Abstract
The evolution of reproductive mode is expected to have profound impacts on the genetic composition of populations. At the same time, ecological interactions can generate close associations among species, which can in turn generate a high degree of overlap in their spatial distributions. Caenorhabditis elegans is a hermaphroditic nematode that has enabled extensive advances in developmental genetics. Caenorhabditis inopinata, the sister species of C. elegans, is a gonochoristic nematode that thrives in figs and obligately disperses on fig wasps. Here, we describe patterns of genomic diversity in C. inopinata. We performed RAD-seq on individual worms isolated from the field across three Okinawan island populations. C. inopinata is about five times more diverse than C. elegans. Additionally, C. inopinata harbors greater differences in diversity among functional genomic regions (such as between genic and intergenic sequences) than C. elegans. Conversely, C. elegans harbors greater differences in diversity between high-recombining chromosome arms and low-recombining chromosome centers than C. inopinata. FST is low among island population pairs, and clear population structure could not be easily detected among islands, suggesting frequent migration of wasps between islands. These patterns of population differentiation appear comparable with those previously reported in its fig wasp vector. These results confirm many theoretical population genetic predictions regarding the evolution of reproductive mode and suggest C. inopinata population dynamics may be driven by wasp dispersal. This work sets the stage for future evolutionary genomic studies aimed at understanding the evolution of sex as well as the evolution of ecological interactions.
Keywords: population genomics, outcrossing, selfing, dispersal, fig wasp, Caenorhabditis
Significance.
Caenorhabditis elegans is a hermaphroditic nematode model system widely used in molecular and developmental genetics, but very little is known about patterns of genomic diversity in female/male Caenorhabditis species. Here, we describe such patterns in the sister species of C. elegans, Caenorhabditis inopinata, a fig-associated nematode that migrates on fig wasps. We find C. inopinata harbors higher patterns of genomic diversity, greater efficacy of selection, more uniform levels of diversity within chromosomes, and lower levels of intrachromosomal linkage disequilibrium than C. elegans. Additionally, we find low levels of population differentiation among island populations of C. inopinata, suggesting widespread migration on its fig wasp vector. Our findings largely confirm the predictions of population genetic theory regarding the evolution of sexual systems, and they set the stage for further work regarding the evolution of ecological interactions.
Introduction
Reproductive systems vary widely across phylogeny (Bell 1982). Self-fertile hermaphrodites propagate via the union of gametes generated by the same individual, and selfing has evolved multiple times independently in eukaryotes (Jarne and Charlesworth 1993; Jarne and Auld 2006). Because such radical changes in sexual reproduction are expected to have profound impacts on patterns of genetic diversity (Glémin et al. 2019), the evolution of selfing has long been of interest to evolutionary biologists. For instance, selfers are expected to harbor less diversity (Haldane 1932), maintain smaller effective population sizes (Pollak 1987), and retain much longer linkage disequilibrium (LD) tracts (Nordborg 2000) than outcrossing species. Empirical population genetic studies have generally supported theoretical predictions (Cutter 2019; Glémin et al. 2019), although there are some exceptions (such as patterns of transposable element loads (Szitenberg et al. 2016; Glémin et al. 2019; Woodruff and Teterina 2020)). However, while there have been a number of studies addressing the genetic consequences of the evolution of selfing (Wright et al. 2007; Cutter et al. 2008; Foxe et al. 2008; Glémin and Muyle 2014; Chen et al. 2017; Cutter 2019), very few have examined the impacts of selfing on the genomic organization of diversity across whole chromosomes (Burgarella et al. 2023; Teterina et al. 2023). Only in recent years have chromosome-level genome assemblies become tractable and cost-effective to complete; such assemblies enabled such comparisons to be explicitly made. How does the evolution of self-fertile hermaphrodism mold chromosomal patterns of diversity?
At the same time, ecological interactions are expected to shape patterns of genomic diversity. Organisms thriving in transient resource patches often have dispersal mechanisms for finding new patches, and phoresy is a common ecological relationship (wherein one species carries another) that facilitates such dispersal. Additionally, the carrier and the carried also frequently differ greatly in size: remora fish disperse on loggerhead sea turtles and whales (O'Toole 2002; Bartlow and Agosta 2020), and tardigrades disperse on birds (Mogle et al. 2018; Bartlow and Agosta 2020). Such interactions require the comigration and colocalization of individuals of different species in space. Thus, interspecific relationships like phoresy have the potential to drive concordant patterns of spatial genetic differentiation among divergent species. That is, because they migrate and live together, it is reasonable to suspect that species in these kinds of relationships may have similar patterns of spatial genetic structure. Indeed, such patterns may not be limited to phoresy but may also extend to other intimate interspecific relationships like those between hosts and parasites/pathogens (Mazé-Guilmo et al. 2016). How do such interactions affect patterns of genetic diversity across vast spatial scales, and do interspecific interactions lead to covariance of genetic diversity among community members inhabiting the same transient resource?
The nematode Caenorhabditis inopinata is well-positioned to address the population genomic consequences of both selfing and comigration. C. inopinata, an outcrossing species, is the closest known relative of the self-fertile hermaphroditic species, C. elegans (Kanzaki et al. 2018). In Caenorhabditis nematodes, self-fertile hermaphroditism evolved three times independently from outcrossing ancestors (including C. elegans) (Sloat et al. 2022). As both species have chromosome-level assemblies (Kanzaki et al. 2018; Yoshimura et al. 2019), these species represent a natural focus of studies aimed at understanding how the evolution of selfing impacts the genomic organization of diversity. Recently, the first description of an outcrossing Caenorhabditis species, Caenorhabditis remanei, was published (Teterina et al. 2023). Here, it was found that C. remanei was ten times more diverse than the hermaphroditic C. elegans, while the intrachromosomal structuring of genetic diversity was conserved among the two species (Teterina et al. 2023). The differences among these two species were largely interpreted to be driven by differences in effective population size (and reproductive mode), and the similarities in patterns of diversity along chromosomes were attributed to similarities in intrachromosomal recombination rate variation among species (Teterina et al. 2023). The examination of an additional outcrosser, one more closely related to C. elegans, affords an opportunity to further clarify the population genomic consequences of reproductive mode evolution in Caenorhabditis nematodes.
Not only does the phylogenetic position of C. inopinata afford natural experiments for understanding the genomic consequences of selfing. The ecological niche of C. inopinata also allows hypotheses regarding the population genomic consequences of comigration to be tested. C. inopinata thrives in fresh Ficus septica figs (Kanzaki et al. 2018; Woodruff and Phillips 2018), which are ephemeral resources that harbor complex communities of wasps (Weiblen 2002), ants (Jandér 2015), moths (Sugiura and Yamazaki 2004), mites (Jauharlina et al. 2012), nematodes (Martin et al. 1973), and fungi (Martinson et al. 2012). While figs famously require fig wasps for pollination (Herre et al. 2008), fig wasps and fig nematodes obligately comigrate. Nematodes travel on pollinating fig wasps in order to colonize and proliferate in new fresh figs (Van Goor et al. 2023). Briefly, the fig wasp life cycle begins when one or more female foundress pollinating fig wasps enter the fig lumen via specialized opening called the ostiole (Weiblen 2002). Figs are not exactly fruits; rather, they are inflorescences with the flowers facing inward (syconia) (Janzen 1979). When the foundress enters, free-living nematodes disembark and reproduce. Concurrently, the foundress pollinates the florets and lays eggs in some of the fig ovules. The foundress dies, and the wasp progeny develop. Male wasp progeny (whose entire lives are restricted to the interior of a single fig) emerge first and mate with female wasps before they hatch. Male wasps cut a hole in the fig, and the females emerge and leave the fig to repeat the life cycle. Before females disperse, nematode dauer juveniles embark on such females to likewise continue their life cycle. Notably, for both fig nematodes and fig wasps, meiosis and syngamy are restricted entirely to the lumen of an individual fig (Weiblen 2002; Van Goor et al. 2023). Fig wasps are also capable of migrating hundreds of kilometers despite their small size (Ahmed et al. 2009). Thus, C. inopinata is well-positioned to understand how interspecific interactions can drive genetic structure among community members of the same transient resource.
Both the evolution of selfing (Pollak 1987) and comigration (Gilbert and Whitlock 2015; Engelbrecht et al. 2016) are expected to influence effective population sizes, and the extent to which these forces interact to shape patterns of genomic diversity remain unclear. The evolution of reproductive mode is also expected to cooccur with changes in recombination rate (Glémin et al. 2019), which itself interacts with effective population size to drive genome-wide patterns of evolutionary change. C. inopinata provides a unique opportunity to examine the influence of the mode of reproduction on patterns of genomic diversity in a system that is closely related to one of the most important model systems in biology (Corsi et al. 2015). Here, we describe chromosome-wide patterns of genomic diversity in C. inopinata. First, we compare these patterns with C. elegans to understand how the evolution of self-fertile hermaphroditism impacts genomic diversity at chromosomal scales. Then, we compare the spatial structuring of C. inopinata island populations with previously published reports of its fig wasp vector to understand how comigration may influence patterns of population differentiation.
Results
The Genomic Landscape of Polymorphism in C. inopinata
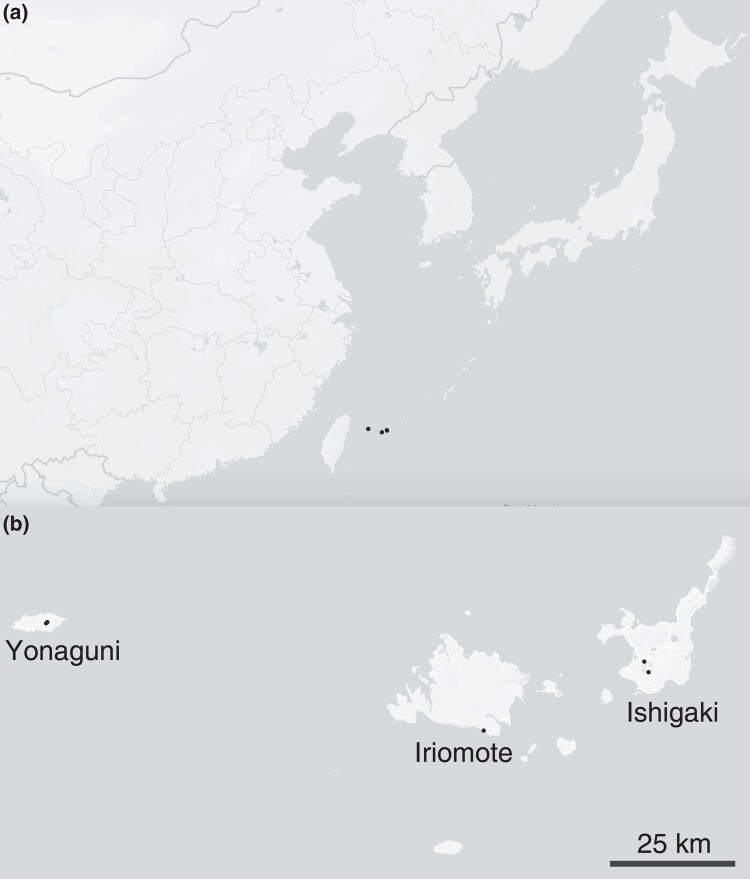
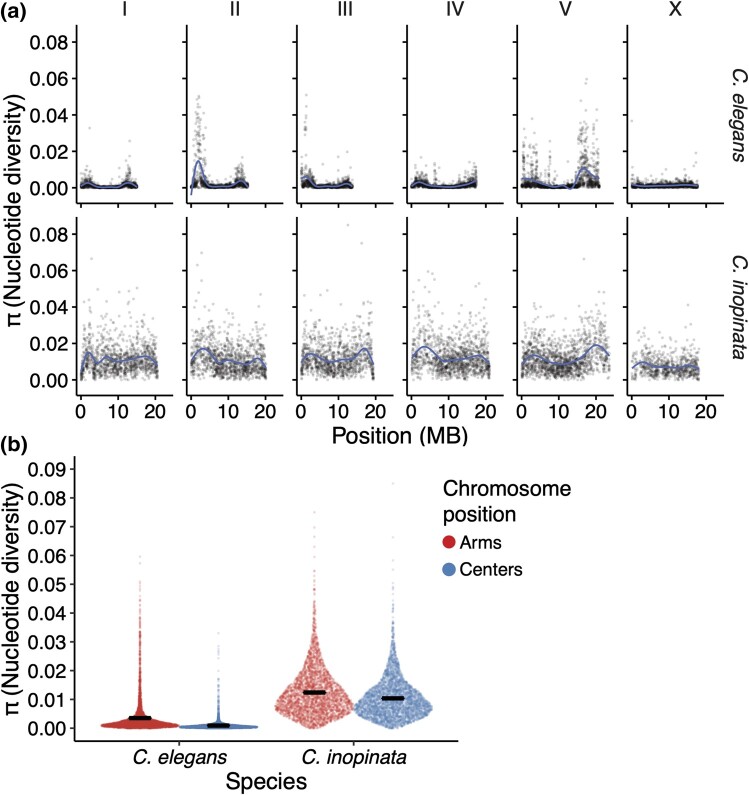
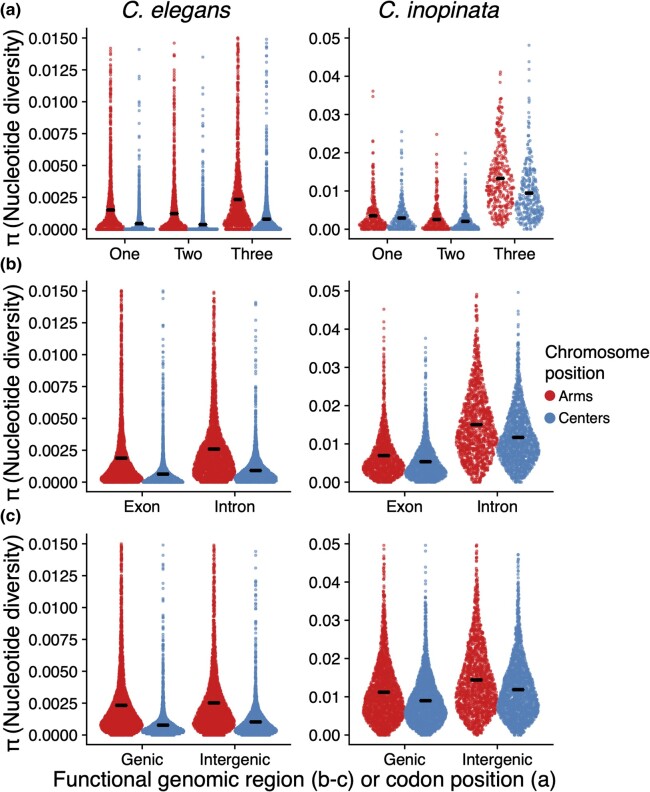
To understand patterns of genetic diversity in C. inopinata, we performed reduced-representation genome sequencing (i.e. bestRAD (Ali et al. 2016)) on 24 individual C. inopinata animals isolated from three islands of the Yaeyama archipelago, the westernmost islands of Okinawa, Japan (Fig. 1; see Sheets 1 and 2 in supplementary table S1, Supplementary Material online for GPS coordinates and sample sizes, respectively). We also retrieved previously published alignment files of C. elegans wild isolates (Cook et al. 2017; Lee et al. 2021; Crombie et al. 2024) to compare genome-wide levels of diversity between C. inopinata and C. elegans. Only alignments associated with the same restriction site we used for RAD in our C. inopinata samples were used for population estimates in C. elegans. After aligning and genotyping C. inopinata sequences, we retained 218,388 biallelic single-nucleotide polymorphism (SNPs) across 195,265 loci of the C. inopinata genome (comprising 4,835,448 bp when including invariant sites; see Sheets 3 to 5 in supplementary table S1, Supplementary Material online for read counts as well as alignment and coverage information). These revealed even coverage across the genome after accounting for variation in restriction site density along C. inopinata chromosomes (supplementary fig. S1, Supplementary Material online). We then found C. inopinata exhibits levels of genome-wide polymorphism that are about five times higher than C. elegans (Fig. 2a; C. inopinata: mean π = 0.011, range = 0.0 to 0.085; C. elegans mean π = 0.0022, range = 0.0 to 0.060 [whole-population mean π across 10 kb genomic windows]). Additionally, genome-wide polymorphism is three times more variable in C. inopinata than in C. elegans (C. inopinata π variance = 6.0 × 10−5; C. elegans π variance = 2.0 × 10−5 [whole-population variance in mean π across 10 kb genomic windows]). As previously reported in C. elegans and other Caenorhabditis species (Koch et al. 2000; Cutter and Payseur 2003; Maydan et al. 2007; Cutter et al. 2009; Andersen et al. 2012; Thomas et al. 2015; Noble et al. 2021; Teterina et al. 2023), polymorphism is elevated on chromosome arms relative to chromosome centers in C. inopinata (Fig. 2b; mean πarms = 0.012, mean πcenters = 0.010). However, the magnitude of elevated diversity on chromosome arms is much lower in C. inopinata (Cohen's d effect size = 0.26) than in C. elegans (Fig. 2b; Cohen's d effect size = 0.61). Additionally, both C. elegans and C. inopinata reveal less enrichment of polymorphism on the arms of the X chromosome (Fig. 2a), although this difference is magnified in C. elegans (autosome Cohen's d effect size = 0.69; X Cohen's d effect size = 0.083) compared with C. inopinata (autosome Cohen's d effect size = 0.27; X Cohen's d effect size = 0.25). Taken together, these results suggest that broad patterns of the genomic organization of polymorphism are conserved between these species despite differences in reproductive mode. But although C. inopinata has much higher levels of polymorphism than the hermaphroditic species C. elegans, C. inopinata is only modestly diverse compared with other outcrossing Caenorhabditis species (supplementary fig. S2, Supplementary Material online; Caenorhabditis brenneri, Caenorhabditis sinica, C. remanei, Caenorhabditis latens, and Caenorhabditis japonica; π = ∼0.01 to 0.16 (Wang et al. 2010; Dey et al. 2012; Dey et al. 2013; Li et al. 2014)).
Fig. 1.
Geographic distribution of genotyped individuals. Twenty-four individual worms were genotyped from five trees across three islands in Okinawa. a) Regional view including major neighboring nations. b) The islands of Ishigaki, Iriomote, and Yonaguni. Two trees sampled on Yonaguni are so close to each other that the points overlap. Maps were generated with Mapbox, OpenStreetMap, and their data sources. © Mapbox, © OpenStreetMap (https://www.mapbox.com/about/maps/; http://www.openstreetmap.org/copyright).
Fig. 2.
The genomic landscape of nucleotide diversity (π) in C. inopinata and C. elegans. a) Nucleotide diversity across 10 kb genomic windows. Lines were fit by LOESS local regression. b) Sina plots (strip charts with points taking the contours of a violin plot) revealing differences among chromosome arms and centers in C. inopinata and C. elegans. Horizontal bars represent means. All C. elegans data for this publication were retrieved from the CaeNDR resource (https://caendr.org/; Crombie et al. 2024).
Patterns of Diversity across Functional Genomic Regions
Patterns of diversity were also assessed across genomic regions harboring putatively variable selective forces. For instance, bases at the third codon position are expected to be under weaker selection than bases at the second codon position due to the degeneracy of the genetic code (Kimura 1977). Additionally, exons are expected to be under stronger selection than introns, and genic regions should be more evolutionarily constrained than intergenic regions. Our data allowed us to look at the extent of genomic diversity across such functional regions in an obligate outcrosser (C. inopinata) and self-fertile hermaphrodite (C. elegans). Additionally, the chromosomal scale of our data allowed us to look at such patterns across putatively high-recombining (“arms”) and low-recombining (“centers”) regions of the genome (Rockman and Kruglyak 2009; Ross et al. 2011; Noble et al. 2021; Stevens et al. 2022).
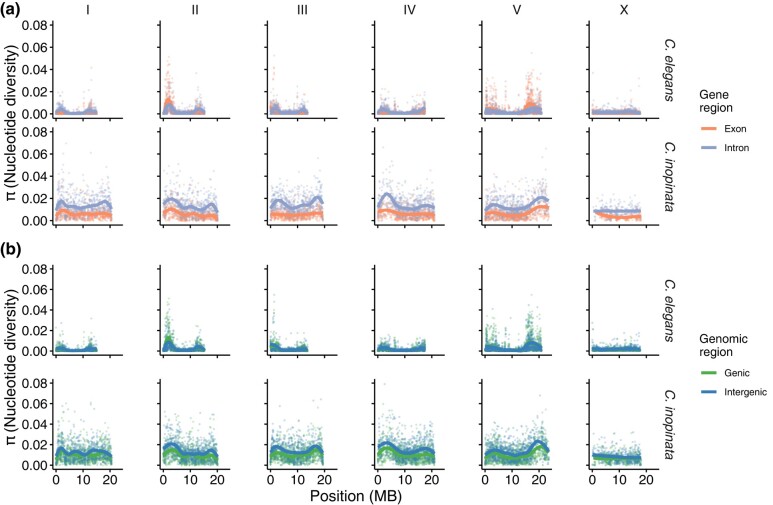
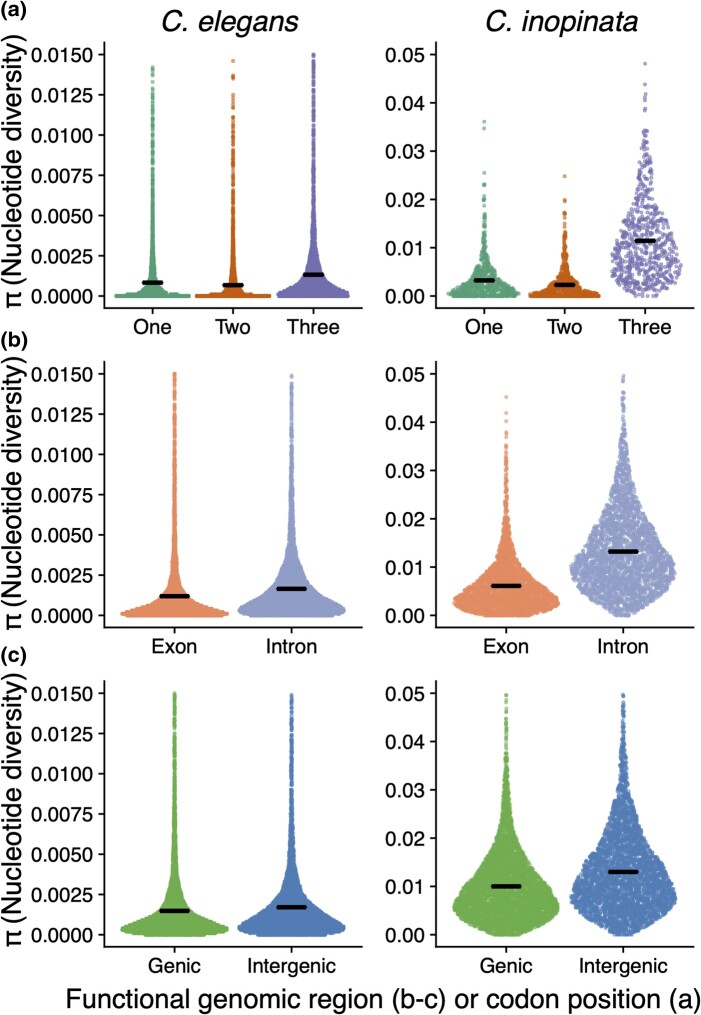
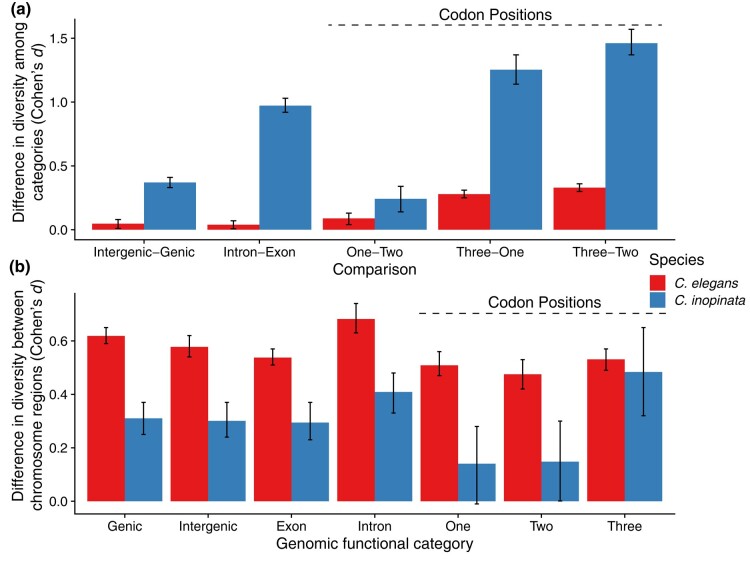
In C. elegans, genetic diversity is only marginally higher in intergenic regions (mean π = 0.0021) than genic regions (mean π = 0.0019; Cohen's d effect size = 0.09; Wilcoxon rank sum test P = 1.3 × 10−24; Figs. 3 and 4; supplementary fig. S3, Supplementary Material online). Similarly, diversity in introns (mean π = 0.0018) is only marginally higher than exon diversity (mean π = 0.0017; Cohen's d effect size = 0.07; Wilcoxon rank sum test P = 1.9 × 10−107; Figs. 3 and 4). In contrast, in C. inopinata, there are substantial differences in diversity among these genomic regions (Figs. 3 and 4). Intergenic diversity (mean π = 0.013) is 30% higher on average than genic diversity (mean π = 0.010) in C. inopinata (Figs. 3 and 4; supplementary fig. S3, Supplementary Material online; Cohen's d effect size = 0.41; Wilcoxon rank sum test P < 2.2 × 10−308). This difference is even greater between introns (mean π = 0.013) and exons (mean π = 0.0062; about double on average; Cohen's d effect size = 1.03; Wilcoxon rank sum test P < 2.2 × 10−308). A similar pattern is seen among codon positions—C. inopinata reveals greater differences in diversity across codon positions (Codon position 3 is 396% more diverse on average than codon position 2) than C. elegans (203%; Figs. 4 and 5). Codon position covaries with genetic code degeneracy, and similar patterns are observed among sites with 0-, 2-, and 4-fold degeneracy (supplementary fig. S4, Supplementary Material online). Synonymous and nonsynonymous variable sites were not explicitly considered because by definition these would not include invariant sites and would then be difficult to compare with the other genomic regions considered above. Moreover, our RAD-seq approach limited the number of these kinds of sites amenable to analysis.
Fig. 3.
Genomic landscapes of diversity across functional positions. a) Genomic landscapes of nucleotide diversity among sites in exons and introns. b) Genomic landscapes of nucleotide diversity among sites in genic and intergenic regions. Plotted are means across 10 kb windows. Solid lines were fit by LOESS local regression.
Fig. 4.
Distributions of diversity across functional positions. Sina plots (strip charts with points taking the contours of a violin plot) reveal differences in nucleotide diversity among various functional genomic categories: a) codon position, b) exons and introns, and c) genic and intergenic regions. Horizontal bars represent means. Here, the tails of the distributions have been clipped to better visualize the differences between functional positions.
Functionally variable genomic regions reveal consistently higher differences in diversity in C. inopinata than C. elegans (Figs. 3 and 4). In contrast, when considering each type of genomic region on its own, all of them reveal greater differences among chromosome arms and centers in C. elegans (Fig. 5). The magnitude of the difference in diversity between chromosome arms and centers is substantial for all such genomic regions in C. elegans (Cohen's d effect size = 0.48 to 0.68). This quantity is smaller in C. inopinata for all functional genomic regions compared with C. elegans (Cohen's d effect size = 0.14 to 0.48; Figs. 5 and 6). This pattern is consistent with the previously described genomic landscapes of diversity (irrespective of functional context; Fig. 2). It is also consistent with patterns of intrachromosomal LD—in C. elegans, alleles in chromosome centers are more likely to cooccur than alleles in chromosome arms (supplementary figs. S5 and S6, Supplementary Material online). C. inopinata reveals no such chromosomal variation in LD, with LD being uniformly low across chromosomes (supplementary figs. S5 and S6, Supplementary Material online). However, LD is higher in C. inopinata (mean r2 for presumably unlinked loci > 2 kb apart is ∼0.075; supplementary fig. S7, Supplementary Material online) than would be expected by the sample size alone (1/n (Hahn 2019) or ∼0.042 to 0.059, depending on the site; supplementary fig. S7, Supplementary Material online). Thus, while C. inopinata reveals higher differences in diversity among putatively variably constrained genomic regions than in C. elegans (Fig. 6a), C. elegans reveals a greater magnitude in the partitioning of diversity across chromosomal regions than C. inopinata (Fig. 6b).
Fig. 5.
Intrachromosomal distributions of diversity across functional positions. Sina plots (strip charts with points taking the contours of a violin plot) reveal differences in nucleotide diversity among various functional genomic categories by chromosome position: a) codon position, b) exons and introns, and c) genic and intergenic regions. Chromosome arms and centers represent the outer and inner half of chromosomes, respectively. Horizontal bars represent means. Here, the tails of the distributions have been clipped to better visualize the differences between functional positions.
Fig. 6.
Effect sizes of diversity differences among functional genomic classes and chromosomal regions by species. a) Differences in nucleotide diversity between functional classes with putative differences in functional constraint. Comparisons labeled “Codon Positions” display differences in nucleotide diversity at respective codon positions in protein-coding sequences. Higher values represent greater differences in diversity in the first functional genomic category compared with the second listed on the x-axis label. b) Differences in nucleotide diversity between chromosome arms and centers at different functional genomic regions or codon positions. Higher values represent greater differences in diversity in chromosome arms compared with chromosome centers for that particular genomic functional category. All values are Cohen's d effect sizes, which are differences in group means divided by the pooled standard deviation. Scale bars represent 95% confidence intervals estimated by 1,000 bootstrap replicates of the data.
C. inopinata Harbors Potentially Elevated Inbreeding and Low Population Differentiation
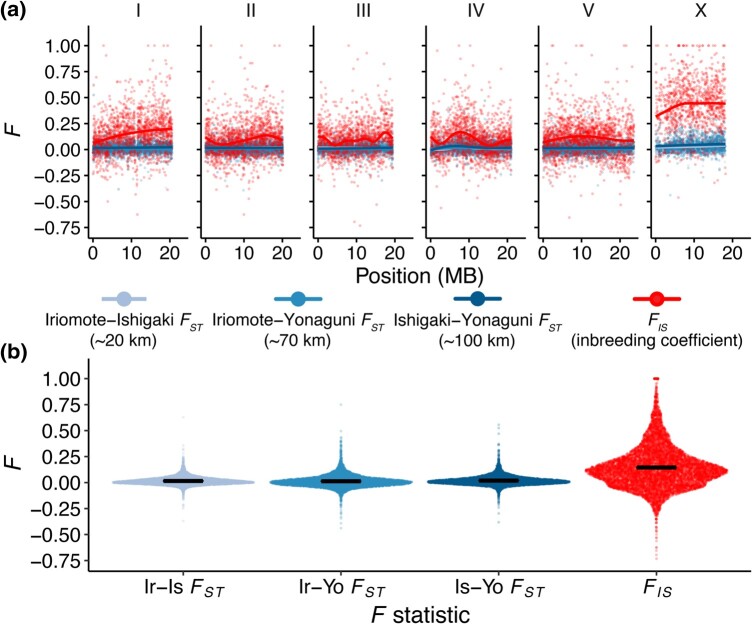
Because C. inopinata is distributed across the islands of Okinawa and Taiwan (Kanzaki et al. 2018; Woodruff and Phillips 2018; Hammerschmith et al. 2020) (and potentially throughout the islands of southeast Asia (Zavodna et al. 2005; Lin et al. 2008; Rodriguez et al. 2017)), we explored the possibility of genetic differentiation among island populations. However, FST is generally low across all island population pairs (Fig. 7; mean pairwise FST = 0.016; SD = 0.049; range = −0.44 to 0.75; Sheet 6 in supplementary table S1, Supplementary Material online). Despite these low values of FST, they are significantly different from 0 (one-sample Wilcoxon rank sum test P < 2.2 × 10−16 for all pairwise comparisons). Additionally, principal components analysis and network phylogenetic approaches do not reveal clusters or clades that correspond with island of origin (supplementary figs. S8 and S9, Supplementary Material online). Discriminant analysis of principal components (Jombart et al. 2010) likewise does not reveal genetic partitioning across islands, plants, or figs across relevant values of K (supplementary fig. S8, Supplementary Material online). These results all suggest C. inopinata migrates frequently among the Yaeyama islands. At the same time, genome-wide estimates of FIS (i.e. the inbreeding coefficient) are much higher and variable than those for FST (Fig. 7; mean FIS = 0.15, SD = 0.20, range = −0.73 to 1.0), suggestive of moderately high levels of mating among close relatives in C. inopinata. We noticed that genomic regions of high FIS were enriched on the C. inopinata X chromosome (Fig. 7; supplementary figs. S10 and S11, Supplementary Material online). Inbreeding coefficients on the C. inopinata X chromosome were nearly four times higher on the X than on autosomes (mean fx/f ratio of 3.96; Fig. 7; supplementary figs. S10, S11, and S13, Supplementary Material online). In C. elegans, these values were about the same (mean fx/f ratio of 1.01; Fig. 7; supplementary figs. S10, S11, and S13, Supplementary Material online). However, C. elegans is a self-fertile hermaphroditic species with very high inbreeding coefficients (supplementary fig. 1, Supplementary Material online), so we also compared interchromosomal inbreeding coefficients in the outcrossing C. remanei (whose whole-genome sequencing population genomic data were recently analyzed and publicly shared (Teterina et al. 2023)). In C. remanei, inbreeding coefficients were slightly elevated on autosomes compared with the X (mean fx/f ratio of 0.87; supplementary figs. S11 and S13, Supplementary Material online), in contrast to the pattern seen in C. inopinata. Because C. inopinata has lower genetic diversity on the X (Fig. 2; supplementary fig. S11, Supplementary Material online), we examined the relationship between nucleotide diversity and inbreeding coefficients both including and excluding the X chromosome (supplementary fig. S12, Supplementary Material online). In C. inopinata, while the X chromosome has a large impact on the negative relationship between FIS and π, this pattern persists even when the X chromosome is excluded (supplementary fig. S12, Supplementary Material online). Additionally, while X chromosomes have similar decreases in diversity compared with autosomes in both C. inopinata and C. remanei (supplementary fig. S11 and S13, Supplementary Material online), C. remanei does not reveal a comparable X chromosome-specific increase in FIS (supplementary fig. S11 and S13, Supplementary Material online). Regardless, these F statistics suggest the unusual cooccurrence of frequent migration with high inbreeding in this species.
Fig. 7.
C. inopinata reveals low genetic differentiation among island populations despite high inbreeding. a) Genomic landscapes of FIS and island population pairwise FST across 10 kb genomic windows. Lines were fit by LOESS local regression. b) Sina plots (strip charts with points taking the contours of a violin plot) revealing the distributions of the data in a). Horizontal bars represent means.
C. inopinata and Its Fig Wasp Carrier Have Potentially Comparable Patterns of Genetic Diversity
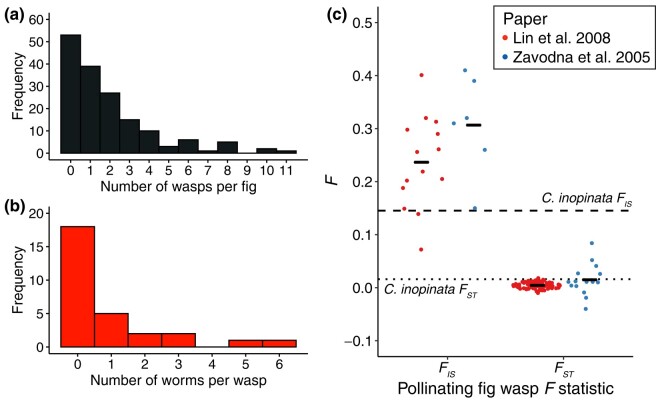
C. inopinata coexists with and is dispersed by the pollinating fig wasp Ceratosolen bisulcatus. We reasoned its patterns of genetic diversity across islands may be explained by its natural history and relationship with fig wasps. We retrieved previous C. inopinata ecological ((Woodruff and Phillips 2018); Fig. 8a and b) and Ceratosolen bisulcatus population genetic ((Zavodna et al. 2005; Lin et al. 2008); Fig. 8c) data to situate the above results in their broader ecological context. Indeed, F. septica figs tend to be pollinated by a small number of pollinating wasps (Fig. 8a; mean = 1.9, range = 0 to 11; N = 162), and most wasps carry only a small number of C. inopinata animals (Fig. 8b; mean = 0.90, range = 0 to 6, N = 29) (Woodruff and Phillips 2018). Moreover, two previous studies of Ceratosolen bisulcatus examined differentiation among populations that both included islands separated by open ocean in Indonesia (Zavodna et al. 2005) and Taiwan (Taiwan and Lanyu Island (Lin et al. 2008)). Both studies revealed low differentiation among populations (mean pairwise FST in Zavodna et al. (2005) = 0.015; mean pairwise FST in Zavodna et al. (2005) = 0.0047). They also observed high inbreeding coefficients in Ceratosolen bisulcatus populations (mean FIS (Zavodna et al. 2005) = 0.31; mean FIS (Lin et al. 2008) = 0.24). Thus, Ceratosolen bisulcatus likewise migrates frequently across islands and exhibits high levels of inbreeding (Zavodna et al. 2005; Lin et al. 2008), mirroring our observations of the worms they carry (Fig. 8c).
Fig. 8.
Low C. inopinata founding populations in figs and the fig wasp vector of C. inopinata has high dispersal and inbreeding. a) The distribution of founding Ceratosolen bisulcatus pollinating fig wasps among pollinated F. septica figs. b) The distribution of C. inopinata worms observed traveling on Ceratosolen pollinating fig wasps. Data in a) and b) are from Woodruff and Phillips (2018). c) The distribution of Ceratosolen bisulcatus FIS and FST statistics from two previous studies (Lin et al. 2008 and Zavodna et al. 2005). For Lin et al. (2008): 1,502 bp of the COI locus plus 15 microsatellites, 15 localities across Taiwan and Lanyu Island used as populations, 86 trees and 219 (COI) or 398 (microsatellites) wasps, FIS data from Table 5 and pairwise FST data from Table 6. For Zavodna et al. (2005): five microsatellites, six localities including four islands across Indonesia used as populations, 74 wasps, FIS data from Table 3 and pairwise FST data from Table 5. Both studies revealed low differentiation among fig wasps on islands separated by open ocean (Lin et al. (2008), 60 km; Zavodna et al. (2005), 40 km). Black horizontal bars represent means. Dotted and dashed lines denote mean island population pairwise FST and mean FIS, respectively, among 10 kb genomic windows in C. inopinata (Fig. 7).
Discussion
Diversity, Selfing, and Outcrossing
Although myriad population genetic studies have been performed in outcrossing Caenorhabditis species using a handful of genetic loci (Graustein et al. 2002; Jovelin et al. 2003; Cutter et al. 2006; Cutter 2008; Jovelin 2009; Wang et al. 2010; Dey et al. 2012, 2013; Gimond et al. 2013; Li et al. 2014), this work examines diversity at a genomic scale in an obligately outcrossing Caenorhabditis species (a recent study of C. remanei also afforded such a comparison (Teterina et al. 2023)). As much genomic work has been performed on selfing Caenorhabditis species, this study allows the opportunity to compare genomic patterns of diversity among species with differing modes of reproduction. Indeed, the population genetic consequences of selfing are expected to be wide-ranging (Cutter 2019). In selfers, polymorphism is expected to be lower, homozygosity higher, and effective population sizes smaller. Moreover, selection is expected to be less efficient, and LD tracts are expected to be longer. All of these patterns emerge from high rates of self-fertilization—despite the existence of meiosis, fertilization occurs largely within the same individual. Selfing inhibits the segregation and recombination of alleles expected under outcrossing, leading to high homozygosity and low levels of effective recombination.
Our results provide a natural test of these expectations of population genetic theory. C. elegans is five times less diverse than C. inopinata (Fig. 2). The diversity among exons and introns, genic and intergenic regions, and codon positions is lower in C. elegans than C. inopinata (Figs. 3 to 6). C. elegans is far more inbred than C. inopinata (supplementary fig. S10, Supplementary Material online), and C. elegans harbors far higher levels of intrachromosomal LD than in C. inopinata (supplementary figs. S5 and S6, Supplementary Material online). Thus, population genetic expectations regarding differences in polymorphism (Fig. 2), the efficacy of selection (Figs. 3 to 6), inbreeding (Fig. 7; supplementary fig. S10, Supplementary Material online), and LD (supplementary figs. S5 and S6, Supplementary Material online) between selfers and outcrossers are aligned with our observations. Indeed, similar results were recently observed in another outcrossing Caenorhabditis species, C. remanei (Teterina et al. 2023). Thus, descriptions of nematode genomic diversity confirm the predictions of population genetic theory regarding the evolution of reproductive modes, consistent with previous studies (Cutter et al. 2009).
Diversity and Recombination
Differences in the extent of intrachromosomal diversity were also observed (Figs. 2 to 6). Specifically, intrachromosomal differences in polymorphism were markedly greater in C. elegans than C. inopinata (Fig. 2). This general chromosome-level pattern has long been noted and has been observed in other selfing species of Caenorhabditis (Koch et al. 2000; Cutter and Payseur 2003; Maydan et al. 2007; Cutter et al. 2009; Andersen et al. 2012; Thomas et al. 2015; Cook et al. 2016, 2017; Lee et al. 2021; Noble et al. 2021). This pattern is likely to be driven by variation in recombination rates within chromosomes, as chromosome centers have far lower recombination rates than chromosome arms in three selfing Caenorhabditis species (Rockman and Kruglyak 2009; Ross et al. 2011; Noble et al. 2021; Stevens et al. 2022). And a similar pattern was recently described in the outcrossing species C. remanei (Teterina et al. 2023). It remains unclear how recombination rate variation is structured in C. inopinata chromosomes. Although the difference is smaller in C. inopinata, chromosome arms are still more diverse than chromosome centers (Fig. 2). Conversely, genomic landscapes of transposable element density and gene density are more uniform in this species compared with its close relatives (Woodruff and Teterina 2020). The cooccurrence of these more uniform landscapes suggests a change in intrachromosomal recombination rates may cause changes in the distribution of genes and transposable elements across chromosomes. This hypothesis could be tested through phylogenetic comparative approaches. There is at least one additional Caenorhabditis species associated with figs (Ficus hispida (Jauharlina et al. 2022)) that is likely to be related to C. inopinata. The characterization of its genomic patterns of diversity, recombination, gene density, and transposable element density could inform when these features evolve and the possibility of recombination-driven changes in genome organization. Moreover, at least two additional Caenorhabditis species have potentially independently evolved uniform transposable element genomic landscapes (Caenorhabditis bovis (Stevens et al. 2020) and C. japonica (Woodruff and Teterina 2020)). Additional chromosome-contiguous assemblies, population genomic surveys, and recombination mapping across the Caenorhabditis genus will inform whether changes in genomic organization is highly correlated with the evolution of intrachromosomal recombination rates (and when these various patterns evolve). Regardless, it is likely that low global effective recombination, high levels of LD in gene-rich chromosome centers, and linked selection (Andersen et al. 2012; Lee et al. 2021) lead to the large disparity in intrachromosomal diversity in C. elegans. Outcrossing, effective global recombination, and perhaps lower differences in intrachromosomal recombination rate may lead to lower differences in intrachromosomal diversity in C. inopinata. Ongoing work in genetic map construction in C. inopinata will inform the role of recombination in molding patterns of genomic diversity in this species.
Population Differentiation, Inbreeding, and the Fig Wasp Life Cycle
Our observations yield the unusual pattern of elevated inbreeding with low differentiation among island populations (Fig. 7). Specifically, inbreeding is not expected to occur if alleles easily disperse, and outbreeding is expected if individuals are freely migrating across large areas. How is this pattern we observe then possible? We propose that the ability of fig wasps to travel long distances, in tandem with low founding populations in individual figs, generates this pattern. That is, the unique life cycles, propagule sizes, and comigration of fig wasps and fig nematodes together drive high inbreeding with low differentiation among islands in both species. Fig wasps can disperse for dozens of kilometers on the wind (Ahmed et al. 2009), even across expanses of open ocean (Zavodna et al. 2005; Lin et al. 2008). C. inopinata nematodes travel on such wasps (Woodruff and Phillips 2018), and their dispersal and life cycle likely depend on this phoretic relationship. However, despite their ability to travel long distances, the reproductive (i.e. mating) phases of both fig wasp and fig nematode life cycles are restricted to the confined lumen of the individual fig (Weiblen 2002; Woodruff and Phillips 2018; Van Goor et al. 2023). And as wasp (and nematode) founding populations are small (Fig. 8a), reproduction is likely to occur between close relatives. Indeed, figs with one foundress wasp are common (Fig. 8a)—in this case, mating necessarily occurs between the siblings who all share one mother. Thus, life cycles, the high dispersal ability of wasps, obligate comigration of nematodes with wasps, and low founding numbers in figs all converge to generate this unusual partitioning of alleles among individuals and islands (Fig. 7). And, both wasps and worms have similar levels of FIS and FST (Fig. 8b), consistent with comigration underlying this pattern. Not all members of the fig microcosm may necessarily share this costructuring—the nonpollinating and pollinating fig wasp species of Ficus hirta harbor variable levels of isolation by distance (Deng et al. 2021). However, our comparisons of fig wasp and fig nematode diversity should be met with caution as we are comparing data sets separated by years, geographic regions, and methodology (Zavodna et al. 2005; Lin et al. 2008). Indeed, we have only sampled a portion of the geographic range of F. septica (Rodriguez et al. 2017), and future work may reveal genetically distinct populations. If these cooccur among fig nematodes and fig wasps, geographic distance may drive such differences, as has been reported in F. hirta and its Valisia pollinators (Tian et al. 2015).
Fig nematode migration may also be influenced by human activity. Although most Indo-Australian Ficus species are not thought to be traditional food sources, a number of fig species have historically been used for dietary and medicinal purposes (Shi et al. 2018). F. septica leaves have been reported as a treatment of illnesses in Papua New Guinea (Holdsworth et al. 1980; Holdsworth and Lacanienta 1981) and the Solomon Islands (Altschul 1973). Beyond historical uses, contemporary human activity may likewise influence nematode migration patterns as the introduction of nonnative Ficus species to new localities does occur (Galias et al. 2018; Peniwidiyanti et al. 2022). Additionally, as F. septica syconia are frequently consumed by bats (Shilton et al. 1999), this interaction may also facilitate fig nematode migration.
We also found the C. inopinata X chromosome to be enriched for these high inbreeding coefficients (Fig. 7; supplementary figs. S10 to S13, Supplementary Material online). Assuming neutrality and unbiased sex ratios, nucleotide diversity on the X is expected to be 75% of that on the autosomes ((Wilson Sayres 2018); although this is not to expected to hold for the self-fertile hermaphrodite, C. elegans). Diversity on the C. inopinata X chromosome is 64% of autosomal nucleotide diversity (supplementary fig. S13, Supplementary Material online). Under random mating, no inbreeding is expected, and as X chromosome inbreeding coefficients depend largely on specific pedigrees (Wright 1933; Crow and Kimura 2009) or specific demographic scenarios (Wang 1999), assigning a null inbreeding hypothesis is difficult. Yet, under inbreeding with no bias in male–female cross directions, the X:autosome inbreeding coefficient ratio is expected to approach 4/3 (1.333…) as the distances between relatives increases (Freire-Maia and Freire-Maia 1961). In C. inopinata, this ratio is far higher (3.96; supplementary fig. S13, Supplementary Material online). As its X:autosome inbreeding coefficient ratio is quite high (supplementary fig. S13, Supplementary Material online), it is possible complex demographic histories (Wang 1999), selection, or other factors may be driving this observation. As X chromosomes are generally expected to be subject to differing evolutionary forces than the autosomes (Vicoso and Charlesworth 2006), any of these could likewise interact to promote asymmetries on the X chromosome in C. inopinata. Most of the individuals we sequenced were female (Sheet 1 in supplementary table S1, Supplementary Material online), so it is unlikely that an excess of males in our sample is connected to this unexpected pattern. This high X:autosome inbreeding coefficient ratio is also unlikely to be a technical artifact as coverage is comparable across autosomes and the X chromosome (supplementary fig. S1, Supplementary Material online). A more likely explanation is that natural populations of C. inopinata harbor unequal sex ratios, which would be expected to generate unequal ratios of X:autosome inbreeding coefficients. Pollinating fig wasps themselves famously modulate their sex ratios depending on the potential for local mate competition (Herre 1985, 1987). Fig nematodes may also do this, which could generate such asymmetrical patterns on the X chromosome. A highly speculative explanation for this observation could be environmental sex determination (ESD)—in fig-associated Parasitodiplogaster and Ficophagus nematodes, sex ratios in syconia were more female-biased than those observed on fig wasps (Van Goor et al. 2022). Moreover, nematode sex ratios in syconia had a far lower variance than expected (Van Goor et al. 2022). If ESD can transform X0 animals into females, ESD could promote high observed X chromosome inbreeding coefficients (as X0 females would appear to be entirely homozygous on the X chromosome), as well as lead to the kinds of demographic scenarios that can increase inbreeding coefficients on the X chromosome (Wang 1999; Crow and Kimura 2009). Sex determination has likely evolved in C. inopinata as her-1, a gene encoding a signaling ligand critical for male fates in C. elegans (Perry et al. 1993), has been disrupted by a transposon insertion in this species (Kanzaki et al. 2018). And, X0-to-female transformants have been observed among Caenorhabditis hybrid broods (Baird 2002), showing such transformations are at least possible in this group. However, ESD has not been reported in the Caenorhabditis genus (to the best of our knowledge), although it has evolved multiple times in nematodes (Haag 2005). Future work involving evolutionary simulations (Haller and Messer 2023) could potentially disentangle these possible causes of asymmetry among the X chromosome and the autosomes. Regardless, future work on fig wasps and nematodes sampled at the same time, in the same places, and interrogated with the same methods will be required to confirm this apparent alignment of their patterns of genetic differentiation.
C. inopinata Diversity in Its Phylogenetic Context
Patterns of chromosome-scale variation were recently described in the outcrossing species, C. remanei (Teterina et al. 2023). How do these results compare? As in the case of C. remanei (Teterina et al. 2023), we found the outcrossing C. inopinata is far more genetically diverse than the selfing C. elegans (Fig. 2). Additionally, like in the previous report (Teterina et al. 2023), we find that diversity is enriched on chromosome arms relative to chromosome centers in both outcrossers (Figs. 2 and 3). And, we likewise find that diversity among functional genomic regions (such as introns and exons) are much greater in outcrossers than selfers (Figs. 3 to 6), indicative of more efficient selection in outcrossing species (Teterina et al. 2023). Qualitatively, our results are then aligned with previous interpretations that levels of genomic diversity are driven by differences in reproductive mode, while conserved distributions along chromosomes are driven by conserved patterns of intrachromosomal recombination rates (Teterina et al. 2023).
Despite this overlap, our work reveals at least one major qualitative difference with that of Teterina et al. (2023). Specifically, intrachromosomal differences in variation were greater in C. remanei (Cohen's d = 1.08) than C. elegans (Cohen's d = 0.62) (Teterina et al. 2023). We find the opposite pattern—the differences in diversity between chromosome arms and centers were greater in C. elegans (Cohen's d = 0.61) than C. inopinata (Cohen's d = 0.26; Figs. 2, 3, and 6). In Teterina et al. (2023), the greater intrachromosomal diversity disparity in outcrossers was attributed to their greater effective population recombination rate. As C. inopinata has far lower intrachromosomal LD than C. elegans, it likely also that it has a much greater effective population recombination rate than this selfing species. Notably, C. inopinata's lower degree of intrachromosomal diversity mirrors its similar lack of structuring of gene density and transposable elements along chromosomes (while C. elegans and C. remanei have similar, structured intrachromosomal distributions of gene number and repeat content) (Woodruff and Teterina 2020). Speculatively, this low intrachromosomal diversity is suggestive of a divergent genomic recombination landscape in C. inopinata. If C. inopinata has a uniform recombination rate among chromosome arms and centers, then this uniformity would be consistent with its more uniform intrachromosomal patterns of diversity, gene density, and repeat content. Ongoing work involving the construction of a C. inopinata genetic map will be invaluable toward testing this possibility.
Beyond intrachromosomal variation, there are a number of quantitative differences between this work and the C. remanei study (Teterina et al. 2023). For instance, our estimate of C. elegans nucleotide diversity (π = 0.0022) is nearly double the Teterina et al. (2023) estimate (π = 0.0012). Teterina et al. (2023) used whole-genome sequencing; examined one Toronto population of C. remanei and one Hawaiian population of C. elegans and used a different computational workflow compared with our study. We used RAD-seq and attempted to capture a broad range of diversity in C. inopinata and C. elegans (i.e. we looked at three island populations of C. inopinata and included the “divergent set” of previously sequenced C. elegans strains (Cook et al. 2017; Lee et al. 2021; Crombie et al. 2024)). We then suspect most of these quantitative differences derive from methodological and sampling (which has been shown to influence biological interpretations (Städler et al. 2009; Cutter et al. 2012)) variation. And as our results are largely qualitatively concordant as discussed above, they appear robust to such changes in methodological approach.
The Caenorhabditis genus is noteworthy in the range of intraspecific polymorphism among its constituent species (Wang et al. 2010; Andersen et al. 2012; Dey et al. 2012, 2013; Gimond et al. 2013; Li et al. 2014; Thomas et al. 2015; Noble et al. 2021) (supplementary fig. S2, Supplementary Material online). This range in polymorphism is driven largely by the independent evolution of self-fertile hermaphroditism in three species (including C. elegans) that have lower levels of polymorphism due to high selfing rates. Conversely, most Caenorhabditis species (including C. inopinata) are obligate outcrossers (Kiontke et al. 2011; Cutter 2015; Kanzaki et al. 2018). A number of these species appear to harbor exceptional levels of diversity (i.e. are “hyperdiverse” with π ≥ 0.05) suggestive of massive population sizes (Cutter et al. 2013). C. inopinata harbors low levels of polymorphism compared with its hyperdiverse outcrossing relatives while being far more diverse than self-fertilizing Caenorhabditis species (supplementary fig. S2, Supplementary Material online). Its modest diversity as a female/male species may be explained by its vector specialist lifestyle. It is thought that specialists should have lower population sizes and more differentiation among populations compared with generalists because there are fewer opportunities for them to thrive and reproduce in space (Li et al. 2014). Notably, C. japonica is another vector specialist that also has lower levels of diversity among outcrossing Caenorhabditis (Li et al. 2014). These comparatively lower levels of diversity in C. inopinata and C. japonica provide evidence for this specialist–generalist variation hypothesis (SGVH (Li et al. 2014)). However, the lack of population differentiation in C. inopinata is inconsistent with the expectations of population fragmentation under the SGVH (Fig. 7). This incongruity could result from the notable migration distances of fig wasps (Ahmed et al. 2009), the limited spatial area of our study, or both. Moreover, there are more known Caenorhabditis vector specialists and generalists whose diversity has not been determined (Cutter 2015; Sloat et al. 2022). Exploring diversity in more Caenorhabditis species and situating them in their phylogenetic context, as well as the geographic range and extent of population differentiation in C. inopinata, will be needed to show if the SGVH holds in this group.
Caveats
It is important to note some potential limitations of this study. The number of individuals sampled per population is relatively small (Nindividuals = 6, 12, and 6 for the islands of Iriomote, Ishigake, and Yonaguni, respectively). These low sample sizes have the potential to pose statistical sampling issues in our estimates of population genetic measures connected to population differentiation. That is, estimates of FST may be inaccurate due to small per-population sample sizes. Indeed, such parameters are largely assumed to be sensitive to sample size (Holsinger and Weir 2009), and the failure to capture rare alleles due to small sample sizes can lead to underestimates of FST (Bhatia et al. 2013). On the other hand, both simulations (Willing et al. 2012) and empirical data (Nazareno et al. 2017) have suggested that these parameters are robust to sample size variation, and if anything, extremely small sample sizes should lead to overestimates of FST (Willing et al. 2012). In this scenario, our conclusions would not be impacted as our estimates of FST are quite low and suggest rampant migration among islands. An overestimate in FST values would then suggest our populations are even less differentiated than reported here. Regardless, additional sampling of these areas, in addition to increased sampling across the known geographic range of F. septica, holds the potential to capture uncharacterized alleles and provide a more accurate picture of genomic variation in C. inopinata.
Additionally, this study characterizes genomic variation via a rarely used combination of linear genomic amplification and RAD-seq. The use of genome amplification must be approached with some caution as the underlying molecular biases are unclear—here, we are assuming all alleles are amplified with equal fidelity. Notably, we see do variation in coverage across the genome, but this variation is readily explained by the distribution of GC content across the genome (i.e., we used EcoRI as a restriction enzyme for RAD-seq, and its recognition sequence is not uniformly distributed along chromosomes; supplementary fig. S1, Supplementary Material online). However, the possibility remains that biases in genome amplification have led to the exclusion of some alleles in our data. Despite the widespread presence of heterozygous calls (indeed, many genomic regions reveal an excess of heterozygotes compared with Hardy–Weinberg expectations; Fig. 7b), we cannot definitively exclude this possibility. Future whole-genome sequencing studies will reveal whether or not biases in linear amplification impact our ability to characterize genomic variation. Similarly, we used RAD-seq to measure genomic diversity. Indeed, we only captured ∼4% of the C. inopinata genome in our dataset (4.8 MB out of a genome assembly size of 123 MB) despite the evenness of coverage across the genome (supplementary fig. S1, Supplementary Material online). Comparisons of the C. elegans pseudo-RAD data with WGS data suggest that RAD-seq may underestimate nucleotide diversity (supplementary fig. S14, Supplementary Material online). Moreover, sampling only 24 C. elegans strains also leads to an underestimate of nucleotide diversity compared with a sample of 330 strains (supplementary fig. S14, Supplementary Material online) (Lee et al. 2021). The limitations of RAD-seq in characterizing patterns of genomic diversity are even more apparent considering known hyperdivergent regions in nematode genomes (Lee et al. 2021; Stevens et al. 2022, 2023). These regions are of particular interest because they are frequently associated with segregating selfish genetic elements (Ben-David et al. 2017, 2021; Noble et al. 2021; Widen et al. 2023). A recent paper characterizing hyperdivergent regions in C. elegans defines such regions as having at least nine consecutive 1 kb windows harboring ≥16 variants or harboring substantially low coverage compared with a genome-wide average (Lee et al. 2021). As we did not assemble genomes de novo, we could not use low read depth as a metric for potential hyperdivergence (in fact, to ensure confidence in genotype calls, we only looked at regions with high coverage and consistent alignment with reference assemblies). Because RAD-seq captures sequences associated with restriction sites, we only rarely captured segregating sites across nine consecutive windows (11 such genomic regions in our C. inopinata data; 18 in the C. elegans pseudo-RAD data; supplementary figs. S15 and S16, Supplementary Material online). None of the regions we captured qualified as hyperdivergent by the definition above using our data alone (supplementary fig. S15, Supplementary Material online). However, although our data cannot be used to discover hyperdivergent regions (as defined by Lee et al. (2021)), nearly all of the previously characterized hyperdivergent regions contain sites captured by our C. elegans pseudo-RAD data (299/312 regions with overlap; 96%). Indeed, hyperdivergent regions were reported to cover a nontrivial fraction of the C. elegans genome (20% (Lee et al. 2021)). As a consequence, a fraction of our C. elegans pseudo-RAD data falls within such regions (14%; 2,105,793 sites). Our C. inopinata RAD-seq data are then likely to include sites in hyperdivergent regions as well, although whole-genome sequencing data will be required to pinpoint these regions. Thus, our diversity measures are surely underestimated. Regardless, whole-genome sequencing of a large and diverse sample of C. inopinata individuals will be required to address the shortcomings of reduced-representation sequencing in characterizing the genomic diversity of populations.
The Promise of Understanding Coevolutionary Patterns
Understanding the causes of spatial genetic structure is a fundamental goal of molecular ecology. Here, we suggest fig nematodes and their fig wasp vectors may share similar patterns of genetic differentiation due to their shared modes of dispersal and their common symbiotic relationship with figs. Interspecific relationships that involve the colocalization and comigration in space of individuals from different species are ubiquitous. Host–pathogen, host–parasite, and phoretic relationships (among others) abound and could lead to similar alignments of genetic diversity among divergent species. Future work examining a larger sample and range of fig nematodes and fig wasps will inform if ecological interactions can drive the alignment genetic structure despite the vast range of geographic distances and body sizes of the species involved.
Methods
Nematode Isolation, Sample Preparation, and Sequencing
Animals were collected from the field in previously described work (Woodruff et al. 2018; Hammerschmith et al. 2022). Briefly, 24 individual C. inopinata animals were isolated from fresh, dissected F. septica figs from three Okinawan islands in May 2016. Live animals were fixed in 100% ethanol and kept at −20 °C for 3 to 11 mo. Fixed animals were then washed three times in phosphate-buffered saline (PBS), and individual worms were transferred to individual tubes. Animals were digested with Proteinase K in 20 μL reactions. The proteinase was heat-inactivated (10 min at 95 °C), and then, half of the reaction was used for linear amplification with the Illustra GenomiPhi V3 amplification kit (GE Lifesciences). DNA was then purified with the Zymo Genomic DNA Clean and Concentrator kit. EcoRI bestRAD libraries were prepared (Ali et al. 2016), and paired-end 150 bp reads were generated with the Illumina Hi-Seq 4000.
Genotyping and Inference of Population Genetic Statistics
Reads were reoriented for processing with Flip2BeRad (https://github.com/tylerhether/Flip2BeRAD), and the first 2 base pairs were removed from all reads with fastx_trimmer (version 0.0.13; options -f 3 -Q 33; http://hannonlab.cshl.edu/fastx_toolkit/) for downstream processing. Reads were then demultiplexed with stacks process_radtags (version 2.0; options -e ecoRI -r -c -q) (Rochette et al. 2019, 2). Reads were aligned to the reference C. inopinata genome assembly (Kanzaki et al. 2018) with gsnap (version 2018 March 25; options --trim-mismatch-score=0 --trim-indel-score=0 --format=sam) (Wu et al. 2016). Unique alignments were then extracted (see align_genotype_pop_gen.sh; all code associated with this work have been deposited in GitHub: https://github.com/gcwoodruff/inopinata_population_genomics_2020).
Genotypes were called with bcftools mpileup call (version 1.9; options mpileup -Ou; options call -c -Ob –ploidy-file –samples-file) (Li 2011); X chromosome sites in males were called as haploid, while female X chromosome and all autosome sites were called as diploid. Alignment files of all samples were then merged with bcftools merge (version 1.9; options --info-rules DP:join, MQ0F:join, AF1:join, AC1:join, DP4:join, MQ:join, FQ:join). Sites with coverage <15× and with <80% of samples having genotype calls were removed with bedtools view (version 1.9; for coverage: options -i “DP> = 15”; for missing genotype calls on autosomes: options -i “COUNT(GT = “mis”) < 5”; for missing genotype calls on X chromosomes: options -i “COUNT(GT = “mis”) < 4”). Biallelic sites were extracted with bedtools view (version 1.9; options: -m2 -M2 -v snps --min-ac 2:minor) and combined with invariant sites to produce a single VCF for inference of population genetics statistics (Quinlan and Hall 2010). Additionally, separate VCF files were generated for estimating population genetics statistics of the X and autosomes.
VCF files were processed with popgenwindows.py (https://github.com/simonhmartin/genomics_general/blob/master/popgenWindows.py) to estimate π (retrieved 2019 May 28; options --windType coordinate -w 10000 -s 10000 -m 160 -f phased) and FST (options --windType coordinate -w 10000 -s 10000 -m 160 --analysis popPairDist -f phased -p ishigaki -p iriomote -p yonaguni --popsFile) in 10 kb genomic windows. The script codingSiteTypes.py was used to extract sites in protein-coding genes by codon positions, and popgenwindows.py was used as above to estimate nucleotide diversity. To estimate nucleotide diversity in other genomic regions (introns, exons, intergenic regions, and genic regions), bedtools intersect (version 2.25.0) (Quinlan and Hall 2010) was used with annotation files to extract regions of VCF files spanning respective genomic regions (site counts across genomic and chromosome regions are included in Sheet 7 in supplementary table S1 and supplementary fig. S17, Supplementary Material online). Stacks populations (Rochette et al. 2019) was used to estimate site FIS (version 2.2; options --sigma 3333 --genepop --structure --phylip), and bedtools map (version 2.25.0; -o mean -c 4) (Quinlan and Hall 2010) was used to initially determine the mean FIS in 50 bp genomic windows. This was done to account for the high number of missing sites resulting from reduced-representation RAD sequencing. After the removal of windows composed only of missing sites, bedtools map (version 2.25.0; -o mean -c 4) (Quinlan and Hall 2010) was used to find mean FIS in 10 kb genomic windows. For comparing π on chromosomes arms and centers between C. inopinata and C. elegans, windows were normalized by chromosome position by setting the median chromosome base pair to 0 and the end chromosome base pairs to 0.5 (as in Woodruff and Teterina (2020)). Chromosome “centers” were defined as those genomic windows with normalized chromosomal position < 0.25 and chromosome “arms” as those with normalized chromosomal position ≥ 0.25.
F statistics data in Ceratosolen bisulcatus were collected from Lin et al. (2008) (Tables 5 and 6) and (Zavodna et al. 2005) (Tables 3 and 5). Natural history data for C. inopinata occupancy on fig wasps and fig wasp foundress number in F. septica figs were communicated in Figs. 5 and 6 and supplementary fig. S1, Supplementary Material online of Woodruff and Phillips (2018). VCFtools (version 0.1.16, option --geno-r2) (Danecek et al. 2011) was used to estimate the correlation among genotypes (i.e. patterns of LD) within each chromosome. Mean values of r2 for each site were used for the estimation of mean r2 across 10 kb genomic windows (with bedtools map as above).
For estimates in C. elegans, alignment files (BAM) of 24 previously whole-genome sequenced C. elegans strains (BRC20067, CB4856, CX11271, CX11276, CX11285, CX11314, DL200, DL226, DL238, ECA246, ECA251, ECA36, ED3017, ED3040, ED3048, ED3049, EG4725, JT11398, JU258, JU775, LKC34, MY16, MY23, and N2) were retrieved from the CaeNDR database (Cook et al. 2017; Lee et al. 2021; Crombie et al. 2024) (retrieved June 2021; version 20210121; https://caendr.org/data/data-release/c-elegans/20210121). These strains were chosen because they included the “divergent set” of 12 strains (CB4856, CX11314, DL238, ED3017, EG4725, JT11398, JU258, JU775, LKC34, MY16, MY23, and N2; version 20210121; https://caendr.org/data/data-release/c-elegans/20210121) plus an additional arbitrarily chosen 12 strains to produce a data set with a sample size comparable with our C. inopinata data. The positions of EcoRI cut sites in the C. elegans reference genome were identified with EMBOSS fuzznuc (version 6.6.0, -pattern “GAATTC” -complement -rformat gff) (Rice et al. 2000). C. elegans alignments located within 332 bp of an EcoRI site were extracted with samtools view (options -b -L) (Danecek et al. 2021). Alignments were then genotyped with bcftools mpileup call and processed as above. All analyses and figures for this paper were generated in the R statistical programming language (R Core Team). All statistics were performed in R (all code used for this work have been deposited in GitHub; https://github.com/gcwoodruff/inopinata_population_genomics_2020). The R packages “ggplot2” (Wickham 2016), “lemon” (Edwards 2020), “ggforce” (Pedersen 2022a), “reshape2” (Wickham 2007), “cowplot” (Wilke 2020), “rstatix” (Kassambara 2023), “vcfr” (Knaus and Grünwald 2017), “adegenet” (Jombart and Ahmed 2011), “plyr,” “cowplot,” and “patchwork” (Pedersen 2022b) were used.
Supplementary Material
Acknowledgments
We thank Anastasia Teterina for offering helpful feedback throughout the development of this work and the preparation of this manuscript. Bill Cresko, Peter Ralph, Andy Kern, and their laboratory members likewise provided helpful comments throughout the development of this work. We thank Waldir Berbel-Filho and Kimberly Moser for providing helpful comments on earlier versions of this manuscript. We thank the University of Oklahoma Biogeography of Behavior Journal Club for their valuable feedback regarding the revision of this manuscript. We thank Tyler Hether for sharing Flip2BeRAD and assisting the processing of bestRAD reads. We thank Erik Andersen and the Caenorhabditis Natural Diversity Resource for sharing C. elegans variant data. We thank the University of Oregon Genomics and Cell Characterization Core Facility (GC3F) for assistance with Illumina sequencing. This work also benefited from access to the University of Oregon high-performance computer, Talapas. Some of the computing for this project was performed at the University of Oklahoma (OU) Supercomputing Center for Education & Research (OSCER) at the OU.
Contributor Information
Gavin C Woodruff, Institute of Ecology and Evolution, University of Oregon, Eugene, OR 97403, USA; Present address: Department of Biology, University of Oklahoma, Norman, OK 73019, USA.
John H Willis, Institute of Ecology and Evolution, University of Oregon, Eugene, OR 97403, USA.
Patrick C Phillips, Institute of Ecology and Evolution, University of Oregon, Eugene, OR 97403, USA.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
G.C.W. and P.C.P. devised and designed the project. G.C.W. collected nematodes, performed bioinformatic analyses, and wrote the first draft of the paper; J.H.W. prepared DNA libraries for sequencing; G.C.W. and P.C.P. revised and prepared the final manuscript.
Funding
This work was supported by funding from the National Institutes of Health to G.C.W. (grant no. 5F32GM115209-03) and to P.C.P. (grant nos. R01GM102511, R01AG049396, and R35GM131838).
Data Availability
FASTQ and BAM files have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA769443. VCF files have been deposited in Figshare (https://figshare.com/projects/C_inopinata_population_genomics_2021/123973). Sample metadata can be found in Sheet 1 in supplementary table S1, Supplementary Material online. All other data and code affiliated with this work have been deposited in GitHub (https://github.com/gcwoodruff/inopinata_population_genomics_2020).
Literature Cited
- Ahmed S, Compton SG, Butlin RK, Gilmartin PM. Wind-borne insects mediate directional pollen transfer between desert fig trees 160 kilometers apart. Proc Natl Acad Sci U S A. 2009:106(48):20342–20347. 10.1073/pnas.0902213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, Miller MR. RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 2016:202(2):389–400. 10.1534/genetics.115.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S von R. Drugs and foods from little-known plants. Cambridge (MA): Harvard University Press; 1973. [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix M-A, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012:44(3):285–290. 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SE. Haldane's rule by sexual transformation in Caenorhabditis. Genetics 2002:161(3):1349–1353. 10.1093/genetics/161.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlow AW, Agosta SJ.. 2020. Phoresy in animals: review and synthesis of a common but understudied mode of dispersal. Bio Rev. 96(1):223–246. 10.1111/brv.12654 [DOI] [PubMed] [Google Scholar]
- Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. New York (NY): Routledge. [Google Scholar]
- Ben-David E, Burga A, Kruglyak L. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science 2017:356(6342):1051–1055. 10.1126/science.aan0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Pliota P, Widen SA, Koreshova A, Lemus-Vergara T, Verpukhovskiy P, Mandali S, Braendle C, Burga A, Kruglyak L. Ubiquitous selfish toxin-antidote elements in Caenorhabditis species. Curr Biol. 2021:31(5):990–1001.e5. 10.1016/j.cub.2020.12.013. [DOI] [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013:23(9):1514–1521. 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C, Brémaud M-F, Von Hirschheydt G, Viader V, Ardisson M, Santoni S, Ranwez V, David J, Glémin S. Mating systems and recombination landscape strongly shape genetic diversity and selection in wheat relatives. bioRxiv 532584. 10.1101/2023.03.16.532584, 20 March 2023, preprint: not peer reviewed. [DOI]
- Chen J, Glémin S, Lascoux M. Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol Biol Evol. 2017:34(6):1417–1428. 10.1093/molbev/msx088. [DOI] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Roberts JP, Andersen EC. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 2017:45(D1):D650–D657. 10.1093/nar/gkw893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Tanny RE, Seo B, Riccardi DD, Noble LM, Rockman MV, Alkema MJ, Braendle C, Kammenga JE, et al. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 2016:204(1):371–383. 10.1534/genetics.116.191148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 2015:200(2):387–407. 10.1534/genetics.115.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie TA, McKeown R, Moya ND, Evans KS, Widmayer SJ, LaGrassa V, Roman N, Tursunova O, Zhang G, Gibson SB, et al. CaeNDR, the Caenorhabditis Natural Diversity Resource. Nucleic Acids Res. 2024:52(D1):D850–D858. 10.1093/nar/gkad887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 2009. An introduction to population genetics theory. Caldwell (NJ): Blackburn Press. [Google Scholar]
- Cutter AD. Multilocus patterns of polymorphism and selection across the x chromosome of Caenorhabditis remanei. Genetics 2008:178(3):1661–1672. 10.1534/genetics.107.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Caenorhabditis evolution in the wild. BioEssays 2015:37(9):983–995. 10.1002/bies.201500053. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Reproductive transitions in plants and animals: selfing syndrome, sexual selection and speciation. New Phytol. 2019:224(3):1080–1094. 10.1111/nph.16075. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Baird SE, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics 2006:174(2):901–913. 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Dey A, Murray RL. Evolution of the Caenorhabditis elegans genome. Mol Biol Evol. 2009:26(6):1199–1234. 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Jovelin R, Dey A. Molecular hyperdiversity and evolution in very large populations. Mol Ecol. 2013:22(8):2074–2095. 10.1111/mec.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol Biol Evol. 2003:20(5):665–673. 10.1093/molbev/msg072. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Wang G-X, Ai H, Peng Y. Influence of finite-sites mutation, population subdivision and sampling schemes on patterns of nucleotide polymorphism for species with molecular hyperdiversity. Mol Ecol. 2012:21(6):1345–1359. 10.1111/j.1365-294X.2012.05475.x. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Wasmuth JD, Washington NL. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 2008:178(4):2093–2104. 10.1534/genetics.107.085787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience 2021:10(2):giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen L, Tian E, Zhang D, Wattana T, Yu H, Kjellberg F, Segar ST. Low host specificity and broad geographical ranges in a community of parasitic non-pollinating fig wasps (Sycoryctinae; Chalcidoidea). J Anim Ecol. 2021:90(7):1678–1690. 10.1111/1365-2656.13483. [DOI] [PubMed] [Google Scholar]
- Dey A, Chan CK, Thomas CG, Cutter AD. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc Natl Acad Sci U S A. 2013:110(27):11056–11060. 10.1073/pnas.1303057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Jeon Y, Wang G-X, Cutter AD. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics 2012:191(4):1257–1269. 10.1534/genetics.112.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. lemon: freshing up your ‘ggplot2’ plots. 2020. https://CRAN.R-project.org/package=lemon.
- Engelbrecht A, Matthee S, du Toit N, Matthee CA. Limited dispersal in an ectoparasitic mite, Laelaps giganteus, contributes to significant phylogeographic congruence with the rodent host, Rhabdomys. Mol Ecol. 2016:25(4):1006–1021. 10.1111/mec.13523. [DOI] [PubMed] [Google Scholar]
- Foxe JP, Dar VU, Zheng H, Nordborg M, Gaut BS, Wright SI. Selection on amino acid substitutions in arabidopsis. Mol Biol Evol. 2008:25(7):1375–1383. 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Maia N, Freire-Maia A. The structure of consanguineous marriages and its genetic implications. Ann Hum Genet. 1961:25(1):29–39. 10.1111/j.1469-1809.1961.tb01494.x. [DOI] [PubMed] [Google Scholar]
- Galias DC, Guerrero JJG, General MA, Bañares EN, Serrano JE. Diversity of tree species within an urban forest fragment in albay province, eastern Philippines. BU R&D Journal. 2018:21(1):33–44. https://www.academia.edu/download/63693023/34-Article_Text-124-1-10-2020060220200620-90000-1el0ma2.pdf. [Google Scholar]
- Gilbert KJ, Whitlock MC. Evaluating methods for estimating local effective population size with and without migration. Evolution 2015:69(8):2154–2166. 10.1111/evo.12713. [DOI] [PubMed] [Google Scholar]
- Gimond C, Jovelin R, Han S, Ferrari C, Cutter AD, Braendle C. Outbreeding depression with low genetic variation in selfing Caenorhabditis nematodes. Evolution 2013:67(11):3087–3101. 10.1111/evo.12203. [DOI] [PubMed] [Google Scholar]
- Glémin S, François CM, Galtier N, Anisimova M. Genome evolution in outcrossing vs. selfing vs. asexual species. Evolutionary genomics: statistical and computational methods. New York (NY): Methods in Molecular Biology Springer; 2019. p. 331–369. 10.1007/978-1-4939-9074-0_11. [DOI] [PubMed] [Google Scholar]
- Glémin S, Muyle A. Mating systems and selection efficacy: a test using chloroplastic sequence data in angiosperms. J Evol Biol. 2014:27(7):1386–1399. 10.1111/jeb.12356. [DOI] [PubMed] [Google Scholar]
- Graustein A, Gaspar JM, Walters JR, Palopoli MF. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 2002:161(1):99–107. 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag ES. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. In The C. elegans research community, WormBook. 2005. 10.1895/wormbook.1.120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Molecular population genetics. New York (NY): Sinaur; 2019. [Google Scholar]
- Haldane JB. 1932. The causes of evolution. Princeton (NJ): Princeton University Press. [Google Scholar]
- Haller BC, Messer PW. SLim 4: multispecies eco-evolutionary modeling. Am Natt. 2023:201(5):E127–E139. 10.1086/723601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmith EW, Woodruff GC, Moser KA, Johnson E, Phillips PC. Opposing directions of stage-specific body shape change in a close relative of C. elegans. BMC Zool. 2022:7(1):38. 10.1186/s40850-022-00131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmith EW, Woodruff GC, Phillips PC. Opposing directions of stage-specific body length change in a close relative of C. elegans. bioRxiv 168039. 10.1101/2020.06.23.168039, 24 June 2020, preprint: not peer reviewed. 10.1101/2020.06.23.168039. [DOI] [PMC free article] [PubMed]
- Herre EA. Sex ratio adjustment in fig wasps. Science 1985:228(4701):896–898. 10.1126/science.228.4701.896. [DOI] [PubMed] [Google Scholar]
- Herre EA. Optimality, plasticity and selective regime in fig wasp sex ratios. Nature 1987:329(6140):627–629. 10.1038/329627a0. [DOI] [Google Scholar]
- Herre EA, Jandér KC, Machado CA. Evolutionary ecology of figs and their associates: recent progress and outstanding puzzles. Annu Rev Ecol Evol Syst. 2008:39(1):439–458. 10.1146/annurev.ecolsys.37.091305.110232. [DOI] [Google Scholar]
- Holdsworth DK, Hurley CL, Rayner SE. Traditional medicinal plants of New Ireland, Papua New Guinea. Q J Crude Drug Res. 1980:18(3):131–139. 10.3109/13880208009065191. [DOI] [Google Scholar]
- Holdsworth D, Lacanienta E. Traditional medicinal plants of the central province of Papua New Guinea. Part II. Q J Crude Drug Res. 1981:19(4):155–167. 10.3109/13880208109070594. [DOI] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 2009:10(9):639–650. 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandér KC. Indirect mutualism: ants protect fig seeds and pollen dispersers from parasites. Ecol Entomol. 2015:40(5):500–510. 10.1111/een.12215. [DOI] [Google Scholar]
- Janzen DH. How to be a fig. Annu Rev Ecol Syst. 1979:10(1):13–51. 10.1146/annurev.es.10.110179.000305. [DOI] [Google Scholar]
- Jarne P, Auld JR. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 2006:60(9):1816–1824. 10.1111/j.0014-3820.2006.tb00525.x. [DOI] [PubMed] [Google Scholar]
- Jarne P, Charlesworth D. The evolution of the selfing rate in functionally hermaphrodite plants and animals. Annu Rev Ecol Syst. 1993:24(1):441–466. 10.1146/annurev.es.24.110193.002301. [DOI] [Google Scholar]
- Jauharlina, Oktarina H, Sriwati R, Sayuthi M, Kanzaki N, Quinnell RJ, Compton SG. Association of fig pollinating wasps and fig nematodes inside male and female figs of a dioecious fig tree in Sumatra, Indonesia. Insects 2022:13(4):320. 10.3390/insects13040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauharlina J, Quinnell RJ, Compton SG, Lindquist EE, Robertson HG. Fig wasps as vectors of mites and nematodes. Afr Entomol. 2012:20(1):101–110. 10.10520/EJC119300. [DOI] [Google Scholar]
- Jombart T, Ahmed I. . adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011:27(21):3070–3071. 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010:11(1):94. 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R. Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis. Mol Biol Evol. 2009:26(10):2373–2386. 10.1093/molbev/msp142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Ajie BC, Phillips PC. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol Ecol. 2003:12(5):1325–1337. 10.1046/j.1365-294X.2003.01805.x. [DOI] [PubMed] [Google Scholar]
- Kanzaki N, Tsai IJ, Tanaka R, Hunt VL, Liu D, Tsuyama K, Maeda Y, Namai S, Kumagai R, Tracey A, et al. Biology and genome of a newly discovered sibling species of Caenorhabditis elegans. Nat Commun. 2018:9(1):3216. 10.1038/s41467-018-05712-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. rstatix: pipe-friendly framework for basic statistical tests. 2023. https://CRAN.R-project.org/package=rstatix.
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 1977:267(5608):275–276. 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kiontke KC, Félix M-A, Ailion M, Rockman MV, Braendle C, Pénigault J-B, Fitch DHA. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011:11(1):339. 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus BJ, Grünwald NJ. vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 2017:17(1):44–53. 10.1111/1755-0998.12549. [DOI] [PubMed] [Google Scholar]
- Koch R, van Luenen HG, van der Horst M, Thijssen KL, Plasterk RH. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 2000:10(11):1690–1696. 10.1101/gr.GR-1471R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Zdraljevic S, Stevens L, Wang Y, Tanny RE, Crombie TA, Cook DE, Webster AK, Chirakar R, Baugh LR, et al. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat Ecol Evol. 2021:5(6):794–807. 10.1038/s41559-021-01435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011:27(21):2987–2993. 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Jovelin R, Yoshiga T, Tanaka R, Cutter AD. Specialist versus generalist life histories and nucleotide diversity in Caenorhabditis nematodes. Proc Biol Sci. 2014:281(1777):20132858. 10.1098/rspb.2013.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R-C, Yeung CK-L, Li S-H. Drastic post-LGM expansion and lack of historical genetic structure of a subtropical fig-pollinating wasp (Ceratosolen sp. 1) of Ficus septica in Taiwan. Mol Ecol. 2008:17(23):5008–5022. 10.1111/j.1365-294X.2008.03983.x. [DOI] [PubMed] [Google Scholar]
- Martin GC, Owen AM, Way JI. Nematodes, figs and wasps. J Nematol. 1973:5(1):77–78. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619964/. [PMC free article] [PubMed] [Google Scholar]
- Martinson EO, Herre EA, Machado CA, Arnold AE. Culture-free survey reveals diverse and distinctive fungal communities associated with developing figs (Ficus spp.) in Panama. Microb Ecol. 2012:64(4):1073–1084. 10.1007/s00248-012-0079-x. [DOI] [PubMed] [Google Scholar]
- Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007:17(3):337–347. 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazé-Guilmo E, Blanchet S, McCoy KD, Loot G, Thrall P. 2016. Host dispersal as the driver of parasite genetic structure: a paradigm lost? Eco Let. 2016:19(3):336–347. 10.1111/ele.12564 [DOI] [PubMed] [Google Scholar]
- Mogle MJ, Kimball SA, Miller WR, McKown RD.. 2018. Evidence of avian-mediated long distance dispersal in American tardigrades. Figure 1: Migration flyways in the Americas, with examples of disjunct distributions of some tardigrades. Figure 2: (A) Blue-gray gnatcatcher (Polioptila caerulea) nest covered with lichen in a black walnut (Juglans nigra) in Wabaunsee County, KS. (B) Ruby-throated hummingbird (Archilochus colubris) nest constructed with lichen in a green ash (Fraxinus pennsylvanica) in Douglas County, KS. C. SAK collecting the blue-gray gnatcatcher nest. Table 1: Density and diversity of tardigrades found in bird nests. Table 2: Tardigrades found on carcasses of recently-dead birds. Peer J. 2018:6:e5035. 10.7717/peerj.5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazareno AG, Bemmels JB, Dick CW, Lohmann LG. Minimum sample sizes for population genomics: an empirical study from an Amazonian plant species. Mol Ecol Resour. 2017:17(6):1136–1147. 10.1111/1755-0998.12654. [DOI] [PubMed] [Google Scholar]
- Noble LM, Yuen J, Stevens L, Moya N, Persaud R, Moscatelli M, Jackson JL, Zhang G, Chitrakar R, Baugh LR, et al. Selfing is the safest sex for Caenorhabditis tropicalis. eLife 2021:10:e62587. 10.7554/eLife.62587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 2000:154(2):923–929. 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole B. 2002. Phylogeny of the species of the superfamily Echeneoidea (Perciformes: Carangoidei: Echeneidae, Rachycentridae, and Coryphaenidae), with an interpretation of echeneid hitchhiking behaviour. Can J Zoo. 2002:80(4):596–623. 10.1139/z02-031 [DOI] [Google Scholar]
- Pedersen T. ggforce: accelerating ‘ggplot2'. 2022a. https://CRAN.R-project.org/package=ggforce.
- Pedersen T. patchwork: the composer of plots. 2022b. https://cloud.r-project.org/web/packages/patchwork/index.html.
- Peniwidiyanti P, Qayim I, Chikmawati T. A study on diversity and distribution of figs (Ficus, Moraceae) in Bogor city, West Java, Indonesia. J Trop Biodivers Biotechnol. 2022:7(2):68516. 10.22146/jtbb.68516. [DOI] [Google Scholar]
- Perry MD, Li W, Trent C, Robertson B, Fire A, Hageman JM, Wood WB. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 1993:7(2):216–228. 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 1987:117:353–360. 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010:26(6):841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/.
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000:16(6):276–277. 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rochette NC, Rivera-Colón AG, Catchen JM. Stacks 2: analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol. 2019:28(21):4737–4754. 10.1111/mec.15253. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009:5(3):e1000419. 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LJ, Bain A, Chou L-S, Conchou L, Cruaud A, Gonzales R, Hossaert-McKey M, Rasplus J-Y, Tzeng H-Y, Kjellberg F. Diversification and spatial structuring in the mutualism between Ficus septica and its pollinating wasps in insular South East Asia. BMC Evol Biol. 2017:17(1):207. 10.1186/s12862-017-1034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Koboldt DC, Staisch JE, Chamberlin HM, Gupta BP, Miller RD, Baird SE, Haag ES. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011:7(7):e1002174. 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mon AM, Fu Y, Zhang Y, Wang C, Yang X, Wang Y. The genus Ficus (Moraceae) used in diet: its plant diversity, distribution, traditional uses and ethnopharmacological importance. J Ethnopharmacol. 2018:226:185–196. 10.1016/j.jep.2018.07.027. [DOI] [PubMed] [Google Scholar]
- Shilton LA, Altringham JD, Compton SG, Whittaker RJ. Old world fruit bats can be long–distance seed dispersers through extended retention of viable seeds in the gut. Proc R Soc Lond B Biol Sci. 1999:266(1416):219–223. 10.1098/rspb.1999.0625. [DOI] [Google Scholar]
- Sloat SA, Noble LM, Paaby AB, Bernstein M, Chang A, Kaur T, Yuen J, Tintori SC, Jackson JL, Martel A, et al. Caenorhabditis nematodes colonize ephemeral resource patches in neotropical forests. Ecol Evol. 2022:12(7):e9124. 10.1002/ece3.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics 2009:182(1):205–216. 10.1534/genetics.108.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Martínez-Ugalde I, King E, Wagah M, Absolon D, Bancroft R, Gonzalez de la Rosa P, Hall JL, Kieninger M, Kloch A, et al. Ancient diversity in host-parasite interaction genes in a model parasitic nematode. Nat Commun. 2023:14(1):7776. 10.1038/s41467-023-43556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Moya ND, Tanny RE, Gibson SB, Tracey A, Na H, Chitrakar R, Dekker J, Walhout AJM, Baugh LR, et al. Chromosome-level reference genomes for two strains of Caenorhabditis briggsae: an improved platform for comparative genomics. Genome Biol Evol. 2022:14(4):evac042. 10.1093/gbe/evac042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Rooke S, Falzon LC, Machuka EM, Momanyi K, Murungi MK, Njoroge SM, Odinga CO, Ogendo A, Ogola J, et al. The genome of Caenorhabditis bovis. Curr Biol. 2020:30(6):1023–1031.e4. 10.1016/j.cub.2020.01.074. [DOI] [PubMed] [Google Scholar]
- Sugiura S, Yamazaki K. Moths boring into Ficus syconia on Iriomote Island, south-western Japan. Entomol Sci. 2004:7(2):113–118. 10.1111/j.1479-8298.2004.00056.x. [DOI] [Google Scholar]
- Szitenberg A, Cha S, Opperman CH, Bird DM, Blaxter ML, Lunt DH. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol Evol. 2016:8(9):2964–2978. 10.1093/gbe/evw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina AA, Willis JH, Lukac M, Jovelin R, Cutter AD, Phillips PC. Genomic diversity landscapes in outcrossing and selfing Caenorhabditis nematodes. PLoS Genet. 2023:19(8):e1010879. 10.1371/journal.pgen.1010879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Wang W, Jovelin R, Ghosh R, Lomasko T, Trinh Q, Kruglyak L, Stein LD, Cutter AD. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 2015:25(5):667–678. 10.1101/gr.187237.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Nason JD, Machado CA, Zheng L, Yu H, Kjellberg F. Lack of genetic isolation by distance, similar genetic structuring but different demographic histories in a fig-pollinating wasp mutualism. Mol Ecol. 2015:24(23):5976–5991. 10.1111/mec.13438. [DOI] [PubMed] [Google Scholar]
- Van Goor J, Herre EA, Gómez A, Nason JD. Extraordinarily precise nematode sex ratios: adaptive responses to vanishingly rare mating opportunities. Proc Biol Sci. 2022:289(1967):20211572. 10.1098/rspb.2021.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor J, Kanzaki N, Woodruff G. How to be a fig nematode. Acta Oecol. 2023:119:103916. 10.1016/j.actao.2023.103916. [DOI] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006:7(8):645–653. 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Wang J. Effective size and F-statistics of subdivided populations for sex-linked loci. Theor Popul Biol. 1999:55(2):176–188. 10.1006/tpbi.1998.1398. [DOI] [PubMed] [Google Scholar]
- Wang G-X, Ren S, Ren Y, Ai H, Cutter AD. Extremely high molecular diversity within the East Asian nematode Caenorhabditis sp. 5. Mol Ecol. 2010:19(22):5022–5029. 10.1111/j.1365-294X.2010.04862.x. [DOI] [PubMed] [Google Scholar]
- Weiblen GD. How to be a fig wasp. Annu Rev Entomol. 2002:47(1):299–330. 10.1146/annurev.ento.47.091201.145213. [DOI] [PubMed] [Google Scholar]
- Wickham H. Reshaping data with the reshape package. J Stat Soft. 2007:21(12):1–20. 10.18637/jss.v021.i12. [DOI] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- Widen SA, Bes IC, Koreshova A, Pliota P, Krogull D, Burga A. Virus-like transposons cross the species barrier and drive the evolution of genetic incompatibilities. Science 2023:380(6652):eade0705. 10.1126/science.ade0705. [DOI] [PubMed] [Google Scholar]
- Wilke C. cowplot: streamlined plot theme and plot annotations for ‘ggplot2'. 2020. https://CRAN.R-project.org/package=cowplot.
- Willing E-M, Dreyer C, van Oosterhout C. Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS One 2012:7(8):e42649. 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres MA. Genetic diversity on the sex chromosomes. Genome Biol Evol. 2018:10(4):1064–1078. 10.1093/gbe/evy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff GC, Phillips PC. Field studies reveal a close relative of C. elegans thrives in the fresh figs of Ficus septica and disperses on its Ceratosolen pollinating wasps. BMC Ecol. 2018:18(1):26. 10.1186/s12898-018-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff GC, Teterina AA. Degradation of the repetitive genomic landscape in a close relative of Caenorhabditis elegans. Mol Biol Evol. 2020:37(9):2549–2567. 10.1093/molbev/msaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff GC, Willis JH, Phillips PC. Dramatic evolution of body length due to postembryonic changes in cell size in a newly discovered close relative of Caenorhabditis elegans. Evol Lett. 2018:2(4):427–441. 10.1002/evl3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Inbreeding and homozygosis. Proc Natil Acad Sci U S A. 1933:19(4):411–420. 10.1073/pnas.19.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Iorgovan G, Misra S, Mokhtari M. Neutral evolution of synonymous base composition in the Brassicaceae. J Mol Evol. 2007:64(1):136–141. 10.1007/s00239-005-0274-1. [DOI] [PubMed] [Google Scholar]
- Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. In: Mathé E, Davis S, editors. Statistical genomics: methods and protocols. New York (NY): Methods in Molecular Biology Springer; 2016. p. 283–334. 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
- Yoshimura J, Ichikawa K, Shoura MJ, Artiles KL, Gabdank I, Wahba L, Smith CL, Edgley ML, Rougvie AE, Fire AZ, et al. Recompleting the Caenorhabditis elegans genome. Genome Res. 2019:29(6):1009–1022. 10.1101/gr.244830.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodna M, Arens P, Van Dijk PJ, Vosman B. Development and characterization of microsatellite markers for two dioecious Ficus species. Mol Ecol Notes. 2005:5(2):355–357. 10.1111/j.1471-8286.2005.00924.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FASTQ and BAM files have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA769443. VCF files have been deposited in Figshare (https://figshare.com/projects/C_inopinata_population_genomics_2021/123973). Sample metadata can be found in Sheet 1 in supplementary table S1, Supplementary Material online. All other data and code affiliated with this work have been deposited in GitHub (https://github.com/gcwoodruff/inopinata_population_genomics_2020).