Abstract
B lymphocytes express the nonclassical class II molecule HLA-DO, which modulates the peptide loading activity of HLA-DM in the endocytic pathway. Binding to HLA-DM is required for HLA-DO to egress from the endoplasmic reticulum (ER). To gain insights into the mode of action of DO and on the role of DM in ER release, we sought to identify DM-binding residues on DO. Our results show that DOα encompasses the binding site for HLA-DM. More specifically, mutation of residue DOα41 on an exposed lateral loop of the α1 domain affects the binding to DM, ER egress, and activity of DO. Using a series of chimeric DR/DO molecules, we confirmed the role of the α chain and established that a second DM-binding region is located C-terminal to the DOα80 residue, most probably in the α2 domain. Interestingly, after mutation of a buried proline (α11) on the floor of the putative peptide-binding groove, HLA-DO remained functional but became independent of HLA-DM for ER egress and intracellular trafficking. Collectively, these results suggest that the binding of HLA-DM to DOα allows the complex to egress from the ER by stabilizing intramolecular contacts between the N-terminal antiparallel β-strands of the DOαβ heterodimer.
Keywords: antigen processing, MHC, class II, HLA-DR
Classical MHC class II proteins are highly polymorphic heterodimers expressed at the surface of antigen-presenting cells where they bind antigenic peptides derived from endocytosed antigens (1). The α and β subunits associate in the endoplasmic reticulum (ER) with the nonpolymorphic invariant chain (Ii), and the complex is sorted to endocytic compartments where Ii is degraded. A small fragment [class II-associated Ii peptide (CLIP)] of the Ii is protected inside the groove and must be released before the binding of antigenic peptides (2). HLA-DM (DM), a nonpolymorphic intracellular chaperone, is responsible for CLIP removal and also for editing the peptide repertoire to favor those of higher class II-binding affinity (3).
Class II-restricted antigen presentation in B lymphocytes is tightly regulated to ensure specificity of the activation process. These cells express another nonclassical class II molecule called HLA-DO (DO; H2-O in mice) that modulates the presentation of antigens in the endocytic pathway (4). Recently, it was shown that transfection of DO in class II transactivator (CIITA)+ cells caused the accumulation of classical class II molecules associated with CLIP (5, 6). These results clearly showed the inhibitory role of DO on class II-restricted antigen presentation.
The precise molecular mechanism by which DO inhibits the catalytic activity of DM remains to be clarified. DO is found mostly in endosomes but is retained in the ER of DM- cells. Interestingly, DO-bound DM is not sequestered there but rather allows the complex to egress from the ER (7). Trafficking of DO/DM to and inside the endocytic pathway is regulated by sorting signals located in the cytoplasmic tails of both molecules (8, 9). The need for DO to access peptide-loading compartments and to modify the subcellular sorting of DM suggests an elaborate modulator role on the antigen-processing rather than a unique inhibitory function (10-13). More recently, it was proposed that the capacity of H2-O to affect, positively or negatively, the presentation of antigens depends on the particular B cell receptor-antigen combination (14).
The mechanism by which DO is retained in the ER and how DM allows for efficient ER egress remains unknown. DO lacks ER-retention motifs such as the cytoplasmic di-basic sequences (15). Pulse-chase and immunoprecipitation experiments revealed that DOα- and β-chains associate early after biosynthesis. However, in the absence of DM, DOα- and β-chains do not show carbohydrate modifications to suggest a passage through the Golgi (7). To gain insights into the role of DM in ER release, we sought to delineate the DM binding site on DO and to characterize the defect in the assembly of DOα- and β-chains.
Materials and Methods
Plasmids and Mutagenesis. pBSDOα.9, pBSDOβ, pBudCE4-A, pBudDOαβ, RSV.5gptDN1, RSV.3DRβ008, and pBudDM are described in refs. 8 and 16. pBudDRβ is the BamHI fragment of RSV.3DRβ008 in pBudCE4-A. pR EP4CIITA and pCDNA3CIITA cDNAs were obtained from J. Ting (University of North Carolina, Chapel Hill). pBudDOα is the BamHI fragment of pBSDOα.9 into the BglII site of pBudCE4-A. pBudDOβ is the BamHI fragment from pBSDOβ into BglII site of pBudCE4-A. The pBudDMY was generated by PCR to modify the cytoplasmic 226-YTPL motif (8). A frameshift past this motif resulted in a longer cytoplasmic tail (226-ATPLLGPIIQKDGTFPRGRIPAARGIH).
A DR18/DOα chimeric cDNA (cDOα) was made by PCR using the DRα cDNA cloned in the BamHI site of pBlueScript (Stratagene) and RSV.5gptDN1 as templates. The SalI-PvuII fragment encompassing the junction was cloned in RSV.5gptDN1. A 2-kbp BamHI fragment was cloned into the BglII site of pBudDRβ to generate pBudDR18/DOα + DRβ. For pBudDR42/DOα + DRβ, pBudDR63/DOα + DRβ, pBudDR77/DOα + DRβ, and pBudDR84/DOα, pBSDOα.9 and pBSDRα were used as templates. The NotI-BstEII fragment including the junction was cloned into pBudDR18/DOα + DRβ. The DOα mutations were created by using pBSDOα.9. PCR products were digested with SalI-PvuII and cloned in pBudDOαβ (pBudDOαP11V/βwt, pBudDOαΔP11/βwt, and pBudDOαA12N/βwt). For other DOα mutations (E41K, F52S, and αEF-KS), a 450-bp SalI-BstEII PCR fragment was simultaneously cloned with a BstEII-XbaI fragment, digested from pBSDOα.9, into the SalI-XbaI sites of pBudCE4-A (pBudDOαE41K, pBudDOαF52S, and pBudDOαEF-KS). A pBSDOβ BamHI fragment was cloned in BglII to generate pBudDOαE41K + βwt, pBudDOαF52S + βwt, and pBudDOαEF-KS + βwt.
Antibodies and Reagents. The following mAbs were used: L243 (DRα) (17), LB3.1 (DRα) (18), CerCLIP.1 (CLIP; Pharmingen) (19), XD5.117 (XD5; DRβ) (20), MaP.DM1 (DM; Pharmingen) (19), Mags.DO5 (21), HKC5 (DOβ) (22), and phycoerythrin (PE)-coupled anti-CD63 (Becton Dickinson). The rabbit sera against DOα and DMβ are described in refs. 8 and 16. Secondary Abs were Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes), biotinylated goat anti-rabbit (BIO/CAN, Montreal), and Texas-red-coupled streptavidin (Amersham Pharmacia Biotech). Biotinylated SEA was from Toxin Technology (Sarasota, FL).
Cell Lines and Transfections. HeLa and HeLa DM.5 cells were provided by R. P. Sekaly (Université de Montréal). HEK 293T cells were obtained from E. Cohen (Université de Montréal). Cells were cultured in DMEM, 10% FBS (Invitrogen), and selective agents. HeLa cells were transfected with FuGENE 6 (Roche Diagnostics) by using 1 μg of each DNA (8). For transient expression, HEK 293T cells were transfected by using calcium phosphate (8).
Flow Cytometry and Fluorescence Microscopy. Cells were harvested, washed, and stained for surface expression. Alternatively, cells were permeabilized in saponin as described in ref. 8. Cells were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson) or transferred on slides by centrifugation. Cells were analyzed by fluorescence microscopy on a Zeiss Axioplan 2 equipped with a Sony (Tokyo) DXC-390P camera.
Immunoprecipitations and Western Blotting. Cells (107) were harvested and lysed in 1% CHAPS or Triton X-100 (8). Immunoprecipitations were performed by using primary Abs bound to protein G coupled to Sepharose 4B (Amersham Pharmacia Biosciences), and samples were analyzed on immunoblots.
Results
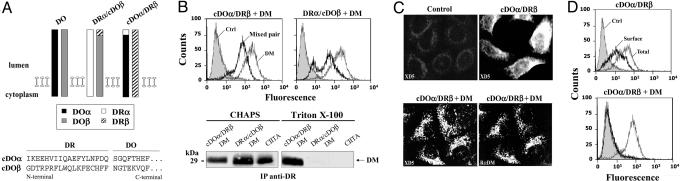
DM Binds to the DOα Chain. First, we determined which chain of DOαβ was responsible for the strong ER interaction with DM by generating mixed heterodimers between DO and DR (Fig. 1A). Such pairing overcomes ER retention but only when using chimeric DO chains (cDO chains) in which the first 18 residues are replaced by those of DR (23). Because DOα shows insertion of one extra amino acid between corresponding residues 9 and 10 of DR (see A Mutation in the Groove of DO Restores ER Egress), position 18 of DRα is thus fused to residue 20 of DOα. The cDOα/DRβ and DRα/cDOβ molecules were stably expressed separately in DM+ HeLa cells (Fig. 1B). To identify the key DM-interacting chain, we took advantage of the fact that DO-DM bonds persist in Triton X-100 and CHAPS whereas DR-DM complexes can only be coprecipitated in CHAPS (7, 24). The weak DR-DM binding was confirmed by immunoprecipitation of DR from CIITA+ HeLa cells (22) after solubilization in Triton X-100 or CHAPS (Fig. 1B). Analysis of mixed DO/DR pairs showed that DM binds DRα/cDOβ only in CHAPS, whereas it binds cDOα/DRβ efficiently in both detergents, suggesting that DOα mediates strong DM binding. These results argue that cDOα/DRβ and DRα/cDOβ associate with DM in a DO- and DR-like fashion, respectively.
Fig. 1.
DOα is responsible for the strong interaction with DM. (A) Mixed pairs between DR and chimeric DO chains. The N-terminal primary sequence of the mature cDOα and cDOβ is shown. (B) HeLa cells stably expressing mixed pairs and DM were permeabilized and stained for class II (XD5 or L243) and DM (Map.DM1). Cells were lysed either in CHAPS or Triton X-100. DRα/cDOβ and cDOα/DRβ mixed pairs were immunoprecipitated with L243 and XD5, respectively. The association with DM was monitored by Western blotting with DMβ-specific rabbit antiserum. HeLa cells transfected with CIITA were used as control, and DR was immunoprecipitated with L243. (C) Permeabilized HeLa cells expressing cDOα/DRβ with or without DM and stained with XD5 and the DM-specific rabbit antiserum (RαDM). Untransfected HeLa cells were used as controls. (D) Surface expression of cDOα/DRβ was monitored by using XD5 in DM+- and DM--transfected HeLa cells (bold line). Total expression was assessed on permeabilized cells (thin line). The intracellular expression of DM is shown in B. Control cells were incubated with the secondary Ab alone (filled histogram).
Because cDOα/DRβ appeared to bind DM as efficiently as DO, we compared the subcellular localization of cDOα/DRβ in DM- and DM+ transfected cells (Fig. 1C). In the absence of DM, the mixed pair is found at the plasma membrane. The diffuse surface staining was expected given the absence of endosomal sorting motifs in DOα and DRβ. However, upon coexpression of DM, staining for cDOα/DRβ revealed punctate structures reminiscent of endosomes. Indeed, the pattern was indistinguishable from the one obtained for DM, suggesting a DM-induced intracellular sequestration (Fig. 1C). The same conclusion was reached from the flow cytometry analysis. Whereas cDOα/DRβ reached the HeLa plasma membrane, the mixed pair was detected in DM+ cells only upon permeabilization (Fig. 1D). Altogether, these results suggest that DM and cDOα/DRβ interacted early in the ER, preventing the mixed pair from taking the default route to the surface. These data point to a major role for DOα in DM binding.
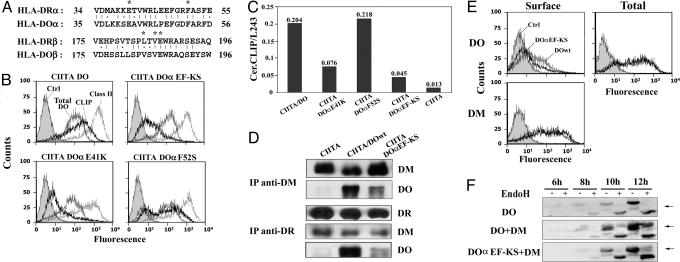
DM Contacts Residue DOαE41. Recently, it was shown that DRαE40, αF51, βL184, βV186, and βE187 are located on the same face and interact with DM (25). These residues are conserved in DO except for the equivalent of DRβL184, which is replaced by a valine (Fig. 2A). To test whether DO also uses this interface for DM binding, we generated mutants in this DR-like binding site on DO. Based on our described results, we concentrated our effort on DOα. The conserved DOα Glu-41 and Phe-52 residues (corresponding to DRαE40 and αF51) were replaced either separately or simultaneously with Lys and Ser residues, respectively. To evaluate their activity, each mutant was cotransfected together with CIITA into HeLa cells, and the level of cell-surface class II-CLIP complexes was determined by flow cytometry. Although CIITA alone slightly up-regulated DO expression, the level of CLIP remained low (22). The ratio of class II-CLIP (CerCLIP.1) to DR (L243) shows that cotransfection of DO strongly inhibits peptide loading (Fig. 2 B and C). In contrast, the double mutant DOαEF-KS no longer inhibits DM function because only low levels of class II-CLIP were generated (Fig. 2 B and C). Staining of permeabilized cells revealed that the observed difference was not due to DO expression levels (Fig. 2B). Next, we found that DOα41 alone is critical for DM inhibition, whereas mutation of α52 had no effect (Fig. 2 B and C). The fact that the conserved glutamate is involved in the binding of DM for both DO and DR suggests possible models for the inhibition of antigen processing by DO (see Discussion). However, the lack of involvement of α52F indicates that DO interacts with DM through a region that is not completely analogous to the one described for DR (26). Moreover, point mutations βV184H or βE187A did not affect the function of DO (data not shown).
Fig. 2.
Mutation of residue DOαE41 affects the interaction with DM. (A) Homology between DR and DO in the regions important for DR-DM association (25). Residues interacting with DM are denoted with asterisks. (B) HeLa cells were stably transfected with CIITA together with DO or DO mutants and purified with magnetic beads coated with L243. Cells were later analyzed after surface staining for DR (L243; gray line) or CLIP (CerCLIP.1; bold line) or after intracellular staining for DO (HKC5; thin line). Control represents untransfected HeLa cells stained with L243 (filled histogram). (C) Transfected HeLa cells were analyzed as above for class II and CLIP cell-surface expression. The histogram shows the ratios of the mean fluorescence values. (D) DM or DR were immunoprecipitated in CHAPS from stably transfected HeLa CIITA cells expressing DO (DOwt) or the DOαEF-KS mutant. Coprecipitating material was analyzed on immunoblots by using Abs for DMβ, DOα, or DRβ. As a control, HeLa cells transfected with CIITA alone were used. (E) 293T cells were transiently transfected with DOαEF-KS (bold line) or wild-type DO (thin line) along with DM lacking its cytoplasmic tyrosine sorting motif (DMY). Surface expression of DO was monitored by using Mags.DO5 Ab (Upper Left). Cells were permeabilized and total expression of DO was assessed by using HKC5 (Upper Right). Control cells were incubated with the secondary Ab alone (filled histogram). Cells were also stained for surface DM by using Map.DM1 (Lower). (F) 293T cells were transfected with DO, DO + DM, or DOαEF-KS + DM and lyzed 6, 8, 10, or 12 h posttransfection. Half of each sample was treated with EndoH and analyzed on immunoblots by using a DOα-specific rabbit antiserum. Arrows indicate the position of glycosylated DOα.
Despite comparable levels of DO and DM expression (Fig. 2B and data not shown), immunoprecipitations showed that DM interacts less efficiently with DOαEF-KS than with wild-type DO (Fig. 2D). However, the DM/DOαEF-KS interaction was not totally abolished, perhaps explaining the low level of class II-CLIP at the surface of these cells (Fig. 2 B and C). Trimolecular DO-DM-DR complexes can be precipitated from B cell lysates as well as from isolated MHC class II compartments (10, 27). DR was immunoprecipitated from the stable transfectants described above and the level of associated DM/DO complexes was determined by Western blotting (Fig. 2D). Results show that both DO and DM are coprecipitated with DR but that the αE41K mutation reduced the amount of DO in the complex.
We confirmed the weak DOαE41K-DM interaction by using transient transfections. We took advantage of the fact that a DM molecule devoid of its cytoplasmic YTPL motif (DMY) is expressed at the cell surface (28, 29) and drags DO to the plasma membrane (Fig. 2E). As expected from the immunoprecipitation data, the DOαEF-KS mutant was expressed at lower levels on the cell surface than wild-type DO. Overall, DO levels were comparable as assessed by staining of permeabilized cells with a mAb specific for the cytoplasmic tail of DOβ (HKC5; Fig. 2E Upper Right). Because DOα and DOβ are encoded on the same plasmid, DOα levels were also identical. Finally, the low surface expression of DOαEF-KS could not be attributed to lower DM expression (Fig. 2E Lower).
To measure DO egress from the ER, we transfected DO and tested for the acquisition of complex carbohydrates over time. In cells expressing DO only, the β chain remained sensitive to EndoH even 12 h posttransfection (Fig. 2F). This finding confirms that DO does not transit through the Golgi in the absence of DM. However, a substantial proportion of the DO pool had acquired EndoH resistance as early as 8 h posttransfection in the presence of DM. On the other hand, DOαEF-KS showed little EndoH resistance at 12 h, confirming its reduced capacity to assemble with DM in the ER.
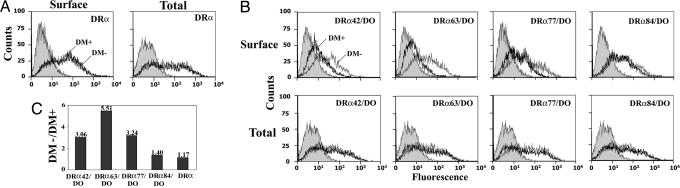
DM Binds Other Regions of DOα. To further characterize the DM-binding region, we generated a series of chimeric DRα/DO molecules. We reasoned that a stepwise increase in the content of DRα should, at some point, abrogate the capacity of DM to interact in the ER and sequester the chimeric molecules in endosomes (Fig. 1). Four more chimeric α-chains were generated to cover most of the α1 domain. Assuming a classical class II-like structure, the DRα40/DO covers two β-strands of the groove floor and most of the two exposed external loops. The DRα63/DO molecule includes DR residues up to about half of the α-helix lining the putative groove. The DR portion in DRα77/DO covers almost the entire α-helix (30).
To assess the interaction with DM, the chimeric molecules were transiently transfected into 293T cells together with DRβ and with or without DM. Surface expression of control DRαβ was unaffected by the presence of DM (Fig. 3A). This was expected because DR takes the default pathway to the surface without Ii, whereas DM is sorted to lysosomes. Also, the DR-DM interactions that might occur in this context would be transient (31). Staining of the permeabilized cells demonstrated that the total amount of class II was comparable in DM+ and DM- cells (Fig. 3A). Fig. 3B shows that all chimeras are efficiently expressed at the surface of DM- cells. However, with DM, the surface expression of DRα42/DO, DRα63/DO, and DRα77/DO was reduced (Fig. 3 B and C). This result suggests that DM interacted in the ER with a substantial fraction of the mixed pairs pool in a DO-like fashion, as shown in Fig. 1 for cDOα/DRβ. Staining of permeabilized cells confirm that these variations in surface expression were not attributable to differences in class II (Fig. 3B) or DM (data not shown) levels. Interestingly, a DRα84/DO construct was not sequestered by DM. However, introduction of the DRα80-84 stretch into DO affected its overall conformation, precluding drawing any conclusions regarding the role of this region in DM binding (see Discussion). Given the fact that residues DRαN78 is conserved in DO (N79) and that mutation of DOα80 does not affect DM binding (32), these results suggest that other DO residues C-terminal to α80 are critical for the interaction with DM.
Fig. 3.
The DM binding site includes a region C-terminal of the DOα1 domain. 293T cells were transiently transfected with DRβ along with DRα (A) or different cDOα-chains (B) in the presence (bold line) or absence (thin line) of DM. The capacity of DM to sequester the various class II molecules was assessed by surface staining with the nonconformational DRβ-specific Ab XD5. Cells were also permeabilized and total expression of the molecules was determined. Control cells were incubated with the secondary Ab alone (filled histogram). (C) Ratios between the XD5 mean fluorescence value obtained for each heterodimer at the surface of DM- and DM+ cells.
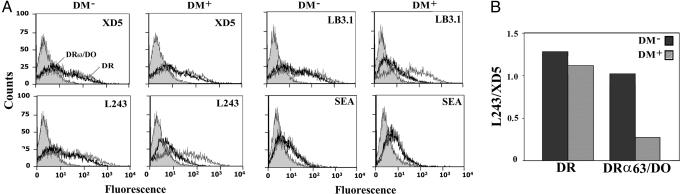
Next, we used the DRα63/DO + DRβ mixed pair to confirm the importance of the α chain in the interaction with DM. On classical class II molecules, the region analogous to the DOα41 loop is part of the epitopes recognized by several mAbs. We hypothesized that these might recognize DRα63/DO and compete for the association with DM. First, we tested the binding of L243 and LB3.1, two DRα-specific monoclonal antibodies recognizing conformational epitopes that include residues DRα18 and DRα39 on the two separate loops beneath the α-helix (33). As a control, we used XD5, which recognizes a linear epitope on the β chain α-helix and which is destroyed by the double mutation βE59A/Q64A (data not shown). DRα63/DO and DR were efficiently recognized by the L243, LB3.1, and XD5 Abs in the absence of DM (Fig. 4A). Interestingly, coexpression of DMY (mutated at its YTPL motif to avoid sequestration) had a dramatic negative effect on the binding of L243 and LB3.1 to the mixed pair but not to DR. The residual Ab staining is most likely because of DRα63/DO molecules that have left the ER before encountering DM. The reduced binding of these α chain-specific Abs in the presence of DM is specific because recognition by XD5 was unaffected. The L243 over XD5 staining ratio for DR was clearly not affected by the presence of DM as compared with the chimeric molecule (Fig. 4B). The inability of L243 and LB3.1 to bind the cell-surface DRα63/DO-DM complex strongly supports our conclusion that DM interacts with the α chain of DO, close to residue E41. Importantly, the lack of L243 binding reflects a DO-like rather than DR-like interaction because this Ab would efficiently bind DR/DM complexes (Fig. 1). This result is in line with our statement made above that DO interacts with DM through a region that is not akin to the interface on DR.
Fig. 4.
HLA-DM competes with DRα-specific antibodies for binding to DRα63/DO. (A) 293T cells were transfected with DRα (thin line) or DRα63/DO (bold line) along with the DRβ chain in absence or presence of DMY. Surface expression was assessed by using the DRβ-specific Ab (XD5), the DRα-specific Abs (L243 or LB3.1), or SEA. Control cells were incubated with the secondary Ab alone or represent class II-negative HeLa cells incubated with SEA (filled histogram). (B) Ratio of the mean fluorescence values obtained for L243 and XD5.
Although the lack of competition between XD5 and DM suggests that the latter does not significantly contact the β1 chain, we confirmed this by analyzing the binding of the superantigen SEA to DRα63/DO + DRβ. SEA binds through coordination of a zinc ion with DRβH81 (23), and our results show that its interaction with the chimeric molecule was as effective in the presence or absence of DMY (Fig. 4A). Collectively, these competition experiments with class II ligands confirm that DM binds to DOα.
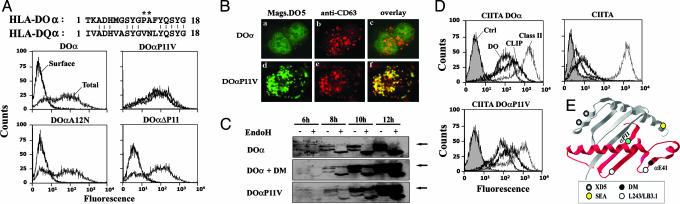
A Mutation in the Groove of DO Restores ER Egress. Next, we addressed the functional importance of the newly identified DM binding site for the maturation of DO. The inability of classical class II molecules to leave the ER has been previously reported in the context of mismatched αβ pairs between various isotypes or haplotypes. In some cases, surface expression was restored after site-directed mutagenesis or by the construction of chimeric molecules, often implicating regions close to the N terminus of either the α or β chain (34-36). Thus, we postulated that the DOαβ interaction was suboptimal and that the binding of DM may stabilize the DO heterodimer. Indeed, introducing the first 18 aa of DR into DO allows the formation of mixed DR/DO pairs (Fig. 1). A sequence alignment of DR, DO, and DQ revealed the presence of an amino acid insertion in both DQα and DOα. This extra Gly residue (αG10) was numbered 9a in I-A (the mouse equivalent of DQ), and the crystal structure revealed that it protrudes in a β bulge conformation outward from the floor of the class II peptide binding groove (37). This bulge maintains the respective arrangement, seen in other class II, of the conserved αβ residues involved in interchain contacts. However, in DOα, this glycine is next to a proline (αP11) that we thought might disrupt the β-sheet and prevent the glycine from bulging (Fig. 5A). This finding is especially interesting given the fact that this proline is close to the α chain loops supporting DM interactions.
Fig. 5.
Mutation of DOαP11 abrogates DM-dependency for ER egress. (A) Alignment between the DOα and DQα N-terminal regions. Mutated residues on DO are denoted with asterisks. 293T cells were transfected transiently with DOαβ-chains (DO) or mutated versions of DOα (DOαP11V, DOαΔP, or DOαA12N) along with DOβ chain. Cells were stained for surface expression of DO with Mags.DO (bold line) or permeabilized and stained with HKC5 (total; thin line). (B) Fluorescence microscopy on permeabilized HEK 293T cells expressing DO (a-c) or DOαP11V mutant (d-f). (C) 293T cells were transfected with DO, DO + DM, or DOαP11V and analyzed as in Fig. 2F.(D) HeLa cells were stably transfected with CIITA alone or together with DO or DOαP11V + DOβ and purified with magnetic beads coated with L243. Cells were subsequently analyzed by flow cytometry after surface staining for DR (L243; gray line) and CLIP (CerCLIP.1; bold line) and after intracellular staining for DO (HKC5; thin line). Control cells represent untransfected HeLa cells stained with L243 (filled histogram). (E) Generic class II structure showing the probable location of key DO residues.
We investigated the importance of this region in DOα by generating a series of mutants (αΔP11, αP11V, αA12N, αY9Q, and αΔP11/A12E). After transfection of these DOα-chains together with DOβ, the transport and subsequent cell-surface expression of DO in the absence of DM was assessed by staining with a DO-specific mAb. Remarkably, DOαP11V + DOβ was efficiently expressed at the cell surface in the absence of DM (Fig. 5A). Surface expression of this DO mutant was also observed in stably transfected HeLa cells (data not shown). None of the other mutations that were introduced in this region (αΔP11, αA12N, αY9Q, αP11/A12E, and βI9Y/A11F) restored surface DO expression in the absence of DM (Fig. 5A and data not shown).
The strong surface expression of DOαP11V suggested that ER egress was highly efficient. DO contains a functional dileucine motif in its β chain cytoplasmic tail (8). This sorting signal should allow DOαP11V to gain access to endosomes, probably after endocytosis. Transfected cells were permeabilized, stained for DO, and analyzed by fluorescence microscopy. The presence of scattered vesicles colocalizing with the endocytic marker CD63 (Fig. 5B) further confirms the overall integrity of DOαP11V mutant and its intrinsic capacity to transport and accumulate in the absence of DM.
To compare the kinetics of maturation between DOαP11V and DO/DM, the acquisition of EndoH resistance by the α-chains was analyzed at various time points after transfection (Fig. 5C). DO was retained in the ER in the absence of DM and, therefore, did not acquire complex sugars. However, the same analysis showed the emergence of EndoH-resistant forms of DOαP11V as early as 8 h posttransfection. Importantly, the kinetic of EndoH resistance was similar for DO molecules coexpressed with DM. Altogether, these results suggest that mutation of the proline to a valine at residue αP11 allows ER egress by correcting a conformation defect in DO.
Finally, we tested the ability of DOαP11V to inhibit CLIP release from classical class II molecules. HeLa CIITA cells were transfected with either DOαP11V or wild-type DO. After selection, the levels of cell-surface class II-CLIP were measured by flow cytometry (Fig. 5D). The results show that both mutant and wild-type DO are functional in terms of their ability to inhibit DM and peptide loading.
Discussion
The regulation of antigen processing and peptide binding to classical class II molecules is a complex process that varies depending on the type of APC or the receptors used for antigen internalization. In B cells, DM is the target of regulatory control through the expression of DO. Although the role and mechanism of action of DM are well characterized, the need for DM modulation in B cells is not well understood.
The crystal structure of DR-DM complexes remains unknown, but models for the mechanism of peptide exchange has been proposed (26, 38). Here, we have identified a region of DO that interacts with DM. As opposed to the results obtained after mutation of the α80-84 stretch (data not shown), mutation of αE41K in the DM-independent DOαP11V molecule (DOαP11V + αE41K) did not affect surface expression (Fig. 6, which is published as supporting information on the PNAS web site). This finding suggests that the reduced interaction between DOαE41K and DM is not the result of a conformation defect. Interestingly, the mouse Gluα41 is polymorphic in H2-O, and CBA/J mice of the k haplotype have a Glu to Ala substitution at position α41 (39). This change most probably affects, to some extent, the interaction with H2-M. The fact that DM covers the DOα-chain may explain why immunization of mice with DO-DM complexes yielded only DOβ-specific mAbs (L.K.D., unpublished work).
The fact that DM mutations affecting DR did not prevent DO-DM association strongly suggests that DO and DR have distinct binding sites (26). A noteworthy point is that DOαE41 is analogous to the DRαE40 residue and both are involved in contacting DM. It will be interesting to see whether this is coincidental or whether both residues interact with the same amino acids on DM. We could envisage that although DO and DR bind to nonoverlapping faces on DM, DO contacts and disengages key DM residues such as DMαF100 that play a role in the destabilization of DR-CLIP bonds (26). At low pH, a conformation change in DO could free the catalytic DM residue.
The discovery that a point mutation renders DO independent of DM for ER egress is of the utmost interest. Karlsson and collaborators showed that even in the absence of DM, the DO α- and β-chains associate and can be coimmunoprecipitated (7). The relative efficiency of DOαβ heterodimer formation but lack of ER egress is reminiscent of results obtained previously with some mismatched αβ class II gene products. Whereas there are clear examples of haplotype-, isotype-, and even species-mismatched heterodimers that can be expressed at the cell surface (40-42), other pairs like EαAβk, DRαAβb, DQ1αAβk, DRαDPβ, and DQ1αDQβw2 could not be detected on transfected cells (35, 36, 43, 44). As for DO in DM- cells, some mismatched αβ classical class II pairs do form in the ER but are not exported beyond the cis-Golgi (43, 44). Although there were doubts in the past that products of the DOA (formerly known as DZ or DN) and DOB genes were part of the same heterodimer, it has become clear from immunoprecipitation and transfection experiments that DOα and DOβ associate (7, 45). The DOA and DOB genes are virtually nonpolymorphic, and it is surprising that they have not evolved to maximize pairing. A selective pressure must exist to explain the instability of DO in the absence of DM. Such safety mechanism might prevent surface expression of DO molecules that could potentially activate autoreactive T cells. Indeed, we have previously shown that DOβ can interact with CD4 (23).
How DM rescues DO from degradation in the ER is a matter of speculation. Classical class II molecules bind calnexin and/or binding luminal protein (BiP) during assembly (46, 47). DM might compete with these chaperones for binding to the loosely associated, most probably aggregated DOα- and β-chains. Another nonmutually exclusive possibility is that DM binding directly affects the conformation of DO to release ER chaperones bound on other regions of the molecule. DO is particularly hydrophobic in its membrane-distal domains (48) and could expose patches recognized by binding luminal protein in the absence of DM. The fact that the DOαP11V mutation restores egress in the absence of DM would favor the latter mechanism. By strongly interacting with the external loops and the α-helix of the DOα1 domain, DM could affect the intra- and/or interchain interactions. Whether DOαP11V binds peptides that could stabilize the groove is currently unknown. Interestingly, DOαP11V binds DM very efficiently as judged by their coimmunoprecipitation in Triton X-100 and the fact that DM sequesters the mutant DO in endosomes (Fig. 7, which is published as supporting information on the PNAS web site). These properties suggest that DOαP11V represents a useful tool to study the function of DO and why free forms must be retained in the ER.
Supplementary Material
Acknowledgments
We thank E. O. Long, J. Ting, and R. P. Sekaly for providing cDNAs and cell lines and E. Mellins and A. Pachine for helpful discussions. F.D. and A.B. were supported by studentships from the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche (Quebec). J.T. holds a Senior Fellowship from the Fonds de la Recherche en Santé du Québec. This work was supported by grants to J.T. from the Canadian Institutes of Health Research and the Cancer Research Society, Inc.
Author contributions: F.D., A.B., A.S., and J.T. designed research; F.D., A.B., and D.A.D. performed research; L.K.D. and A.S. contributed new reagents/analytic tools; F.D., A.B., D.A.D., L.K.D., and J.T. analyzed data; and F.D., A.B., L.K.D., A.S., and J.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; Ii, invariant chain; CLIP, class II-associated Ii peptide; CIITA, class II transactivator; cDO, chimeric DO.
References
- 1.Cresswell, P. (1994) Annu. Rev. Immunol. 12, 259-293. [DOI] [PubMed] [Google Scholar]
- 2.Riberdy, J. M., Newcomb, J. R., Surman, M. J., Barbosa, J. A. & Cresswell, P. (1992) Nature 360, 474-477. [DOI] [PubMed] [Google Scholar]
- 3.Kropshofer, H., Hammerling, G. J. & Vogt, A. B. (1997) Immunol. Today 18, 77-82. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X. & Jensen, P. E. (2004) Immunol. Res. 29, 19-28. [DOI] [PubMed] [Google Scholar]
- 5.Denzin, L. K., Sant'Angelo, D. B., Hammond, C., Surman, M. J. & Cresswell, P. (1997) Science 278, 106-109. [DOI] [PubMed] [Google Scholar]
- 6.Van Ham, S. M., Tjin, E. P. M., Lillemeier, B. F., Gruneberg, U., Van Meijgaarden, K. E., Pastoors, L., Verwoerd, D., Tulp, A., Canas, B., Rahman, D., et al. (1997) Curr. Biol. 7, 950-957. [DOI] [PubMed] [Google Scholar]
- 7.Liljedahl, M., Kuwana, T., Fung-Leung, W. P., Jackson, M., Peterson, P. A. & Karlsson, L. (1996) EMBO J. 15, 4817-4824. [PMC free article] [PubMed] [Google Scholar]
- 8.Brunet, A., Samaan, A., Deshaies, F., Kindt, T. J. & Thibodeau, J. (2000) J. Biol. Chem. 275, 37062-37071. [DOI] [PubMed] [Google Scholar]
- 9.van Lith, M., van Ham, M., Griekspoor, A., Tjin, E., Verwoerd, D., Calafat, J., Janssen, H., Reits, E., Pastoors, L. & Neefjes, J. (2001) J. Immunol. 167, 884-892. [DOI] [PubMed] [Google Scholar]
- 10.Kropshofer, H., Vogt, A. B., Thery, C., Armandola, E. A., Li, B. C., Moldenhauer, G., Amigorena, S. & Hammerling, G. J. (1998) EMBO J. 17, 2971-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Ham, M., van Lith, M., Lillemeier, B., Tjin, E., Gruneberg, U., Rahman, D., Pastoors, L., van Meijgaarden, K., Roucard, C., Trowsdale, J., et al. (2000) J. Exp. Med. 191, 1127-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perraudeau, M., Taylor, P. R., Stauss, H. J., Lindstedt, R., Bygrave, A. E., Pappin, D. J., Ellmerich, S., Whitten, A., Rahman, D., Canas, B., et al. (2000) Eur. J. Immunol. 30, 2871-2880. [DOI] [PubMed] [Google Scholar]
- 13.Brocke, P., Armandola, E., Garbi, N. & Hammerling, G. J. (2003) Eur. J. Immunol. 33, 411-421. [DOI] [PubMed] [Google Scholar]
- 14.Alfonso, C., Williams, G. S., Han, J. O., Westberg, J. A., Winqvist, O. & Karlsson, L. (2003) J. Immunol. 171, 2331-2337. [DOI] [PubMed] [Google Scholar]
- 15.Teasdale, R. D. & Jackson, M. R. (1996) Annu. Rev. Cell Dev. Biol. 12, 27-54. [DOI] [PubMed] [Google Scholar]
- 16.Faubert, A., Samaan, A. & Thibodeau, J. (2002) J. Biol. Chem. 277, 2750-2755. [DOI] [PubMed] [Google Scholar]
- 17.Lampson, L. A. & Levy, R. (1980) J. Immunol. 125, 293-299. [PubMed] [Google Scholar]
- 18.Gorga, J. C., Knudsen, P. J., Foran, J. A., Strominger, J. L. & Burakoff, S. J. (1986) Cell. Immunol. 103, 160-173. [DOI] [PubMed] [Google Scholar]
- 19.Denzin, L. K., Robbins, N. F., Carboy-Newcomb, C. & Cresswell, P. (1994) Immunity 1, 595-606. [DOI] [PubMed] [Google Scholar]
- 20.Radka, S. F., Machamer, C. E. & Cresswell, P. (1984) Hum. Immunol. 10, 177-188. [DOI] [PubMed] [Google Scholar]
- 21.Glazier, K. S., Hake, S. B., Tobin, H. M., Chadburn, A., Schattner, E. J. & Denzin, L. K. (2002) J. Exp. Med. 195, 1063-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalil, H., Deshaies, F., Bellemare-Pelletier, A., Brunet, A., Faubert, A., Azar, G. A. & Thibodeau, J. (2002) Tissue Antigens 60, 372-382. [DOI] [PubMed] [Google Scholar]
- 23.Thibodeau, J., Lavoie, P. M., Samaan, A., Corre, J. P., Sekaly, R. P. & Cazenave, P. A. (1998) Mol. Immunol. 35, 885-893. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson, F., Thomas, C., Neefjes, J. & Trowsdale, J. (1996) Immunity 4, 87-96. [DOI] [PubMed] [Google Scholar]
- 25.Doebele, C. R., Busch, R., Scott, M. H., Pashine, A. & Mellins, D. E. (2000) Immunity 13, 517-527. [DOI] [PubMed] [Google Scholar]
- 26.Pashine, A., Busch, R., Belmares, M. P., Munning, J. N., Doebele, R. C., Buckingham, M., Nolan, G. P. & Mellins, E. D. (2003) Immunity 19, 183-192. [DOI] [PubMed] [Google Scholar]
- 27.Hammond, C., Denzin, L. K., Pan, M., Griffith, J. M., Geuze, H. J. & Cresswell, P. (1998) J. Immunol. 161, 3282-3291. [PubMed] [Google Scholar]
- 28.Marks, M. S., Roche, P. A., van Donselaar, E., Woodruff, L., Peters, P. J. & Bonifacino, J. S. (1995) J. Cell Biol. 131, 351-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindstedt, R., Liljedahl, M., Peleraux, A., Peterson, P. A. & Karlsson, L. (1995) Immunity 3, 561-572. [DOI] [PubMed] [Google Scholar]
- 30.Brown, J. H., Jardetzky, T. S., Gorga, J. C., Stern, L. J., Urban, R. G., Strominger, J. L. & Wiley, D. C. (1993) Nature 364, 33-39. [DOI] [PubMed] [Google Scholar]
- 31.Denzin, L. K. & Cresswell, P. (1995) Cell 82, 155-165. [DOI] [PubMed] [Google Scholar]
- 32.van Lith, M., van Ham, M. & Neefjes, J. (2002) Immunogenetics 54, 591-595. [DOI] [PubMed] [Google Scholar]
- 33.Fu, X.-T. & Karr, R. W. (1994) Hum. Immunol. 39, 253-260. [DOI] [PubMed] [Google Scholar]
- 34.Sant, A. J., Braunstein, N. S. & Germain, R. N. (1987) Proc. Natl. Acad. Sci. USA 84, 8065-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karp, D. R., Teletski, C. L., Jaraquemada, D., Maloy, W. L., Coligan, J. E. & Long, E. O. (1990) J. Exp. Med. 171, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechler, R. I., Sant, A. J., Braunstein, N. S., Sekaly, R., Long, E. O. & Germain, R. N. (1990) J. Immunol. 144, 329-333. [PubMed] [Google Scholar]
- 37.Fremont, D. H., Monnaie, D., Nelson, C. A., Hendrickson, W. A. & Unanue, E. R. (1998) Immunity 8, 305-317. [DOI] [PubMed] [Google Scholar]
- 38.Stratikos, E., Mosyak, L., Zaller, D. M. & Wiley, D. C. (2002) J. Exp. Med. 196, 173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson, L. & Peterson, P. A. (1992) J. Exp. Med. 176, 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Germain, R. N. & Quill, H. (1986) Nature 320, 72-75. [DOI] [PubMed] [Google Scholar]
- 41.Norcross, M. A., Raghupathy, R., Strominger, J. & Germain, R. N. (1986) J. Immunol. 137, 1714-1717. [PubMed] [Google Scholar]
- 42.Lotteau, V., Teyton, L., Burroughs, D. & Charron, D. (1987) Nature 329, 339-341. [DOI] [PubMed] [Google Scholar]
- 43.Sant, A. J., Hendrix, L. R., Coligan, J. E., Maloy, W. L. & Germain, R. N. (1991) J. Exp. Med. 174, 799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwok, W. W., Kovats, S., Thurtle, P. & Nepom, G. T. (1993) J. Immunol. 150, 2263-2272. [PubMed] [Google Scholar]
- 45.Douek, D. C. & Altmann, D. M. (1997) Int. Immunol. 9, 355-364. [DOI] [PubMed] [Google Scholar]
- 46.Anderson, K. S. & Cresswell, P. (1994) EMBO J. 13, 675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonnerot, C., Marks, M. S., Cosson, P., Robertson, E. J., Bikoff, E. K., Germain, R. N. & Bonifacino, J. S. (1994) EMBO J. 13, 934-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Servenius, B., Rask, L. & Peterson, P. A. (1987) J. Biol. Chem. 262, 8759-8766. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.