Abstract
It is well established that exonic sequences contain regulatory elements of splicing that overlap with coding capacity. However, the conflict between ensuring splicing efficiency and preserving the coding capacity for an optimal protein during evolution has not been specifically analyzed. In fact, studies on genomic variability in fields as diverse as clinical genetics and molecular evolution mainly focus on the effect of mutations on protein function. Synonymous variations, in particular, are assumed to be functionally neutral both in clinical diagnosis and when measuring evolutionary distances between species. Using the cystic fibrosis transmembrane conductance regulator (CFTR) exon 12 splicing as a model, we have established that about one quarter of synonymous variations result in exon skipping and, hence, in an inactive CFTR protein. Furthermore, comparative splicing evaluation of mammalian sequence divergences showed that artificial combinations of CFTR exon 12 synonymous and nonsynonymous substitutions are incompatible with normal RNA processing. In particular, the combination of the mouse synonymous with the human missense variations causes exon skipping. It follows that there are two sequential levels at which evolutionary selection of genomic variants take place: splicing control and protein function optimization.
Keywords: composite exonic regulatory elements of splicing, exonic splicing regulatory sequences, molecular evolution, synonymous variations
Since the first original observations that nucleotide changes in the coding regions can affect normal and alternative cellular pre-mRNA processing (1, 2), extensive evidence has accumulated that exonic variants may affect pre-mRNA splicing (3-6). Mutations within exons are responsible of aberrant splicing profiles of pre-mRNA in several human disease genes, including ataxia telangiectasia mutated (ATM) (7), SMN2 (8-10), BRCA1 (11), neurofibromin 1 (NF-1) (12), cystic fibrosis transmembrane regulator (CFTR) (13-15) genes and in some viral systems such as HIV-1 (16). Nonsense, missense, and even synonymous mutations can induce aberrant skipping of the mutant exon producing nonfunctional proteins. Because direct analysis of mRNA is not routinely performed in the diagnostic field, it is possible that a high amount of exonic mutations may unexpectedly affect splicing, as reported in ATM (7), CFTR (13-15), and NF-1 (12) genes.
In the pre-mRNA of higher eukaryotes, the correct identification of exonic sequences from the larger introns requires conserved discrete sequence elements located at the 5′ and 3′ splice sites and the branch point. In addition, exonic splicing elements that interact with several classes of positive and negative trans-acting splicing regulatory factors contribute significantly to constitutive and alternative splicing regulation (3, 17). The sequence composition of exonic splicing regulatory sequences is highly degenerated (3, 18-21), and their effect on splicing may be significantly affected by the context (13, 14) and/or depend on the formation of RNA secondary structures (22, 23). The application of computational strategies indicates that exonic splicing regulatory sequences may be remarkably common in the exons of higher eukaryotes (20, 24, 25).
Notwithstanding the proven presence of exonic regulatory elements of splicing that overlap with coding capacity, a great many studies on genomic variability in fields as diverse as clinical genetics and molecular evolution mainly focus on the effect of mutations on protein function. In fact, the potential effect of synonymous and nonsynonymous substitutions on the splicing efficiency has never been systematically analyzed.
In clinical diagnosis, synonymous variations have been routinely classified as innocuous polymorphisms and are assumed to be functionally neutral. A well known exception is for synonymous changes that directly affect the splice sites, such as those that create a consensus splice site inside the exon (26, 27) or occur in the last nucleotide of the exon (28).
In molecular evolution, the presumed neutrality of synonymous variations is the key assumption in the measurement of the evolutionary distances between species through the ratio of the rate of nonsynonymous (Ka) and synonymous (Ks) substitutions. Low Ka/Ks ratios indicate purifying selection, values of 1 suggest neutral evolution, and greater values indicate positive selection. However, recent comparative studies between human and chimpanzee transcripts have shown that a significant proportion of silent sites in protein coding regions are under purifying selection (29) and that, in humans, the average substitution rate of silent sites is lower in functional genes than in pseudogenes (30). These statistical analyses indicate the need of reconsidering the apparent neutrality of synonymous mutations.
In the human CFTR exons 9 and 12, we have previously defined the composite exonic regulatory elements of splicing (CERES) (13, 14). They are short, context-dependent 6- to 8-nt sequences in which natural and site-directed mutants may increase or decrease the exon splicing efficiency (14, 31, 32). Skipping of these CFTR exons removes a highly conserved region encoding part of the first nucleotide binding fold of CFTR, rendering the protein nonfunctional (33). Depending on the extent of exon skipping and thus on the residual levels of normal transcript, splicing mutations in CFTR may result in severe cystic fibrosis or less severe phenotypes, such as male sterility resulting from congenital absence of vas deferens, bronchiectasia, or pancreatitis (14, 31, 32). In particular, because even mild CFTR defects cause male sterility, splicing CFTR mutations with partial exon skipping may have an evolutionary disadvantage and may thus be expected to be eliminated by negative selection.
Here, we have extensively evaluated the contribution of exonic synonymous and nonsynonymous changes on the splicing efficiency along the whole mammalian CFTR exon 12. Our results indicate that synonymous changes cannot evolve freely, because they are significantly constrained by splicing requirement, and that the composition of exons during evolution is selected to ensure accurate and efficient splicing, an essential requirement to maintain optimal protein-coding capacity.
Materials and Methods
GenBank Accession Numbers for CFTR Exon 12 Nucleotide Sequences. Accession numbers and Taxa are listed in parentheses: human (GenBank accession no. AJ574986, Homo sapiens), baboon (GenBank accession no. AF162413, Papio hamadryas anubis); chimpanzee (GenBank accession no. AC090112, Pan troglodytes); lemur (GenBank accession no. AC123543, Lemur catta); horse (GenBank accession no. AC124906, Equus caballus); pig (GenBank accession no. AY585334, Sus scrofa); cow (GenBank accession no. M76128, Bos taurus); sheep (GenBank accession no. AF022453, Ovis aries); mouse (GenBank accession no. AF162137, Mus musculus); rat (GenBank accession no. M89906, Rattus norvegicus); cat (GenBank accession no. AC091382, Felis catus); and dog (GenBank accession no. AC091119, Canis familiaris); ferret (GenBank accession no. S82688, Mustela putorius furo).
Hybrid Minigene Constructs. The natural and artificial point mutations were introduced in the previously described WTB minigene (14) between the AccI and BamHI sites, which were substituted with the appropriate AccI-BamHI cassettes created by PCR-mediated site-directed mutagenesis and verified by sequencing. The sequences of the oligonucleotides are available upon request.
Analysis of the Hybrid Minigene Expression. Transient transfection of Hep3B cells, RNA extraction, RT-PCR, and quantitation of PCR products were done as described (14). PCRs were optimized to remain in the exponential range of amplification and products were routinely fractionated in 1.5% (wt/vol) agarose gel. For quantitation of the PCRs, [α-32P]dCTP was included in the PCR mixture, the products were loaded on 5% denaturing polyacrylamide/8 M urea gel, dried, and exposed to a Cyclone Instant Imager. The counts of each splicing band were corrected by the number of C/G present in the PCR product sequence.
Results
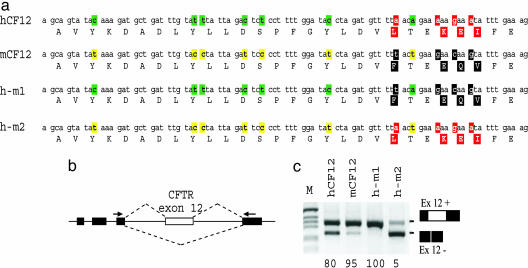
Splicing Constraint of Synonymous and Nonsynonymous Differences Between Human and Mouse CFTR Exon 12. To evaluate the contribution of exonic synonymous and nonsynonymous site divergences on the splicing efficiency along the whole exon, we have functionally compared human and mouse CFTR exon 12 sequences. The former presents a variable degree of aberrant exon 12 skipping, whereas the mouse is constitutively spliced in vivo (14, 33). Although the rodent lineage is considered to be closer to human than to several other nonprimate mammals (34, 35), the mouse CFTR exon 12 has seven synonymous and four nonsynonymous substitutions. The latter are located at the 3′ portion of the exon (Fig. 1a). To evaluate the role of the exonic sequence differences in splicing regulation, the mouse synonymous or nonsynonymous changes were introduced separately in the context of the human sequences by using a previously reported hybrid minigene system (14, 36) (Fig. 1b). The analysis of the splicing products derived from transfection experiments followed by RT-PCR amplification showed that human and mouse CFTR exon 12 minigenes (hCF12 and mCF12) generated 80% and 95%, respectively, of transcripts containing exon 12 and that the four mouse nonsynonymous substitutions in the human context (h-m1) produced a completely processed exon (Fig. 1c). On the contrary and unexpectedly, the h-m2 minigene, which has the seven mouse synonymous substitutions and thus the same amino acid coding potential of the normal human exon, showed a severe splicing defect with nearly complete exon skipping (only 5% of the mature mRNA included the exon; Fig. 1c). Thus, the combination of the mouse synonymous variations with the human missense mutations is incompatible with normal RNA processing, indicating that synonymous and nonsynonymous changes are under selective constraint to maintain inclusion of the exon in mRNA. To exclude the presence of species-specific splicing factors eventually responsible of the differential exon inclusion, the human-mouse minigenes were transfected in a mouse-derived cell line (3T3). The splicing patterns obtained were identical to those seen in the human cell lines (data not shown).
Fig. 1.
Comparative analysis of human and mouse CFTR exon 12 sequences evaluated for splicing efficiency. (a) Comparison of human (hCF12) and mouse (mCF12) CFTR exon 12 nucleotide sequences and amino acids (in one letter code). Synonymous (green for human and yellow for mouse) and nonsynonymous (in red or black for human and mouse, respectively) differences are highlighted. h-m1 and h-m2 are human-mouse hybrid exons: h-m1 contains the human-specific synonymous along with and the mouse-specific nonsynonymous substitutions; h-m2 contains the mouse-specific synonymous along with and the human-specific nonsynonymous substitutions. (b) Schematic representation of the hybrid minigene used in transient transfection splicing assays. Minigene exons and introns are indicated as black boxes and lines, respectively. Exon 12 sequences (white box) were tested for splicing efficiency by using specific primers (arrows). Dotted lines show the two CFTR exon 12 alternative splicing possibilities. (c) RT-PCR products from transfection experiments. RNA splicing variants corresponding to exon 12 inclusion and exclusion are shown. M is the 1-kb marker. Numbers at the bottom indicate the percentage of exon inclusion. Transfection experiments in NIH 3T3 mouse cells lines showed comparable changes in the splicing efficiency.
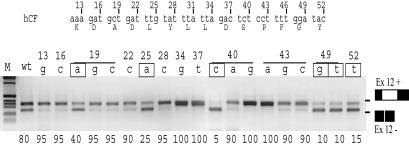
Synonymous Sites Are Under Selective Pressure to Maintain the Exon Inclusion. The severe splicing defect (5% of exon inclusion) observed in the h-m2 minigene, which has the same amino acid coding potential of the normal human exon, indicates that synonymous sites, presumed to evolve neutrally, may have an unexpected critical effect on splicing. To mimic the natural evolution at synonymous sites and to establish their effect on splicing, we analyzed the exon splicing efficiency of single synonymous changes introduced in the human CFTR exon 12 (Fig. 2). The synonymous nucleotides between position 13 and 52 of the human CFTR exon 12 were individually substituted, and the resulting minigenes were evaluated for splicing efficiency. We tested 19 synonymous changes over 22 possible sites. Compared with the normal human exon that generated ≈80% of transcript containing exon 12 (Fig. 2, wt), most mutations produced a small increase in exon inclusion, whereas six changes, 19A, 25A, 40C, 49G, 49T, and 52T, induced exon skipping (Fig. 2). In two positions (19 and 40), the effect on splicing depended on the nucleotide substitution: 19A and 40C caused an increase in exon skipping, whereas 19G, 19C, 40G, and 40A increased the exon inclusion. Thus, the probability of inducing exon skipping with a single synonymous substitution in CFTR exon 12 is ≈30%. This means that synonymous variations in CFTR exon 12 cannot evolve neutrally, but that they are significantly constrained by splicing requirements. A similar splicing effect of synonymous changes is also found in human NF-1 exon 37 (M. Baralle, personal communication)
Fig. 2.
Synonymous mutations in human CFTR exon 12 induce exon skipping. (Upper) The nucleotide and amino acid composition (in one letter code) of part of the human CFTR exon 12 (hCF). Third nucleotide positions are numbered according to their location in the exon. (Lower) RT-PCR products from splicing assay from 19 of 22 possible single synonymous changes between position 13 and 52 of the human CFTR exon 12. The position and the nucleotide substitution analyzed are indicated above each lane. The percentages of exon inclusion are reported at the bottom of each lane. The six synonymous substitutions that induce significant exon skipping are boxed.
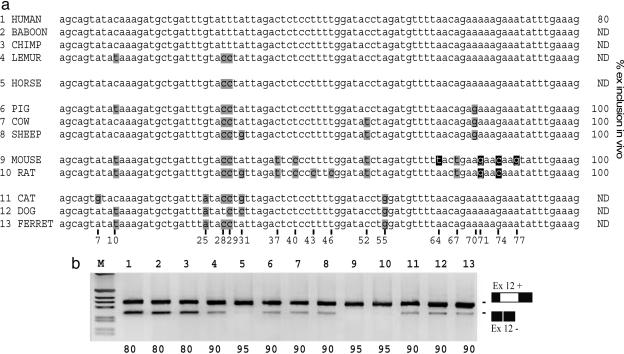
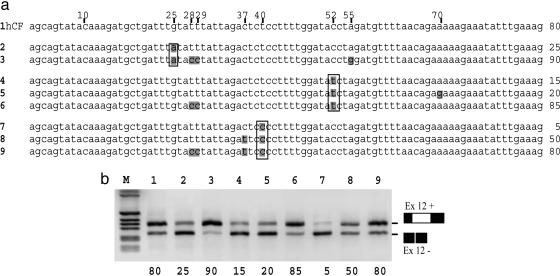
Mammalian CFTR Exon 12 Divergences Analyzed for Splicing Efficiency. Mammalian CFTR exon 12 sequence comparison (Fig. 3a) showed that synonymous substitutions that cause human CFTR exon 12 skipping are present in other species (25A in carnivores, 52T in sheep, and both 40C and 52T in rodents). Nevertheless, these exons are efficiently spliced both in the natural context and in the minigene system (Fig. 3). To follow up this observation, we created, by site-directed mutagenesis, artificial combinations of natural synonymous substitutions and analyzed their splicing pattern using the minigene system (Fig. 4). The results of this study clearly showed that the exonic context in which these mutations are embedded determines the splicing efficiency. Indeed, the 25A substitution does not induce exon skipping when associated with the carnivores-specific C28, C29, and G55 combination (Fig. 4b, compare lanes 2 and 3). In the same manner, 52T is compensated by C28, C29 (Fig. 4b, compare lane 4 with lane 6), but not by G70 (compare with lane 5), and 40C by the combination C28, C29, and 37T (Fig. 4b, lanes 7 and 9), but only partially by 37T alone (lane 8).
Fig. 3.
Comparative analysis of mammalian CFTR exon 12 sequences evaluated for splicing efficiency. (a) Mammalian CFTR exon 12 sequences grouped into primates, (human, baboon, chimpanzee, and lemur), perissodactyla (horse), certiodactyla (pig, cow, and sheep), rodents (mouse and rat), and carnivora (cat, dog, and ferret). Compared to human sequences, synonymous (black) and nonsynonymous (white) differences are highlighted and numbered according to their position in the exon. Sequence comparison shows some degree of lineage-specific conservation in the exon. Nonsynonymous sites are specific for rodents. Among the synonymous changes, T28/T29 is a recent acquisition of the human/Pan/gorilla group, T37, C40, and T67 are rodent-specific, and A25 and G55 are exclusive for carnivores. The remaining synonymous changes at positions 10, 31, and 52 occur in more than one lineage, C43 and C46 are only present in rat, and only G7 is present in cat. Numbers on the right indicate the percentage of CFTR exon 12 inclusion in vivo. ND, not done. (b) Splicing efficiency of mammalian exons analyzed by RT-PCR products from transfection experiments. RNA splicing variants correspond to exon 12 inclusion and exclusion. M is the 1-kb marker. Percentage of exon inclusion is indicated at the bottom.
Fig. 4.
Synonymous substitutions affect the splicing efficiency according to the mammalian exonic context. (a) Sequences of CFTR exon 12 highlighting the artificial composition of synonymous substitutions introduced in hybrid minigenes by site-directed mutagenesis. The three synonymous changes that in the human context induce exon skipping (25A, 40C, and 52T) are boxed. Numbers on the right of each sequence indicate the percentage of exon inclusion as determined by minigene splicing assay. For clarity, these percentages are also reported under the image in b that shows corresponding RT-PCR products from splicing assay. The RNA splicing variants that correspond to exon 12 inclusion and exclusion are indicated. M is the 1-kb marker.
Discussion
Genomic variation is the substrate of evolution, and novel coding alleles are generated by selection of specific mutations that confer particular advantages to protein function. However, there is now a further consideration to be made because of the presence of splicing regulatory elements overlapping with the amino acid code. In fact, before any genomic variant can be the subject of selection at the protein level, it has to be compatible with the splicing process. Indeed, mutations that cause exon skipping will not be included in the mature message and, even if they may have improved protein function, they would never be the substrate of positive selection. These observations have important implications in both the clinical genetics and molecular evolution fields.
The fact that any new genomic variation has to be compatible with the pre-mRNA splicing process has to be kept in mind when analyzing apparently neutral polymorphisms in diagnostic genomic scans, which may be erroneously classified as innocuous. Here, we describe systematic analysis of the role of synonymous variants on the splicing efficiency. Our results show that a high proportion (one in four) of them may induce CFTR exon 12 skipping reducing normal transcript below 25%, and hence may be a disease causing mutation (Fig. 2). It remains to be established whether exonic regulatory elements that make splicing vulnerable to synonymous point mutations are restricted to peculiar exons or are a general feature of mammalian genes. At least this phenomenon seems to be present in constitutive or alternative spliced exons that have weak splice sites. In fact, roughly one quarter of the synonymous changes studied in CFTR exon 9 (13), CFTR exon 12 (this paper), or NF-1 exon 37 (M. Baralle, personal communication) produce a splicing defect. It is also possible that missense mutations, such those found at the 3′ end of mouse CFTR exon 12 (Fig. 1), might create a dominant enhancer effect and thus allow for more flexibility and diversity in other parts of the exon.
Our results show clearly that several natural mammalian synonymous and nonsynonymous substitutions outside their species-specific context induce exon skipping. The effect on the splicing efficiency of these substitutions indicates that exonic sequences are significantly constrained by splicing requirements that in turn restrict evolutionary productive genome variability. Strikingly, in the human-mouse comparison, the functional splicing analysis of the evolutionary divergences revealed that the combination of mouse synonymous substitutions with the human nonsynonymous mutations is not compatible with proper splicing (Fig. 1).
It is then reasonable to conclude that, because of the presence of cis-acting splicing regulatory elements embedded in the exonic sequence, the evolution of mammalian exons is conditioned by two sequential selection processes. The first is the pressure to maintain the exon included in the final mRNA and the second is the selection of optimal protein function. This means that synonymous and nonsynonymous changes must not induce exon skipping leading to nonfunctional proteins, otherwise they will be discarded by negative selection. Only when exon inclusion is preserved can novel amino acid sequences be generated by selection of nonsynonymous changes that confer particular advantages to protein function.
Previous comparative studies between human and chimpanzee transcripts, have shown that 39% of silent sites in protein coding regions are under purifying selection (29) and that, in humans, the average substitution rate of silent sites is 30% lower in functional genes than in pseudogenes (30). Here, we provide experimental evidence that a significant amount of substitutions at synonymous sites cannot evolve freely because they encode splicing regulatory sequences. It is comforting that the percentage of synonymous substitutions involved is comparable to that suggested to be under purifying selection in comparative inter- and intraspecies studies (29, 30),
One method of evolutionary rate measurement is given by the ratio between the rate of change of nonsynonymous (Ka) and synonymous (Ks) substitutions. Because our results show that synonymous mutations are not strictly neutral but constrained by splicing requirement, the Ka/Ks ratios may yield inaccurate estimates of evolutionary rates. These findings are consistent with the unexpected low Ks observed in certain genomic coding regions (37, 38). Therefore, the presumed Ks evolutionary neutrality should be reevaluated in the light of splicing requirements.
Amino acid mutations with a presumed slightly deleterious effect on protein function segregate in the human population (29) and are responsible for human protein polymorphisms. However, the fate of these changes may not simply be elimination by purifying selection as previously suggested (29), but tolerated to preserve proper exon inclusion. An even more interesting extension of this hypothesis is that, because of strong splicing constraints on a fraction of the exons, suboptimal protein function might be tolerated to allow for the persistence of sequences that are essential for exon inclusion.
Acknowledgments
We thank Rodolfo Garcia and Marco Baralle for critical reading of the manuscript and Cristiana Stuani for technical assistance. This work was supported by the Telethon Onlus Foundation (Italy), Associazione Italiana per la Ricerca sul Cancro, and the Fondo Investimenti per la Ricerca di Base RBNE01W9PM.
Author contributions: F.P. designed research; M.R. performed research; F.E.B. analyzed data; and F.P. wrote the paper.
Abbreviation: CFTR, cystic fibrosis transmembrane regulator.
References
- 1.Mardon, H. J., Sebastio, G. & Baralle, F. E. (1987) Nucleic Acids Res. 15, 7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed, R. & Maniatis, T. (1986) Cell 46, 681-690. [DOI] [PubMed] [Google Scholar]
- 3.Cartegni, L., Chew, S. L. & Krainer, A. R. (2002) Nat. Rev. Genet. 3, 285-298. [DOI] [PubMed] [Google Scholar]
- 4.Faustino, N. A. & Cooper, T. A. (2003) Genes Dev. 17, 419-437. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Blanco, M. A., Baraniak, A. P. & Lasda, E. L. (2004) Nat. Biotechnol. 22, 535-546. [DOI] [PubMed] [Google Scholar]
- 6.Pagani, F. & Baralle, F. E. (2004) Nat. Rev. Genet. 5, 389-396. [DOI] [PubMed] [Google Scholar]
- 7.Teraoka, S. N., Telatar, M., Becker-Catania, S., Liang, T., Onengut, S., Tolun, A., Chessa, L., Sanal, O., Bernatowska, E., Gatti, R. A., et al. (1999) Am. J. Hum. Genet. 64, 1617-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorson, C. L. & Androphy, E. J. (2000) Hum. Mol. Genet. 9, 259-265. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni, L. & Krainer, A. R. (2002) Nat. Genet. 30, 377-384. [DOI] [PubMed] [Google Scholar]
- 10.Kashima, T. & Manley, J. L. (2003) Nat. Genet. 34, 460-463. [DOI] [PubMed] [Google Scholar]
- 11.Liu, H. X., Cartegni, L., Zhang, M. Q. & Krainer, A. R. (2001) Nat. Genet. 27, 55-58. [DOI] [PubMed] [Google Scholar]
- 12.Ars, E., Serra, E., Garcia, J., Kruyer, H., Gaona, A., Lazaro, C. & Estivill, X. (2000) Hum. Mol. Genet. 9, 237-247. [DOI] [PubMed] [Google Scholar]
- 13.Pagani, F., Buratti, E., Stuani, C. & Baralle, F. E. (2003) J. Biol. Chem. 278, 26580-26588. [DOI] [PubMed] [Google Scholar]
- 14.Pagani, F., Stuani, C., Tzetis, M., Kanavakis, E., Efthymiadou, A., Doudounakis, S., Casals, T. & Baralle, F. E. (2003) Hum. Mol. Genet. 12, 1111-1120. [DOI] [PubMed] [Google Scholar]
- 15.Aznarez, I., Chan, E. M., Zielenski, J., Blencowe, B. J. & Tsui, L. C. (2003) Hum. Mol. Genet. 12, 2031-2040. [DOI] [PubMed] [Google Scholar]
- 16.Caputi, M. & Zahler, A. M. (2002) EMBO J. 21, 845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291-336. [DOI] [PubMed] [Google Scholar]
- 18.Schaal, T. D. & Maniatis, T. (1999) Mol. Cell. Biol. 19, 261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulter, L. R., Landree, M. A. & Cooper, T. A. (1997) Mol. Cell. Biol. 17, 2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. (2002) Science 297, 1007-1013. [DOI] [PubMed] [Google Scholar]
- 21.Wang, Z., Rolish, M. E., Yeo, G., Tung, V., Mawson, M. & Burge, C. B. (2004) Cell 119, 831-845. [DOI] [PubMed] [Google Scholar]
- 22.Buratti, E., Muro, A. F., Giombi, M., Gherbassi, D., Iaconcig, A. & Baralle, F. E. (2004) Mol. Cell. Biol. 24, 1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratti, E. & Baralle, F. E. (2004) Mol. Cell. Biol. 24, 10505-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, X. H. & Chasin, L. A. (2004) Genes Dev. 18, 1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo, G., Hoon, S., Venkatesh, B. & Burge, C. B. (2004) Proc. Natl. Acad. Sci. USA 101, 15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard, I. & Beckmann, J. S. (1995) Nat. Genet. 10, 259. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., Park, W. J., Pyeritz, R. E. & Jabs, E. W. (1995) Nat. Genet. 9, 232-233. [DOI] [PubMed] [Google Scholar]
- 28.Akli, S., Chelly, J., Mezard, C., Gandy, S., Kahn, A. & Poenaru, L. (1990) J. Biol. Chem. 265, 7324-7330. [PubMed] [Google Scholar]
- 29.Hellmann, I., Zollner, S., Enard, W., Ebersberger, I., Nickel, B. & Paabo, S. (2003) Genome. Res. 13, 831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustamante, C. D., Nielsen, R. & Hartl, D. L. (2002) Mol. Biol. Evol. 19, 110-117. [DOI] [PubMed] [Google Scholar]
- 31.Buratti, E., Dork, T., Zuccato, E., Pagani, F., Romano, M. & Baralle, F. E. (2001) EMBO J. 20, 1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buratti, E., Brindisi, A., Pagani, F. & Baralle, F. E. (2004) Am. J. Hum. Genet. 74, 1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaney, S. J., Rich, D. P., Thomson, S. A., Hargrave, M. R., Lovelock, P. K., Welsh, M. J. & Wainwright, B. J. (1993) Nat. Genet. 4, 426-431. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, W. J., Eizirik, E., Johnson, W. E., Zhang, Y. P., Ryder, O. A. & O'Brien, S. J. (2001) Nature 409, 614-618. [DOI] [PubMed] [Google Scholar]
- 35.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788-793. [DOI] [PubMed] [Google Scholar]
- 36.Pagani, F., Buratti, E., Stuani, C., Bendix, R., Dork, T. & Baralle, F. E. (2002) Nat. Genet. 30, 426-429. [DOI] [PubMed] [Google Scholar]
- 37.Hurst, L. D. & Pal, C. (2001) Trends Genet. 17, 62-65. [DOI] [PubMed] [Google Scholar]
- 38.Orban, T. I. & Olah, E. (2001) Trends Genet. 17, 252-253. [DOI] [PubMed] [Google Scholar]