Abstract
The p53 tumor suppressor protein is a master regulatory transcription factor that coordinates cellular responses to DNA damage and cellular stress. Besides mutations in p53, or in proteins involved in the p53 response pathway, genetic variation in promoter response elements (REs) of p53 target genes is expected to alter biological responses to stress. To identify SNPs in p53 REs that may modify p53-controlled gene expression, we developed an approach that combines a custom bioinformatics search to identify candidate SNPs with functional yeast and mammalian cell assays to assess their effect on p53 transactivation. Among ≈2 million human SNPs, we identified >200 that seem to disrupt functional p53 REs. Eight of these SNPs were evaluated in functional assays to determine both the activity of the putative RE and the impact of the candidate SNPs on transactivation. All eight candidate REs were functional, and in every case the SNP pair exhibited differential transactivation capacities. Additionally, six of the eight genes adjacent to these SNPs are induced by genotoxic stress or are activated directly by transfection with p53 cDNA. Thus, this strategy efficiently identifies SNPs that may differentially affect gene expression responses in the p53 regulatory pathway.
Keywords: bioinformatics, single nucleotide polymorphism, yeast, regulatory sequences, gene expression
Human genetic diversity plays a major role in determining susceptibility to stress. Numerous examples of functional polymorphisms that have demonstrable effects on stress response phenotype have been described (1, 2). In general, these polymorphisms consist of variation in protein coding sequences, although examples of variation in regulatory sequences are also reported (3). Genome-scale sequencing has led to the discovery of millions of human SNPs (4). Most are expected to have little functional consequence. Identifying functional SNPs, including those in regulatory regions, among the vast number of uncharacterized SNPs and evaluating their potential impact on human health are formidable challenges.
The p53 tumor suppressor protein is a master regulator of a prominent transcriptional network that can control the fate of cells in response to stress. By coordinating expression of many target genes, p53 directs cells into cell-cycle arrest or apoptosis after DNA damage or other perturbations (5, 6). Of note, p53 mutations are associated with nearly half of all cancers (7).
Sequence-specific transcription factors such as p53 can interact with many related chromosomal binding sites, known as response elements (REs), to modulate transcription. Among p53-responsive genes, stress-induced transactivation varies broadly, attributable in part to sequence variation in individual p53 binding sites (8). p53-mediated transactivation in a yeast model system revealed large variations (up to 1,000-fold) in transactivation potential among 44 RE sequences despite close similarity to the proposed p53 consensus (9, 10).
These observations suggest that substantial diversity in transcription can result from polymorphisms in p53 REs, possibly leading to variability in cellular stress responses. SNPs are the most common type of genetic variation, with an estimated density of 1-3 SNPs per 1,000 bases (11, 12). Based on the length of p53 REs (≈20 bases), a few such polymorphisms might be expected in known p53 target REs. Both genomic analysis and microarray expression studies suggest thousands of additional p53 targets exist (8, 13). Therefore, the actual number of polymorphisms at p53 REs could be substantial.
We developed an approach for selecting SNPs from the dbSNP database that is likely to result in functional differences between putative p53 RE alleles. Candidate REs are identified based on relationship to p53 RE consensus sequence, assessment of the impact of mismatches to consensus, and proximity to transcriptional start sites. This search revealed numerous putative polymorphic REs in the vicinity of candidate p53 responsive genes. Functional analysis of a subset of these polymorphic REs in our yeast-based system demonstrated strong differences in p53 transactivation capacities for the different SNP alleles. Additionally, when candidate SNPs were examined in reporter constructs in human cell lines, pronounced allelic differences in p53-dependent gene expression were revealed. Polymorphisms in p53 REs are thus a potential source of variation in stress responses between individuals. Furthermore, genome-wide bioinformatics searches, combined with functional analyses, can identify potential regulatory SNPs from among millions of candidates, thereby facilitating discovery of functional variation.
Materials and Methods
Bioinformatics. Bioinformatics programs were written in perl and included some elements of bioperl (www.bioperl.org). SNP data were from the National Center for Biotechnology Information (NCBI, May 12, 2003). Genome assembly and gene annotation data were from the European Molecular Biology Laboratory (EMBL, March 17, 2003). sql analysis was performed in mysql (www.mysql.org).
Yeast Strains and p53 Transactivation. Genomic p53 RE reporter constructs were generated by using the Delitto Perfetto site-specific mutagenesis system (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site) (14). Wild-type human p53 was expressed under the inducible GAL1-10 promoter from pTSG-p53, a centromeric TRP1-selectable expression vector. The ADE2-based transactivation assay was performed on plates with low adenine (5 mg/liter) to allow color detection (9). Luciferase assays were performed under conditions of constitutive p53 expression in strains containing plasmid pTSAd-p53 (15).
Plasmids for Transfections. Reporter plasmids containing the SNP REs, the p21WAF RE, or the AIP1 RE upstream of the firefly luciferase reporter gene were generated in a pGL3 vector (Promega). pRL-SV40, a Renilla reniformis luciferase plasmid (Promega) was a control for transfection efficiency. The expression plasmids pCMV-p53WT and pCMV-G279E (10) encode the indicated human p53 protein controlled by the cytomegalo-virus promoter.
Human Cells. Human SaOS-2 osteosarcoma [American Type Culture Collection (ATCC) HTB no. 85], RKO colon carcinoma (ATCC CRL#2577), and HT1080 fibrosarcoma cell line (ATCC CCL no. 121) were grown in McCoy's A5 medium supplemented with 10% FBS and 100 μg/ml penicillin and streptomycin. Lymphoblastoid cells (GM15223, Coriell Cell Repositories, Camden, NJ) were grown in RPMI medium 1640 with 15% FBS, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B. Cells were incubated at 37°C with 5% CO2.
Genotyping. A panel of 72 individuals (Asian, European, and African ancestry, 24 each) and the first 90 individuals from the Polymorphism Discovery Resource were genotyped for the eight SNPs. The ARHGEF7 and SCGB1D2 SNPs were not detected in these populations, so an additional 292 individuals (181 European and 111 African ancestry) were genotyped. Probe-based genotyping assays (Assays-by-Design, Applied Biosystems) were run on an Applied Biosystems Prism HTS 7900.
Transfection and Luciferase Assays in Human Cell Lines. p53 wild-type RKO cells (2 × 105) were cultured in 24-well plates, 18 h before transfection. Cells at 80% confluence were transfected with FuGENE 6 (Roche Molecular Biochemicals) following the manufacturer's instructions. A mixture of 100 ng of pGL3 reporter plasmid containing the RE-SNP and 75 ng of pRL-SV40 with FuGENE 6 was added to each well. After 30 h, the cells were UV-irradiated to induce p53 responses.
For luciferase assays in the p53-null SaOS-2 cell line, 1 × 105 cells were seeded in 24-well plates 24 h before transfection. The cells were cotransfected with 100 ng of pGL3 reporter plasmid, 25 ng of pCMV-p53WT expression vector, and 75 ng of pRL-SV40. Total plasmid DNA per well was adjusted to an equal level by adding the empty vector pCMV-Neo.
Cell cultures were harvested 48 h after transfection, and luciferase activities were assessed by using the Dual-luciferase assay system (Promega) according to the manufacturer's protocol. Firefly luciferase activity was normalized with Renilla luciferase. Experiments were performed at least twice in triplicate.
Gene Expression Analysis. For RKO, HT1080, and SaOS2 cells, media were removed, and cells were washed twice with PBS. Cells were covered with a thin layer of PBS and exposed to 254 nm of UV light (UVC) at a dose of 20 J/m2. Fresh medium was added, and cells were harvested at indicated intervals. Unexposed control cells were treated similarly.
For real-time RT-PCR assays, RKO and HT1080 were seeded at 3 × 105 cells per well in a six-well plate 1 day before irradiation. For SaOS-2 cells, 2 × 105 cells were cultured in a six-well plate for 24 h. SaOS2 cells were transiently transfected in the presence of FuGENE 6 with 2 μg of pCMV-Bam-Neo vector alone, with vector carrying the p53 gene with the inactivating mutation G279E, or with vector carrying the wild-type p53 gene. After UV-irradiation or transfection, cells were grown for an additional 24 h before harvesting.
Total RNA was isolated from RKO, HT1080, and SaOS-2 cell lines with TRIzol reagent (GIBCO/BRL) according to the manufacturer's protocol. Extracted RNA was cleaned with Qiagen RNeasy columns (Qiagen, Valencia, CA). One microgram was used for reverse transcription and amplification using TaqMan Reverse Transcription Reagents and gene-specific primer-probe sets (Assays on Demand, Applied Biosystems). Reactions were carried out in 96-well plates by using an Applied Biosystems PRISM 7700 Sequence Detection System.
For lymphoblastoid cells, 3 × 106 cells were pelleted and resuspended in PBS at a density of 5 × 105 cells per ml before exposure. Cells were exposed to 15 J/m2 UV, returned to conditioned media, and grown for 16 h before RNA harvesting. RNA was extracted by using an Applied Biosystems 6100 following the manufacturer's protocol (Applied Biosystems). cDNA was generated by using the First-strand Synthesis system (Invitrogen). Probe-based expression assays were performed on an Applied Biosystems Prism HTS 7900 (Assays-on-Demand, Applied Biosystems).
Results
Identification of Functional SNPs in Putative p53 REs. We used a combination of custom bioinformatics applications and in vivo functional analyses to identify putative polymorphic REs that might mediate differential transactivation by p53 (Fig. 3 and Table 2, which are published as supporting information on the PNAS web site). The strategy is divided into three phases: (i) computational identification of SNPs within putative p53 REs, (ii) SNP validation, and (iii) functional evaluation in yeast and human cells.
The consensus p53 RE sequence is RRRCWWGYYY-N-RRRCWWGYYY, where N is 0-13 bases (16, 17). Although the consensus was derived from in vitro binding studies and functional analysis, these assays do not address strength of binding, actual transactivation capacity, or the impact of chromatin on protein-DNA interactions. We used data from direct in vivo measurements of transactivation at various REs in a quantitative yeast model system to inform our search algorithms (9). This in vivo system can identify single base changes in p53 RE sequences that dramatically affect transactivation (10), and we have used it to evaluate candidate SNPs. In human cells, chromatin structure and promoter organization may modulate p53-induced transactivation at REs (18, 19); however, chromosomal context is likely to be similar for paired SNP alleles, which should allow a direct comparison of single base changes.
We developed rules for identifying SNPs at sites where only one allele is likely to support strong p53 transactivation. Because these rules are conservative, there are likely to be many more functional sites in the genome, both polymorphic and nonpolymorphic, and recent surveys support this view (13). Common features of strong binding sites, deduced in part from our previous analysis of known mammalian p53 REs, include the following: (i) most functional REs have a spacer length of 0 or 1 base between the two 10-nt half sites (10); (ii) although most p53 REs contain non-consensus bases and the number of mismatches does not directly correlate with transactivation capacity, typically fewer than four total mismatches and no more than three mismatches per 10-nt half site are found in active REs; (iii) a change in the conserved C or G in the RE dramatically affects transactivation; (iv) within the WW motif, AT provides the strongest transactivation, and nonconsensus changes have a strong negative effect (9); (v) mismatches at the R and Y bases have a greater negative impact on transactivation the closer they occur to the central CWWG. Most bona fide p53 REs fall within a few thousand base pairs of transcriptional start sites (9). Thus, we limited our study to REs positioned within 2,500 bp of gene start sites. The search rules and the number of candidates identified at each step are shown in Fig. 3.
Identification of SNPs in Putative p53 REs. Approximately 2.3 million uniquely mapped SNPs from the National Center for Biotechnology Information's dbSNP (build 114) database were selected for analysis. The computational search for putative REs was performed by using a series of progressively more restrictive rules. Initially, 7,161 SNPs with the following features were selected: the sequence surrounding the SNP matched the p53 consensus binding site with four or fewer mismatches; the SNP was close to reported or computationally identified genes (within 5 kb); and the selected RE preserved core C and G positions (CWWG) in the consensus alignment of both adjacent dimers in at least one allele. Of note, none of the candidate SNPs overlapped known REs. Although it is tempting to speculate that SNPs are excluded from functional REs, perhaps as a consequence of conservative evolutionary pressure, the current data are insufficient to address this hypothesis. Given the number of bona fide sites detected by our search strategy (37 of 41 validated REs examined) and the estimated density of reported SNPs at the time of our study (1-3 per kb), it is not possible to establish that polymorphisms are globally underrepresented at established functional REs.
From the initial set, we selected 680 candidate SNPs located within 2.5 kb of named genes. We then restricted the candidate pool to 231 potential REs that maintained well defined core binding sites (at least three of four “W” positions maintained in the core). Of these candidates, 81 REs had three or fewer total mismatches, and ≈40 were predicted to exhibit significant differences in p53 transactivation between alleles. Eight SNP pairs were chosen for functional evaluation. Final candidate selection was based in part on the known or suspected function of RE-associated genes, in that preference was given to genes with functions related to p53-mediated biological pathways (e.g., cell cycle control and DNA repair). None of the eight selected REs were adjacent to previously identified p53 responsive genes.
The eight candidate RE SNPs selected for further evaluation are described in Table 1. For each SNP, the alignment of the adjacent flanking sequence to the p53 RE consensus is shown, along with the position and identity of the polymorphism. Allele frequencies were measured directly in two ethnically diverse human lymphoblast cell line DNA collections (see Materials and Methods). For the reported SNP site upstream of the SCGB1D2 and ARHGEF7 genes, only one allele was observed, suggesting either a very low minor allele frequency or that the original dbSNP submission was in error (dbSNP contains ≈20% false-positive entries) (20, 21). Of the genes identified as candidates for further analysis, DCC, SEI1, SCGB1D2, and ARHGEF7 are either known or suspected tumor suppressor genes or have predicted roles in cell cycle control. ADAR2, EOMES, RRM1, and TLR8 play diverse roles in fundamental processes (RNA editing, T cell differentiation, nucleotide metabolism, and pathogen response, respectively). Although these candidate genes are not known to have a role in the development of exposure-induced disease such as cancer, most have a plausible relationship to p53-mediated signaling or stress response pathways.
Table 1. SNPs at candidate p53 response elements.
| Associated | Alignment | Allele | ||||
|---|---|---|---|---|---|---|
| gene | Offset | SNP ID | RRRCWWGYYYNRRRCWWGYYY | frequencies | Mismatches | Reported gene function* |
| ADAR2 | −2214 | 2838769 | GGACAAGTTg-AAACTT*CaC | G: 0.747 | 2 | Adenine deaminase acting on RNA; contributes to post-transcriptional modification of multiple RNAs |
| A: 0.253 | ||||||
| ARHGEF7 | −1255 | 1658728 | AAACATGTCa-*cACTTGCTT | G: 1.000† | 2 | Rho guanine nucleotide exchange factor (GEF) 7 |
| T: 0.000 | ||||||
| DCC | −2297 | 934345 | *AGCATGTTC-AcACAAGCCa | G: 0.671 | 2 | Deleted in colorectal carcinoma; apoptotic inducer and proposed tumor suppressor gene |
| C: 0.329 | ||||||
| EOMES | −838 | 3806624 | GGGCcTGTCT-cAACT*GCCC | T: 0.270 | 2 | Human Eomesodermin homolog; T-box transcription factor with essential roles in development |
| C: 0.730 | ||||||
| RRM1 | −1472 | 1465952 | GGG*ATGTgC-AttCAAGTTT | C: 0.115 | 3 | Subunit of the human ribonucleotide reductase complex; two other subunits of this complex (RRM2, P53R) interact with p53 |
| T: 0.885 | ||||||
| SCGB1D2 | −445 | 2232945 | GGtCTTGTTT-AGACTT*CTC | G: 1.000† | 1 | Lipophilin protein associated with secreted mammoglobin; possible diagnostic marker for breast cancer |
| A: 0.000 | ||||||
| SEI1 | −141 | 14716 | GGGCTT*agg-GcGCATGCCC | G: 0.047 | 4 | CDK4-binding protein and antagonist of the p 16 protein, which inhibits CDK4; putative oncogene. |
| C: 0.953 | ||||||
| TLR8 | −1077 | 3761624 | AGGCAAGaTg-AAACAT*TCa | G: 0.437 | 3 | Toll-like receptor 8; Toll-like receptors regulate innate immune responses by binding ligands from pathogens |
| A: 0.563 |
SNPs and RE structures are shown for candidate binding sites identified by bioinformatics analysis. Associated genes are those located near candidate REs. Offset indicates the position of the SNP relative to the reported start of transcription for the associated gene, and SNP ID numbers are dbSNP RefSNP IDs. Sequences are aligned to the consensus response element (column 4, top), with spacer regions denoted by -. Mismatches against the consensus sequence are shown as lowercase in the alignments, and the position of the SNP is indicated by *. Allele frequencies are reported as predicted strong over predicted weak allele. Mismatches are reported for the allele that best matches the consensus binding site.
See Discussion for more extensive discussion of gene functions.
For SNPs associated with ARHGEF7 and SCGB1D2, only one allele was detected in the test set (n = 365).
Analysis of Putative p53 RE SNPs for Transactivation Capacity in Yeast. To establish both the functionality of the REs and the impact of the single nucleotide polymorphisms, we determined relative transactivation capacities by using two yeast-based reporter assays (10). Two groups of isogenic p53 reporter strains, each with a distinct RE SNP upstream of either a luciferase or an ADE2 reporter, were constructed by using Delitto Perfetto in vivo site-directed mutagenesis (14). In the quantitative luciferase assay, there is moderate constitutive expression of p53 (under control of the ADH1 promoter). The ADE2-based visual assay is performed at various levels of p53 expression, an approach that has allowed detection of subtle functional differences among p53 REs as well as among p53 mutant alleles.
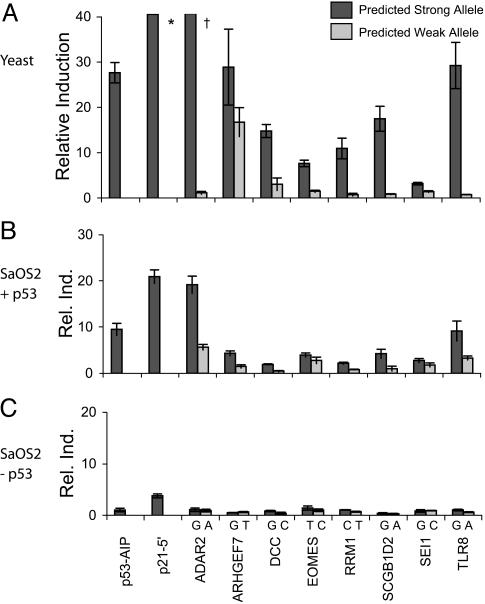
Except for the ARHGEF7 and SEI1 REs, we observed strong differences in p53 transactivation between pairs of RE-SNPs using the luciferase reporter assay (Fig. 1A). Comparable results were observed with the ADE2 assay (data not shown). In every case, the predicted strong allele (i.e., the one that best fit the rules described above) was more efficient in driving luciferase expression than the predicted weak allele. Even SNPs at the poorly conserved first “R” position of the consensus showed some difference in transactivation between alleles (ARHGEF7 and DCC).
Fig. 1.
Transactivation capacity of wild-type p53 toward SNP alleles. The ability of SNP RE alleles to support transactivation by p53 was examined under isogenic conditions in yeast and SAOS2 cells (lacking p53). REs were placed upstream of minimal promoters and luciferase reporter genes. (A) Transactivation capacity in yeast. Each strain contained an integrated luciferase construct under the control of a modified cyc1 promoter that included one RE allele. p53 was expressed by using a constitutive ADH1 promoter from a single copy stable plasmid (pTSAd-p53). Luciferase activity for the predicted strong and predicted weak allele for each RE SNP is presented as relative induction between p53-expressing cells and control cells lacking p53. (B) Transactivation capacity in mammalian cells. SaOS2 cells were transiently cotransfected with vectors containing p53 under the control of a CMV promoter. The luciferase reporter is downstream of a minimal SV40 promoter (pGL3-promoter vector) that contains one of the RE alleles. Luciferase activity of each RE SNP was measured relative to the empty pGL3-promoter vector in the absence or presence of p53. The nucleotide below each bar indicates the specific SNP allele. The nonpolymorphic AIP1 and P21-5′ p53 REs are positive controls. Data represent the averages and standard deviations for three experiments. *, Value is 289 ± 16.2; †, value is 105 ± 16.0.
Different levels of transactivation activity were observed among the functional REs, but none reached the level of the p21-5′ element, previously found to be one of the most p53-responsive of human REs (9). Although the responsiveness for ADAR2, ARHGEF7, and TLR8 was comparable with AIP1, the other elements were less active, particularly EOMES and SEI-1. However, several p53-REs, such as those from the mdm2, bax, igf-bp3, c-fos, Xpc, and p48 promoters, are weak when examined in yeast as single tetramer binding sites (9, 22). Notably, the results with the EOMES RE corroborate our observations of the strong impact of mismatches in the CWWG motif.
Differential Transactivation Capacity of Polymorphic REs in Human Cells. To assess the potential of the polymorphic REs to respond differentially to p53-induced transactivation in human cells, oligonucleotides corresponding to the eight pairs of REs were cloned into a luciferase reporter plasmid carrying a minimal eukaryotic promoter. Luciferase activity levels were determined 48 h after cotransfection of SaOS2 (p53-null) cells with the RE reporter plasmid and a p53-expressing plasmid (pC53-SN3) (23). As shown in Fig. 1B, differences in p53-induced luciferase expression were observed between the RE alleles for six of the pairs, with the anticipated stronger allele yielding a greater response in each case. For TLR8 and ADAR2, the activity of the strong alleles was comparable with the activity mediated by the AIP and p21-5′ REs, respectively. For the EOMES and SEI-1 pairs, the differences were small, but consistent with results observed in yeast. In the absence of p53, there was no induction, suggesting that only p53 can activate the REs, unlike the case with the control p21-5′ RE, which showed limited induction in the absence of p53 (Fig. 1C).
Wild-type p53 RKO cells were also transiently transfected with the different reporter plasmids. To induce endogenous p53, RKO cells were UV-irradiated (20 J/m2) 24 h after transfection, and luciferase activity was measured 18 h later. Overall, results were qualitatively similar to those described for SaOS2. Most (seven of eight) of the predicted stronger alleles support transactivation, and there were significant differences between alleles for five of the RE SNPs (Fig. 4, which is published as supporting information on the PNAS web site).
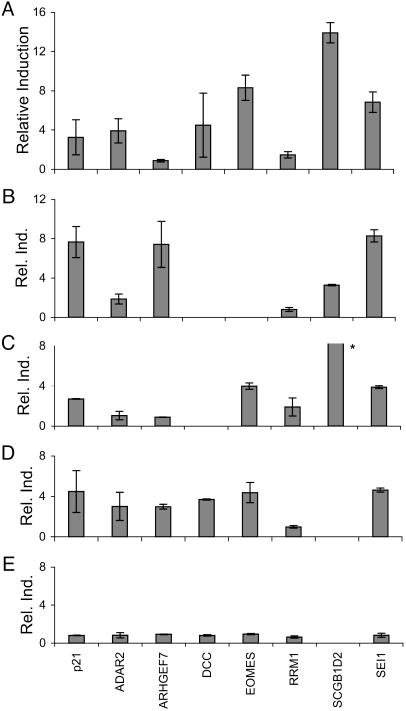
p53-Dependent Stress Responses in Endogenous Human Genes Containing Polymorphic REs. None of the eight RE-associated genes described above were known to be induced in response to p53 activation. We used real-time RT-PCR to measure p53-mediated expression changes of these genes in human RKO, HT1080, and lymphoblast cell lines. Methods for assaying p53-mediated expression in human cells include induction of endogenous p53 by genotoxic stress and expression of p53 from a transfected plasmid; we observed changes in candidate gene expression with both methods. Cells were exposed to a DNA-damaging but nonapoptotic dose of UVC radiation (UV, Fig. 2). Among the eight genes, five were UV inducible in at least one cell type. Notably, large inductions (>3-fold) were observed for the SEI-1, EOMES, and SCGB1D2 genes in HT1080 cells, for ARHGEF7, SEI-1, and SCGB1D2 in RKO, and for EOMES, SEI-1, and SCGB1D2 in lymphoblastoid cells. The observed inductions equaled or exceeded levels obtained with the p21 and AIP1 genes, which are highly p53-responsive upon UV treatment (24).
Fig. 2.
Exposure-induced expression of candidate genes in human cells. Expression of genes adjacent to candidate RE SNPs was measured by real-time RT-PCR in cultured human cells after exposure to 254 nm of UV light (UV) or upon exogenous addition of p53. Expression is normalized to GAPDH levels, and values represent fold change versus unexposed cells (inductions) or versus vector control (transfections). (A and B) UV induction in RKO (A) and HT1080 (B) cells. Cells were exposed to 20 J/m2 UV and grown for 24 h before measuring gene expression. (C) UV induction in GM15223 (PDR004) lymphoblastoid cells. Cells were exposed to 15 J/m2 UV and grown for 16 h before measuring gene expression. (D and E) Induction of endogenous gene expression by transient transfection of exogenous wild-type p53 protein. SaOS-2 cells (p53-null) were transiently transfected with a plasmid expressing wild-type p53 (D) or the transactivation-deficient mutant G279E (E) and cultured for 24 h. *, Value is 307 ± 1.83. Expression was undetectable before induction. Fold change was calculated versus the measured limit of detection for the assay.
We also measured p53-induced expression of candidate genes after transfection of a p53-expressing plasmid into p53-null SaOS2 cells. Substantial transactivation (>3-fold) of ADAR2, DCC, ARHGEF7, EOMES, and SEI1 was observed with the functional p53 plasmid (Fig. 2D), but not with a plasmid containing the transactivation-defective mutant G279E. The level of induction observed for some of these genes in SaOS-2 cells was comparable with that observed for p21.
Of the genes examined, six exhibited increased expression after p53 induction in at least one cell type. RRM1 was not induced in any cell type, whereas TLR8 expression could not be measured (TLR8 mRNA was not detectable in any of the cell lines examined). For the remaining genes, in addition to differences in the p53 RE sequences, variation in induction between cell lines may correspond to differences in other cis- or trans- acting factors (19), unlike the situation with reporter plasmids. Nonetheless, these data show that the majority of the candidate genes examined are bona fide p53-responsive genes in these cell lines. Although it is possible that the candidate REs are not specifically responsible for the observed expression changes, the transfection assays establish that the sites are strongly functional. Given the functional spacing of the REs relative to the genes (as compared with established p53 targets), their demonstrated activity, and the p53 responsiveness of the candidate genes, it is likely that the candidate polymorphic REs are involved directly in p53 transactivation.
Discussion
Identification of Functional Variation in the p53 Regulatory Network. Polymorphisms in regulatory sequences can have important phenotypic consequences (3). Our approach to identifying functionally meaningful SNPs in the REs of a large regulatory network merges experimentally based estimation of transactivation consequences of SNPs, computational identification of REs, and direct cellular evaluation of allelic differences. We suggest that this approach can be generally applied to the analysis of genetic variation at sequence-specific binding sites for transcription factors and other DNA binding proteins in human chromosomal DNA.
The computational identification of SNPs that alter p53-dependent gene expression was developed from rules derived from literature analysis and our previous studies with yeast model systems. Our subsequent functional analyses strongly support the assignment of several of the newly identified REs as bona fide p53 target sequences and the inclusion of the adjacent genes as new direct targets in the p53 transcription network. For these candidate genes, the observed allelic differences could contribute to variability in gene expression after exposure. The candidate SNPs, therefore, may represent genetic risk factors in environmentally influenced diseases such as cancer.
SNPs in regulatory sequences represent an important class of genetic variation, with implications for disease phenotypes (3). For example, an SNP in the AP4 binding site upstream of the macrophage migratory inhibitory factor gene was found to associate with disease risk in juvenile idiopathic arthritis (25). Similarly, an SNP in a putative C/EBPβ binding site near the FasL gene alters basal expression by at least 2-fold, and one allele is associated with systemic lupus erythematosus in African Americans (26). In the p53 pathway itself, recent work has demonstrated that an SNP in the MDM2 promoter can perturb SP1 transcription factor binding and thereby influence MDM2 levels, leading to attenuation of p53 activity (27). This regulatory SNP is also associated with accelerated tumor formation in human cancers. In these examples, candidate disease genes were identified first, and SNPs in transcription factor binding sites were subsequently characterized. In contrast, our study uses a systematic genomic search for regulatory SNPs, a relatively unexplored class of functional genetic variation in humans. More specifically, we have used this approach to identify functional polymorphisms in a motif that defines a set of genes in a master regulatory network.
Biological Role of Genes with Polymorphic REs. Few of the candidate genes described in this study have been directly investigated for involvement in either p53-mediated stress response or susceptibility to exposure-induced disease. We have established, however, that most of the genes are responsive to p53 signals. Many of the genes play significant roles in development, cell-cycle regulation, or DNA repair processes that are often aberrantly regulated in tumor cells. ADAR2 is a member of a family of adenosine deaminases acting on RNA (ADAR proteins) that is important in posttranscriptional regulation of a variety of genes (28). The ADAR proteins may participate in developmental pathways mediated by p53 homologs such as p63 and p73, or may govern the modification of unidentified transcripts that are involved in stress response. SCGB1D2 is a lipophilin protein that associates with secreted mammoglobin and is a marker for breast cancer (29). SEI-1 antagonizes the function of p16, an inhibitor of the cyclin-D/CDK4 complex (30). Disruption of SEI-1 regulation could therefore exert a powerful effect on cellular proliferation. EOMES (Eomesodermin) is a T-box transcription factor that plays an essential role in development and is also a major determinant of T cell differentiation (31, 32). EOMES may play a role in governing immune surveillance, or may also be a target for p53 homologs involved in human development. DCC (deleted in colorectal carcinoma) is a tumor suppressor gene highly correlated with human colorectal tumorigenesis. It also acts as a transmembrane receptor for netrin-1, an important secreted mediator of nervous system development (33). ARHGEF7, also known as βPIX, was found to be overexpressed in >90% of breast tumors and may play a role in tumor invasion by regulating changes in the actin cytoskeleton of cancer cells (34). Although it is not clear what roles TLR8, a toll-like receptor involved in innate immune response, may play in the p53 stress response network, this protein seems to play a key role in detecting single-stranded viral RNAs, and may have other undescribed functions in immune surveillance (35). Interestingly, all of the TLR proteins contain putative p53 REs upstream of their promoter regions (36).
Establishing a direct biological consequence for the SNPs presented here would in most cases require sensitive assays specifically tailored to the biological role of the candidate gene under investigation. Additionally, model system investigations of p53 occupancy at polymorphic promoters is complicated by numerous factors related to both cell biology and assay limitations (discussed below). Studies of genotype/phenotype relationship in human cells are substantially limited by the lack of isogenic cell line models that allow direct comparisons of SNP alleles. Epstein-Barr virus-transformed lymphoblastoid cell lines, which are available from a large number of individuals and represent many genotypes of interest, are a tempting model for investigating gene-environment interactions. However, recent studies have suggested that these cell lines become immortal through a series of poorly characterized transformations that include both p53 mutations and aneuploidy (37). In keeping with this idea, we observed substantial variation in p53-induced expression between lymphoblastoid cell lines for well known target genes that lack SNPs in their p53 REs (e.g., p21) (data not shown). The impact of SNPs could also be examined at the level of promoter occupancy by p53 or RNA polymerase by using chromatin immunoprecipitation, or at the level of mRNA synthesis in cases where other SNPs allow direct measurement of expressed allelic imbalance. Because there seem to be limitations in these approaches (as suggested for p53 binding) (19) and variation in both the magnitude and direction of expressed allelic imbalance across cell lines (38), especially for relatively small differences in expression, we chose an isogenic transfection model to compare the activity of polymorphic REs. This approach enabled us to demonstrate clear differences in the potential of RE SNPs to influence transactivation under controlled conditions. Establishing a relationship between particular RE alleles and disease risk, however, will likely require epidemiological investigation in humans.
This study supports and extends the concept of master genes of diversity applied initially to p53 (10). Diversity can arise through functional mutations in a master regulator that change both the spectrum and intensity of downstream gene expression responses. The present observations demonstrate that diversity can also result from variation in REs that are contained in individual genomes. This diversity could be an important factor in governing environmental responses and the potential for disease. The combination of diversity in REs, along with diversity in a master regulatory gene, could greatly broaden the disease consequences of specific p53 mutations. Thus, we suggest that the impact of identical cancer-associated functional p53 mutations could differ between individuals because of polymorphism in their genomic REs.
Supplementary Material
Acknowledgments
We thank Dr. Jack Taylor and Dr. Alex Merrick (National Institute of Environmental Health Science) for providing insightful reviews.
Author contributions: D.J.T., A.I., and D.M. designed research; D.J.T., A.I., D.M., G.S.P., and M.R.C. performed research; D.J.T., A.I., D.M., and F.S. contributed new reagents/analytic tools; D.J.T., A.I., D.M., and G.S.P. analyzed data; D.A.B. and M.A.R. were pricipal investigators; and D.J.T., A.I., and D.M. wrote the paper.
Abbreviation: RE, response element.
References
- 1.Kelada, S. N., Eaton, D. L., Wang, S. S., Rothman, N. R. & Khoury, M. J. (2003) Environ. Health Perspect. 111, 1055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, D. A., Taylor, J. A., Paulson, D. F., Robertson, C. N., Mohler, J. L. & Lucier, G. W. (1993) J. Natl. Cancer Inst. 85, 1159-1164. [DOI] [PubMed] [Google Scholar]
- 3.Hudson, T. J. (2003) Nat. Genet. 33, 439-440. [DOI] [PubMed] [Google Scholar]
- 4.Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M. & Sirotkin, K. (2001) Nucleic Acids Res. 29, 308-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko, L. J. & Prives, C. (1996) Genes Dev. 10, 1054-1072. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 7.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607-614. [DOI] [PubMed] [Google Scholar]
- 8.Zhao, R., Gish, K., Murphy, M., Yin, Y., Notterman, D., Hoffman, W. H., Tom, E., Mack, D. H. & Levine, A. J. (2000) Genes Dev. 14, 981-993. [PMC free article] [PubMed] [Google Scholar]
- 9.Inga, A., Storici, F., Darden, T. A. & Resnick, M. A. (2002) Mol. Cell. Biol. 22, 8612-8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Resnick, M. A. & Inga, A. (2003) Proc. Natl. Acad. Sci. USA 100, 9934-9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao, Z., Fu, Y. X., Hewett-Emmett, D. & Boerwinkle, E. (2003) Gene 312, 207-213. [DOI] [PubMed] [Google Scholar]
- 12.Packer, B. R., Yeager, M., Staats, B., Welch, R., Crenshaw, A., Kiley, M., Eckert, A., Beerman, M., Miller, E., Bergen, A., et al. (2004) Nucleic Acids Res. 32, D528-D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cawley, S., Bekiranov, S., Ng, H. H., Kapranov, P., Sekinger, E. A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A. J., et al. (2004) Cell 116, 499-509. [DOI] [PubMed] [Google Scholar]
- 14.Storici, F., Durham, C. L., Gordenin, D. A. & Resnick, M. A. (2003) Proc. Natl. Acad. Sci. USA 100, 14994-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inga, A., Storici, F. & Resnick, M. A. in Yeast as a Tool in Cancer Research, eds. Heitman, J. & Nitiss, J. L. (Springer, New York), in press.
- 16.el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1, 45-49. [DOI] [PubMed] [Google Scholar]
- 17.Tokino, T., Thiagalingam, S., el-Deiry, W. S., Waldman, T., Kinzler, K. W. & Vogelstein, B. (1994) Hum. Mol. Genet. 3, 1537-1542. [DOI] [PubMed] [Google Scholar]
- 18.Espinosa, J. M. & Emerson, B. M. (2001) Mol. Cell 8, 57-69. [DOI] [PubMed] [Google Scholar]
- 19.Espinosa, J. M., Verdun, R. E. & Emerson, B. M. (2003) Mol. Cell 12, 1015-1027. [DOI] [PubMed] [Google Scholar]
- 20.Marth, G., Yeh, R., Minton, M., Donaldson, R., Li, Q., Duan, S., Davenport, R., Miller, R. D. & Kwok, P. Y. (2001) Nat. Genet. 27, 371-372. [DOI] [PubMed] [Google Scholar]
- 21.Reich, D. E., Gabriel, S. B. & Altshuler, D. (2003) Nat. Genet. 33, 457-458. [DOI] [PubMed] [Google Scholar]
- 22.Qian, H., Wang, T., Naumovski, L., Lopez, C. D. & Brachmann, R. K. (2002) Oncogene 21, 7901-7911. [DOI] [PubMed] [Google Scholar]
- 23.Kern, S. E., Pietenpol, J. A., Thiagalingam, S., Seymour, A., Kinzler, K. W. & Vogelstein, B. (1992) Science 256, 827-830. [DOI] [PubMed] [Google Scholar]
- 24.Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., Nishimori, H., Tamai, K., Tokino, T., Nakamura, Y. & Taya, Y. (2000) Cell 102, 849-862. [DOI] [PubMed] [Google Scholar]
- 25.Donn, R. P., Shelley, E., Ollier, W. E. & Thomson, W. (2001) Arthritis Rheum. 44, 1782-1785. [DOI] [PubMed] [Google Scholar]
- 26.Wu, J., Metz, C., Xu, X., Abe, R., Gibson, A. W., Edberg, J. C., Cooke, J., Xie, F., Cooper, G. S. & Kimberly, R. P. (2003) J. Immunol. 170, 132-138. [DOI] [PubMed] [Google Scholar]
- 27.Bond, G. L., Hu, W., Bond, E. E., Robins, H., Lutzker, S. G., Arva, N. C., Bargonetti, J., Bartel, F., Taubert, H., Wuerl, P., et al. (2004) Cell 119, 591-602. [DOI] [PubMed] [Google Scholar]
- 28.Bass, B. L. (2002) Annu. Rev. Biochem. 71, 817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter, D., Douglass, J. F., Cornellison, C. D., Retter, M. W., Johnson, J. C., Bennington, A. A., Fleming, T. P., Reed, S. G., Houghton, R. L., Diamond, D. L. & Vedvick, T. S. (2002) Biochemistry 41, 6714-6722. [DOI] [PubMed] [Google Scholar]
- 30.Sugimoto, M., Nakamura, T., Ohtani, N., Hampson, L., Hampson, I. N., Shimamoto, A., Furuichi, Y., Okumura, K., Niwa, S., Taya, Y. & Hara, E. (1999) Genes Dev. 13, 3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura, N., Nakashima, K., Ueno, M., Kiyama, H. & Taga, T. (1999) Brain Res. Dev Brain Res. 115, 183-193. [DOI] [PubMed] [Google Scholar]
- 32.Pearce, E. L., Mullen, A. C., Martins, G. A., Krawczyk, C. M., Hutchins, A. S., Zediak, V. P., Banica, M., DiCioccio, C. B., Gross, D. A., Mao, C. A., et al. (2003) Science 302, 1041-1043. [DOI] [PubMed] [Google Scholar]
- 33.Forcet, C., Ye, X., Granger, L., Corset, V., Shin, H., Bredesen, D. E. & Mehlen, P. (2001) Proc. Natl. Acad. Sci. USA 98, 3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn, S. J., Chung, K. W., Lee, R. A., Park, I. A., Lee, S. H., Park, D. E. & Noh, D. Y. (2003) Cancer Lett. 193, 99-107. [DOI] [PubMed] [Google Scholar]
- 35.Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., Lipford, G., Wagner, H. & Bauer, S. (2004) Science 303, 1526-1529. [DOI] [PubMed] [Google Scholar]
- 36.Hoh, J., Jin, S., Parrado, T., Edington, J., Levine, A. J. & Ott, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto, M., Tahara, H., Ide, T. & Furuichi, Y. (2004) Cancer Res. 64, 3361-3364. [DOI] [PubMed] [Google Scholar]
- 38.Pastinen, T., Sladek, R., Gurd, S., Sammak, A., Ge, B., Lepage, P., Lavergne, K., Villeneuve, A., Gaudin, T., Brandstrom, H., et al. (2004) Physiol. Genomics 16, 184-193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.