Abstract
Plant pathogenic Pseudomonas syringae deliver type III effector proteins into the host cell, where they function to manipulate host defense and metabolism to benefit the extracellular bacterial colony. The activity of these virulence factors can be monitored by plant disease resistance proteins deployed to “guard” the targeted host proteins. The Arabidopsis RIN4 protein is targeted by three different type III effectors. Specific manipulation of RIN4 by each of them leads to activation of either the RPM1 or RPS2 disease resistance proteins. The type III effector AvrRpt2 is a cysteine protease that is autoprocessed inside the host cell where it activates RPS2 by causing RIN4 disappearance. RIN4 contains two sites related to the AvrRpt2 cleavage site (RCS1 and RCS2). We demonstrate that AvrRpt2-dependent cleavage of RIN4 at RCS2 is functionally critical in vivo. This event leads to proteasome-mediated elimination of all but a membrane-embedded ≈6.4-kDa C-terminal fragment of RIN4. One or more of three consecutive cysteines in this C-terminal fragment are required for RIN4 localization; these are likely to be palmitoylation and/or prenylation sites. AvrRpt2-dependent cleavage at RCS2, and release of the remainder of RIN4 from the membrane, consequently prevents RPM1 activation by AvrRpm1 or AvrB. RCS2 is contained within the smallest tested fragment of RIN4 that binds AvrB in vitro. Thus, at least two bacterial virulence factors target the same domain of RIN4, a ≈30-aa plant-specific signature sequence found in a small Arabidopsis protein family that may be additional targets for these bacterial virulence factors.
Keywords: disease resistance, plant immune system
Plant disease resistance (R) genes control the plant immune response upon pathogen recognition. R proteins initiate cellular events that efficiently limit pathogen reproduction (1). Most plant R proteins defined to date possess a central nucleotide binding (NB) domain together with C-terminal leucine-rich repeats (LRR), and hence are termed NB-LRR proteins. NB-LRR proteins are specific in that each is activated by a particular pathogen-encoded molecule. These are polymorphic across the pathogen population. The proteins from Gram-negative plant-pathogenic bacteria that trigger NB-LRR action are type III effector proteins delivered into plant cells through the type III secretion system (2). These are called avirulence (Avr) proteins because their presence renders a given pathogen strain avirulent on a resistant plant genotype (R). Avr proteins, however, can also contribute to pathogen virulence on susceptible plant genotypes (r).
An Avr ligand-R receptor model could explain the genetic specificity of disease resistance, although experimental support for this model is limited. Rather, accumulating evidence suggests that Avr proteins from pathogenic bacteria can be recognized indirectly by their action on one or more host targets (1, 3, 4). In this model, type III effector proteins manipulate host targets, thereby contributing to pathogen virulence. The plant NB-LRR protein, if present, senses the outcome of this manipulation, activating host defenses. This “guard hypothesis” explains the action of several NB-LRR proteins, including the two discussed here, RPM1 (resistance to P. syringae pv. maculicola) and RPS2 (resistance to P. syringae) (5-7).
RPM1 interacting protein 4 (RIN4) is a small (211 aa) protein that, along with RPM1 and RPS2, is localized to the plasma membrane (8-10). RIN4 associates with both RPM1 and RPS2 in yeast two-hybrid and coimmunoprecipitation experiments from plant extracts (9, 11, 12). RPM1 is activated by either of the unrelated AvrRpm1 or AvrB type III effector proteins. Both AvrRpm1 and AvrB are acylated in the host cell after type III secretion system delivery and hence are targeted to the plasma membrane (13). Infection of both RPM1 and rpm1 plants with P. syringae expressing either AvrB or AvrRpm1 leads to phosphorylation of RIN4. RIN4 phosphorylation is correlated with RPM1 activation (9). RIN4 is also targeted by a third type III effector, AvrRpt2, which encodes a cysteine protease (12). Delivery of AvrRpt2 into plant cells results in its autoprocessing and activation (14), which in turn leads to RIN4 disappearance and consequent activation of RPS2 (11, 12). Thus, three different type III effectors use at least two different mechanisms to manipulate RIN4, and thereby regulate the activation of two different NB-LRR proteins. In the absence of RIN4, RPS2 is activated to an extent that is lethal (9), whereas RPM1 is weakly activated, suggesting that RIN4 has a formal negative regulatory effect on both RPS2 and RPM1 (15). Despite its importance in regulation of RPM1 and RPS2, RIN4 is dispensable for the virulence function of AvrRpm1 and AvrRpt2 in disease-susceptible rin4 rpm1 rps2 plants, suggesting the existence of additional host cellular targets (15).
Delivery of AvrRpt2 interferes with AvrRpm1- or AvrB-dependent activation of RPM1 (16), suggesting that these three type III effectors compete for a common factor required for RPM1 function. The isolation of RIN4 offers the opportunity to test directly the hypothesis of whether AvrRpt2-dependent cleavage and subsequent elimination of RIN4 from the plasma membrane interferes with AvrRpm1- or AvrB-dependent activation of RPM1. We also examine the requirements for RIN4 cleavage by AvrRpt2 and the means by which RIN4 is localized to the plasma membrane.
Materials and Methods
Bacterial Inoculations. Hypersensitive response (HR) and growth restriction assays were as described (15).
Site-Directed PCR-Mediated Mutagenesis. Mutations in RIN4 (At3g25070) were generated by PCR-based site-directed mutagenesis using PFU turbo high-fidelity polymerase (Strategene). Complementary PCR forward and reverse primers were used for mutagenesis of RIN4 (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for details).
Transgenic Plants. RIN4 mutant derivatives were cloned into binary vector pTA7002 (17). Agrobacterium tumefaciens (GV3101) carrying pTA7002 with each RIN4 derivative were vacuum-infiltrated into flowering Arabidopsis rin4 rps2 RPM1 or rin4 rps2 rpm1 plants (15). Basta (AgroEvo, Frankfurt, Germany) was sprayed on 1-week-old T1 seeds to select for transgenic progeny. All analyses of transgenic plants were carried out by using Basta-resistant T2 lines treated with 20 μM dexamethsone (DEX) (9).
Protein. Two to three leaves of 2 cm2 were ground in sucrose buffer (20 mM Tris, pH 8.0/0.33 M sucrose/1 mM EDTA/1× Sigma protease inhibitor mixture) and centrifuged at 2,000 × g, and the supernatant was collected as the total protein extract. Half of the total extract was centrifuged at 20,000 × g, resulting in soluble fraction and microsomal membrane fractions. Protein concentrations of total, soluble, and microsomal membrane fractions were equalized by using 6× loading buffer (120 nM Tris, pH 8.6/50% glycerol/6% SDS/2% 2-mercaptoethanol). Samples were separated by 12% SDS/PAGE and transferred onto nitrocellulose membranes for Western detection (ECL kit; Amersham Pharmacia Biosciences). Anti-RIN4 sera (9) was used at 1:5,000 in 1× TTBS (1 M Tris, pH 8.0/5 M NaCl/0.05% Tween 20/1% milk powder). The proteasome inhibitor clasto-lactacysteine β-lactone (Calbiochem; 20 μM in 1% DMSO) was hand-inoculated into leaves.
2-Bromopalmitic Acid (2-BPA) Treatment of Arabidopsis Plants. Leaves of 3-week-old rin4 rps2 RPM1 plants were hand-inoculated with 2-BPA (Sigma-Aldrich) at 100 μM in 1% DMSO. Plants were sprayed with 20 μM DEX 1 h after 2-BPA treatment. Leaves were harvested 24 h later and fractionated into total, soluble, and membrane fractions as described above. Proteins were separated by 12% SDS/PAGE for Western blot analysis.
In Vitro AvrB-RIN4 Binding Assays. AvrB was cloned from P. syringae pv. glycinea with an N-terminal histidine tag and transformed into BL21CodonPlus (DE3)-RIL cells (Stratagene). Expressed protein was purified on high-trap chelating columns according to the manufacturer's instructions (Amersham Pharmacia Biosciences; see Supporting Materials and Methods for details). RIN4 fragments were amplified by PCR, subcloned as N-terminal GST-fusion proteins by using Gateway pDEST-15 vector (Invitrogen), and transformed into BL21CodonPlus (DE3)-RIL cells (Stratagene). Fusion proteins were purified by using Glutathione Sepharose 4B (Amersham Pharmacia Biosciences; see Supporting Materials and Methods for details).
For native gel electrophoresis, AvrB was mixed with a purified GST-RIN4 fragment, incubated on ice, and run on a Homogeneous 12.5 PhastGel (Amersham Pharmacia Biosciences) by using a Phast System (Amersham Pharmacia). For gel filtration analysis, isolated RIN4142-179 peptide was mixed with AvrB, and the mixture was separated by using a calibrated hand-poured XK 16/70 Superdex 75 (Amersham Pharmacia) column equilibrated in 20 mM Hepes (pH 7.5), 150 mM NaCl, and 2 mM DTT. Five-milliliter fractions were collected and analyzed by SDS/PAGE (see Supporting Materials and Methods for details).
Results
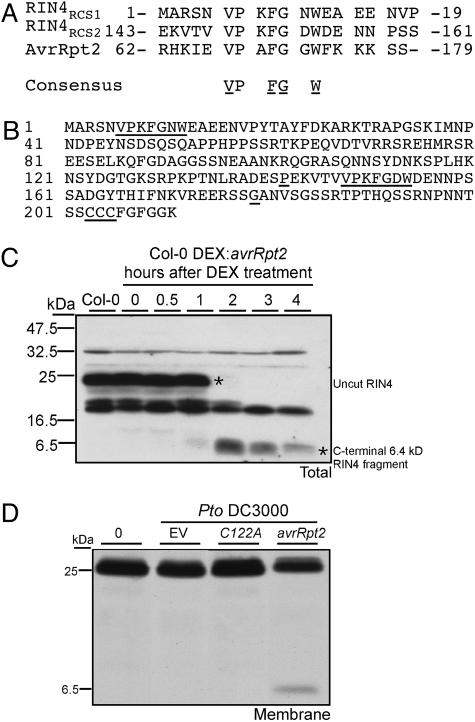
AvrRpt2 Cleaves RIN4. AvrRpt2 has a cysteine protease cleavage site similar to the catalytic core of staphopains (CA clan) cysteine proteases (12). AvrRpt2 is amino-terminally processed in plant cells into 7- and 21-kDa fragments; the latter is the protease (14). Mutations in the conserved catalytic core of AvrRpt2 prevented autoproteolysis and both RIN4 degradation and RPS2 activation (12). RIN4 contains two sites related to the AvrRpt2 autocleavage site (Fig. 1A), termed RCS (AvrRpt2 cleavage site) 1 (V6-W12) and RCS2 (V148-W154) (Fig. 1B and ref. 18). Using GST-RIN4 fusion proteins, we demonstrated that the presence of catalytically active AvrRpt2 and an unknown factor present in Arabidopsis extracts led to the appearance of appropriately sized RIN4 fragments (data not shown and Supporting Materials and Methods).
Fig. 1.
RIN4 contains two cysteine protease cleavage sites. (A) A consensus cysteine protease cleavage site is present in both RIN4 and AvrRpt2. Block mutations to alanine discussed below were made in the underlined residues, V-FG-W. (B) RIN4 amino acid sequence features: RCS1 (V6-W12), RCS2 (V148-W154), and a putative palmitoylation site at C203-C205 are underlined. The fragment from P142 to G179 (end points underlined) is discussed in the Fig. 6 legend. (C) Overexpression of proteolytically active AvrRpt2 leads to RIN4 cleavage and accumulation of a C-terminal RIN4 fragment. Col-0 transgenic plants conditionally overexpressing AvrRpt2 were treated with 20 μM DEX, and total extracts were prepared from leaves harvested at the indicated time points indicated (hours post-DEX) for SDS/PAGE and Western blot with anti-RIN4 sera. (D) Delivery of enzymatically active AvrRpt2 from P. syringae specifically results in accumulation of a 6.4-kDa membrane-bound RIN4 fragment, consistent with cleavage at RCS2. Col-0 plants were infected with 5 × 107 colony-forming units/ml of Pto DC3000 carrying either avrRpt2 or a protease dead mutant avrRpt2C122A. Protein samples from the membrane microsomal fraction were tested by protein blot with anti-RIN4 sera at 0, 4, and 8 h postinfection.
A C-Terminal, AvrRpt2-Dependent Cleavage Product of RIN4 Is Membrane-Localized. Our in vitro data suggested that both RIN4 RCS1 and RCS2 (Fig. 1 A and B) can be cleaved by AvrRpt2. We reasoned that cleavage of RIN4 by AvrRpt2 might lead to the disappearance of some or all of the RIN4 products from the plant microsomal membrane fraction (hereafter membrane). Cleavage at RCS1 would result in 22.2- and 1.2-kDa fragments, cleavage at RCS2 would result in fragments of 17 and 6.4 kDa, and cleavage at both RCS1 and RCS2 would result in fragments of 15.9, 6.4, and 1.2 kDa. Conditional overexpression (using a DEX-inducible transgene; see Materials and Methods) of WT AvrRpt2 led to RIN4 cleavage and the accumulation over time of a 6.4-kDa RIN4 fragment, consistent with the predicted RIN4 C-terminal fragment remaining after cleavage at RCS2 (Fig. 1C). The same-sized RIN4 fragment was seen exclusively in the membrane fraction after delivery of WT AvrRpt2 to plant cells from P. syringae starting at 4 h postinoculation. This fragment did not appear after the delivery of a catalytically inactive AvrRpt2 C122A mutant (Fig. 1D). Appearance of this 6.4-kDa RIN4 fragment suggests that at least RCS2 is cleaved in vivo and further suggests that the C-terminal cleavage product remains membrane localized. A faint 16-kDa fragment was also observed in the soluble fraction, but only when leaves were pretreated with proteasome inhibitors (data not shown). It is likely that the N-terminal cleavage fragment of 1.2 kDa would not be detected in these experiments.
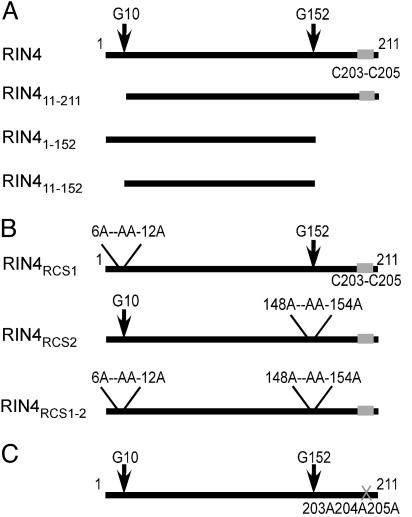
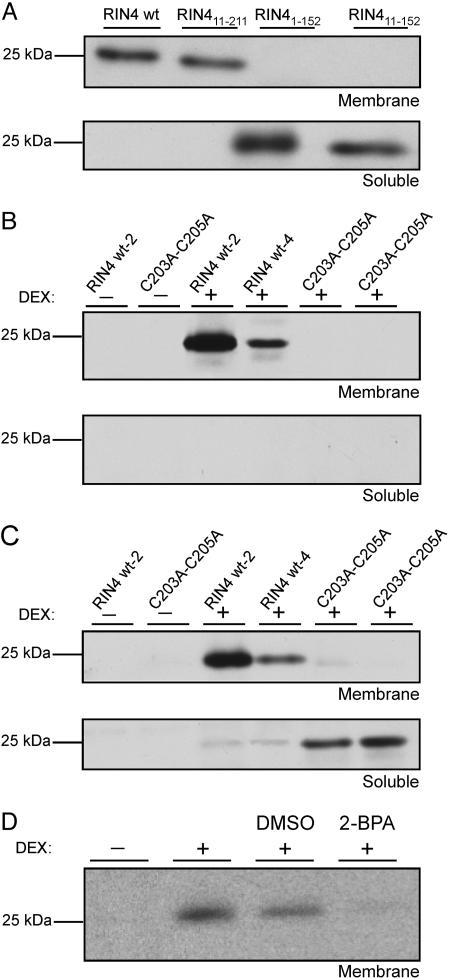
The C-Terminal Region of RIN4 Determines Its Localization by Three Cysteine Residues That Are Likely Palmitoylation and/or Prenylation Sites. To analyze the functional consequences of RIN4 proteolysis at either RCS1 or RCS2, we made three sets of RIN4 mutant derivatives. We mimicked RIN4 cleavage at RCS1 with RIN411-211, cleavage at RCS2 with RIN41-152, and cleavage at both RCS1 and RCS2 with RIN411-152 (Fig. 2A). Transgenic plants conditionally expressing these derivatives were made in rin4 rps2 RPM1. A WT C terminus was required for proper RIN4 localization, because RIN41-152 and RIN411-152 were found only in the soluble fraction, and RIN411-211 localized only to the membrane fraction (Fig. 3A). The conditional RIN4 expression levels were not significantly different from WT RIN4 in any experiments (data not shown, but see Fig. 4B).
Fig. 2.
RIN4 mutant derivative constructs used to test the effect of RIN4 proteolysis on RPM1 activation and AvrRpt2 virulence function. Two sets of conditionally expressed constructs to express mutant RIN4 proteins are shown. (A) The 211-aa RIN4 protein is schematically shown at the top. Putative AvrRpt2 cleavage sites follow G10 and G152. Putative palmitoylation sites at C203, C204, and C205 are denoted by a gray box. RIN4 derivatives that mimic the putative products of AvrRpt2-mediated cleavage were constructed: the larger product from N-terminal cleavage, termed RIN411-211, the product ending at the C-terminal cleavage site, RIN41-152, or the internal product of both cleavage events, RIN411-152. (B) Alanine RIN4 block mutants that alter AvrRpt2-mediated cleavage events at either the N-terminal cleavage site, termed RIN4RCS1, the C-terminal cleavage site, RIN4RCS2, or both, RIN4RCS1-2 (see Fig. 1 for consensus sequences mutated to alanine). (C) Alanine block mutation (gray X) of the RIN4 putative palmitoylation site, C203A/C204A/C205A, generates RIN4C203AC204AC205A.
Fig. 3.
RIN4 is localized to the membrane by a putative palmitoylation site at C203/C204/C205. (A) Membrane localization requires the RIN4 C-terminal region. The RIN4 derivatives that mimic AvrRpt2 cleavage (Fig. 2), RIN411-211, RIN41-152, and RIN411-152, were conditionally expressed in transgenic rin4 rps2 RPM1 plants (20 μM DEX treatment) for 24 h. Protein samples from the microsomal and soluble fractions were analyzed for presence of the respective RIN4 fragments. (B and C) The three possible cysteine palmitoylation sites, C203, C204, and C205 are required for RIN4 localization membrane. Conditionally expressed transgenes encoding WT RIN4 or the RIN4C203AC204AC205A block mutant (Fig. 2) were constructed in rin4 rps2 RPM1 plants. Two transgenic lines for each construct were tested for localization of RIN4 either in the absence (B) or presence (C) of 20 μM clasto-lactacysteine β-lactone (see Materials and Methods). (D) A palmitoylation inhibitor diminishes the amount of RIN4 targeted to the membrane. 2-BPA (100 μM) was infiltrated 1 h before DEX treatment of transgenic rin4 rps2 RPM1 plants conditionally expressing WT RIN4, and samples were taken 24 h later for protein blots.
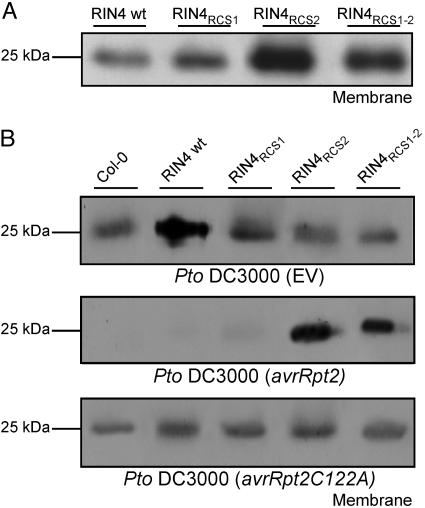
Fig. 4.
In vivo cleavage site of RIN4 by AvrRpt2 at RCS2 is critical. (A) Mutation of the AvrRpt2 cleavage sites in RIN4 does not alter their proper localization to the membrane. Membrane and soluble protein fractions were prepared from leaves 24 h after 20 μM DEX treatment of transgenic rin4 rps2 RPM1 plants conditionally expressing the proteins listed at the top (see Fig. 2). (B) AvrRpt2-dependent cleavage at RIN4RCS2 is required for elimination of RIN4 from the membrane. Plants were treated with DEX as in A and inoculated 24 h later with 5 × 105 colony-forming units/ml of Pto DC3000(EV), Pto DC3000(avrRpt2), or Pto DC3000 (avrRpt2C122A). Leaf samples were collected for microsomal membrane protein extraction 12 h after infection.
The C-terminal region of RIN4 contains possible palmitoylation and/or prenylation sites at C203, C204, and C205 (Fig. 1B). Posttranslational acylation of any of these cysteines could drive membrane association and/or integration of RIN4 into lipid microdomains within the plasma membrane (19). We created the RIN4C203AC204AC205A mutant (Fig. 2). Membrane and soluble fractions from two independent rin4 rps2 RPM1 transgenic lines conditionally expressing either WT RIN4 or RIN4C203AC204AC205A were analyzed. As expected, WT RIN4 is in the membrane fraction (Fig. 3 B and C). We could not detect any RIN4C203AC204AC205A in these extracts unless we inoculated a proteasome inhibitor at the time of DEX induction (compare Fig. 3 B with C). Thus, C203, C204, and C205 are required for RIN4 localization, and RIN4C203AC204AC205A is rapidly degraded via the proteasome.
We used a palmitoylation inhibitor, 2-BPA (Materials and Methods; ref. 19), to test whether palmitoylation of RIN4 contributes to its membrane localization. Infiltration of 2-BPA into leaves of transgenic plants before DEX treatment significantly reduced RIN4 accumulation in the membrane fraction, as compared with untreated or mock-treated leaves (Fig. 3D). Further, 2-BPA treatment eliminated RIN4 accumulation from the soluble fraction (data not shown). This result suggests that one or more of the cysteines at C203, C204, and C205 are palmitoylated, this acylation is responsible for efficient RIN4 localization, and full-length RIN4 is destabilized in the absence of acylation. Despite their genotype, the rin4 rps2 RPM1 plants used for our transgenics do not generate an AvrRpm1-driven HR because diminution of RIN4 levels leads to proportional reduction in RPM1 accumulation (9). Consistent with these findings, DEX-treated RIN4C203AC204AC205A plants did not express an RPM1-dependent HR after inoculation with DC3000(avrRpm1), whereas DEX-treated RIN4 plants did (data not shown).
RIN4 Mutants in RCS1 and RCS2 Retain Proper Membrane Localization, and AvrRpt2 Cleavage at RCS2 Is Required for RIN4 Disappearance. We ultimately wanted to test whether AvrRpt2-dependent cleavage of RIN4 from the plasma membrane perturbs RPM1 activation by AvrRpm1. First, we conditionally expressed RIN4 mutants that have alanine substitutions at either RCS1 or RCS2 or both (Figs. 1 A and B and 2) in rin4 rps2 RPM1 transgenic plants. We did not change the conserved RCS proline to avoid possible structural disruption. Mutation of the RCS sites did not alter membrane localization of these RIN4 derivatives, because all three accumulated only in the membrane fraction (Fig. 4A). We inoculated DEX-treated plants expressing WT RIN4, RIN4RCS1, RIN4RCS2, or RIN4RCS1-2 with either Pseudomonas syringae pv. tomato (Pto) DC3000(EV), Pto DC3000(avrRpt2), or Pto DC3000(avrRpt2C122A). AvrRpt2 caused disappearance of both the endogenous RIN4 in Col-0 and the slightly higher levels of WT RIN4 achieved with the conditional expression system (Fig. 4B). AvrRpt2 also drove the disappearance of RIN4RCS1. By contrast, AvrRpt2 did not trigger disappearance of RIN4RCS2 and RIN4RCS1-2 from the membrane (Fig. 4B). As expected, the enzymatic activity of AvrRpt2 is required for these events (Fig. 4B). Combined with data from Fig. 3, our results demonstrate that (i) AvrRpt2 action at RCS2 is critical in vivo for RIN4 elimination and (ii) cleavage at RCS2 releases RIN4 from the membrane, allowing subsequent proteasome-dependent elimination of the resulting fragments.
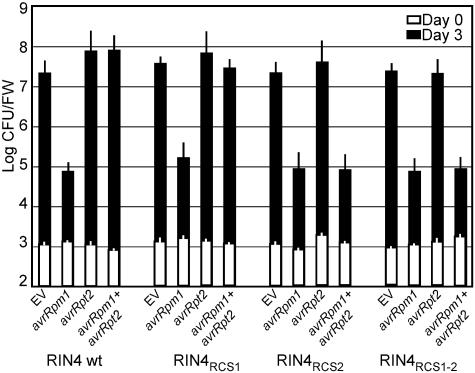
AvrRpt2-Dependent Cleavage of RCS2 Is Required to Interfere with RPM1 Activation. Because the RIN4RCS1, RIN4RCS2, and RIN4RCS1-2 derivatives were properly localized, we addressed whether they supported RPM1 activation. Both RPM1-dependent restriction of bacterial growth and RPM1-mediated HR were restored by each RCS mutant (Fig. 5 and Fig. 7, which is published as supporting information on the PNAS web site). Thus, RCS mutation does not alter AvrRpm1 (or AvrB, data not shown) activation of RPM1. Inoculation of plants expressing WT RIN4 with Pto DC3000(avrRpm1 plus avrRpt2) neither resulted in restricted growth (Fig. 5B), nor elicited RPM1-mediated HR (Fig. 7) as expected (16). AvrRpt2 also blocked these AvrRpm1-dependent responses in RIN4RCS1 plants (Fig. 5). However, AvrRpt2 could not block either of the two AvrRpm1-dependent responses in RIN4RCS2- or RIN4RCS1-2-expressing plants (Fig. 5). Similar results were obtained for AvrB-dependent responses (data not shown). We conclude that AvrRpt2 cleavage of RIN4 at RCS2 is required to interfere with RPM1 activation by either AvrRpm1 or AvrB.
Fig. 5.
AvrRpt2-dependent cleavage at RIN4RCS2 blocks activation of RPM1 by AvrRpm1. Plants expressing RIN4 and RCS mutant derivatives listed at the bottom were treated with DEX (as in Fig. 4) to induce expression of WT RIN4 and RCS mutant derivatives listed below and then inoculated with 5 × 105 colony-forming units/ml of Pto DC3000-expressing genes listed. Bacterial growth was measured 0 (empty bars) and 3 days (filled bars) postinoculation. Error bars represent standard deviation from three samples. The values from the experiment are representative of two additional replicates.
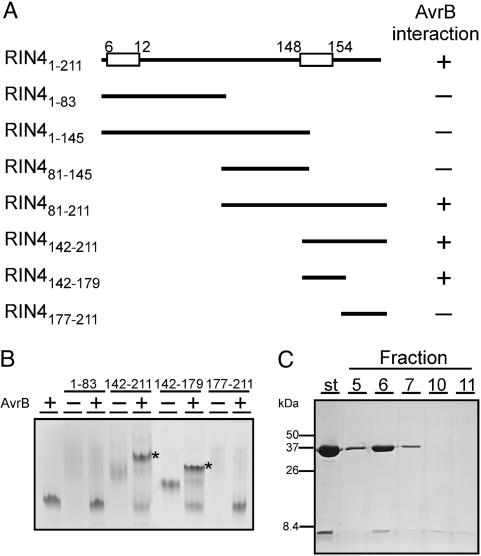
The AvrB Interacting Domain of RIN4 Overlaps with the C-Terminal Cysteine Protease Cleavage Site. AvrRpm1 and AvrB associate with RIN4, resulting in its phosphorylation. AvrRpm1 coimmunoprecipitates with RIN4, but direct interaction with RIN4 has not been demonstrated. AvrB, however, interacts with RIN4 in yeast two-hybrid assays, suggesting direct interaction (9). We mapped the region of RIN4 that interacts directly with AvrB by using epitope-tagged RIN4 deletion derivatives (Fig. 6A) purified from Escherichia coli extracts (see Materials and Methods). Each purified RIN4 derivative was incubated with purified AvrB and analyzed on native PAGE gels. We observed a molecular weight band higher than AvrB alone only when WT RIN4, RIN4142-211, or RIN4142-179 (Fig. 6B, indicated by *) were incubated with AvrB. The AvrB and RIN4142-179 mixture was loaded onto a calibrated native gel filtration column, and fractions were collected and run on SDS/PAGE. We observed a protein complex of ≈44 kDa, near the expected molecular mass of 40.4 kDa, consistent with an AvrB and RIN4142-179 complex in fractions 5-7 (Fig. 6C).
Fig. 6.
The RIN4 domain that interacts with AvrB overlaps RCS2. (A) RIN4 deletion mutants were constructed and expressed in E. coli (see Materials and Methods) to define a region sufficient for interaction with AvrB in vitro. The location of RCS1 and RCS2 are indicated by boxes on the RIN4 sequence. (B) Purified AvrB and RIN4 deletion derivatives were incubated together before being run on native 12.5% PAGE gels. Addition of either RIN4142-211 or RIN4142-179 fragments with AvrB resulted in slower band migration (indicated by *). (C) Confirmation of the RIN4142-179 fragment as the AvrB interacting domain was made by native gel filtration and subsequent SDS/PAGE of the relevant fractions. St, starting mixture of AvrB and RIN4142-179. No other fractions contained either protein.
Discussion
The Arabidopsis protein RIN4 is targeted by at least three bacterial virulence factors. One is AvrRpt2, a cysteine protease (12). AvrRpt2-dependent disappearance of RIN4 is required for activation of the RPS2 NB-LRR class disease resistance protein (10, 11). AvrRpm1 and AvrB have no easily predicted biochemical function, yet their presence leads to accumulation of phosphorylated RIN4 and RPM1 activation. Thus, three unrelated type III effector proteins target RIN4 by at least two different mechanisms. Manipulation of RIN4 initiates the activation of two different NB-LRR disease resistance proteins, RPM1 and RPS2. In disease-susceptible plants, the same RIN4 modifications occur, presumably to manipulate host responses regulated by RIN4. AvrRpt2 and AvrRpm1 also have additional targets besides RIN4 in the plant cell (15). The present work was aimed at understanding (i) whether the RCS sequences were cleaved in vivo by AvrRpt2, (ii) how RIN4 was localized, and (iii) whether in vivo cleavage of RCS sequences is the means by which AvrRpt2 interferes with AvrRpm1- or AvrB-mediated activation of RPM1 (16).
Two AvrRpt2 cleavage sites, RCS1 near the N terminus and RCS2 near the C terminus, are found in RIN4 (Fig. 1) (18, 20). RCS1 and RCS2 can be cleaved in vitro by AvrRpt2 in the presence of an unidentified eukaryotic host factor (data not shown and ref. 14). AvrRpt2 drives RIN4 disappearance from total extracts (10, 11). We observed the AvrRpt2-driven accumulation of a small membrane-localized RIN4 fragment. This finding is consistent with cleavage at RCS2 and retention of the remaining C-terminal RIN4 fragment in the membrane. We did not clearly observe RIN4 fragments of sizes consistent with cleavage at either RCS1 or both RCS1 and RCS2 in vivo, unless we coinoculated a proteasome inhibitor. Hence, these presumably soluble fragments might be very short-lived.
We demonstrated RIN4 is tethered to the membrane via probable palmitoylation at C203, C204, and/or C205. Alternatively, the cluster of three cysteines near the C-terminal end of a protein may lead to both palmitoylation and prenylation (19). RIN4C203AC204AC205A did not accumulate in the soluble fraction unless we included a proteasome inhibitor. In contrast, RIN4 derivatives truncated at residue 152 did accumulate in the soluble fraction. Hence, nonacylated RIN4 is rapidly directed to the proteasome, perhaps because it is misfolded, by signals that include sequences between residues 153 and 211.
The RIN4RCS2 and RIN4RCS1-2 mutations abolished AvrRpt2-dependent disappearance of RIN4 from the membrane, whereas the RIN4RCS1 mutation did not. Thus, RCS2 is the key in vivo site for AvrRpt2 action. RIN4RCS1, RIN4RCS2, and RIN4RCS1-2 were properly localized and were competent for AvrRpm1- or AvrB-mediated activation of RPM1, suggesting that neither RCS sequence per se is required for those activities. Both the RIN4RCS2 and RIN4RCS1-2 mutations abolished the ability of AvrRpt2 to inhibit RPM1 activation by AvrRpm1 or AvrB. RCS1 mutation had no effect. Hence, the observation that AvrRpt2 can interfere with AvrRpm1- or AvrB-mediated activation of RPM1 is clarified: AvrRpt2 cleaves RIN4 at RCS2, releasing it from the membrane, disrupting proper RPM1 localization and accumulation. This cleavage blocks the ability of AvrRpm1 or AvrB to activate RPM1. Consistent with these data, we found that AvrB interacts with a small RIN4 142-179 fragment containing RCS2. There was no interaction with various RIN4 fragments containing RCS1. Strikingly, then, at least two type III effectors have evolved to target not only the same host protein, but the same subdomain within that protein.
The targeting of multiple, related host factors by a type III effector may contribute to efficient virulence, compared to the targeting of a single host protein. We discovered a small family of ≈11 Arabidopsis proteins that share with RIN4 a conserved “nitrate-induced” (NOI) domain (pfam05627.3) containing both the RCS and the AvrB binding domain of RIN4. The NOI domain is conserved in small families of otherwise unrelated proteins in both monocots and dicots, suggesting that it has a conserved, plant-specific function. The Arabidopsis proteins (Table 1, which is published as supporting information on the PNAS web site) contain either one or two (NOI10 and NOI11) RCS consensus sequences; we predict that most, but not all, will be cleaved by AvrRpt2. NOI10 and NOI11 are clearly related by duplication. The RCS2 sequence from each deviates significantly from the consensus, suggesting that these will not be AvrRpt2 cleavage sites. Similarly, some, but not all, are predicted to bind AvrB. We find possible palmitoylation or prenylation sequences in all 11 NOI proteins, supporting our contention that these proteins are all acylated into the host plasma membrane.
We propose that the NOI domain proteins have conserved functions in regulation of plant defense responses and that these are targets for at least AvrRpt2, AvrRpm1, and AvrB. For example, an NOI-containing protein from Nicotiana benthamiana (orthologous to At5g55850; NOI4) was required for HR induced through the NB-LRR protein Prf (21). Furthermore, RIN4 overexpression suppresses basal defense responses, and the elimination of RIN4 (in a rin4 rpm1 rps2 triple mutant) leads to enhanced basal defense responses (22). Manipulation of RIN4 and other NOI-containing proteins at the highly conserved NOI domain by AvrRpt2, AvrRpm1, and AvrB may modulate the plant immune system. Because the RCS consensus and the domain that binds AvrB are largely conserved across the small NOI family, we speculate that the conserved sequences are required for the ancestral RIN4 and NOI domain function.
Cleavage of RIN4 by AvrRpt2 at RCS2 removes most of RIN4 from the membrane, presumably altering interactions with, and regulation of, both RPM1 and RPS2. For example, when RIN4 levels are decreased, RPS2 and RPM1 are activated, and levels of RPM1 are proportionally diminished (9, 15). Both AvrRpm1 and AvrB require myristoylation for their localization at the plasma membrane and function (13). The membrane localized, enzymatically active fragment of AvrRpt2 also carries an N-terminal myristoylation site, G72. Both RPM1 and RPS2 require the glycosylphosphatidylinositol-anchored NDR1 protein for their function (23, 24). Acylated proteins are commonly found in membrane subdomains, or lipid rafts. Thus, it is increasingly likely that a set of acylated proteins assembled into a lipid microdomain plays a key role in activation of the plant immune system. This finding would be consistent with type III effectors from animal pathogens that target lipid microdomains for their function in manipulating host cell physiology (25, 26).
Supplementary Material
Acknowledgments
We thank Youssef Belkhadir for excellent discussions and comments on the manuscript. This work was supported by National Science Foundation Arabidopsis 2010 Project Grant IBN-0114795 (to J.L.D.) and National Institutes of Health Grant GM-65533 (to J.S.). H.-S.K. receives partial support from the Cottrell Foundation, and D.D. was a Fellow of the Natural Science and Engineering Research Council of Canada.
Author contributions: H.-S.K., D.D., A.U.S., J.S., and J.L.D. designed research; H.-S.K., D.D., A.U.S., and P.P. performed research; H.-S.K., D.D., A.U.S., P.P., J.S., and J.L.D. analyzed data; and H.-S.K., D.D., and J.L.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Avr, avirulence; HR, hypersensitive response; Pto, Pseudomonas syringae pv. tomato; RPM1, resistance to P. syringae pv. maculicola; RPS2, resistance to P. syringae; RIN4, RPM1 interacting protein 4; NB, nucleotide binding; LRR, leucine-rich repeats; DEX, dexamethsone; 2-BPA, 2-bromopalmitic acid; RCS, AvrRpt2 cleavage site; NOI, nitrate-induced.
References
- 1.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826-833. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R. & Collmer, A. (2004) Annu. Rev. Phytopathol. 42, 385-414. [DOI] [PubMed] [Google Scholar]
- 3.van der Biezen, E. A. & Jones, J. D. G. (1998) Trends Biochem. Sci. 23, 454-456. [DOI] [PubMed] [Google Scholar]
- 4.Van der Hoorn, R. A., De Wit, P. J. & Joosten, M. H. (2002) Trends Plant Sci. 7, 67-71. [DOI] [PubMed] [Google Scholar]
- 5.Grant, M. R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R. W. & Dangl, J. L. (1995) Science 269, 843-846. [DOI] [PubMed] [Google Scholar]
- 6.Bent, A. F., Kunkel, B. N., Dahlbeck, D., Brown, K. L., Schmidt, R., Giraudat, J., Leung, J. & Staskawicz, B. J. (1994) Science 265, 1856-1860. [DOI] [PubMed] [Google Scholar]
- 7.Mindrinos, M., Katagiri, F., Yu, G.-L. & Ausubel, F. M. (1994) Cell 78, 1089-1099. [DOI] [PubMed] [Google Scholar]
- 8.Boyes, D. C., Nam, J. & Dangl, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 15849-15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackey, D., Holt, B. F., III, Wiig, A. & Dangl, J. L. (2002) Cell 108, 743-754. [DOI] [PubMed] [Google Scholar]
- 10.Axtell, M. J. & Staskawicz, B. J. (2003) Cell 112, 369-377. [DOI] [PubMed] [Google Scholar]
- 11.Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R. & Dangl, J. L. (2003) Cell 112, 379-389. [DOI] [PubMed] [Google Scholar]
- 12.Axtell, M. J., Chisholm, S. T., Dahlbeck, D. & Staskawicz, B. J. (2003) Mol. Microbiol. 49, 1537-1546. [DOI] [PubMed] [Google Scholar]
- 13.Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R. T., Katagiri, F. & Dangl, J. L. (2000) Cell 101, 353-363. [DOI] [PubMed] [Google Scholar]
- 14.Mudgett, M. & Staskawicz, B. (1999) Mol. Microbiol. 32, 927-941. [DOI] [PubMed] [Google Scholar]
- 15.Belkhadir, Y., Nimchuk, Z., Hubert, D. A., Mackey, D. & Dangl, J. L. (2004) Plant Cell 16, 2822-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritter, C. & Dangl, J. L. (1996) Plant Cell 8, 251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoyama, T. & Chua, N. H. (1997) Plant J. 11, 605-612. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. A. & Takemoto, D. (2004) Curr. Opin. Immunol. 16, 48-62. [DOI] [PubMed] [Google Scholar]
- 19.Smotrys, J. E. & Linder, M. E. (2004) Annu. Rev. Biochem. 73, 559-587. [DOI] [PubMed] [Google Scholar]
- 20.Chisholm, S. T., Dahlbeck, D., Krishnamurthy, N., Day, B., Sjolander, K. V. & Staskawicz, B. J. (2005) Proc. Natl. Acad. Sci. USA 102, 2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., Rathjen, J. P., Bendahmane, A., Day, L. & Baulcombe, D. C. (2003) EMBO J. 22, 5690-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, M.-G., da Cunha, L., Belkhadir, Y., DebRoy, S., Dangl, J. L. & Mackey, D. (2005) Cell, in press. [DOI] [PubMed]
- 23.Coppinger, P., Repetti, P. P., Day, B., Dahlbeck, D., Mehlert, A. & Staskawicz, B. J. (2004) Plant J. 40, 225-237. [DOI] [PubMed] [Google Scholar]
- 24.Century, K. S., Holub, E. B. & Staskawicz, B. J. (1995) Proc. Natl. Acad. Sci. USA 92, 6597-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triantafilou, M., Miyake, K., Golenbock, D. T. & Triantafilou, K. (2002) J. Cell Sci. 115, 2603-2611. [DOI] [PubMed] [Google Scholar]
- 26.Lafont, F., Tran Van Nhieu, G., Hanada, K., Sansonetti, P. & van der Goot, F. G. (2002) EMBO J. 21, 4449-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.