Abstract
Mutations in mitochondrial DNA (mtDNA) cause a variety of relatively rare human diseases and may contribute to the pathogenesis of other, more common degenerative diseases. This stimulates interest in the capacity of mitochondria to repair damage to mtDNA. Several recent studies have shown that some types of damage to mtDNA may be repaired, particularly if the lesions can be processed through a base excision mechanism that employs an abasic site as a common intermediate. In this paper, we demonstrate that a combination of enzymes purified from Xenopus laevis mitochondria efficiently repairs abasic sites in DNA. This repair pathway employs a mitochondrial class II apurinic/apyrimidinic (AP) endonuclease to cleave the DNA backbone on the 5′ side of an abasic site. A deoxyribophosphodiesterase acts to remove the 5′ sugar-phosphate residue left by AP endonuclease. mtDNA polymerase γ fills the resulting 1-nucleotide gap. The remaining nick is sealed by an mtDNA ligase. We report the first extensive purification of mtDNA ligase as a 100-kDa enzyme that functions with an enzyme-adenylate intermediate and is capable of ligating oligo(dT) strands annealed to poly(rA). These properties together with preliminary immunological evidence suggest that mtDNA may be related to nuclear DNA ligase III.
Mitochondrial DNA (mtDNA) encodes a small set of 13 proteins that are critically important for aerobic metabolism. In the last several years, deletions in mtDNA and point mutations in mitochondrial tRNA and protein-encoding genes have been shown to cause a variety of human diseases. These diseases are commonly referred to with abbreviations such as MELAS, MERRF, LHON, NARP, and KSS/CPEO (reviewed in references 17 and 55). Affected individuals frequently exhibit neurological and/or muscular symptoms. These diseases are, fortunately, rare, since mtDNA is highly polyploid and a high proportion of defective genomes is required for penetrance. However, there have been suggestions that mtDNA damage or polymorphism may be a contributing factor for a number of more common age-related disorders, including Parkinson’s disease and type II diabetes mellitus (56, 57). Understanding the role of mtDNA mutations in these human diseases requires an appreciation of the mechanisms whereby the cell can protect the integrity of its mtDNA genomes.
In an early study, Clayton et al. (4) found that pyrimidine dimers in mtDNA were not actively repaired in cultured mouse cells. Other groups subsequently showed that certain carcinogen DNA adducts accumulated to high levels in mtDNA and were not repaired efficiently (2, 36). These investigations suggested that mitochondria lacked the ability to repair bulky lesions in their DNA. To date, there has been no clear demonstration that vertebrate cell mitochondria possess the sort of nucleotide excision repair processes that handle such bulky lesions in bacterial DNA or eukaryotic nuclear DNA. It has been speculated that mtDNA molecules bearing bulky adducts may be diluted as undamaged molecules continue to replicate or may be targeted for destruction.
Recent reports have documented repair of some types of mtDNA damage, including purine base alkylation (19, 33, 38, 41, 46) and some classes of damage induced by cis-platinum (18), bleomycin (47), or 4-nitroquinoline oxide (50). In addition, mtDNA may be a frequent target of oxidative damage, since superoxide radicals have been shown to be generated as a by-product of oxidative metabolism in mitochondria (48). The content of one oxidative product, 8-oxo-dG, has been reported to occur with a severalfold-higher frequency in mtDNA than in nuclear DNA and to increase with age (31, 43). Although some studies have failed to observe a high steady-state level of 8-oxo-dG in mtDNA (11, 12), DNA backbone damage produced by oxidative stress can be documented and is subject to repair (9, 12). Many of these repairable lesions in mtDNA may be processed by a base excision repair (BER) mechanism that should be maintained to permit repair of the frequent spontaneous base loss that occurs in any DNA (23, 24).
Very little is known concerning the enzymatic machinery for BER in mitochondria. Mitochondria contain a uracil DNA glycosylase (UDG) that can initiate repair of U residues misincorporated during DNA replication. The mitochondrial form of the enzyme is an alternate product of the cellular UDG gene (35, 49). The existence of this glycosylase strongly suggests that vertebrate cell mitochondria are likely to contain enzymes capable of complete repair of abasic sites. Other damage-specific glycosylases and endonucleases have been studied but are less well understood (6, 53). A partial purification of mitochondrial apurinic/apyrimidinic (AP) endonucleases has been reported (52), although no definitive characterization has been published to our knowledge. Although a β-like polymerase has been reported for the protozoan Crithidia (54), the only DNA polymerase identified in mitochondria in higher eukaryotes is DNA polymerase γ (pol γ), which is the replicative polymerase of mitochondria. The ability of this polymerase to participate in repair reactions has received little attention beyond the demonstration that the associated 3′→5′ exonuclease is capable of editing replication errors (10, 14, 15, 25, 37). Although an mtDNA ligase must exist to participate in mtDNA replication, the literature contains only a single brief report of the existence of this activity, with no molecular characterization of the enzyme (21).
In this paper, we report the first complete reconstitution of base excision repair of abasic sites in DNA by using highly purified enzymes prepared from mitochondria. As part of this effort, we provide a more complete description of the mtDNA ligase than has been available previously.
MATERIALS AND METHODS
Oligonucleotide and DNA substrates for repair reactions.
Duplex oligonucleotide substrates with two different sequence contexts were used for standard AP endonuclease assays, as listed in Table 1. Oligonucleotides 31F and 32F contained a synthetic analog of an abasic site, 3-hydroxy-2-hydroxymethyltetrahydrofuran (also referred to as tetrahydrofuran or, within sequences, as F). Oligonucleotide 31F, used for Fig. 2, was 5′ labeled with polynucleotide kinase and [γ-32P]ATP and annealed to its complement (with A opposite F). Alternatively, oligonucleotide 32U or 32F, used for Fig. 4, were either 5′ labeled, as for 31F, or 3′ labeled by incubation of duplex oligonucleotides with the Klenow fragment of DNA pol I and [α-32P]dATP. In repair reactions in Fig. 1, unlabeled oligonucleotide duplex 32U was used after pretreatment with 2.5 U of UDG (Boehringer Mannheim) for 90 min at 37°C in a buffer containing 50 mM Tris (pH 8.0), 2 mM dithiothreitol (DTT), and 100 μg of bovine serum albumin per ml. The covalently closed circular repair substrate was prepared as described previously (28), except that the oligonucleotide contained a U residue. Control templates were prepared by using an oligonucleotide containing a T residue in place of the U residue. Covalently closed circular DNA (cccDNA) substrates were prelabeled on the 5′ side of the lesion 5 nucleotides away from the U residue, or on the 3′ side 10 nucleotides away from the U residue, as described previously (28) and illustrated in Fig. 8. DNA substrates were pretreated before the repair reaction with UDG as described above. Where indicated, cccDNA was also pretreated with AP endonuclease as described below.
TABLE 1.
Oligonucleotide substrates with U residues or abasic site analogs
| Name | Sequence |
|---|---|
| 31F | 5′-CACTCGGCCTTTFCTACTTTCCTCTCCATTT |
| GTGAGCCGGAAAAGATGAAAGGAGAGGTAAA | |
| 32F | 5′-CATGGGCCGACATGAFCAAGCTTGAGGCCAAG |
| GTACCCGGCTGTACTAGTTCGAACTCCGGTTCT | |
| 32U | 5′-CATGGGCCGACATGAUCAAGCTTGAGGCCAAG |
| GTACCCGGCTGTACTAGTTCGAACTCCGGTTCT |
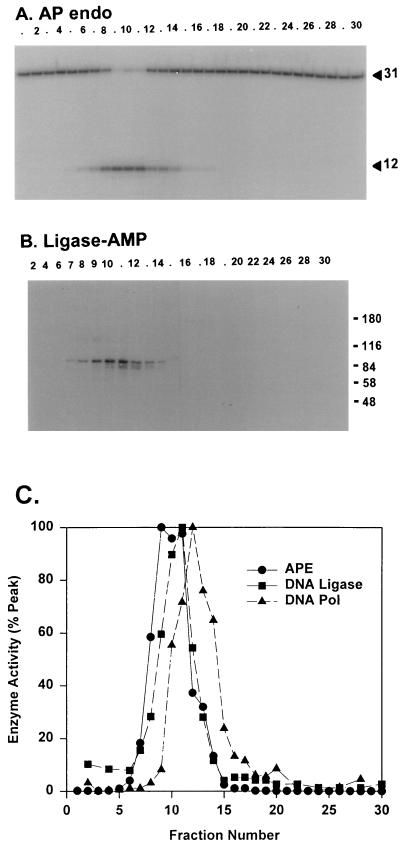
FIG. 2.
DNA pol γ, DNA ligase, and AP endonuclease activities coelute from S Sepharose. (A) S Sepharose column fractions were assayed for AP endonuclease (AP endo) by cleavage of a 5′-32P-labeled 31-bp oligonucleotide containing a synthetic abasic site to generate a 12-nucleotide product that was detected by electrophoresis on a denaturing polyacrylamide-urea gel. Numbers on the right are sizes in nucleotides. (B) Fractions were assayed for DNA ligase by the production of 32P-AMP-ligase detected by autoradiography following SDS-PAGE. Numbers on the right are molecular masses in kilodaltons. (C) Elution profiles for these two enzymes and DNA pol γ, normalized to 100% activity in the fraction containing maximal activity for each enzyme. The fraction numbers are offset slightly with respect to the column fractions used in Fig. 1, which was generated with a different enzyme preparation. APE, AP endonuclease.
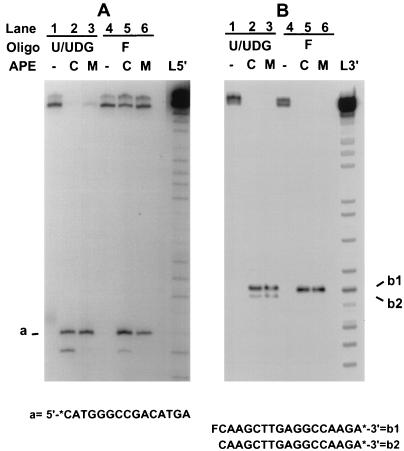
FIG. 4.
The mitochondrial AP endonuclease cleaves to generate 3′-OH and 5′ deoxyribosephosphate termini. Two 32-mer oligonucleotides containing either a U residue or a tetrahydrofuran residue at position 16 were labeled at the 5′ end (A) or 3′ end (B) as described in Materials and Methods. Oligonucleotide (Oligo) duplexes containing U residues were pretreated with UDG. Oligonucleotides were then treated with X. laevis nuclear AP endonuclease (APE) (lanes C) or the mitochondrial AP endonuclease (lanes M). Reaction mixtures for panel B were treated with NaBH4 following the enzyme incubation as described previously (28). The products were ethanol precipitated, boiled in formamide loading solution, and analyzed by electrophoresis on a 20% polyacrylamide gel containing 8 M urea. Lanes L5′ and L3′ show purine cleavage products (30) as marker ladders generated by partial depurination with formic acid (pH 2) and piperidine treatment of 5′- or 3′-labeled wild-type oligonucleotides containing a T or F at position 16 for L5′ and L3′, respectively. It should be noted that the Maxam-Gilbert chemistry leaves 5′ and 3′ phosphate residues adjacent to the position of nucleoside loss (30). For each panel, all samples were originally run on the same gel. Lane L5′ in panel A was from a longer autoradiographic exposure than the rest of the lanes.
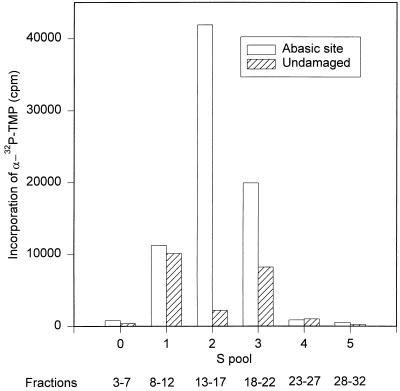
FIG. 1.
A crude fraction containing DNA pol γ supports damage-specific incorporation into DNA. Two duplex oligonucleotide substrates, one containing a U residue at position 16 and the other containing T as an undamaged control, were prepared as described in Materials and Methods. The duplex containing U was preincubated with UDG immediately before the repair reaction to generate an abasic site. Repair reaction mixtures (40 μl) contained 1 pmol of oligonucleotide and 3 μl of pooled fractions obtained by chromatography of a mitochondrial extract on S Sepharose. Incorporation of [α-32P]TMP into oligonucleotides was assayed following trichloroacetic acid precipitation. The peak of DNA pol γ activity in this column eluate was in fraction 15.
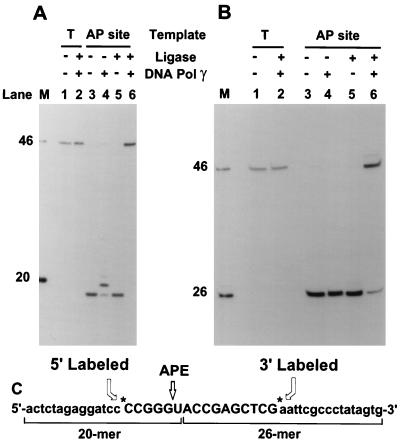
FIG. 8.
Repair of abasic sites by mitochondrial enzymes. Two cccDNAs were used in repair reactions. One contained a U residue (as shown in panel C) and was pretreated with UDG and mitochondrial AP endonuclease (APE) to create a specific nicked abasic site (AP site). The control DNA contained a T residue in place of the U residue. Each type of DNA was labeled either on the 5′ side (A) or on the 3′ side (B). Repair reactions were conducted as described in Materials and Methods. DNA samples were treated with HinfI prior to analysis on a 20% polyacrylamide–urea gel. Individual lanes are described in Results. Markers (lanes M) were generated by cleavage of the T-containing control DNA substrates with HinfI (46-mer) or HinfI plus RsaI (20-mer in panel A, 26-mer in panel B). Labeled DNA fragments were detected by autoradiography.
DNA pol γ.
DNA pol γ was purified as described previously (13, 32) following incorporation of [α-32P]TMP by using oligo(dT) · poly(rA) as a primer-template. The S Sepharose column fractions containing the peak of DNA pol γ activity were adjusted to 1.2 M ammonium sulfate and loaded on a Poros PH hydrophobic interaction chromatography column. The flowthrough fraction was processed as described below to recover AP endonuclease and DNA ligase (see Fig. 3).
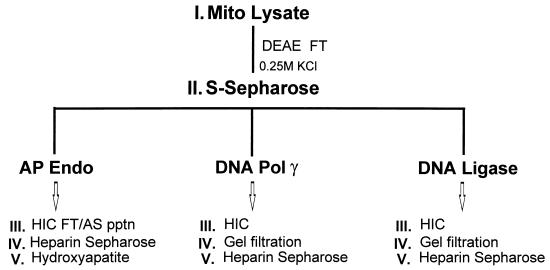
FIG. 3.
Scheme for purification of mitochondrial DNA repair activities. Mito, mitochondrial; FT, flowthrough; Endo, endonuclease; HIC, hydrophobic interaction chromatography; AS pptn, ammonium sulfate precipitation.
DNA ligase.
For routine purposes, DNA ligase was detected by monitoring the mechanism-based adenylation of enzyme. One microliter of enzyme fraction was incubated in a 10-μl reaction mixture containing 20 mM Tris (pH 8.0), 40 mM NaCl, 5 mM MgCl2, 5 mM DTT, 8% glycerol, 0.02% Triton X-100, and 2 μCi of [α-32P]ATP. Reaction mixtures were incubated at 25°C for 15 min, and reactions were stopped by addition of an equal volume of sodium dodecyl sulfate (SDS) sample loading buffer (16). Proteins were resolved by electrophoresis on an 8 or 10% polyacrylamide gel containing SDS (16) and detected by autoradiography or direct Phosphorimager analysis of the dried gel. In some cases, proteins were precipitated with trichloroacetic acid prior to electrophoresis. An alternative procedure to assay DNA ligase involved incubation of enzyme with substrates prepared by annealing 10 pmol of 5′-32P-oligo(dT)40 annealed to 1 μg of either poly(dA) or poly(rA) in reaction mixtures containing DNA ligase buffer and 1 mM ATP. Ligation products were analyzed by electrophoresis on a 10% polyacrylamide gel containing 8 M urea and detected by autoradiography or Phosphorimager analysis.
mtDNA ligase was prepared from the same fractions of the S Sepharose column eluate used to prepare DNA pol γ as described above. mtDNA ligase was observed to flow through the Poros PH column at 1.2 M ammonium sulfate. The flowthrough fraction from this column was adjusted to 1.8 M ammonium sulfate by addition of an appropriate volume of saturated ammonium sulfate, filtered, and loaded on a 4.6- by 100-mm Poros HP hydrophobic interaction column (Perceptive Biosystems) at a flow rate of 2 ml/min. Bound proteins were eluted with a 15-ml gradient of 1.8 to 0 M ammonium sulfate in a buffer containing 20 mM Tris (pH 8.4), 0.5 mM EDTA, and 2 mM DTT. These and all column buffers described below contained a standard cocktail of protease inhibitors, including 0.5 mM benzamidine HCl, 1 μM pepstatin A, and 2 μg each of leupeptin, antipain, E64 {N-[N-(l-3-trans-carboxirane-2-carbonyl)-l-leucyl]-agmatine}, and aprotinin per ml. Fractions containing DNA ligase adenylation activity were pooled and applied to a Superdex 200 HiLoad (16/60) gel filtration column equilibrated with a buffer containing 0.4 M NaCl, 5% glycerol, 20 mM Tris (pH 8), 2 mM DTT, 0.5 mM EDTA, 0.02% Triton X-100, 100 μg of acetylated gelatin per ml, and the standard protease inhibitor mix. Fractions containing DNA ligase adenylation activity were diluted approximately fivefold with gel filtration buffer lacking NaCl and applied to a 1-ml heparin Sepharose column (HiTrap; Pharmacia). Bound proteins were eluted with 15 ml of gel filtration buffer lacking gelatin but containing a gradient of KCl increasing linearly from 50 mM to 1 M. At this stage and other intermediate stages, fractions were stored at −80°C after addition of one-half volume of storage buffer containing 75% glycerol, 20 mM Tris (pH 8), 2 mM DTT, 0.02% Triton X-100, and the standard protease inhibitors plus 0.2 mM phenylmethylsulfonyl fluoride.
Nuclear DNA ligase I was purified from the postmitochondrial supernatant of Xenopus ovary homogenates. This fraction was centrifuged at 100,000 × g, and proteins were precipitated from the resulting supernatant with 62% saturated ammonium sulfate as described previously (5). DNA ligase I was purified by S Sepharose, hydroxylapatite, hydrophobic interaction, gel filtration, and heparin Sepharose chromatography. DNA ligase I was detected in each column eluate as a polypeptide of 180 kDa by using the adenylation assay described above for mtDNA ligase.
Human DNA ligase III was prepared from a HeLa cell nuclear extract by using phosphocellulose and mono-S column chromatography as described previously (44). A DNA ligase III-specific rabbit antiserum was generated against a peptide coupled to keyhole limpet hemocyanin. The antigen peptide sequence, CMFEKLERARATTKK, is conserved in the published sequence of human DNA ligase III and in a partial cDNA sequence for Xenopus DNA ligase III (37a).
Mitochondrial AP endonuclease.
The standard AP endonuclease assay was performed with a 10-μl reaction mixture with 500 fmol of tetrahydrofuran-containing oligonucleotide (either 31F or 32F [Table 1]) as the substrate in a buffer containing 20 mM Tris (pH 8.0), 20 mM KCl, 2 mM MgCl2, 2 mM DTT, 20 μM zinc acetate, 10% glycerol, 0.05% Triton X-100, and 10 μg of bovine serum albumin per ml. In some cases, to provide excess substrate, the oligonucleotide concentration was increased to 2 pmol per reaction mixture. Reaction mixtures were incubated for 20 min at 37°C, and reactions were stopped by addition of an equal volume of loading solution containing 95% HCONH2, 25 mM Tris (pH 8.4), 25 mM EDTA, and 50 μg each of bromophenol blue and xylene cyanol per ml. An aliquot of each reaction mixture was subjected to electrophoresis on a 15% polyacrylamide gel containing 8 M urea. AP endonuclease activity was measured by the conversion of 5′-labeled oligonucleotide 32F to a 15-mer or of 5′-labeled oligonucleotide 31F to a 12-mer as monitored by autoradiography or direct Phosphorimager analysis. One unit of enzyme activity was defined as the amount required to cleave 10 fmol of substrate in a standard reaction.
AP endonuclease activity was purified from the same S Sepharose fractions used as a source of DNA ligase. AP endonuclease was found to flow through the Poros HP column, which removed most proteins with greater hydrophobic character. AP endonuclease was recovered by precipitation with 80% saturated ammonium sulfate. The ammonium sulfate precipitate was resuspended in 20 mM Tris (pH 8)–2 mM DTT and dialyzed for 3 h against a solution containing 40% glycerol, 20 mM Tris (pH 8), 5 mM DTT, 100 mM NaCl, 0.02% Triton X-100, and standard protease inhibitors to generate fraction III. AP endonuclease was subjected to chromatography on a 1-ml heparin Sepharose HiTrap column as described for mtDNA ligase. Fractions containing AP endonuclease activity were combined and applied to a 1-ml HAP-Ultrogel column (IBF Biotechnics, Inc.) equilibrated with 10 mM KPi (pH 6.8)–50 mM HEPES (pH 7.5)–5% glycerol–2 mM DTT–protease inhibitors. Proteins were eluted with a 15-ml gradient of 10 to 600 mM KPi. In one case (see Table 2) chromatography on a 1-ml double-stranded DNA cellulose (Sigma) column was used as an alternative to hydroxylapatite for the final purification step. In this case, proteins were applied to the column in buffer containing 40 mM KCl, 20 mM Tris (pH 8), 2 mM DTT, 0.1 mM EDTA, 8% glycerol, and protease inhibitors. Bound proteins were eluted with a 16-ml gradient of KCl increasing from 50 mM to 1 M. Fractions containing peak enzyme activity from either the double-stranded DNA or hydroxylapatite column were mixed with enzyme storage buffer as described for mtDNA ligase and were considered equivalent sources of fraction V AP endonuclease activity. When the properties of the mitochondrial AP endonuclease were compared to those of the cellular AP endonuclease, the latter enzyme was purified as described previously (28).
TABLE 2.
Purification of X. laevis mitochondrial AP endonucleasea
| Fraction | Step | Vol (ml) | Protein (mg) | Activity (U/μl) | Total activity (U) | Sp act (U/mg) |
|---|---|---|---|---|---|---|
| I | Lysate | 182 | 1,320 | NDb | ||
| II | S Sepharose | 30 | 25.8 | 8 | 240,000 | 9,300 |
| III | Ammonium sulfate precipitate | 1.8 | 1.20 | 19 | 34,200 | 28,500 |
| IV | Heparin Sepharose | 8 | ND | 5.26 | 42,070 | |
| V | Double-stranded DNA | 4 | ND | 3.9 | 15,694 |
Note that double-stranded DNA cellulose rather than hydroxylapatite was used as the final column in this preparation. AP endonuclease activity was detectable in the crude lysate but not in the flowthrough of the S Sepharose column. Due to the presence of contaminating activities, the enzyme activity was not quantified at this stage.
ND, not determined.
Borohydride trapping of deoxyribophosphodiesterase.
Borohydride trapping of the Schiff base intermediate in the AP lyase reaction employed a duplex oligonucleotide substrate essentially identical to oligonucleotide 32U (Table 1) except that the top strand consisted of two oligonucleotides. An unlabeled 15-mer (CATGGGCCGACATGA) and a 5′-phosphorylated labeled 17-mer (32P-UCAAGCTTGAGGCCAAG) were annealed to the lower strand of oligonucleotide 32U. Following treatment with UDG, this duplex oligonucleotide would contain a labeled 2′-deoxyribose 5′-phosphate (5′-dRP) residue adjacent to a nick in the top strand. Two microliters of DNA pol γ or mtDNA ligase was incubated with 50 fmol of labeled oligonucleotide in a 20-μl reaction mixture containing 10 mM HEPES (pH 8), 1 mM MgCl2, 2 mM DTT, 50 μg of gelatin per ml, 0.1 U of UDG (HK-UNG; Epicentre Technologies), and 20 mM freshly prepared NaBH4 for 30 min at 25°C. Ten microliters of a solution containing 20 mM CaCl2, 20 mM HEPES (pH 8), and 20 μg of micrococcal nuclease per ml was added, and incubation was continued for 10 min at 37°C. Reactions were stopped by addition of an equal volume of sample loading buffer (16), and the mixtures were boiled. Proteins were subjected to electrophoresis on a 10% polyacrylamide gel containing SDS. The gel was fixed in 10% methanol–10% acetic acid, dried, and exposed to X-ray film or to a Phosphorimager plate to detect label transferred to protein.
Stage 2 of repair reaction.
Stage 1 of repair reactions involved pretreatment with UDG to expose an abasic site followed by treatment with mitochondrial AP endonuclease to cleave adjacent to the lesion. For stage 2 of the repair reaction, 200 fmol of prelabeled DNA substrate was incubated for 30 min at 25°C in a 40-μl reaction mixture containing 80 mM NaCl, 8 mM MgCl2, 25 μM zinc acetate, 20 mM Tris (pH 8.0), 2 mM DTT, 10% glycerol, 1 mM ATP, 20 μM (each) four deoxynucleoside triphosphates, and 100 μg of bovine serum albumin per ml. Reaction mixtures included 2 μl of both DNA pol γ and mtDNA ligase except when either enzyme was specifically omitted. Reactions were stopped by addition of 160 μl of 0.3 M sodium acetate in Tris-EDTA buffer containing 10 μg of glycogen per ml as a carrier and extracted with 1:1 phenol-CHCl3 followed by ether. Nucleic acids were precipitated with ethanol, resuspended, treated with restriction endonuclease HinfI, and reprecipitated. Samples were resuspended in formamide loading solution and analyzed by electrophoresis on a 15% polyacrylamide gel containing 8 M urea. 32P-labeled fragments were detected by autoradiography or by Phosphorimager analysis.
RESULTS
A crude fraction of mitochondrial enzymes supports DNA synthesis on templates containing abasic sites.
We first asked whether a crude enzyme fraction containing mtDNA pol γ could selectively incorporate DNA precursors into an oligodeoxynucleotide template containing a single abasic site. A duplex oligodeoxynucleotide containing a U · A base pair was pretreated with UDG to expose a single abasic site. Repair of this lesion should require incorporation of a TMP residue. A control oligodeoxynucleotide duplex was prepared with a T residue base paired to A in place of the abasic site. In order to suppress label incorporation due to end labeling of the annealed oligonucleotide, the sequences were designed to lack T residues near the 3′ ends of both strands. As a source of potential repair activity, we used pooled fractions from a standard S Sepharose column procedure used as the first step for purification of Xenopus ovary mtDNA pol γ. This has also been used as the first chromatographic step for purification of mitochondrial single-stranded-DNA binding protein, mitochondrial RNA polymerase, and transcription factors mtTFB and mtTFA. Approximately 700 mg of protein in a cleared Triton X-100 lysate of ovary mitochondria was loaded onto a 20-ml S Sepharose column. Bound proteins were eluted with a 6-column-volume gradient from 50 to 700 mM KCl and collected into 32 fractions. Samples of fractions were pooled to test for damage-specific DNA synthesis. We observed significant damage-dependent incorporation into oligonucleotides by pool 2, which contained DNA pol γ (Fig. 1). This incorporation is consistent with repair of the abasic site but does not provide proof of a complete repair reaction, since it is possible that it represents strand displacement synthesis by DNA polymerase. At a minimum, this result requires the presence of AP endonuclease to generate a 3′-OH terminus to support DNA synthesis by DNA pol γ.
Detection of mitochondrial AP endonuclease and DNA ligase.
Previous experiments have shown that mammalian mitochondria contain AP endonuclease and DNA ligase activities. We assayed S Sepharose fractions for AP endonuclease activity by monitoring cleavage of a 5′-labeled oligonucleotide adjacent to a synthetic analog of an abasic site in DNA. To test for DNA ligase, we incubated samples of column fractions with [α-32P]ATP to assay the mechanism-based formation of a DNA ligase adenylate. These assays showed clear single peaks of activity for both enzymes overlapping the peak of DNA pol γ activity (Fig. 2). At this time, we have no evidence that these enzymes coelute from this column as a complex. It is interesting that the adenylated mtDNA ligase has a molecular mass of approximately 100 kDa and that the predominant nuclear DNA ligase in Xenopus oocytes, the 180-kDa DNA ligase I, was not detected in the mitochondrial enzyme preparation.
The mitochondrial AP endonuclease is a class II enzyme.
We developed a general strategy for further purification of mitochondrial AP endonuclease and DNA ligase, as shown in Fig. 3. One goal of this approach was to purify both of these proteins as well as DNA pol γ from the same S Sepharose fractions. Thus, the choice of chromatography steps was influenced by the need to recover multiple activities at several stages in purification. The peak fractions of DNA pol γ prepared by this protocol contain two major polypeptides, the catalytic 137-kDa subunit and a smaller subunit of 43 kDa. This preparation has been used in a number of previous studies (32, 40, 59).
The single peak of mitochondrial AP endonuclease observed in Fig. 2 appears to represent essentially all detectable mitochondrial AP endonuclease, since we consistently found that all of the AP endonuclease activity in the crude mitochondrial lysate bound to S Sepharose. Further purification of the AP endonuclease through three additional highly resolving steps resulted in a nearly homogeneous preparation (Table 2). However, the final yield was only 6.5% of the initial activity, so we cannot rule out the possibility that there is a selective loss of one form of AP endonuclease.
To determine the specificity of the mitochondrial AP endonuclease, we compared the products of action of this enzyme and the major nuclear AP endonuclease on oligonucleotide substrates containing a natural AP site (a U residue excised with UDG) or a synthetic AP site, known as a tetrahydrofuran residue (F). We have previously shown that the major Xenopus nuclear AP endonuclease is a class II enzyme antigenically related to the human HAP1 or APE enzyme (7, 28, 45). With 5′-labeled oligonucleotide substrates, the mitochondrial and nuclear AP endonucleases produced identical products with a 3′-OH terminus characteristic of a class II enzyme (species a in Fig. 4A). However, it is apparent in Fig. 4A that the nuclear enzyme preparation contains detectable 3′ exonuclease that is absent in the mitochondrial enzyme preparation. The mitochondrial and nuclear AP endonucleases also produced identical products upon cleavage of 3′-labeled oligonucleotides containing a natural AP site (compare lanes 2 and 3 of Fig. 4B). The mobility of the 3′ product, labeled b1 in Fig. 4B, is similar to that of the cleavage product obtained when the oligonucleotide contained a synthetic AP site or tetrahydrofuran residue. When a natural AP site was used, a fraction of the initial cleavage product underwent β-elimination to generate species b2. We conclude that the mitochondrial AP endonuclease, like the nuclear enzyme, is a class II AP endonuclease. The properties of the highly purified mitochondrial AP endonuclease appear to be similar to those of the mouse mitochondrial AP endonuclease reported by Tomkinson et al. (52).
Further characterization of mtDNA ligase.
We purified the 100-kDa mtDNA ligase, as outlined in Fig. 3 and described in Materials and Methods, by using the enzyme adenylation assay. To confirm that the final enzyme fraction was active as a DNA ligase, we tested its ability to ligate oligonucleotides annealed to homopolymer templates. One of the classical assays employed to discriminate between various eukaryotic DNA ligase activities is to test the ability of an enzyme to join oligo(dT) annealed to poly(rA) (22). The results in Fig. 5 show that the protein we purified as a potential mtDNA ligase by using the adenylation assay is active as a DNA ligase and is capable of using the oligo(dT) · poly(rA) substrate. As a control, we purified the Xenopus laevis nuclear DNA ligase I and confirmed that it does not utilize this substrate, as has been shown for DNA ligase I from other sources (Fig. 5). This suggests that the 100-kDa mtDNA ligase is not a breakdown product of DNA ligase I.
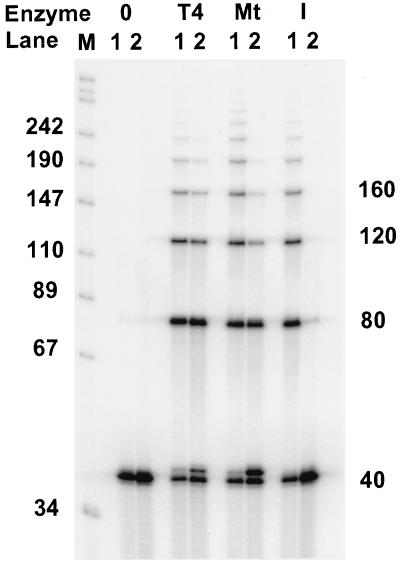
FIG. 5.
mtDNA ligase joins oligo(dT)40 strands annealed to either poly(dA) or poly(rA). Oligo(dT)40 annealed to either poly(dA) or poly(rA) (lanes 1 or 2, respectively) was incubated either without enzymes (0) or with T4 DNA ligase (T4), mtDNA ligase (Mt), or X. laevis DNA ligase I (I) as described in Materials and Methods. Products were precipitated with ethanol and analyzed by electrophoresis on an 8% polyacrylamide–urea gel. Ligation results in a ladder of increasing numbers of 40-mers. Mobility markers (in kilodaltons) consisting of labeled MspI fragments of pUC18 are shown in lane M. The apparent 41-mer in samples incubated with T4 DNA ligase or mtDNA ligase most likely represents oligo(dT)40 modified by addition of AMP by DNA ligase.
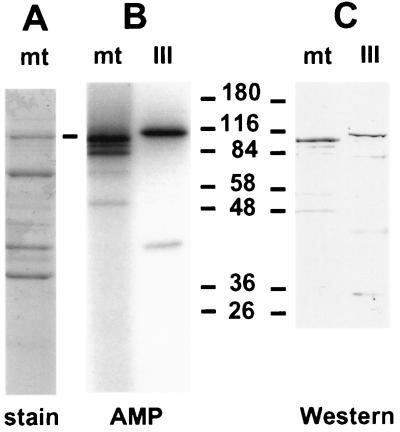
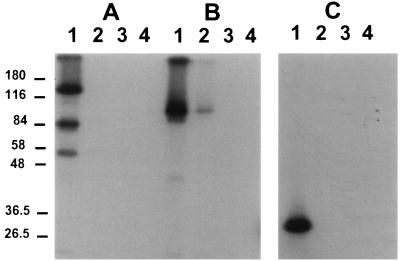
The size of the Xenopus mtDNA ligase and its activity on an oligo(dT) · poly(rA) template suggested that it may be a form of the amphibian homolog of mammalian DNA ligase III or IV (3, 26, 58) (see Discussion). However, we were not aware of any previous reports of either of these DNA ligases in Xenopus. We used degenerate oligonucleotides to amplify portions of both DNA ligase III and IV by using cDNA prepared from Xenopus ovary mRNA. Although the sequencing of these cDNAs is still continuing, we were able to identify peptide sequences conserved between Xenopus and human DNA ligase III and absent from DNA ligase I or IV (37a). We raised a polyclonal rabbit antiserum against one DNA ligase III-specific peptide, CMFEKLERARATTKK. This serum was used to probe a Western blot of a protein gel containing Xenopus mtDNA ligase and human DNA ligase III. Figure 6 shows that this serum reacted with both DNA ligases, which suggests that the Xenopus mtDNA ligase is related to DNA ligase III. The Xenopus mtDNA ligase identified by either adenylation or immunoblotting migrates slightly more rapidly than human nuclear DNA ligase III on the gel. Figure 6 also shows that our current preparation of mtDNA ligase is highly purified, since the immunoreactive band is a major protein species. The mtDNA ligase has not yet been purified to homogeneity, although this work is in progress.
FIG. 6.
mtDNA ligase is related to DNA ligase III. (A) Coomassie blue stain of a 10% polyacrylamide–SDS gel analysis of the mtDNA ligase preparation. (B) Autoradiogram of a gel containing mtDNA ligase and human DNA ligase III following adenylation of proteins with [α-32P]ATP. (C) Immunoblot of mtDNA ligase and human DNA ligase III, probed with a 1:15,000 dilution of antiserum directed against a peptide sequence conserved between human and Xenopus DNA ligase III and developed with alkaline phosphatase-conjugated goat anti-rabbit serum. Numbers indicate molecular masses in kilodaltons.
Repair of abasic sites in DNA by highly purified mitochondrial enzymes.
The main purpose of our present experiments is to determine whether DNA pol γ, mtDNA ligase, and mitochondrial AP endonuclease are capable of repairing abasic sites in DNA. We confirmed that all three repair enzymes were free of the other activities (Fig. 7). We then studied repair of single abasic sites embedded in the same cccDNA as used in our previous studies of AP site repair with nuclear enzymes (27, 28). These substrates for repair offer several advantages. The lesion is contained in high-molecular-weight DNA, so that binding of repair proteins is not limited by the small size of an oligonucleotide substrate. The abasic site that serves as a target for repair is precisely positioned in a context that allows prelabeling of the substrate either 5 nucleotides upstream or 10 nucleotides downstream of the unique U residue (Fig. 8). Cleavage of the initial cccDNA with HinfI generated a 46-mer containing the original oligonucleotide. HinfI cleavage of intermediates in the repair reaction was used to monitor the 5′- and 3′-end groups surrounding the original damaged residue at each stage in the repair reaction.
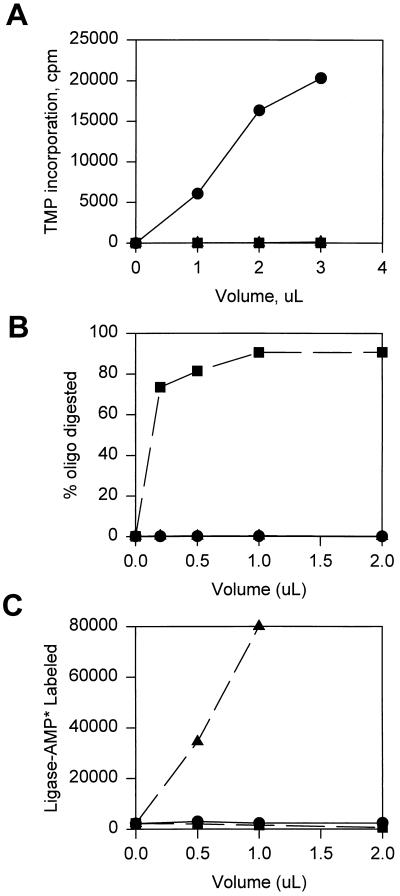
FIG. 7.
Purity of mitochondrial repair enzymes. Final preparations of DNA pol γ (•), mtDNA ligase (▴), and AP endonuclease (▪) were assayed for DNA polymerase (A), AP endonuclease (B), and DNA ligase adenylation (C) by using standard assays described in Materials and Methods. oligo, oligonucleotide.
Repair reactions were conducted in two stages. In the first stage, substrates were treated with UDG to generate abasic sites, followed by mitochondrial AP endonuclease to generate a 3′-OH terminus on the 5′ sides of lesions. Cleavage with HinfI showed that the substrates containing U residues were quantitatively nicked in the first-stage reaction to generate products of the expected sizes (lanes 3 of Fig. 8A and B for 5′- and 3′-labeled substrates, respectively). Control substrates containing a T residue in the oligonucleotide were not incised (Fig. 8, lanes 1).
Figure 8 shows the products of HinfI digestion of molecules processed in the second stage of the repair reaction. Figure 8A shows the products of molecules labeled on the 5′ side of the lesion. Incubation with only DNA pol γ produced primarily a 1-nucleotide extension of the 5′-labeled fragment as a repair intermediate (Fig. 8A, lane 4). This intermediate comigrated on a gel with the 20-mer marker generated by combined treatment of wild-type DNA with HinfI and RsaI. Incubation with only mtDNA ligase did not result in resealing of the DNA strand (Fig. 8A, lane 5). Incubation of the incised substrates with both DNA pol γ and mtDNA ligase provided efficient repair of the plasmids, as evidenced by the conversion of the HinfI fragment to an intact 46-mer (Fig. 8A, lane 6).
The progress of repair reactions was also monitored for plasmids labeled on the 3′ side of the lesion (Fig. 8B). HinfI digestion of the initial product of incision adjacent to the abasic site resulted in a fragment that comigrated with a 26-mer marker generated by HinfI/RsaI digestion of a control plasmid (Fig. 8B, lane 3). The incised AP site templates were efficiently repaired in a reaction that required both DNA pol γ and mtDNA ligase (Fig. 8B, lane 6). Additional control experiments showed that both 5′- and 3′-labeled substrates lacking damage, i.e., containing a T residue in place of the U residue, were not affected by the second stage of the repair reaction in the presence of DNA pol γ and mtDNA ligase (Fig. 8, lanes 2).
The efficient repair of AP sites in under 30 min under our in vitro conditions requires removal of 5′-dRP groups at a rate higher than that of the spontaneous loss of this labile end group. We tested mitochondrial repair enzymes for reactivity with a 5′-32P-dRP residue in an oligonucleotide substrate by using a borohydride-trapping procedure. This assay detects AP lyase activity by reducing the Schiff base intermediate formed during the attack of lysine or an N-terminal amino acid on the dRP residue (8, 29, 34, 39, 51). With this assay, we detected AP lyase activity in both the 137-kDa catalytic subunit of DNA pol γ and the 100-kDa mtDNA ligase (Fig. 9). Some labeling of proteolytic degradation products of DNA pol γ was also observed in Fig. 9. DNA pol γ is known to be highly susceptible to proteolysis (13, 15, 25). For both of these mitochondrial proteins, as well as for the positive control, Escherichia coli formamidopyrimidine glycolase (FPG) protein, cross-linking was dependent on the treatment of the U-containing oligonucleotide with UDG and on the presence of a strong reducing agent, such as NaBH4 or NaBH3CN.
FIG. 9.
DNA pol γ and mtDNA ligase contain AP lyase activity. DNA pol γ (A), mtDNA ligase (B), and, as a positive control, recombinant E. coli 8-oxo-guanine glycosylase (FPG protein) (C) (51) were tested for their ability to form a Schiff base as a presumptive intermediate in the AP lyase mechanism as described in Materials and Methods. Proteins were incubated with a 5′-32P-labeled, 5′-U-containing oligonucleotide that was either pretreated with UDG (lanes 1 and 2) or mock incubated without UDG (lanes 3 and 4). Incubations contained either 20 mM NaBH4 (lanes 1 and 3) or 20 mM NaCl (lanes 2 and 4). Proteins were fractionated by SDS-PAGE, and radioactivity labeled proteins were detected by autoradiography of the dried gel. The gel mobilities of molecular mass standards (in kilodaltons) are shown on the left.
DISCUSSION
Abasic site repair with mitochondrial proteins.
We have reconstituted a complete system for repair of abasic sites in DNA by using mitochondrial enzymes. We emphasize that this is a robust repair reaction capable of repairing a large fraction of input DNA molecules (Fig. 8). At the inception of this work, we expected that AP site repair should require at least mitochondrial AP endonuclease and DNA ligase in addition to DNA pol γ. This presented several obstacles, since neither of these accessory proteins had been well characterized in mitochondria and the ability of DNA pol γ to participate in short patch repair reactions had not been established. The following will review the current understanding of the roles of these enzymes in base excision repair in mitochondria.
DNA pol γ is a repair polymerase.
Since DNA pol γ is the only DNA polymerase identified in vertebrate mitochondria, most prior studies of this enzyme had considered it mainly as a replicative polymerase. Randahl et al. (42) demonstrated that HeLa DNA pol γ is capable of filling small gaps in a DNA substrate. However, this work did not employ precisely defined substrates, and a substantial fraction of poorly ligatable products was observed. In our preliminary experiments, we found that repair is optimized by the use of conditions that minimize strand displacement synthesis. Thus, our standard repair reactions employ a relatively high MgCl2 concentration and a reduced temperature that is physiological for Xenopus. It may be that additional accessory factors exist to reduce strand displacement replication in vivo. Our results provide a clear demonstration that DNA pol γ has the ability to participate in an efficient short patch repair process.
Identification of mtDNA ligase as a form of DNA ligase III.
Previous efforts to catalog eukaryotic DNA ligases have largely ignored mtDNA ligase. The only report of mtDNA ligase prior to our work (21) also predated the discovery that vertebrate genomes encode a variety of DNA ligases. The X. laevis mtDNA ligase has a mass of ∼100 kDa on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and is capable of using oligo(dT) · poly(rA) as a substrate. We consider that this is a legitimate mtDNA ligase, since the much larger and more abundant DNA ligase I (1, 20) is not detected in the mitochondrial lysate. This provides the ideal control to indicate that the 100-kDa DNA ligase in the mitochondrial fraction is not a nuclear contaminant.
Since the size of mtDNA was similar to that of human DNA ligase III or ligase IV, we raised antisera against peptides from the Xenopus homologs of both of these proteins. The anti-DNA ligase III antiserum reacted with the 100-kDa adenylated polypeptide in the mtDNA ligase preparation (Fig. 6), suggesting that the mtDNA ligase may be an alternative product of the DNA ligase III gene. Work is in progress to obtain protein sequence information on mtDNA ligase as an additional test of its relationship with DNA ligase III. It is interesting that analysis of cDNA clones of human (3, 58) and mouse (26) DNA ligase III reveal a potential mechanism whereby the DNA ligase III gene could supply mtDNA ligase. Both cDNA sequences encode upstream, in-frame AUG codons that were considered by the authors who reported the cDNA sequences to occur in sequence contexts unfavorable for translation initiation. The N-terminal sequences of the hypothetical polypeptides that would be produced by translation initiation at the upstream AUG sequences of mouse and human DNA ligase III are highly conserved and contain potential mitochondrial transit peptide sequences. Thus, a low rate of translation initiation from an upstream AUG codon in the DNA ligase III mRNA may be sufficient to supply the requirement for mtDNA ligase. Alternative translation initiation at the downstream AUG could provide the nuclear fraction of DNA ligase III. The human DNA ligase IV cDNA shows a similar potential mitochondrial targeting sequence upstream from the published AUG (58). One study in which indirect immunofluorescence was used to localize DNA ligase III in mouse cells did not identify this enzyme in mitochondria (26). If a form of mtDNA ligase III is directed to mitochondria in these cells, it is possible that the quantity of mitochondrial enzyme was below the limits of detection. Further experiments are required to study the relationship between mtDNA ligase and nuclear DNA ligase III in mammals and amphibians and to determine whether a form of DNA ligase IV may be directed to mitochondria in some organisms.
Which mitochondrial enzymes provide deoxyribophosphodiesterase activity?
Figure 9 shows that both DNA pol γ and mtDNA ligase react in a borohydride-trapping reaction that is routinely used to identify AP lyases (8, 34, 39, 51). The specificity of this assay may be questioned, since many enzymes can be labeled to some extent with this procedure, and there is no a priori assurance that an enzyme active in the borohydride-trapping reaction is an active AP lyase. However, our control experiments have shown that a number of proteins, including the basic mitochondrial proteins mitochondrial single-stranded-DNA binding protein and transcription factor mtTFA, as well as other DNA metabolic enzymes such as E. coli DNA ligase and Xenopus type I topoisomerase, do not react in the borohydride-trapping procedure (data not shown). The data shown in Fig. 9 are presented at this time only to identify mtDNA ligase and DNA pol γ as potential candidates for the AP lyase involved in BER in mitochondria. For both enzymes, additional experiments will be required to determine whether the apparent AP lyase activity revealed by borohydride-trapping experiments is actually required for repair. Since both DNA pol γ and mtDNA ligase bind to nicks in DNA, it may be that both enzymes contain appropriately positioned lysine residues capable of promoting β-elimination of a dRP residue at a rate sufficient to facilitate repair.
ACKNOWLEDGMENTS
We thank J. Tchou for the gift of E. coli FPG protein and S. Klein and R. Perez-Jannotti for cDNA sequence information that permitted generation of the antipeptide antiserum. We thank A. P. Grollman for critical review of the manuscript.
This research was supported by the National Institute of Environmental Health Sciences.
REFERENCES
- 1.Aoufouchi S, Prigent C, Theze N, Philippe M, Thiebaud P. Expression of DNA ligases I and II during oogenesis and early development of Xenopus laevis. Dev Biol. 1992;152:199–202. doi: 10.1016/0012-1606(92)90171-c. [DOI] [PubMed] [Google Scholar]
- 2.Backer J M, Weinstein I B. Mitochondrial DNA is a major cellular target for dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Tomkinson A E, Ramos W, Mackey Z B, Danehower S, Walter C A, Schultz R A, Besterman J M, Husain I. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton D A, Doda J N, Friedberg E C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cozzarelli N R, Gerrard S P, Schlissel M, Brown D D, Bogenhagen D F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 6.Croteau D L, apRhys C M J, Hudson A K, Dianov G L, Hansford R G, Bohr V A. An oxidative damage-specific endonuclease from rat liver mitochondria. J Biol Chem. 1997;272:29338–27344. doi: 10.1074/jbc.272.43.27338. [DOI] [PubMed] [Google Scholar]
- 7.Demple B, Herman T, Chen D S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodson M L, Michaels M L, Lloyd R S. Unified catalytic mechanism for DNA glycosylases. J Biol Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 9.Driggers W J, LeDoux S P, Wilson G L. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;288:22042–22045. [PubMed] [Google Scholar]
- 10.Gray H, Wong T W. Purification and identification of subunit structure of the human mitochondrial DNA polymerase. J Biol Chem. 1992;267:5835–5841. [PubMed] [Google Scholar]
- 11.Hegler J, Bittner D, Boiteux S, Epe B. Quantification of oxidative DNA modifications in mitochondria. Carcinogenesis. 1993;14:2309–12. doi: 10.1093/carcin/14.11.2309. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi Y, Linn S. Purification of all forms of HeLa mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J Biol Chem. 1995;270:7950–7956. doi: 10.1074/jbc.270.14.7950. [DOI] [PubMed] [Google Scholar]
- 13.Insdorf N F, Bogenhagen D F. DNA polymerase gamma from Xenopus laevis. I. The identification of a high molecular weight catalytic subunit by a novel DNA polymerase photolabeling procedure. J Biol Chem. 1989;264:21491–21497. [PubMed] [Google Scholar]
- 14.Insdorf N F, Bogenhagen D F. DNA polymerase gamma from Xenopus laevis. II. A 3′-5′ exonuclease is tightly associated with the DNA polymerase activity. J Biol Chem. 1989;264:21498–21503. [PubMed] [Google Scholar]
- 15.Kunkel T A, Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-γ. J Biol Chem. 1988;263:4450–4459. [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Larsson N-G, Clayton D A. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 18.LeDoux S P, Wilson G L, Beecham E J, Stevnsner T, Wassermann K, Bohr V A. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 19.LeDoux S P, Patton N J, Avery L J, Wilson G L. Repair of N-methylpurines in the mitochondrial DNA of xeroderma pigmentosum group D cells. Carcinogenesis. 1993;14:913–917. doi: 10.1093/carcin/14.5.913. [DOI] [PubMed] [Google Scholar]
- 20.Lepetit D, Thiebaud P, Aoufouchi S, Prigent C, Guesne R, Theze N. The cloning and characterization of a cDNA encoding Xenopus laevis DNA ligase I. Gene. 1996;172:273–277. doi: 10.1016/0378-1119(96)00175-8. [DOI] [PubMed] [Google Scholar]
- 21.Levin C J, Zimmerman S B. A DNA ligase from mitochondria of rat liver. Biochem Biophys Res Commun. 1976;69:514–520. doi: 10.1016/0006-291x(76)90551-9. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Barnes D E. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 24.Loeb L A. Apurinic sites as mutagenic intermediates. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 25.Longley M J, Mosbaugh D W. Properties of the 3′ to 5′ exonuclease associated with porcine liver DNA polymerase γ. J Biol Chem. 1991;266:24702–24711. [PubMed] [Google Scholar]
- 26.Mackey Z B, Ramos W, Levin D S, Walter C A, McCarrey J R, Tomkinson A E. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol Cell Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto Y, Bogenhagen D F. Repair of a synthetic abasic site involves concerted reactions of DNA synthesis followed by excision and ligation. Mol Cell Biol. 1991;11:4441–4447. doi: 10.1128/mcb.11.9.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto Y, Kim K, Bogenhagen D F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 30.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Mecocci P, MacGarvey U, Kaufman A E, Koontz D, Shoffner J M, Wallace D C, Beal M F. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 32.Mikhailov V S, Bogenhagen D F. Effects of Xenopus laevis mitochondrial single-stranded DNA binding protein on primer-template binding and 3′→5′ exonuclease activity of DNA polymerase γ. J Biol Chem. 1996;271:18939–18946. doi: 10.1074/jbc.271.31.18939. [DOI] [PubMed] [Google Scholar]
- 33.Myers K A, Saffhill R, O’Connor P J. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9:285–92. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 34.Nash H M, Bruner S D, Scharer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 35.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus T A, Skorpen F, Krokan H E. Nuclear and mitochondrial uracil-DNA glycosylase are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niranjan B G, Bhat N K, Avadhani N G. Preferential attack of mitochondria DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- 37.Olson M W, Kaguni L S. 3′-5′ exonuclease in Drosophila mitochondrial DNA polymerase. J Biol Chem. 1992;267:23136–23142. [PubMed] [Google Scholar]
- 37a.Perez-Janotti, R., S. Klein, and D. F. Bogenhagen. Unpublished data.
- 38.Pettepher C C, LeDoux S P, Bohr V A, Wilson G L. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- 39.Piersen C E, Prasad R, Wilson S H, Lloyd R S. Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 40.Pinz K, Shibutani S, Bogenhagen D. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J Biol Chem. 1995;270:9202–9206. doi: 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- 41.Pirsel M, Bohr V A. Methyl methanesulfonate adduct formation and repair in the DHFR gene and in mitochondrial DNA in hamster cells. Carcinogenesis. 1993;14:2105–2108. doi: 10.1093/carcin/14.10.2105. [DOI] [PubMed] [Google Scholar]
- 42.Randahl H, Elliott G C, Linn S. DNA repair reactions by purified HeLa DNA polymerases and exonucleases. J Biol Chem. 1988;263:12228–12234. [PubMed] [Google Scholar]
- 43.Richter C, Park J-W, Ames B N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robins P, Lindahl T. DNA ligase IV from HeLa cell nuclei. J Biol Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 45.Robson C N, Hickson I D. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satoh M S, Huh N-H, Rajewsky M F, Kuroki T. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem. 1988;263:6854–6856. [PubMed] [Google Scholar]
- 47.Shen C C, Wertelecki W, Driggers W J, LeDoux S P, Wilson G L. Repair of mitochondrial DNA damage induced by bleomycin in human cells. Mutat Res. 1995;337:12–23. doi: 10.1016/0921-8777(95)00008-8. [DOI] [PubMed] [Google Scholar]
- 48.Shigenaga M D, Hagen T M, Ames B N. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slupphaug G, Markussen F H, Olsen L C, Aasland R, Aarsaether N, Bakke O, Krokan H E, Helland D E. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–84. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyderwine E G, Bohr V A. Gene- and strand-specific damage and repair in Chinese hamster ovary cells treated with 4-nitroquinoline 1-oxide. Cancer Res. 1992;52:4183–4189. [PubMed] [Google Scholar]
- 51.Tchou J, Grollman A P. The catalytic mechanism of Fpg protein. J Biol Chem. 1995;270:11671–11677. doi: 10.1074/jbc.270.19.11671. [DOI] [PubMed] [Google Scholar]
- 52.Tomkinson A E, Bonk R T, Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 53.Tomkinson A E, Bonk R T, Kim J, Bartfield N, Linn S. Mammalian mitochondrial endonuclease activities specific for ultraviolet-irradiated DNA. Nucleic Acids Res. 1990;18:929–935. doi: 10.1093/nar/18.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torri A, Englund P. A DNA polymerase beta in the mitochondrion of the trypanosomatid Crithidia fasciculata. J Biol Chem. 1995;270:3495–3497. doi: 10.1074/jbc.270.8.3495. [DOI] [PubMed] [Google Scholar]
- 55.Wallace D C. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 56.Wallace D C. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 57.Wallace D C. Mitochondrial DNA variation in human evolution, degenerative disease, and aging. Am J Hum Genet. 1995;57:201–223. [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Y-F, Robins P, Carter K, Caldecott K, Pappin D J C, Yu G-L, Wang R-P, Shell B K, Nash R A, Schar P, Barnes D E, Haseltine W A, Lindahl T. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye F, Carrodeguas J A, Bogenhagen D F. The γ subfamily of DNA polymerases: cloning of a developmentally regulated cDNA encoding Xenopus laevis mitochondrial DNA polymerase γ. Nucleic Acids Res. 1996;24:1481–1488. doi: 10.1093/nar/24.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]