, complex patterns during embryogenesis (25). Gene disruption of Tcf-1 yields a severe T-cell developmental defect as the only phenotypic abnormality (41). Mutation of Lef-1 results in abnormalities in the development of various skin appendages, teeth, and the trigeminal nucleus but leaves the immune system intact (40).
As described below, recent studies have led to the surprising realization that Tcf proteins mediate Wingless/Wnt signaling in insects and vertebrates (22, 39). In the course of these studies, several novel Tcf proteins were cloned from diverse organisms, i.e., dTCF (or Pangolin) from Drosophila melanogaster (4, 39) and XTcf-3 from Xenopus laevis (19). In addition, a novel human family member, hTcf-4, was found to be expressed in colonic epithelium and malignancies derived therefrom (16).
The Wingless signaling pathway establishes segment polarity in Drosophila, while the ectopic expression of components of the Wnt signaling cascade induces axis duplication in Xenopus. A striking symmetry exists between these two pathways (reviewed in reference 15). Wingless/Wnt signaling is assumed to reprogram gene expression in responding cells. Despite the identification of multiple components of this signal transduction pathway, the mechanism by which the reprogramming occurs remained a mystery, and until very recently, β-catenin represented the most-downstream component known in the Wnt signal transduction pathway (reviewed in reference 28). β-Catenin and its Drosophila homolog Armadillo play dual roles in cell-cell adhesion and in signaling. Both proteins occur in adherent junctions in a complex with α-catenin and cadherin homologs, and both accumulate inside cells in response to Wingless/Wnt signals.
In a yeast two-hybrid screen, Lef-1 was cloned with β-catenin as a bait (2). In a reciprocal experiment, we cloned β-catenin by using human Tcf-1 as a bait (19). It was subsequently shown that murine Lef-1 (mLef-1) and XTcf-3, an embryonically expressed member of the Tcf family in Xenopus, bind to β-catenin upon microinjection in embryos. The complex accumulates in the nucleus. We further showed that XTcf-3 in isolation binds DNA but does not activate transcription of target genes, while the interaction with β-catenin endows XTcf-3 with potent transactivational properties (19). The biological relevance of the interaction between β-catenin and Tcf proteins was tested by ectopic expression of engineered forms of XTcf-3 and Lef-1 in Xenopus embryos. Injection of β-catenin into early Xenopus embryos induces a secondary axis (8), mimicking the effect of Wnt activation. Ectopic expression of a dominant-negative form of XTcf-3, lacking the region required for β-catenin binding, blocks β-catenin’s ability to cause axis duplication and, more importantly, blocks formation of the endogenous axis (19). In concordance with these findings, injection of mLef-1 induces the formation of a secondary axis in Xenopus embryos (2, 11).
The maternally expressed Drosophila Tcf family member, dTCF/Pangolin, was subsequently demonstrated to play a key role in Wingless signaling (4, 29, 39). dTCF binds a canonical Tcf DNA motif and interacts with the β-catenin homolog Armadillo. Mutations in dTCF and expression of a dominant-negative dTCF transgene or an mLef-1 transgene cause Wingless-like phenotypes and affect expression of the Wingless target genes engrailed and Ultrabithorax. In accordance with the biochemical data, genetic epistasis experiments position dTCF downstream of Armadillo in the Wingless cascade.
It is likely that early Wnt-driven developmental signals during embryogenesis in the mouse are mediated by transcription factors of the Tcf family. However, the available knockout data for mLef-1 and mTcf-1 are not consistent with a role for these two factors in Wnt signaling in the early embryo. This would suggest the involvement of other Tcf proteins in early murine development. We have now cloned full-length cDNAs of two additional murine members of the Tcf family, determined their expression patterns, and provided evidence for their potential involvement in Wnt signaling.
MATERIALS AND METHODS
Cloning of mTcf-3 and mTcf-4.
Mouse 11- and 15-day embryo cDNA libraries (Clontech, Palo Alto, Calif.) were screened at low stringency with a mixed probe derived from HMG boxes of XTcf-3, chicken Tcf (chTcf), and hTcf-1. Positive clones were subcloned in pBluescript SK and sequenced.
In situ hybridization.
Dissected embryos were fixed 4 to 18 h in 4% paraformaldehyde, dehydrated in ethanol and butanol, and embedded in paraffin. Embryos were sectioned at 6-μm thickness and mounted on 3-aminopropyl-triethoxy-silane-coated slides. Slides were deparaffined, digested with proteinase K, acetylated with acetic anhydride dissolved in triethanolamine, dehydrated, and hybridized overnight at 55°C. Subsequently, high-stringency washing was performed in 50% formamide–2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) 10 mM dithiothreitol (DTT) at 65°C. Slides were treated with RNase A, rinsed in 2× SSC–0.1× sodium dodecyl sulfate, dehydrated, and dipped in Ilford emulsion G.5. After drying, the slides were exposed for 1 to 10 days at 4°C, developed in Kodak D-19, and fixed with Kodak LX-24.
RNA probes.
DNA fragments encoding amino acids (aa) 270 to 342 for mTcf-3 and aa 258 to 328 for mTcf-4 were subcloned into pBluescript SK, constructs were linearized, and radiolabeled antisense RNA probes were generated from these templates, using T7 RNA polymerase and [35S]UTP (Stratagene, La Jolla, Calif.). Both constructs covered unique DNA sequences. Probes were used for hybridization at 105 cpm/μl.
RNA analysis.
Total RNA from different tissues and cell lines was isolated by the method of Chomczynski and Sacchi (7). Approximately 15 μg of total RNA was loaded per lane and separated on an agarose gel containing 2% formaldehyde. The gel was blotted onto a nitrocellulose membrane and probed with 32P-labeled probes. Probes were the same as for the in situ analysis. Poly(A)+ RNA was isolated from total RNA by using Oligo-dT Dynal beads (Dynal, Oslo, Norway). Approximately 2 μg of poly(A)+ RNA was loaded per lane.
Yeast two-hybrid analysis.
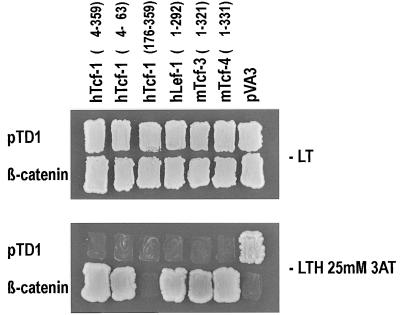
All TCF constructs were created by fusing the regions encoding the indicated amino acids to the GAL4 binding domain of pMD4: hTcf-1 constructs, aa 4 to 359, 4 to 63, and 176 to 359; hLef-1, aa 1 to 292; mTcf-3, aa 1 to 321; and mTcf-4, aa 1 to 331. pVA3 encodes a murine p53–GAL4 DNA-binding domain hybrid in pGBT9 (Clontech). These constructs were transformed into the MATa Saccharomyces cerevisiae strain HF7C (Clontech). β-Catenin was inserted in frame with the GAL4 activation domain in pGADrx (Stratagene). pTD1 encodes simian virus 40 large T antigen in pGAD3F (Clontech); both were transformed into Y187 (MATα) (Clontech). HF7C cells were mated to Y187 cells. The resulting cells were grown on nonselective (Leu- and Trp-deficient) or selective (lacking Leu, Trp, and His and containing 25 mM 3-aminotriazole) medium. β-Galactosidase filter assays were performed as described for the Matchmaker two-hybrid system (Clontech).
Generation of vectors.
The vectors for generating cell lines with a tetracycline-repressible Wnt-1 cDNA were kindly provided by H. Bujard (10) and modified as follows: pUHD15-1 was digested with BamHI and EcoRI (blunted) to obtain a 1-kb fragment containing the tetracycline-sensitive transactivator. This fragment was cloned into the StuI site of pHyTCX to generate pHyTCtTA (pHyTCX was generated from pLNCX [GenBank name SYNMMLPLN3] by John Murphy, who replaced the neomycin resistance gene with the hygromycin B resistance gene). pUHD10-3 was digested with XhoI and EcoRI to release a 0.46-kb fragment containing a minimal cytomegalovirus promoter with heptamerized Tetracycline operators. This fragment was cloned into a 6.5-kb EcoRI-digested and partially XhoI-digested fragment of pROSAbgal (provided by P. Soriano [32]). The resulting vector was named pROTX. An EcoRI fragment containing the Wnt-1 cDNA (from V101) was cloned into the EcoRI site of pROTX to generate pROTWnt-1.
Generation of the 2-69-23 cell line.
C57MG cells carrying the Wnt-1 gene under the control of the tetracycline-repressible promoter were generated as follows: C57MG cells were first infected with virus obtained from pROTWnt-1-transfected PE501 cells and selected in G418 until colonies appeared. Clones with a flat morphology were subsequently infected with virus obtained from PE501 cells transfected with pHyTCtTA and selected with hygromycin B in the presence of 5 μg of tetracycline per ml. Individual colonies were isolated and screened for the ability to induce Wnt-1 overexpression in the absence of tetracycline.
Wnt-1 Western blotting.
Cells were washed in phosphate-buffered saline, lysed in ice-cold lysis buffer (1% Triton X-100, 50 mM Tris-HCl [pH 8.0], 150 mM NaCl) containing protease inhibitors (1 mM Pefabloc SC [Boehringer Mannheim], 1 mM phenylmethylsulfonyl fluoride leupeptin [1 μg/ml], aprotinin [2 μg/ml], pepstatin [1 μg/ml] [Sigma]), and incubated on ice for 20 min. Insoluble material was removed by centrifugation at 20,000 × g for 10 min at 4°C. The protein concentration was determined by the method of Bradford (Bio-Rad protein assay). Equal amounts of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein was transferred to nitrocellulose. Wnt-1 immunoblots were blocked in 1% bovine serum albumin in Tris-buffered saline plus 0.1% Tween 20 (TBST) and incubated in a 1:1,000 dilution of the monoclonal anti-Wnt-1 antibody Mc123 (raised to a Wnt-1 peptide spanning aa 200 to 212) (3).
Cell culture and Wnt-1 induction.
C57MG cells were grown in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal calf serum, 2 mM glutamine, penicillin, streptomycin, and 10 mg of bovine pancreatic insulin (Sigma) per ml. C57MG cells with a tetracycline-repressible Wnt-1 gene expression (cell line 2-69-23) were grown in complete medium supplemented with G418 (400 μg/ml; Boehringer), hygromycin B (100 μg/ml; Boehringer), and tetracycline (50 ng/ml; Sigma). For induction studies, cells were washed with phosphate-buffered saline, cultivated for additional 24 h in tetracycline-free medium, and then harvested.
Gel retardation assays.
Assays were performed as described previously (16). Extracts were prepared from intact nuclei of induced and control C57MG cells. Isolated nuclei were washed extensively prior to lysis. The anti-β-catenin antibody was purchased from Transduction Laboratories (Lexington, Ky.). Binding reaction mixtures contained 3 μg of nuclear protein, 0.5 ng of probe, and 100 ng of poly(dI-dC) in 25 μl of binding buffer (60 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). Samples were incubated for 20 min at room temperature, antibody was added, and samples were incubated for another 20 to 30 min and subjected to nondenaturing polyacrylamide gel electrophoresis.
CAT assays and luciferase assays.
C57MG and 2-69-23 cells (induced or noninduced) (2 × 105 of each cell type) were transfected with luciferase reporter construct pTOPFLASH or pFOPFLASH (16), using LipofectAmine reagent (Gibco). Cells were harvested after 24 h and lysed in 1 mM DTT–1% Triton X-100–15% glycerol–25 mM Tris (pH 7.8)–8 mM MgCl2. Luciferase activity was determined on a Lumac/3M biocounter. Plasmid pCATCONTROL (Promega) was used as an internal control. Chloramphenicol acetyltransferase (CAT) values were determined as pristane-xylene-extractable radiolabeled, butyrylated chloramphenicol (37).
RESULTS
Cloning of mTcf-3 and mTcf-4.
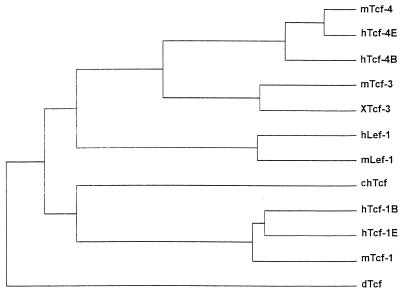
To isolate Tcf-like genes from mice, low-stringency screening of several embryonic cDNA libraries was performed with a mixed probe consisting of the HMG boxes of XTcf-3 (19), chTcf (5), and hTcf-1 (37). All efforts yielded cloned sequences derived from two different Tcf homologs. The amino acid sequences directly N-terminal to the HMG boxes were aligned with the corresponding regions encoded by the human genomic fragments of hTCF-3 and hTCF-4 (6). This identified the two novel genes as the murine orthologs of these partially characterized human genes. No full-length mammalian Tcf-3 homolog has been cloned previously. In the course of the present study, we isolated full-length cDNAs for human Tcf-4 (16), which was 98% identical to mTcf-4 at the amino acid level. An alignment of mTcf-3, hTcf-4, mTcf-4, and XTcf-3 is given in Fig. 1. An evolutionary tree was constructed for the available Drosophila and vertebrate Tcf sequences by using the program Clustal (Fig. 2). mTcf-3 clustered with XTcf-3, while mTcf-4 and hTcf-4 clustered separately. The overall match between mTcf-3 and XTcf-3 is 75%. Both proteins align well in the regions C terminal to the HMG box (65% identity); the other three mammalian homologs contain less conserved C termini. We therefore believe that mTcf-3 and XTcf-3 are encoded by orthologous genes.
FIG. 1.
Sequence comparison of mTcf-4, hTcf-4, mTcf-3, and XTcf-3. The highly conserved N-terminal β-catenin interaction domain and the HMG box region are underlined. hTcf-4 is 98% identical to mTcf-4 at the amino acid level; the overall match between mTcf-3 and XTcf-3 is 75%; in particular, they align in the region C terminal to the HMG box (65% identity), which provides a unique signature for individual Tcf factors.
FIG. 2.
Phylogenetic relationship among members of the Tcf/Lef gene family based on aligned amino acid sequences. Relative branch lengths are drawn proportional to the number of inferred substitutions per site.
Two regions of conservation between mTcf-3, mTcf-4, and other members of the family were of particular interest: (i) the N terminus, which in XTcf-3, hTcf-1 and -4, Lef-1, and dTCF/Pangolin (2, 4, 11, 16, 19, 39) constitutes the β-catenin interaction domain, and (ii) the HMG box DNA-binding domain, which has >90% identity between individual members of the Tcf family. This predicted that mTcf-3 and -4 would bind to Tcf consensus DNA motifs and would interact with β-catenin.
Predominantly embryonic expression of mTCF-3 and mTCF-4.
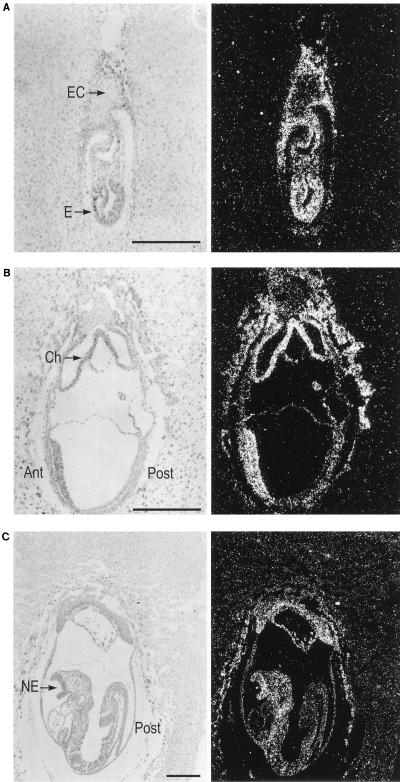
We next determined the embryonic expression pattern of mTcf-3 and mTcf-4. As depicted in Fig. 3A for a longitudinal section of an embryonic day 6.5 (E6.5) embryo, strong expression of mTcf-3 was observed throughout the embryo proper. In addition, all extraembryonic tissues with the exception of the ectoplacental cone and the decidua were positive. By E7.5, expression levels were declining, with highest signals remaining in the anterior part of the embryo. The posterior part, including the primitive streak, showed lower expression levels (Fig. 3B). At E8.5, the head region still showed expression, especially in the anterior neurectoderm. Expression had almost disappeared in the posterior part of the embryo (Fig. 3C). From E10.5 onward, mTcf-3 expression became undetectable (not shown).
FIG. 3.
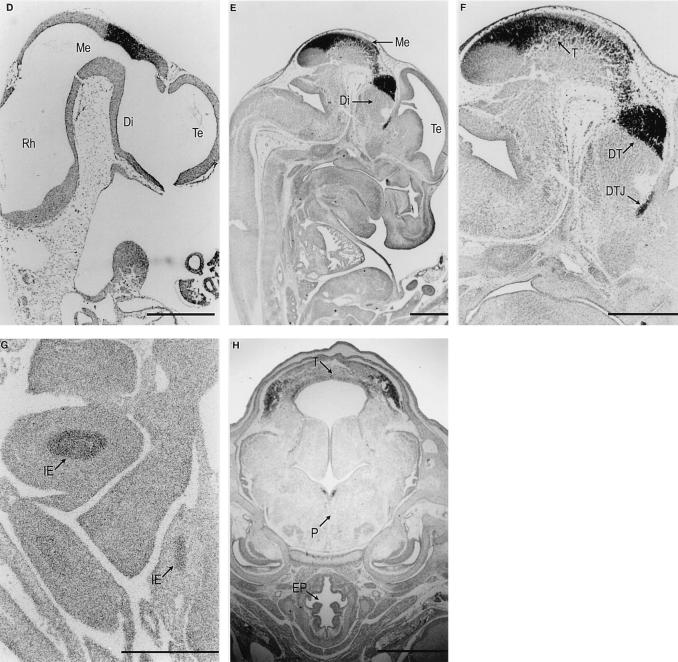
In situ hybridization analysis of mTcf-3 (A to C) and mTcf-4 (D to H) expression on murine embryo sections. mTcf-3 was detected in E6.5 (A), E7.5 (B), and E8.5 (C) embryos. Bright-field (left) and the accompanying dark-field (right) pictures of the longitudinal sections are shown. Embryo proper (E), ectoplacental cone (EC), chorion (Ch), and neuroectoderm (NE) are indicated. mTcf-4 expression was first observed at E10.5 (D) in the central nervous system. The expression remains restricted at E13.5 (E). (F) Detail of the head region; (G) detail of the intestine at E13.5; (D to G) parasagittal sections; (H) coronal section of the head at E16.5. Rhombencephalon (Rh), mesencephalon (Me), diencephalon (Di), telencephalon (Te), tectum (T), dorsal thalamus (DT), di-telencephalic junction (DTJ), intestinal epithelium (IE), pons (P), and esophagus (EP) are indicated. The bar in panel G represents 0.1 mm; bars in other panels represent 0.3 mm.
mTcf-4 expression was undetectable on E6.5, E7.5, and E8.5 (not shown) and was first observed at E10.5 in the roof of the diencephalon and in the anterior part of the mesencephalon, with a sharp posterior boundary (Fig. 3D). At all later embryonic stages, high-level expression was mostly restricted to the central nervous system. At E13.5, mTcf-4 mRNA was expressed in the roof of the mesencephalon (the tectum) and in the dorsal thalamus. The expression was sharply demarcated from the remainder of the thalamus complex by the mTcf-4-negative zona limitans intrathalamica. High-level expression was also detected at the ditelencephalic junction (Fig. 3E and F). At E13.5, a relatively low level of mTcf-4 mRNA was observed in the intestinal epithelium (Fig. 3G). The intestinal epithelium was the only embryonic tissue with detectable levels of mTcf-4 expression outside the central nervous system. This expression pattern remained essentially unchanged at E16.5. On coronal sections at E16.5, mTcf-4 mRNA was also detected in the pons cerebri (Fig. 3H).
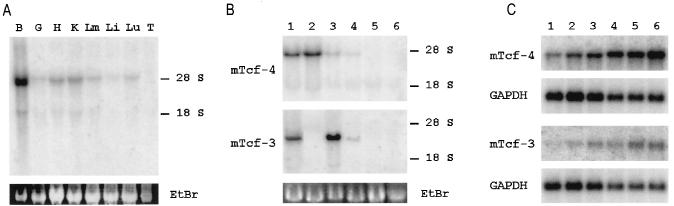
Expression of mTcf-3 and mTcf-4 was also analyzed by Northern blotting on a panel of mouse embryonic (E18.5) and adult tissues and on several selected cell lines. As expected, no mTcf-3 expression was observed in any of the embryonic or adult tissue RNA samples (not shown). By contrast, high-level expression was detected in E14 embryonic stem cells and in the mammary epithelial cell line C57MG, while low-level expression occurred in NIH 3T3 fibroblasts (Fig. 4B). In line with the in situ data, high-level mTcf-4 expression was observed in embryonic brain; all other tissues were essentially negative (Fig. 4A), as were all adult tissues tested (not shown). hTcf-4 is highly expressed in different cell lines derived from colon adenocarcinomas, and hTcf-4 mRNA is clearly detected by in situ analysis in human colon epithelium (16). By in situ analysis, we noticed a low-level mTcf-4 expression in the intestinal epithelium at E13.5 (Fig. 3G). To document expression in the adult gut, we performed Northern blot analysis on poly(A)+ RNA purified from different regions of the adult mouse intestine. As shown in Fig. 4C, mTcf-4 is detected in all parts of the mouse intestine, with a clear gradient along the rostrocaudal axis. The relative values indicate a 10-fold increase of mTcf-4 expression in the distal colon compared to the duodenal part of the small intestine. A similar pattern of expression, albeit at much lower levels, was detected for mTcf-3 (Fig. 4C). Among the tested cell lines, high mTcf-4 expression was observed in the C57MG and PC12 pheochromocytoma cell lines; embryonic stem cells and NIH 3T3 fibroblasts expressed relatively low levels of mTcf-4 mRNA (Fig. 4B).
FIG. 4.
Northern blot analysis of mTcf-4 and mTcf-3 expression. (A) Tissue-specific mTcf-4 expression in mouse embryo at E18.5. The analysis was performed on total RNA isolated from brain (B), gut (G), heart (H), kidney (K), limbs (Lm), liver (Li), lung (Lu), and thymus (T). (B) mTcf-4 and mTcf-3 expression in C57MG cells (lane 1), PC12 cells (lane 2), embryonic stem cells (lane 3), 3T3 fibroblasts (lane 4), RBL-5 T-lymphocyte cells (lane 5), and NS-1 B lymphocyte cells (lane 6). The positions of 18S and 28S rRNAs are shown. EtBr, ethidium bromide stain. (C) Northern blot analysis of poly(A)+ RNA isolated from different parts of the intestine. Lanes: 1, duodenum; 2, jejunum; 3, ileum; 4, cecum; 5, proximal colon; 6, distal colon. GAPDH, control hybridization with a GAPDH probe.
mTcf-3 and mTcf-4 interact with β-catenin.
We next wished to obtain molecular support for the involvement of the two Tcf proteins in Wnt/β-catenin signaling. The interaction between other Tcf members and β-catenin occurs between the N terminus of the former and the Armadillo repeat of the latter. We used the yeast two-hybrid system to demonstrate the interaction. As depicted in Fig. 5, two-hybrid interactions were readily detectable between the two novel mTcf proteins and β-catenin, and were comparable with the previously reported two-hybrid interactions between β-catenin and Tcf-1 (19) and between β-catenin and Lef-1 (2).
FIG. 5.
Two-hybrid mating assay for the interaction of Tcfs and β-catenin. Baits (indicated above the lanes; pVA3 encodes p53) and preys were transformed in MATa and MATα yeast strains, respectively, and mated with each other. As shown by growth on selective plates (lower panel) or β-galactosidase assay (not shown), all Tcfs (but not the N-terminally truncated hTcf-1 or simian virus 40 large T) interact specifically with β-catenin and not with a control p53 protein (pVA3). All matings grow on nonselective plates (upper panel). −LT, medium lack Leu and Trp; −LTH 25mM 3AT, medium lacking Leu, Trp, and His and containing 25 mM 3-aminotriazole.
Wnt-1 expression induces nuclear β-catenin–Tcf complexes and activates transcription of Tcf reporter genes.
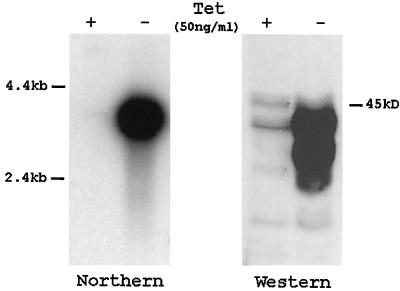
Despite compelling evidence implying Tcf family members as effectors of Wnt signaling, no direct experimental proof has linked Wnt stimulation with the induction of β-catenin–Tcf interactions or with induced Tcf reporter gene transcription. The expression of mTcf-3 and mTcf-4 in the Wnt-1-responsive mammary epithelial C57MG cells allowed us to establish such a direct link. C57MG cells do not express the Wnt-1 gene (23). Upon stimulation by Wnt-1 protein, C57MG cells show morphological transformation and altered growth characteristics (3, 14, 44). Transient expression of Wnt-1 in C57MG cells results in increased stability of β-catenin (26). As shown recently, Wnt-1 stimulation reduces ubiquitination and subsequent degradation of β-catenin via the ubiquitin-proteasome pathway (1). In order to study an effect of Wnt-1 expression on Tcf/β-catenin-mediated transcription, we generated C57MG cells carrying the Wnt-1 gene under the control of a tetracycline-repressible promoter. The resulting 2-69-23 cell line produces a limited amount of Wnt-1 in the repressed state and shows a strong induction of Wnt-1 mRNA and protein within 24 h after removal of tetracycline (Fig. 6).
FIG. 6.
Inducible expression of Wnt-1 in C57MG cells. (A) Northern blot analysis of Wnt-1 RNA in C57MG cells (clone 2-69-23) carrying a Wnt-1 transgene under the control of a tetracycline (Tet)-repressible promoter. Poly(A)+ RNA was isolated from cells cultured in the presence of tetracycline (50 ng/ml) or for 24 h in the absence of the drug. The Wnt-1 transcript was detected by blot hybridization after gel electrophoresis using a 32P-labeled probe for Wnt-1. A longer exposure detects the presence of the major Wnt-1 transcript in tetracycline-treated cells. Wnt-1 expression was quantitated with the model 300A computing densitometer (Molecular Dynamics), and basal levels were found to be 50 times lower than the derepressed levels (data not shown). A probe for neomycin was used to confirm the presence of equal levels of RNA in each lane (not shown). (B) Western blot analysis of 2-69-23 cells. Equal amounts of Triton X-100-soluble protein from cells cultured in the presence of tetracycline (50 ng/ml) or for 24 h in the absence of the drug were analyzed by immunoblotting with anti-Wnt-1 antibody.
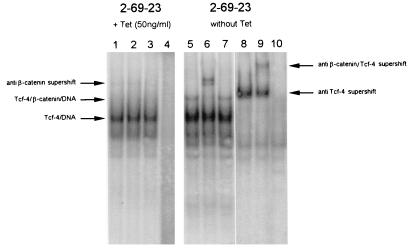
Using a gel retardation-supershift assay, we have previously demonstrated the constitutive presence of a nuclear β-catenin–hTcf-4 complex in APC−/− colon carcinoma cells (16) and in colon carcinoma cells with dominant-positive mutations in β-catenin (21). We now applied the same assay to determine whether Wnt stimulation could induce the formation of such complexes. We performed gel retardation with nuclear extracts from 2-69-23 cells stably transfected with a tetracycline-repressible Wnt-1 expression vector. We readily obtained a Tcf-DNA complex from both uninduced and induced cells, using an optimal Tcf binding motif as probe. The complex could be supershifted with a monoclonal antibody recognizing human and mouse Tcf-4 (Fig. 7). Wnt-1 induction resulted in the appearance of an additional band, which could be further supershifted with a β-catenin antibody (Fig. 7). The anti-Tcf-4 antibody used for the gel retardation/supershift assay does not cross-react with other mouse Tcf proteins (1a). The completeness of the supershift indicated that Tcf-4 was the only Tcf protein present in the extracts. Our attempts to prepare nuclear or whole-cell extracts showing DNA binding activity specific for the Tcf-3 protein were not successful. We concluded that the Tcf-3 protein most likely remained associated with insoluble nuclear components and is not eluted from nuclei by conventional extraction procedures. A similar feature was observed for Tcf-1 protein (38).
FIG. 7.
Wnt-1 induces a Tcf–β-catenin complex in C57MG cells, as determined by a gel retardation assay performed with nuclear extracts from 2-69-23 cells growing in the presence or absence (Wnt-1 induction) of tetracycline (Tet; 50 ng/ml). Samples in lanes 1 and 5 were incubated under standard conditions. Anti-β-catenin antibody (0.2 μg) was added to the samples in lanes 2, 6, and 9. A control antibody (human CD4; 0.2 μg) was added to the samples in lanes 3 and 7. Anti-Tcf-4 antibody (affinity purified, 0.2 μg) was added to samples in lanes 8 and 9. Samples in lanes 4 and 10 were incubated with a probe mutated in the Tcf binding site.
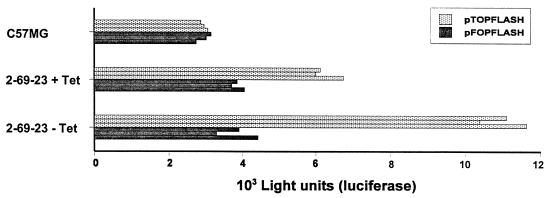
To test if Wnt stimulation could activate transcription from Tcf target genes, we performed transient transfections in C57MG and 2-69-23 cells with the Tcf reporter plasmid pTOPFLASH or the mutant negative control plasmid pFOPFLASH (16). In the original C57MG cell line, no differences in activities of these two reporters were detected (Fig. 8). In repressed 2-69-23 cells, a small induction of Tcf reporter gene transcription was already observed, likely the consequence of leaky Wnt-1 expression. Wnt induction resulted in a threefold stimulation of the Tcf reporter gene transcription compared to negative control plasmid (Fig. 8). Taken together, these data provided formal proof that Wnt stimulation indeed induces the formation of nuclear Tcf–β-catenin complexes and activates transcription from Tcf target genes.
FIG. 8.
Wnt-1 activates Tcf reporter construct in C57MG cells. C57MG cells and 2-69-23 cells growing in the presence or absence of tetracycline (Tet) were transfected with 1 μg of the indicated luciferase reporter construct and 1 μg of plasmid pCATCONTROL as an internal control. Cells were harvested after 24 h, and luciferase and CAT values were determined. All experiments were done in triplicate; triplicate values are given.
DISCUSSION
This report describes the characterization of two murine members of the Tcf family. Based on their expression profiles and the physical and functional interactions with the Wnt effector β-catenin, mTcf-3 and mTcf-4 are potential candidates to mediate Wnt-driven developmental decisions in the mouse. Taking advantage of the expression of the Tcf genes in Wnt-1-responsive C57MG cells, we provide direct evidence that Wnt-1 stimulation induces the formation of nuclear complexes between Tcf family members and β-catenin, which in turn can activate transcription from Tcf reporter genes.
mTcf-3 in the early embryo.
Based on sequence comparisons, the mTcf-3 cDNA clone is most closely related to the maternally expressed Xenopus Tcf family member, XTcf-3. XTcf-3 is broadly expressed in the Xenopus embryo (19). Like XTcf-3, mTcf-3 is ubiquitously expressed in the gastrulating embryo. At E7.5, mTcf-3 expression is localized to the anterior part of the embryo and then gradually disappears. An earlier study showed an opposite pattern of Tcf-1 and Lef-1 expression, with highest levels of both mRNAs in the posterior part of the embryo (25). Thus, three Tcf family members are expressed in partially overlapping patterns during early postimplantation development.
Expression patterns of mouse Wnt genes have been studied in great detail (9, 18, 20, 27, 30, 34, 36, 43). Three Wnt family members, Wnt-3a, Wnt-5a, and Wnt-5b, are expressed in discrete spatial domains in the primitive streak. At early primitive-streak stages (E6.5), Wnt-5a mRNA is detected only in mesodermal cells; Wnt-5b mRNA is localized in epiblast and mesoderm within the streak. By the extended-streak stages (E7.5), Wnt-3a is widely expressed within the streak. In contrast to Wnt-5a and Wnt-5b expression, which is posteriorly localized, Wnt-3a expression extends more anteriorly, suggesting that Wnt-3a may regulate the formation of embryonic mesoderm arising from the anterior primitive streak. Indeed, the phenotype of Wnt-3a-deficient mice indicates that Wnt-3a regulates dorsal mesoderm fate and is required for generation of embryonic mesoderm (34). Tcf-1, Lef-1, and Tcf-3 very likely mediate signaling of one or more of the Wnt genes discussed above. Tcf-1 and Lef-1 completely overlap in their early embryonic expression, implying functional redundancy. Based on its expression pattern, we believe that mTcf-3 may perform a distinct function in early murine embryogenesis, mediating signaling of a Wnt factor, possibly Wnt-3a in the anterior regions of the gastrulating embryo. This function is likely equivalent to the role that XTcf-3 plays in Xenopus axis specification.
mTcf4 and the embryonic brain.
During embryogenesis, mTcf-4 displays a highly specific pattern of expression. It appears much later in development than do the other three family members, and its expression is essentially restricted to the di- and mesencephalon and the intestinal epithelium. Strikingly, the area of mTcf-4 expression in embryonic brain also expresses high levels of Lef-1 (25, 40), suggesting a possible redundancy between Tcf-4 and Lef-1 in midbrain development. At least seven Wnt family members are expressed in largely overlapping regions within the central nervous system. Expression patterns of the Wnt-1, -3, and -3a genes show a temporal and spatial overlap in di- and mesencephalon with that of Tcf-4 and Lef-1 in this region. Therefore, the latter two Tcf genes are likely candidates to mediate signaling of these Wnt factors. Wnt-1, essential for midbrain development as demonstrated by gene knockout (17, 35), is expressed at E10.5 in the dorsal wall of the diencephalon, and its expression continues along the dorsal wall of the midbrain. Additional sites of Wnt-1 expression are detected in the lateral wall of the midbrain close to its junction with hindbrain and ventral wall of midbrain and diencephalon. At E10.5 and at later stages, the Wnt-3 gene is broadly expressed in the dorsal part of the neural tube, with a rostral boundry in the dorsal thalamus. The anterior edge of the boundary is very sharp, clearly separating the Wnt-3-positive dorsal thalamus from the Wnt-3-negative remainder of diencephalon. The same sharp boundary of expression was observed for Tcf-4 and Lef-1. Like Wnt-3, the Wnt-3a gene is expressed in the dorsal part of the neural tube. Expression is mostly observed in the dorsal midline up to the diencephalon. In the hindbrain and in the diencephalon, midline expression bifurcates. Rostrally, the area of Wnt-3a expression extends into the medial walls of the telencephalon. Within the diencephalon, Wnt-3a expression marks the boundary between the dorsal and ventral thalamus.
Null mutations in four Wnt genes have been described (17, 20, 33–35). It is of obvious interest to compare the phenotypes of Wnt knockout mice with those of Tcf/Lef knockout mice. Only the disruption of the Wnt-1 gene yields a defect in midbrain development. The phenotype of the Wnt-1 and Wnt-3a compound mutant mice has demonstrated clear redundancy of these genes in the regulation of dorsal neural tube differentiation (12). Despite the evidence for the involvement of Tcf factors in Wingless/Wnt signaling (22), the single-knockout phenotypes of Tcf-1 and Lef-1 genes have not implied a role for these two genes in Wnt-1-driven development of the midbrain (17, 35), Wnt-3a-regulated somite and tailbud formation (34), Wnt-4-dependent kidney tubulogenesis (33), or Wnt-2-regulated development of the placental tissues (20). However, as mentioned earlier, the expression patterns of Tcf-1 and Lef-1 are virtually completely overlapping, suggestive of functional redundancy. Analysis of Tcf-1/Lef-1 double-knockout mice is ongoing.
mTcf-4 in the intestine.
We recently found hTcf-4 to be expressed in human colonic epithelial cells and cancers derived therefrom (16). The available evidence indicates that loss of the tumor suppressor protein APC or gain-of-function mutations in β-catenin leads to uncontrolled transcriptional activity of Tcf target genes and, as a consequence, to cellular transformation. The mTcf-4 expression in the embryonic intestinal epithelium suggests a physiological role for the gene in this tissue, possibly in the context of Wnt signaling events that control epithelial homeostasis. In contrast to the brain-specific expression, the intestinal mTcf-4 expression persists to the adult age. mTcf-4 mRNA in mouse intestine is not restricted to the colon but is also detected in the small intestine. Interestingly, mTcf-4 mRNA shows a gradient with increased levels along the rostrocaudal axis. The same gradient (albeit at much lower levels) was also detected for mTcf-3. The biological relevance of this gradient is currently not obvious.
Extensive cloning efforts in our lab, as well as searches of expressed sequence tags (EST) databases, have failed to identify yet other members of the Tcf family in mammals. We therefore believe that the full complement of mammalian Tcf proteins has now been cloned. Ongoing knockout experiments of the individual Tcf genes as well as crosses between the knockout mice will reveal the unique and redundant functions of individual Tcf family members and the roles they may play in the various Wnt-driven developmental decisions.
ACKNOWLEDGMENTS
We thank both Harold Varmus and Roel Nusse, in whose labs the cells with tetracycline-responsive Wnt-1 expression were established and characterized. We are grateful to Laura van ’t Veer for two hybrid vectors, Herman van der Putte for cDNA libraries, Bas Defize for helpful discussion, and the members of the Destree and Clevers labs for discussions and for carefully reading the manuscript.
This work was supported by PIONIER and program grants from NWO-GMW to H.C.
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Barker, N. Unpublished data.
- 2.Behrens J, von Kries J P, Kuehl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown A M, Wildin R S, Prendergast T J, Varmus H E. A retrovirus vector expressing the putative mammary oncogen int-1 causes partial transformation of a mammary epithelial cell line. Cell. 1986;46:1001–1009. doi: 10.1016/0092-8674(86)90699-9. [DOI] [PubMed] [Google Scholar]
- 4.Brunner E, Peter O, Schweizer L, Basler K. Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 5.Castrop J, Hoevenagel R, Young J R, Clevers H C. A common ancestor of the mammalian transcription factors TCF-1 and TCF-1α/LEF-1 expressed in chicken T cells. Eur J Immunol. 1992;22:327–1330. doi: 10.1002/eji.1830220531. [DOI] [PubMed] [Google Scholar]
- 6.Castrop J, van Norren K, Clevers H C. A gene family of HMG box factors with homology to TCF-1. Nucleic Acids Res. 1992;20:611. doi: 10.1093/nar/20.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Funayama N, Fagotto F, McCrea P, Gumbiner B M. Embryonic axis induction by the armadillo repeat domain of β-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavin B J, McMahon J A, McMahon A P. Expression of multiple novel Wnt-1/int-1-related genes during fetal and adult mouse development. Genes Dev. 1991;4:2319–2332. doi: 10.1101/gad.4.12b.2319. [DOI] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 12.Ikeya M, Lee S M, Johnson J E, McMahon A P, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 13.Joyner A L. Engrailed, Wnt and Pax genes regulate midbrain-hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- 14.Jue S F, Bradley R S, Rudnicki J A, Varmus H E, Brown A M. The mouse Wnt-1 gene can act via a paracrine mechanism in transformation of mammary epithelial cells. Mol Cell Biol. 1992;12:321–328. doi: 10.1128/mcb.12.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klymkowsky M W, Parr B. The body language of cells: the intimate connection between cell adhesion and behavior. Cell. 1995;83:5–8. doi: 10.1016/0092-8674(95)90226-0. [DOI] [PubMed] [Google Scholar]
- 16.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Sci. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 17.McMahon A P, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 18.McMahon A P, Joyner A L, Bradley A, McMahon J A. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- 19.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 20.Monkley S J, Delaney S J, Pennisi D J, Christiansen J H, Wainwright B J. Targeted disruption of the Wnt2 gene results in placentation defects. Development. 1996;122:3343–3353. doi: 10.1242/dev.122.11.3343. [DOI] [PubMed] [Google Scholar]
- 21.Morin P J, Sparks A, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 22.Nusse R. A versatile transcriptional effector of Wingless signaling. Cell. 1997;89:321–323. doi: 10.1016/s0092-8674(00)80210-x. [DOI] [PubMed] [Google Scholar]
- 23.Olson D J, Papkoff J. Regulated expression of Wnt family members during proliferation of C57mg mammary cells. Cell Growth Differ. 1994;5:197–206. [PubMed] [Google Scholar]
- 24.Oosterwegel M, van de Wetering M, Dooijes D, Klomp L, Winoto A, Georgopoulos K, Meijlink F, Clevers H. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-ɛ and T cell receptor α enhancers. J Exp Med. 1991;173:1133–1142. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 26.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parr B A, Shea M J, Vassileva G, McMahon A P. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- 28.Peifer M. Cell adhesion and signal transduction: the Armadillo connection. Trends Cell Biol. 1995;5:224–229. doi: 10.1016/s0962-8924(00)89015-7. [DOI] [PubMed] [Google Scholar]
- 29.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling imputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 30.Roelink H, Nusse R. Expression of two members of the Wnt family during mouse development—restricted temporal and spatial patterns in the developing neural tube. Genes Dev. 1991;5:381–388. doi: 10.1101/gad.5.3.381. [DOI] [PubMed] [Google Scholar]
- 31.Salinas P C, Nusse R. Regional expression of the Wnt-3 gene in the developing mouse forebrain in relationship to diencephalic neuromeres. Mech Dev. 1990;39:151–160. doi: 10.1016/0925-4773(92)90042-i. [DOI] [PubMed] [Google Scholar]
- 32.Soriano P, Friedrich G, Lawinger P. Promoter interactions in retrovirus vectors introduced into fibroblasts and embryonic stem cells. J Virol. 1991;65:2314–2319. doi: 10.1128/jvi.65.5.2314-2319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark K, Vainio S, Vassileva G, McMahon A P. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 34.Takada S, Stark K L, Shea M J, Vassileva G, McMahon J A, McMahon A P. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:74–89. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 35.Thomas K R, Capecchi M R. Targeted disruption of the murine int-1 protooncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 36.Travis A, Amsterdam A, Belanger C, Grosschedl R. Lef-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 37.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T cell-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generates TCF-1 protein isoforms with differential transcription control properties. Mol Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo co-activates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 40.van Genderen C, Okamura R M, Farinas I, Quo R G, Parslow T G, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1 deficient mice. Genes Dev. 1994;8:2691–2704. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 41.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald H R, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 42.Waterman M L, Fischer W H, Jones K A. A thymus-specific member of the HMG protein family regulates the human T cell receptor Cα enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson D G, Bailes J A, McMahon A P. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell. 1987;50:79–88. doi: 10.1016/0092-8674(87)90664-7. [DOI] [PubMed] [Google Scholar]
- 44.Wong G T, Gavin B J, McMahon A P. Differential transformation of mammary epithelial cells by Wnt Genes. Mol Cell Biol. 1994;14:6278–6286. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]