Abstract
Amyloid diseases encompass >20 medical disorders that include amyloid protein A (AA) amyloidosis, Alzheimer's disease, and type 2 diabetes. A common feature of these conditions is the selective organ deposition of disease-specific fibrillar proteins, along with the sulfated glycosaminoglycan, heparan sulfate. We have generated transgenic mice that overexpress human heparanase and have tested their susceptibility to amyloid induction. Drastic shortening of heparan sulfate chains was observed in heparanase-overproducing organs, such as liver and kidney. These sites selectively escaped amyloid deposition on experimental induction of inflammation-associated AA amyloidosis, as verified by lack of material staining with Congo Red, as well as lack of associated polysaccharide, whereas the same tissues from control animals were heavily infiltrated with amyloid. By contrast, the spleens of transgenic mice that failed to significantly overexpress heparanase contained heparan sulfate chains similar in size to those of control spleen and remained susceptible to amyloid deposition. Our findings provide direct in vivo evidence that heparan sulfate is essential for the development of amyloid disease.
Keywords: inflammation, transgenic mice, endo-glucuronidase, Congo red staining
Amyloid diseases are characterized by deposition of disease-specific fibrillar proteins (amyloid) in various organs, leading to loss of function and to associated clinical symptoms. It has been proposed that heparan sulfate (HS) may facilitate formation of the nidus and/or protofilament around which amyloid fibrillogenesis takes place and impart stability to the amyloid fibril in vivo (1, 2). This notion was supported by the marked increase in fibrillogenesis in vitro exerted by heparin/HS on various amyloidogenic polypeptides, including Aβ and phosphorylated tau (Alzheimer's disease), α-synuclein (Parkinson's disease), islet amyloid polypeptide (IAPP) (type 2 diabetes), and β2-microglobulin (chronic hemodialysis-related amyloid). Aβ fibrillogenesis is precluded by agents that block Aβ:HS interactions (3). The in vivo evidence is circumstantial, primarily observations of codistributed HS proteoglycan (HSPG), notably perlecan, and amyloidogenic peptide in amyloid fibrils (2).
Heparanase is a mammalian endo-β-d-glucuronidase that cleaves HS at a limited number of sites. Cloning of the human heparanase cDNA by several groups suggests that a single dominant HS-degrading endoglycosidase is expressed in mammalian cells (4, 5). Our recent generation of transgenic mice overexpressing heparanase in various tissues revealed that the enzyme plays a role in diverse processes such as embryonic implantation, mammary gland morphogenesis, hair follicle growth, and tissue repair (6). In the present study, the differential expression of heparanase has been exploited to assess the role of HS in amyloid protein A (AA) amyloid generation in vivo. The results demonstrate that overexpression of heparanase, resulting in fragmentation of HS chains, affords protection against amyloidosis.
Materials and Methods
Animals and Amyloid Induction. The homozygous mouse strain overexpressing human heparanase (hpa-tg) and the respective control (ctr) mice (C57BL background) were generated as described (6) and maintained in the animal facility of the Biomedical Center, Uppsala University. The animal experiments were performed in compliance with Swedish legislation for animal welfare (approval number C176/2). Amyloidosis was induced as described (3). Briefly, amyloid enhancing factor (AEF), prepared as AA amyloid fibrils (7), was administered i.p. (200 μg per mouse, 200 μl, n = 6 hpa-tg and n = 6 ctr male mice, 10 weeks old). Immediately after the administration of AEF, 0.5 ml of AgNO3 (2% solution) was injected s.c. into the loose tissue of the back, between the shoulder blades. Mice were killed by cervical dislocation 7 days after commencement of the induction protocol. Spleens, livers, and kidneys were dissected from each animal, fixed overnight in a solution containing 96% ethanol, 1% glacial acetic acid, and 3% distilled water, and stored in 70% ethanol until processed for histological analysis.
Histochemical Analyses. Tissues in 70% ethanol were dehydrated by using standard procedures and embedded in paraffin. Sections of 8-10 μm were stained with Congo red (8) to detect amyloid deposition and with sulfated Alcian blue (SAB) (9) to detect sulfated glycosaminoglycans. The percent tissue area occupied by birefringent Congo red-positive staining in polarized light was determined as described (3) by image analysis by using a program and apparatus from mcid m2 Imaging Research (St. Catherine's, ON, Canada). All comparisons were made after calibrating the apparatus against a set of standard spleen sections containing AA amyloid.
Immunohistochemical Detection of Heparanase. The analysis was performed as described with minor modifications (6, 10, 11). Briefly, 8- to 10-μm sections were deparaffinized and rehydrated. The tissue was then denatured for 3 min in a microwave oven in citrate buffer (0.01 M, pH 6.0). Blocking steps included successive incubations in 3% H2O2 in methanol and 5% goat serum. Tissue sections were incubated with anti-human heparanase antibodies (mAb 130) or with DMEM supplemented with 3.3% horse serum as control, followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibodies (The Jackson Laboratory). Color was developed by using Zymed AEC substrate kit (Zymed) for 10 min, followed by counter staining with Mayer's hematoxylin. The monoclonal mouse anti-human heparanase antibodies (mAb 130) are directed against the C terminus of the 50-kDa heparanase subunit and were produced as described (11). These antibodies do not recognize the mouse heparanase and were kindly provided by InSight Ltd (Rehovot, Israel).
In Vivo Radiolabeling and Purification of HS. Hpa-tg and ctr mice (male, 10 weeks old) were injected i.p. with 0.5 mCi of Na35SO4 (1 Ci = 37 GBq) (Amersham Pharmacia Biosciences) and maintained for 2 h with free access to water and food. The animals were killed by cervical dislocation, and various organs (liver, kidney, spleen, lung, heart, and brain) were dissected. The heparan sulfate was isolated as described (12). Briefly, the organs were cut into small pieces and homogenized in Tris·HCl (50 mM, pH 7.4) extraction buffer containing 4 M urea and 1% Triton X-100 on ice. The homogenates were incubated at 4°C overnight with mild agitation and centrifuged at 2,800 × g for 15 min. The supernatant was mixed with 4 M NaOH to a final concentration of 0.5 M and incubated at 4°C overnight. After neutralization with HCl, the samples were adjusted to 100 mM salt concentration by dilution with the Tris·HCl extraction buffer and applied on a DEAE-Sephacel column (2 ml) equilibrated in the same buffer. Columns were washed with acetate buffer (50 mM, pH 4.5) containing 4 M urea until there was no detectable radioactivity in the effluent. Elution was carried out by using the acetate buffer containing 1.5 M NaCl and 4 M urea, pH 4.5. The eluted radiolabeled material was pooled and desalted on a PD-10 column (Amersham Pharmacia Biosciences) in 10% ethanol and lyophilized. The samples were treated with chondroitinase ABC (1 unit/ml) (Seikagaku, Tokyo) at 37°C overnight, and the free HS chains were recovered by purification on a 1-ml DEAE-Sephacel column connected to an HPLC system (12).
Analysis of HS. Purified metabolically 35S-labeled free HS chains were desalted on a PD-10 column (10% ethanol) and lyophilized. For identification of the products, samples of 5,000 cpm were incubated with bacterial heparinase (1 milliunit) and heparitinase (1 milliunit) (Seikagaku) in 100 μl of 50 mM Tris·HCl buffer (pH 7.2) in the presence of 1.6 mM CaCl2 and 0.005% BSA for 4 h at 37°C, and the digests were analyzed by gel chromatography on a Superose-12 column. For size analysis, samples (≈ 5,000 cpm in a volume of 100 μl) were applied to a similar column (Amersham Pharmacia Biosciences) equilibrated in Tris·HCl buffer (50 mM, pH 7.4) containing 1 M NaCl and 0.1% Triton X-100. Effluent fractions of 0.5 ml were collected and analyzed after addition of OptiPhase HiSafe 3 in a Beckman scintillation counter (12).
Northern Blot Analysis. One hpa-tg and one ctr mouse (10 weeks old) were killed by cervical dislocation. The organs were dissected and frozen immediately in liquid nitrogen. Total RNA was prepared from the different organs by using either TRIzol or the SV Total RNA Isolation System (Promega) according to the manufacturer's instructions. Total RNA (25 μg, estimated by OD260) was separated by electrophoresis in a 1.2% denaturing agarose gel containing 0.66 M formaldehyde. After blotting onto a Hybond NX membrane (Amersham Pharmacia Biosciences) according to standard procedures, the RNA was hybridized with a purified PCR fragment, corresponding to nucleotides 175-734 of the mouse heparanase cDNA, which was labeled with [32P]dCTP by using the Ready-To-Go kit (Amersham Pharmacia Biosciences). Hybridization was carried out at 68°C in ExpressHyb solution (Clontech) as described in the user's manual, with extensive prehybridization (4-16 h). To check the loaded amount of RNA, the blots were reprobed with a MTN β-actin probe from Clontech. The blots were exposed onto Phosphor-Imager screens and scanned in a Fuji scanner. Image analysis was carried out by using image gauge.
Results
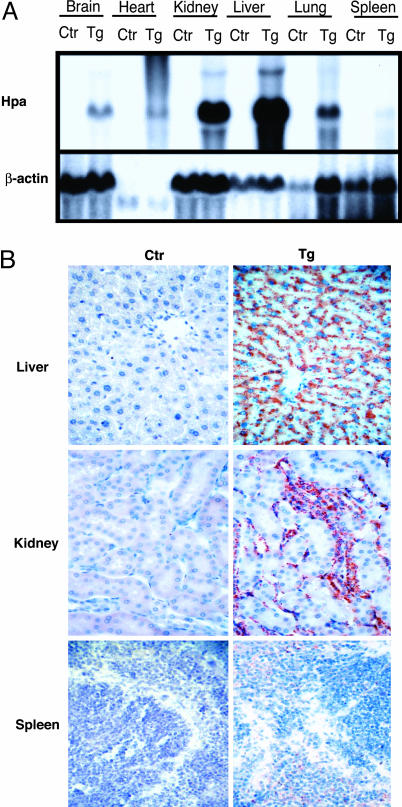
Expression of Heparanase mRNA and Protein. Despite a marked reduction in overall HS chain length, the heparanase transgenic (hpa-tg) mice seemed healthy, were fertile, and had a normal life span (6). Heparanase overexpression showed consistent organ selectivity, and this trait was exploited in the present study to investigate the pathophysiological role of HS in AA amyloidogenesis in vivo. The expression pattern of heparanase transcripts, determined by Northern blot analysis (Fig. 1A), was found to correlate well with immunohistochemical staining of the heparanase protein (Fig. 1B) and with enzyme activity measurements in various organs (6). Although liver and kidney showed the highest expression levels, spleen of the hpa-tg mice differed only marginally from that of the ctr mice and showed little or no expression of human heparanase (Fig. 1 A and B). The amounts of mRNA obtained from heart were substantially smaller than those from other organs, judging from the low levels of β-actin signal, yet clearly revealed overexpression in the hpa-tg mouse.
Fig. 1.
Heparanase mRNA and protein in different organs of hpa-tg vs. ctr mice. (A) Total RNA was hybridized with a 560-bp fragment of the mouse heparanase cDNA (Upper) and reprobed with a MTN β-actin probe (Lower). Overexpression of human heparanase is apparent in all organs analyzed, although at markedly different levels. Only a slight overexpression was noted in the spleen. Hpa, heparanase. (B) Immunohistochemical staining with antihuman heparanase antibody (mAb 130). Abundant heparanase protein expression (reddish color) is seen in the liver and kidney, but not in the spleen, of the hpa-tg mice (magnification ×40).
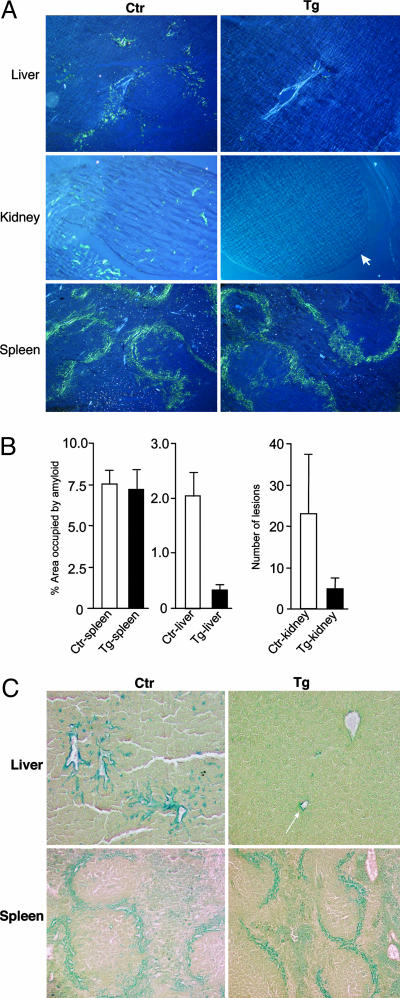
Amyloid and Glycosaminoglycan Deposition in Different Organs. A widely used experimental model for amyloid induction (3) was applied to hpa-tg and ctr mice of the same genetic background. Mice (six hpa-tg and six ctr) received a single i.p. injection of amyloid-enhancing factor (AEF), likely the nidus for AA amyloid (13) along with a s.c. injection of AgNO3 as an inflammatory stimulus (see Materials and Methods). Seven days later, the organs were collected and examined for amyloid deposition by staining with Congo red. Representative sections of liver, kidney, and spleen (i.e., the preferred predilection targets in AA amyloidosis) are shown in Fig. 2A. In ctr mice, substantial amyloid deposits were found around the central veins of the liver, in the renal papillae, and in the perifollicular areas of the spleen. In contrast, the hpa-tg mice showed virtually no amyloid in the kidney or the liver, whereas the perifollicular splenic amyloid seemed similar in hpa-tg and ctr mice. These observations were substantiated by image analysis, which showed the mean spleen area occupied by amyloid in both wild-type and transgenic groups of mice to be ≈7.5% (Fig. 2B). In contrast, the mean liver areas occupied by AA amyloid in the wild-type (≈2.0% of area) and transgenic groups (≈0.4%) differed considerably (P = 0.0013). A similar 4- to 5-fold difference was noted in the kidneys. The renal amyloid was localized exclusively to the papillae (the only site of kidney amyloidogenesis at this early stage of induction), where the number of lesions per high-powered field were 23.1 ± 14.0 and 4.9 ± 2.3 in the wild-type and transgenic animals, respectively (Fig. 2B). Comparison of Congo red staining for amyloid (Fig. 2 A) with immunohistochemical staining for heparanase (Fig. 1B), in adjacent serial tissue sections, revealed an inverse correlation between heparanase overexpression and amyloid formation. The amyloid-resistant organs (liver and kidney) of the hpa-tg mice displayed abundant heparanase, as opposed to the very low expression levels in the amyloid-susceptible spleen of the same animals. Similar results were seen in all six animals of each group.
Fig. 2.
Visualization of amyloid and glycosaminoglycan deposition in different organs of hpa-tg vs. ctr mice. (A) Sections stained for amyloid with Congo red, viewed under crossed-polars, show representative areas of liver (Top), kidney (Middle), and spleen (Bottom). Seven days after amyloid induction, ctr mice had developed amyloidosis (green-stained deposits) in all three tissues (Left), whereas the liver of hpa-tg mice showed barely detectable amyloid, and the kidney of hpa-tg mice showed no detectable amyloid deposition (Top Right and Middle Right). In contrast, the hpa-tg spleen (Bottom Right) showed as abundant amyloid as the ctr tissue. The light-blue objects in the liver sections are blood vessels. (Middle Right) The arrow indicates the margin of the renal papilla (magnification ×115). (B) Image analysis of amyloid deposition, after staining with Congo red. Ctr, control; Tg, heparanase transgenic. (C) Sections stained with SAB reveal codeposition of glycosaminoglycans in amyloid deposits in the liver (Upper) and spleen (Lower) of hpa-tg vs. ctr mice. Both the liver (area around the central vein and adjacent liver parenchyma) and spleen (perifollicular area) of ctr mice (Left) are strongly stained. Essentially no staining is detected in the hpa-tg liver (Upper Right; arrow indicates small focus of AA amyloid) as opposed to an intense staining of the hpa-tg spleen (Lower Right), similar to that of the ctr spleen (magnification ×115).
Codeposition of AA amyloid and HS may also be demonstrated by staining tissue sections with SAB, a dye used to visualize sulfated glycosaminoglycans (9). Although liver from amyloid-induced ctr mice showed abundant SAB staining, primarily around central veins and extending into the liver sinusoids (Fig. 2C), sites coinciding with Congo red staining (Fig. 2 A), the transgenic hpa-tg liver was almost devoid of SAB-positive material (Fig. 2C). Spleen sections, on the other hand, stained strongly with SAB in perifollicular patterns (Fig. 2C) matching the deposition of AA amyloid (Fig. 2A), and with no discernible difference between ctr and hpa-tg tissue. The amyloid-associated SAB-staining material of mouse spleen was previously identified as HS by its sensitivity to nitrous acid treatment (14, 15) and to HS lyase digestion, but not to chondroitinase ABC digestion (16). We conclude that the fragmented HS resulting from heparanase overexpression failed to support amyloidogenesis, hence secondary associated accumulation of polysaccharide.
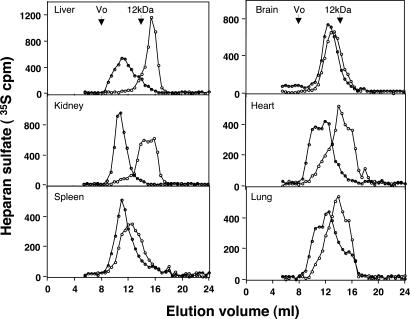
Molecular Size of HS Extracted from Different Organs. The predicted effect of heparanase overexpression on HS molecular size was verified by in vivo labeling of the polysaccharide with [35S]sulfate, followed by isolation of HS from the various organs and gel chromatography (see Materials and Methods). Notably, the total amounts of 35S-labeled HS, relative to the wet weight of each organ, did not differ significantly between hpa-tg and ctr mice (data not shown). Gel chromatography, however, revealed a pronounced reduction in the average chain length of [35S]HS isolated from organs (i.e., liver and kidney) in which heparanase was markedly overexpressed (Fig. 3). Thus, liver HS was reduced in size from an average ≈35 kDa (ctr) to ≈3 kDa (hpa-tg), and renal HS was degraded to products largely <10 kDa. In contrast, HS in spleen and brain, both organs with minor expression of transgenic heparanase (Fig. 1), was only marginally affected, as indicated by the essentially overlapping elution patterns of HS derived from hpa-tg and ctr mice (Fig. 3).
Fig. 3.
Molecular size of HS purified from hpa-tg and ctr organs. HS samples, labeled in vivo with 35S were isolated from the indicated organs as described in Materials and Methods. The labeled polysaccharides were susceptible to digestion with heparin lyases (data not shown). Analysis by gel chromatography on a Superose-12 column showed a pronounced reduction in the size of HS extracted from hpa-tg (open circles) vs. ctr (filled circles) liver and kidney. A significant decrease in the size of HS was noted in the heart and lung, as compared with a marginal decrease in the spleen and brain of the hpa-tg vs. ctr mice. Arrows mark the peak elution position of a standard 12-kDa heparin.
Discussion
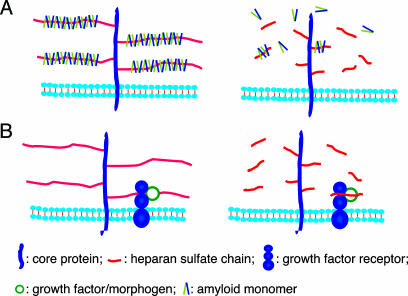
Application of hpa-tg mice in the present study provided straightforward evidence for an essential role of HS in AA amyloidosis. The variable overexpression of heparanase in different organs provided a reliable internal control. Unlike most organs, the spleen essentially escaped excessive heparanase action, retained almost full-size HS chains, and remained susceptible to amyloid development. The lack of a significant amyloid deposition in the liver and kidney, both expressing high levels of heparanase, can therefore be confidently ascribed to the marked decrease in the molecular size of HS observed in these tissues. Interestingly, the spleen of control mice is the first organ affected by AA amyloid, followed by the liver and then the kidney. Also, a higher amount of AA amyloid is deposited in spleen vs. the liver (17). Formation of amyloid fibrils/plaques seems to proceed through intermediate stages of polymerization that are thought to depend on interactions with HS structures (1). We propose that this process critically depends on the macromolecular state of the HS constituent, and that a minimal chain length is a prerequisite for efficient fibril polymerization and deposition in vivo (Fig. 4A). Free HS oligosaccharides generated by heparanase can presumably bind amyloid monomers, which are then unable to polymerize and assemble into larger aggregates. Conversely, infusion in rat brain of perlecan-carrying full-size HS chains was shown to promote deposition of Aβ amyloid, the hallmark of Alzheimer's disease (18). These observations point toward a therapeutic potential for oligosaccharides with the appropriate structure. Indeed, low-molecular weight heparin, with a molecular size (≈5 kDa) similar to the HS fragments extracted from heparanase-overexpressing organs, was found to prevent AA and Aβ-peptide fibril formation in vitro, and to arrest the progression of AA amyloidosis in mice (19). Further, agents (such as small-molecule sulfonates and sulfates) that inhibit the binding of HS to amyloid precursor protein/peptide are effective anti-amyloid compounds both in vivo and in vitro (3, 20). We also demonstrated that degradation of HS chains, through exposure to bacterial heparinase III, markedly inhibits conversion of cellular prion protein (PrPc) into an abnormal conformer (PrPSc) in cultured neuroblastoma cells (21). A similar effect was obtained in response to inhibition of HS chain elongation and/or sulfation. Moreover, partial restoration of PrPSc deposition was induced by exogenously added HS (21), further emphasizing the contributory involvement of HS in amyloid disorders. Precise characterization of HS/protein interactions at the molecular level may provide clues to the generation of drugs against amyloid diseases.
Fig. 4.
Proposed models of HS function in normal and heparanase-overexpressing tissues. (A) Polyvalent interaction. Efficient polymerization and deposition of amyloid peptides is promoted by intact HS (Left), but not by fragments of HS generated upon cleavage of HS by heparanase (Right). (B) Monomeric/oligomeric interactions with proteins. HS side chains of a cell-surface HS proteoglycan bind a ligand (e.g., growth factor, morphogen) and its receptor. Complex formation (here shown to involve a monomeric receptor only) is promoted by the intact HS chain (Left) as well as by HS fragments released by heparanase (Right).
Our findings have bearing on the structure-function relationships of HS in development and homeostasis. Targeted disruption of genes encoding various enzymes involved in HS biosynthesis has demonstrated that HS is essential for normal embryonic development (12, 22-24). The multiple regulatory functions of HS are generally ascribed to interactions with a variety of bioactive proteins, including growth factors and their receptors, morphogens, cytokines, enzymes, and extracellular-matrix proteins (25, 26). Molecular studies suggest that the majority of such interactions can be accommodated within a 10-mer saccharide sequence (25, 26). A putative 10-mer “functional unit” of an HS chain would have a Mr of ≈3,000, thus approaching the size of HS fragments extracted from several organs of the hpa-tg mice (Fig. 3). The relatively modest phenotypic alterations observed in these mice (6) could therefore conceivably reflect the functional competence of such fragments (Fig. 4B), in accord with crystallography data (27, 28). The same fragments, however, are too short to promote assembly of amyloid aggregates that are critical to the pathogenesis of amyloid diseases.
Finally, it is recalled that HS has been implicated in other major diseases involving amyloid formation, such as Alzheimer's disease, type 2 diabetes, and Parkinson's disease. We predict that these conditions may be alleviated by means of appropriately depolymerized heparin/HS-related compounds. Indeed, treatment of APP-transgenic mice (a model of Alzheimer's disease) with low-molecular-weight heparin (Mr ≈ 5,000) attenuated plaque formation and improved neuropathology of the animals (19, 29). It is important to establish whether Aβ aggregation in Alzheimer's disease, as well as amyloid generation in other conditions, depends on HS chain length as shown here for AA amyloidosis. Current studies on mice that overexpress a secreted form of heparanase in the brain may provide information to verify this prediction.
Acknowledgments
We thank Mr. Lee Boudreau for able technical assistance. This work was supported by Swedish Research Council Grant 32X-15023, Swedish Cancer Society Grant 4708-B02-01XAA, European Commission Grant QLK3-CT-2002-02049, Swedish Foundation for Strategic Research Grant A303:156e, Polysackaridforskning AB (Uppsala), the Center for the Study of Emerging Diseases (CSED), Israel Science Foundation Grant 532/02, Canadian Institutes of Health Research Grant MOP-3153, and a Detweiler Traveling Fellowship from the Royal College of Physicians and Surgeons of Canada.
Author contributions: J.-P.L., U.L., and R.K. designed research; M.L.E.G., F.G., and X.Z. performed research; E.Z. and S.M. contributed new reagents/analytic tools; and J.-P.L., R.K., I.V., and U.L. wrote the paper.
Abbreviations: AA, amyloid protein A; HS, heparan sulfate; ctr, control; hpa-tg, heparanase transgenic; SAB, sulfated Alcian blue.
References
- 1.Snow, A. D., Willmer, J. & Kisilevsky, R. (1987) Lab. Invest. 57, 687-698. [PubMed] [Google Scholar]
- 2.Kisilevsky, R. & Fraser, P. E. (1997) Crit. Rev. Biochem. Mol. Biol. 32, 361-404. [DOI] [PubMed] [Google Scholar]
- 3.Kisilevsky, R., Lemieux, L. J., Fraser, P. E., Kong, X., Hultin, P. G. & Szarek, W. A. (1995) Nat. Med. 1, 143-148. [DOI] [PubMed] [Google Scholar]
- 4.Vlodavsky, I. & Friedmann, Y. (2001) J. Clin. Invest. 108, 341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parish, C. R., Freeman, C. & Hulett, M. D. (2001) Biochim. Biophys. Acta 1471, M99-M108. [DOI] [PubMed] [Google Scholar]
- 6.Zcharia, E., Metzger, S., Chajek-Shaul, T., Aingorn, H., Elkin, M., Friedmann, Y., Weinstein, T., Li, J. P., Lindahl, U. & Vlodavsky, I. (2004) FASEB J. 18, 252-263. [DOI] [PubMed] [Google Scholar]
- 7.Baltz, M., Caspi, D., Hind, C., Feinstein, A. & Pepys, M. (1986) in Amyloidosis, eds. Glenner, G., Osserman, E., Benditt, E., Calkins, E., Cohen, A. & Zucker-Franklin, D. (Plenum, New York), pp. 115-121.
- 8.Puchtler, H., Sweat, F. & Levine, M. (1962) J. Histochem. Cytochem. 10, 355-364. [Google Scholar]
- 9.Lendrum, A. C., Slidders, W. & Fraser, D. S. (1972) J. Clin. Pathol. 25, 373-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedmann, Y., Vlodavsky, I., Aingorn, H., Aviv, A., Peretz, T., Pecker, I. & Pappo, O. (2000) Am. J. Pathol. 157, 1167-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlodavsky, I., Friedmann, Y., Elkin, M., Aingorn, H., Atzmon, R., Ishai-Michaeli, R., Bitan, M., Pappo, O., Peretz, T., Michal, I., et al. (1999) Nat. Med. 5, 793-802. [DOI] [PubMed] [Google Scholar]
- 12.Li, J.-P., Gong, F., Hagner-McWhirter, Å., Forsberg, E., Åbrink, M., Kisilevsky, R., Zhang, X. & Lindahl, U. (2003) J. Biol. Chem. 278, 28363-28366. [DOI] [PubMed] [Google Scholar]
- 13.Kisilevsky, R., Gruys, E. & Shirahama, Y. (1995) Amyloid 2, 128-133. [Google Scholar]
- 14.Snow, A. D. & Kisilevsky, R. (1985) Lab. Invest. 53, 37-44. [PubMed] [Google Scholar]
- 15.Snow, A. D., Bramson, R., Mar, H., Wight, T. N. & Kisilevsky, R. (1991) J. Histochem. Cytochem. 39, 1321-1330. [DOI] [PubMed] [Google Scholar]
- 16.Elimova, E., Kisilevsky, R., Szarek, W. A. & Ancsin, J. B. (2004) FASEB J. 18, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 17.Kisilevsky, R., Boudreau, L. & Foster, D. (1983) Lab. Invest. 48, 60-67. [PubMed] [Google Scholar]
- 18.Snow, A. D., Sekiguchi, R., Nochlin, D., Fraser, P., Kimata, K., Mizutani, A., Arai, M., Schreier, W. A. & Morgan, D. G. (1994) Neuron 12, 219-234. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, H., Yu, J. & Kindy, M. S. (2001) Mol. Med. 7, 517-522. [PMC free article] [PubMed] [Google Scholar]
- 20.Kisilevsky, R., Szarek, W. A., Ancsin, J., Bhat, S., Li, Z. & Marone, S. (2003) J. Mol. Neurosci. 20, 291-297. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Zaken, O., Tzaban, S., Tal, Y., Horonchik, L., Esko, J. D., Vlodavsky, I. & Taraboulos, A. (2003) J. Biol. Chem. 278, 40041-40049. [DOI] [PubMed] [Google Scholar]
- 22.Bullock, S. L., Fletcher, J. M., Beddington, R. S. & Wilson, V. A. (1998) Genes Dev. 12, 1894-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, X., Wei, G., Shi, Z., Dryer, L., Esko, J. D., Wells, D. E. & Matzuk, M. M. (2000) Dev. Biol. 224, 299-311. [DOI] [PubMed] [Google Scholar]
- 24.Ringvall, M., Ledin, J., Holmborn, K., van Kuppevelt, T., Ellin, F., Eriksson, I., Olofsson, A. M., Kjellen, L. & Forsberg, E. (2000) J. Biol. Chem. 275, 25926-25930. [DOI] [PubMed] [Google Scholar]
- 25.Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J. & Zako, M. (1999) Annu. Rev. Biochem. 68, 729-777. [DOI] [PubMed] [Google Scholar]
- 26.Esko, J. D. & Lindahl, U. (2001) J. Clin. Invest. 108, 169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiGabriele, A. D., Lax, I., Chen, D. I., Svahn, C. M., Jaye, M., Schlessinger, J. & Hendrickson, W. A. (1998) Nature 393, 812-817. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrini, L. (2001) Curr. Opin. Struct. Biol. 11, 629-634. [DOI] [PubMed] [Google Scholar]
- 29.Bergamaschini, L., Rossi, E., Storini, C., Pizzimenti, S., Distaso, M., Perego, C., De Luigi, A., Vergani, C. & De Simoni, M. G. (2004) J. Neurosci. 24, 4181-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]