Abstract
Gene activation by NF-κB/Rel transcription factors is modulated by synergistic or antagonistic interactions with other promoter-bound transcription factors. For example, Sp1 sites are often found in NF-κB-regulated genes, and Sp1 can activate certain promoters in synergism with NF-κB through nonoverlapping binding sites. Here we report that Sp1 acts directly through a subset of NF-κB binding sites. The DNA binding affinity of Sp1 to these NF-κB sites, as determined by their relative dissociation constants and their relative efficiencies as competitor DNAs or as binding site probes, is in the order of that for a consensus GC box Sp1 site. In contrast, NF-κB does not bind to a GC box Sp1 site. Sp1 can activate transcription through immunoglobulin kappa-chain enhancer or P-selectin promoter NF-κB sites. p50 homodimers replace Sp1 from the P-selectin promoter by binding site competition and thereby either inhibit basal Sp1-driven expression or, in concert with Bcl-3, stimulate expression. The interaction of Sp1 with NF-κB sites thus provides a means to keep an elevated basal expression of NF-κB-dependent genes in the absence of activated nuclear NF-κB/Rel.
A multitude of gene-specific transcription factors which activate or repress transcription of their target genes in a combinatorial fashion through their individual binding sites have been identified. Synergistic protein-protein and protein-DNA interactions among these proteins lead to a functional cross-coupling that allows a high degree of complexity, since the single regulators are individually regulated (see reference 58 for a review).
Members of the NF-κB/Rel family are involved in the transcriptional regulation of a number of cellular and viral genes. Various hetero- and homodimers are formed between the five mammalian subunits p50, p52, p65 (RelA), c-Rel, and RelB, which bind with different affinities to a group of related NF-κB DNA binding sites with the consensus sequence GGGRNNYYCC (17, 67). These proteins share a stretch of 300 amino acids, termed the Rel homology domain (RHD), which is highly conserved among NF-κB/Rel proteins, including the viral Rel homolog v-Rel and the insect developmental and immune control proteins Dorsal and Dif (see references 61 and 65 for reviews). The RHD is required entirely for DNA binding, whereas only its C-terminal part is required for dimerization. Only mammalian p65, c-Rel, and RelB contain transcription activation domains in their unique sequences located carboxy terminally to the RHD; p50 and p52 lack such domains. It is assumed that homodimers of the latter repress transcription and that they require nuclear cofactors, such as Bcl-3, to form ternary complexes which can activate transcription. Bcl-3 belongs to the IκB gene family, which in vertebrates comprises IκBα, IκBβ, IκBɛ, and the precursor proteins for p50 and p52, p105, and p100, respectively. The cytoplasmic IκBs bind to dimerized NF-κB factors and thereby block their nuclear translocation until signaling processes lead to induced IκB degradation and nuclear translocation of NF-κB (see references 2, 4, 51, 61, and 65 for reviews).
All NF-κB/Rel members have distinct physiological functions, as evident from the outcome of gene targeting experiments in mice (2). Both differential regulation by IκBs in the cytoplasm and differential interaction with other transcription factors in the nucleus are likely to establish specificity in the NF-κB system. It is assumed that NF-κB/Rel dimers exert specific functions through nonidentical DNA binding site preferences as well as through individual interactions with other promoter-bound gene-specific or basal factors (27, 53). As one example, Sp1 elements are often found in the enhancers or promoters of NF-κB-regulated genes, including those for human immunodeficiency virus type 1 (HIV-1), intracellular adhesion molecule 1, vascular adhesion molecule 1, or granulocyte-macrophage colony-stimulating factor 1 (23, 41, 55, 56). Further examples are the promoters for the cellular genes encoding δ opioid receptor, interleukin-2 (IL-2) receptor alpha chain (IL-2Rα), manganese superoxide dismutase, NF-κB2, tissue factor 1, preprogalanin, monocyte chemoattractant protein 1, c-Rel, and melanoma growth-stimulating activity (1, 3, 24, 33, 36, 47, 48, 60, 62, 64).
The ubiquitous transcription factor Sp1 contains a three-zinc-finger DNA binding domain and four transactivation domains (9, 10, 25) and binds to GC-rich sites (6, 8). BTEB, Sp3, and Sp4 are related to Sp1 and share a highly conserved zinc finger domain (18, 19, 22). Accordingly, Sp1-type GC (GGGGCGGGC) or GT (GGGTGTGGC) box DNA binding sites are also bound by these proteins.
In the HIV-1 long terminal repeat (LTR), the actions of NF-κB and Sp1 are highly cooperative, involving effects on the DNA binding of both factors to their adjacent binding sites, resulting in increased transcriptional activation (45). Furthermore, binding of Sp1 with either p50 or p65 induces establishment of the nucleosomal arrangement of HIV-1 LTR DNA (44). A direct protein-protein association between Sp1 and NF-κB could be demonstrated even in the absence of DNA. This interaction between Sp1 and NF-κB requires the zinc finger region of Sp1 and an N-terminal part of the Rel homology domain of p65, i.e., the DNA binding domains of both factors (46). It has been shown that most vertebrate NF-κB/Rel factors interact with Sp1. v-Rel increases transcription from promoters containing Sp1 sites by physically interacting with Sp1 (54). The interaction of v-Rel with Sp1 required the N-terminal 147 amino acids of v-Rel. The interaction domain in Sp1 was found to be in the N-terminal region of Sp1 containing transactivation domains. In this study, an Sp1 mutant, lacking the zinc finger region, specifically bound to v-Rel, c-Rel, p50, p52, and p65 (RelA) (54). At another level of functional cross talk, Sp1 regulates the p65 promoter, which, unlike the other genes encoding NF-κB/Rel proteins, is not subject to autoregulation (59).
Although Sp1 is widely regarded as a housekeeping-type nuclear transcription factor with constitutive activity and ubiquitous occurrence, both its expression and activity are subject to regulation. The expression levels of Sp1 vary dramatically during embryonal development and differentiation (49). Several reports indicate modification of Sp1 activity in response to cellular signaling. It has been shown that okadaic acid treatment induces strong phosphorylation of Sp1 in the nucleus (63), and dephosphorylation of Sp1 by protein phosphatase 1 is involved in glucose activation of acetyl coenzyme A carboxylase gene transcription (12). Furthermore, cytomegalovirus infection enhances the Sp1 activation function on the NF-κB–p65 gene promoter (66). Gamma interferon induces phosphorylation and thereby increases DNA binding activity of Sp1 (50).
We provide evidence for an unexpected level of functional coupling between Sp1 and NF-κB. Sp1 binds with high affinity to a subset of NF-κB DNA binding sites and thereby activates transcription. The interactions of Sp1 and NF-κB with NF-κB sites are mutually exclusive. Sp1 may serve to provide an increased basal expression of NF-κB-dependent genes in the absence of nuclear NF-κB. At elevated levels, Sp1 may also compete for the binding site and block access to some NF-κB heteromers but not to others, and thus it may play a role in the determination of gene-specific functions of various NF-κB homo- and heterodimers which have distinct affinities for various NF-κB binding sites.
MATERIALS AND METHODS
Cell culture.
Adherent HeLa cells were grown in Dulbecco’s modified Eagle’s medium (BRL/GIBCO)–1 mM sodium pyruvate–100 U of penicillin per ml–100 μg of streptomycin per ml–10% fetal calf serum. Cells were stimulated with 1 to 25 ng of tumor necrosis factor alpha (TNF-α; Biomol) per ml where indicated. Drosophila SL2 cells were cultured with Schneider’s medium (GIBCO)–10% fetal bovine serum–10 U of penicillin-streptomycin per ml at 25°C without CO2 in tightly closed flasks.
Plasmids and recombinant proteins.
For prokaryotic expression, the pET system (Novagen) was used. pETp50(443) (amino acids 18 to 443) (30) was expressed in Escherichia coli BL21(DE3)pLysS and purified as described previously (40). Recombinant Sp1, purified from HeLa cells infected with recombinant vaccinia virus containing the human Sp1 cDNA, was obtained from Promega. For luciferase reporter experiments, p309LUC and pmkB309LUC, containing P-selectin wild-type and mutant promoters, respectively (42), were provided by R. P. McEver (University of Oklahoma Health Sciences Center). The Sp1 reporter plasmid pSp1HL was described by Hirano et al. (21). In brief, six tandem Sp1 binding sites (KpnI-EcoRI sites) and the −45 to +83 fragment of the HIV-1 promoter (EcoRI-HindIII sites) were inserted into KpnI-HindIII sites of PGV-B (Toyo Ink Co., Tokyo, Japan). The oligonucleotide sequences for the Igκ–NF-κB reporter plasmids were CAGTTGAGGGGACTTTCCCAGATCTAGTTGAGGGGACTTTCCCAG-3′ (tandem immunoglobulin kappa chain [Igκ] enhancer sequence) (2× κB-Luc) and CAGTTGAAGGGACTTTCCCAGATCTAGTTGAAGGGACTTTCCCAG-3′ (2× κB-m1 Luc). These oligonucleotides were annealed to their complementary counterparts and replaced into KpnI/EcoRI sites of pSp1HL.
The pPacSp1 plasmid for Sp1 expression was kindly provided by G. Suske (University of Marburg, Marburg, Germany). pPacSp1ΔC (amino acids 83 to 611) and pPacSp1DBD (amino acids 612 to 778) were constructed from pPacSp1 by PCR using the appropriate primers. To construct vectors for expression of p50 and p65 in SL2 cells (pPacp50 and pPacp65), the Sp1 cDNA insert was removed from pPacSp1 by BamHI-XhoI cleavage and replaced by the PCR-amplified complete cDNA inserts of pECEp50 or pECEp65 (40) with BamHI-XhoI ends. For pPacBcl-3, the Bcl-3 cDNA insert from pcDNABcl-3 (40) was isolated by HindIII-XhoI cleavage. The fragment was cloned into the pPac vector via a BamHI-HindIII linker.
Antibodies.
Rabbit anti-p65, anti-c-Rel, and anti-Sp1 antisera (sc-109, sc-272, and sc-59, respectively) were from Santa Cruz Biotechnology Inc. Rabbit anti-p50 serum was obtained from Rockland Inc.
Preparation of nuclear extracts.
The cells were washed and resuspended in buffer A, and 0.125% Nonidet P-40 was added. The cells were left for 5 min on ice and centrifuged at 1,000 × g for 10 min. The pellet was treated with buffer C for 15 min to yield the nuclear extract as described previously (39).
Silver staining of recombinant proteins.
Recombinant proteins were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE). Gels were preincubated in the fixing solution (50% methanol, 12% acetic acid) for 30 min with gentle shaking and then washed 4 times with distilled H2O (dH2O). Gels were incubated in 0.8% AgNO3 for 15 min with gentle shaking and then washed three times with dH2O. After treatment with a aqueous solution containing 0.005% sodium citrate and 0.02% formaldehyde, gels were incubated in 10% acetic acid, washed with dH2O, and dried.
Immunoblots.
Forty-microgram aliquots of nuclear extract were separated by SDS-PAGE. Prior to transfer, gels were equilibrated in ice-cold blotting buffer (25 mM Tris-HCl [pH 8.3], 0.01% SDS, 20% methanol). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). All Western blots were analyzed by chemiluminescence (Tropix) as described previously (29).
Electrophoretic mobility shift assay (EMSA).
The following oligonucleotides were annealed to complementary strands, resulting in double-stranded probes with 5′-AATT and AGCT-3′ overhanging ends on the top strand (Sp1) or with 5′-GATC overhanging ends on both strands (all other probes): Sp1 (5′-aattACCGGGCGGGCGGGCTACCGGGCGGGCTagct), P-selectin (5′-gatcCGAAGGGGGTGACCCCTTGCC), IL-6 (5′-gatcCTCAAATGTGGGATTTTCCCATGAGTCT), Igκ (5′-gatcCAACAGAGGGGACTTTCCGAGGCCATCTG), mutant Igκ (5′-gatcCAACAGAGGGGACTTTCCGAGGCCATCTG), IL-2Rα (5′-gatcCGGCAGGGGAATCTCCCTCTCC), H2K (5′-gatcCAGGGCTGGGGATTCCCCATCTCCACAGG), HIV proximal site (5′-gatcTCCGCTGGGGACTTTCCAGG), HIV distal site (5′-gatcAAGGGACTTTCCGCTGCAGA), and IL-2 (5′-gatcCTAACAAAGAGGGATTTCACCTACAT). The quantitated probes were 32P labeled with Klenow enzyme by fill-in reaction.
Standard DNA binding reactions were performed with nuclear extracts or recombinant protein in 20 μl of binding buffer [20 mM HEPES (pH 8.4), 60 mM KCl, 4% Ficoll, 5 mM dithiothreitol (DTT), 1 μg of bovine serum albumin (BSA), 2 μg of poly(dI-dC)] for 20 min at 30°C. The reaction mixtures were loaded onto 4% nondenaturing polyacrylamide gels containing 1× Tris-borate-EDTA. Gels were run at 250 V for 1 h, dried, and visualized by autoradiography. The standard binding reaction was modified for the experiments shown in Fig. 2B and C as indicated. Radioactivity on the membrane was quantified with a BAS-2000 bioimaging analyzer (Fujix, Tokyo, Japan).
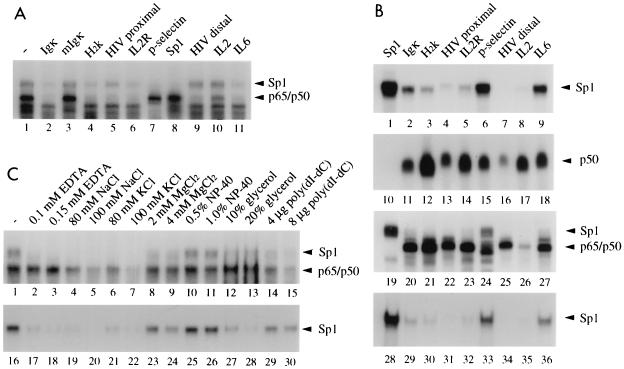
FIG. 2.
Differential interaction of Sp1 with various NF-κB binding sites. (A) Complexes formed between the Igκ probe and nuclear extracts (5 μg of protein/lane) of TNF-α (1 ng/ml, 30 min)-stimulated HeLa cells were challenged without (lane 1) or with (lanes 2 to 11) 50-fold molar amounts of competitor oligonucleotides containing the indicated NF-κB DNA binding sites or an Sp1 binding site. The positions of Sp1 and p65-p50 DNA complexes are indicated; free DNA is not shown. (B) Oligonucleotides containing various NF-κB binding sites or an Sp1 binding site, as indicated, were used as radiolabeled probes in gel retardation assays either with affinity-purified Sp1 (1 footprint-producing unit/lane) (lanes 1 to 9 and 28 to 36), with bacterially expressed p50 (20 ng/lane) (lanes 10 to 18), or with nuclear extract proteins (5 μg/lane) of TNF-α (10 ng/ml, 30 min)-stimulated HeLa cells (lanes 19 to 27). Either NF-κB buffer conditions [20 mM HEPES (pH 8.4), 60 mM KCl, 5 mM DTT, 1 μg of BSA, 2 μg of poly(dI-dC), 4% Ficoll] (lanes 1 to 27) or Sp1 buffer conditions [10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 0.1 mM ZnCl2, 0.2 μg of poly(dA-dT), 10% glycerol] (lanes 28 to 36) were used. The positions of complexes are indicated. Free DNA is not shown. (C) Effect of buffer components on Sp1 or NF-κB complex formation with the IL-6 gene NF-κB site. Either nuclear extracts of TNF-α-stimulated HeLa cells (lanes 1 to 15) or purified Sp1 (lanes 16 to 30) were used in EMSA with the indicated components included in the standard NF-κB binding buffer. NP-40, Nonidet P-40.
Determination of dissociation constants.
KD values were determined by EMSAs as described by Meisterernst et al. (35). Double-stranded oligonucleotide probes were end labeled with [α-32P]dATP and Klenow DNA polymerase. The DNA binding reactions were performed in the presence of poly(dI-dC) for 20 min at 30°C followed by EMSA analysis. The counts/minute values of gel areas corresponding to complexed and free DNA were determined with the BAS-2000 bioimaging analyzer.
Shift-Western blotting.
DNA binding reaction mixtures contained 10,000 cpm of 32P-labeled Igκ probe and 10 μg of nuclear extract protein in 20 mM HEPES (pH 8.4)–60 mM KCl–4% Ficoll–5 mM DTT–1 μg of BSA–2 μg of poly(dI-dC) in a total volume of 10 μl. After 15 min at 30°C, the reaction mixtures were loaded onto 5% nondenaturing acrylamide gels. Shift-Western blotting was performed as described previously (13). In brief, after electrophoretic separation, protein-DNA complexes were transferred onto stacked nitrocellulose and PVDF membranes. The radiolabeled probe that bound to the PVDF membrane was detected by autoradiography, whereas the proteins in the complexes that bound to the nitrocellulose membrane were detected by immunoblotting.
Transient transfection and luciferase assay.
SL2 cells were transfected by calcium phosphate precipitation. Cells were plated in 60-mm-diameter collagen-coated dishes at a density of 5 × 105 cells/ml (5 ml) 24 h before transfection. Aliquots of 0.25 ml of 250 mM CaCl2, containing 16 μg of plasmid DNA were added drop by drops to an equal volume of 2× HeBS (16 g of NaCl, 0.7 g of KCl, 0.4 g of Na2HPO4, 2 g of dextrose, 10 g of HEPES [pH 7.1] [all quantities per liter]) in 12-well plates that were gently rocked by hand. After 30 min at room temperature, the suspension of calcium phosphate complexes was gently pipetted into the 60-mm-diameter dish. After 48 h, cells were harvested by vigorous tapping, centrifuged at 1,000 × g, washed twice with phosphate-buffered saline, and resuspended in a lysis buffer for the luciferase assay system (Toyo Ink). Cells were sometimes prepared by freeze-thawing as an additional step, because cells were not destroyed by lysis buffer alone, followed by the protocol of the luciferase assay system.
RESULTS
Sp1 specifically interacts with a subgroup of NF-κB binding sites.
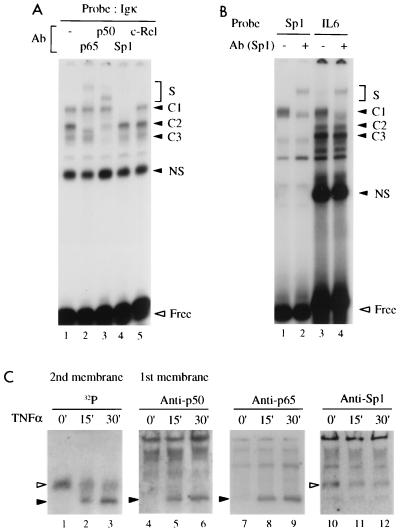
A DNA binding complex that migrated more slowly than the NF-κB DNA complex was occasionally detected in EMSAs with NF-κB binding site probes and nuclear extracts of HeLa or Jurkat cells. For example, the expected NF-κB complexes formed with an Igκ probe in TNF-α-treated HeLa cells could be supershifted with either the anti-p65 (RelA) or anti-p50 antibody (Fig. 1A, lanes 1 to 3, complex C2, heterodimeric NF-κB) or only with anti-p50 antiserum (complex C3, p50 homodimers), whereas an additional complex (C1) remained refractory to antisera raised against p50, p65, and c-Rel (lanes 2, 3, and 5). We have assayed this complex with a panel of antibodies against known transcription factors and found that an anti-Sp1 antibody specifically inhibited C1 (lane 1 versus lane 4). Similarly, complex C1 was also detected with a probe containing the κB site of the IL-6 gene, using the same extract, and was supershifted with the anti-Sp1 antibody (Fig. 1B, lanes 3 and 4). Complexes C2 and C3 were sensitive to anti-p50 and anti-p65 antibodies (not shown). An Sp1 binding site probe containing a consensus GC box and with the same length as the IL-6 probe showed an Sp1-DNA complex migrating like the C1 complex formed with the IL-6 probe (lanes 1 and 2). As expected, this complex was supershifted with the anti-Sp1 antibody (lanes 1 and 2). It is further noteworthy that the efficiencies of Sp1 complex formation with both probes were similar (compare C1 in lanes 1 and 3). To further confirm that Sp1 binds to the Igκ NF-κB binding site, a DNA binding reaction was subjected to shift-Western blotting analysis (13) (see Materials and Methods for details). Nuclear extracts of untreated or TNF-α-stimulated cells yielded C1 and C2 complexes on the DNA blot (Fig. 1C, lanes 1 to 3, open and solid arrowheads, respectively). Only C2 was induced by TNF-α (lanes 2 and 3), whereas C1 activity was weakened after stimulation. On the protein blots, the inducible complex could be identified as heterodimeric NF-κB by blotting with either the anti-p50 or anti-p65 antibody (lanes 4 to 9). Only the upper complex reacted with the anti-Sp1 antibody (lanes 10 to 12). Thus, both cellular Sp1 and NF-κB bind to the Igκ site. The reduction of Sp1 activity upon activation of NF-κB (Fig. 1C) suggests competition for the same binding site.
FIG. 1.
Cellular Sp1 binds to Igκ and IL-6 NF-κB DNA binding sites. (A) Complexes formed between nuclear extract proteins (5 μg) of TNF-α (1 ng/ml, 30 min)-stimulated HeLa cells and an Igκ probe were analyzed by EMSA. The reaction mixtures contained either no (−) antibody (Ab) (lane 1) or antiserum directed against p65, p50, Sp1, or c-Rel (lanes 2 to 5), as indicated. The reactivities with the antibodies identify C1 as Sp1, C2 as p65-p50, and C3 as p50-p50. S, supershifted complex; NS, nonspecific. (B) Complexes formed with an Sp1 (lanes 1 and 2) or IL-6 gene (lanes 3 and 4) NF-κB binding site probe without (lanes 1 and 3) or with (lanes 2 and 4) Sp1 antibody. C1, C2, and C3 contain Sp1, p65-p65, and p50-p65 (not shown), respectively. (C) Shift-Western blotting of complexes formed with the Igκ probe and nuclear extracts of adherent HeLa cells stimulated with TNF-α (10 ng/ml) for the indicated times. EMSA gels were sandwich blotted with a nitrocellulose membrane and then with a PVDF membrane, which were then analyzed by immunoblotting and autoradiography, respectively, to separately visualize bound proteins and labeled DNA of the complexes. The DNAs of two differently migrating complexes (open and solid arrowheads) (lanes 1 to 3) whose intensities were decreased and induced, respectively, upon TNF-α stimulation, were detected. The slower complex (open arrowhead) was detected by an Sp1 antibody (lanes 10 to 12), and the faster one (solid arrowhead) was detected by both p50 and p65 antibodies (lanes 4 to 9). Free DNA is not shown.
To assess whether Sp1 would recognize the conserved NF-κB binding site motif or a part of it, complexes formed with the Igκ binding site probe were challenged with competitor DNAs containing various NF-κB binding sites with natural flanking sequences (Fig. 2A). Both Sp1 and p50-p65 complexes formed with nuclear extract from TNF-α-stimulated HeLa cells were competed by an unlabeled Igκ but not by a mutant Igκ oligonucleotide that differed by only one base in the quadruplet G of the binding site motif (lane 1 compared to lanes 2 and 3). Furthermore, the Sp1-Igκ complex was efficiently competed by oligonucleotides containing the NF-κB binding sites of the major histocompatibility complex H2K gene, the IL-2Rα promoter, the P-selectin promoter, the consensus Sp1 GC box, and the IL-6 gene promoter (lanes 4, 6 to 8, and 11). The HIV-1 LTR proximal site competed less efficiently, and both the distal LTR site and the IL-2 site were inactive (lanes 5, 9, and 10). As expected, all oligonucleotides except the Sp1 and P-selectin competitors (lanes 7 and 8) competed for NF-κB complex formation. The P-selectin site is known to bind p50-p65 with much lower affinity than p50 homodimers (42). Next, the various Sp1 and NF-κB binding sites were used as radiolabeled probes and assayed with purified Sp1 from HeLa cells, bacterial p50, or nuclear extract from TNF-α-stimulated HeLa cells (Fig. 2B). Purified Sp1 strongly bound to the Sp1 GC box and to the P-selectin and IL-6 gene NF-κB binding sites (lanes 1, 6, and 9) and with lower efficiency to the Igκ, H2K, HIV proximal, and IL-2Rα sites but not to the HIV distal or IL-2 site (lanes 2 to 5, 7, and 8). Recombinant p50 bound with comparable affinities to most NF-κB binding sites, as expected, but with reduced affinity to the HIV distal site and not to the Sp1 binding site oligonucleotide (lanes 10 to 18). When testing nuclear extract from TNF-α-stimulated HeLa cells, we found strong complex formation between Sp1 and the Sp1 GC box or the P-selectin and IL-6 binding sites (lanes 19, 24, and 27). Strongly impaired binding, however, was observed in this experiment with the other NF-κB binding sites, possibly due to binding site competition of Sp1 with NF-κB p50-p65. The latter bound efficiently to most NF-κB sites, except for the IL-2 site, but not to the Sp1 site (lanes 19 to 27).
The binding affinity of Sp1 to NF-κB sites was dependent on the buffer conditions. When we used a buffer in the binding reaction previously used for Sp1 (31), the affinity to NF-κB sites compared to the GC box sequence was reduced (lanes 28 to 36 versus lanes 1 to 9), although the P-selectin and IL-6 sites were still bound efficiently (lanes 33 and 36). We tested the effects of various buffer components on complex formation of cellular Sp1 and p50-p65 in nuclear extracts or purified Sp1 with the IL-6 promoter site. In contrast to NF-κB, cellular or purified Sp1 was strongly affected by EDTA (Fig. 2C, lanes 1 to 3 and 16 to 18), MgCl2 (lanes 8, 9, 23, and 24), or glycerol (lanes 12, 13, 27, and 28). Both factors were sensitive to elevated salt concentrations (lanes 1 and 16 versus lanes 4 to 7 and 19 to 22). This sensitivity to various buffer components may explain why Sp1 was not detected in previous studies with NF-κB binding site probes.
Sp1 and NF-κB interact with the same DNA binding site in a mutually exclusive fashion.
The ability of Sp1 to bind to several NF-κB binding sites suggests that the conserved NF-κB motif is recognized by Sp1. This possibility is also supported by the fact that a one-base mutation in the Igκ site eliminates binding of Sp1 (Fig. 2A). Since NF-κB almost wraps around the contacted DNA (16, 38), little of the conserved DNA binding site is exposed to allow excess amounts of other proteins to the same site simultaneously. Therefore, it is likely that NF-κB and Sp1 compete for the common binding site.
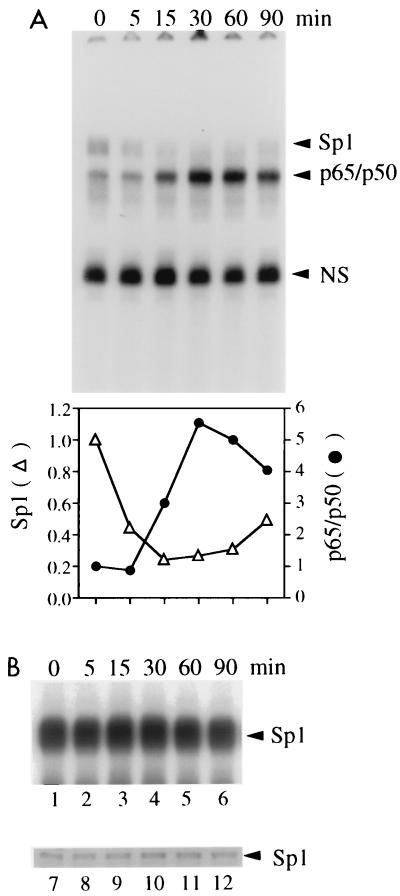
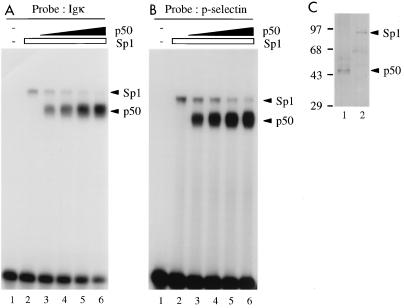
In fact, the Sp1 complex formed on the IL-6 probe with nuclear extracts from HeLa cells was replaced by NF-κB p50-p65 after induction of the latter with TNF-α (Fig. 3A). This loss of Sp1 complexes with the NF-κB site was not due to effects of TNF-α on Sp1 DNA binding affinity or protein amounts (Fig. 3B) and thus should be due to binding site competition. Similarly, upon incubation of the Igκ (Fig. 4A) or P-selectin (Fig. 4B) site with purified Sp1 and increasing concentrations of recombinant p50, Sp1 complexes were diminished (lanes 2 to 6). Competition on the Igκ site was more efficient than that on the P-selectin site, suggesting a higher relative affinity of Sp1 for the latter, consistent with its higher affinity for the P-selectin site observed in Fig. 2A and B. The protein amounts used for Sp1 and p50 were in comparable order, as shown by silver staining (Fig. 4C).
FIG. 3.
Mutually exclusive interaction of Sp1 or NF-κB with the same site. (A) Gel retardation assay with the IL-6 probe and nuclear extracts of adherent HeLa cells stimulated with TNF-α (10 ng/ml) for the indicated times. The increase of NF-κB complex formation after induction and the concomitant decrease of Sp1 complex formation are also visualized by quantitation of the counts/minute of each complex with a BAS2000 phosphorimaging analyzer in arbitrary units (bottom). NS, nonspecific. (B) The nuclear extracts used for panel A were assayed for Sp1 complex formation with an Sp1 DNA probe in a gel retardation assay (lanes 1 to 6; only complexes are shown) and for Sp1 protein amounts in a Western blot (lanes 7 to 12).
FIG. 4.
Competition of Sp1 by p50 homodimers on Igκ (A) or P-selectin (B) NF-κB binding sites, determined by EMSA using purified Sp1 protein from HeLa cells (1 footprint-producing unit (fpu)/lane) (A and B, lanes 2 to 6) and increasing amounts of recombinant p50 (0, 5, 10, 20, and 40 ng/lane) (lanes 2 to 6, respectively). Lanes 1 in panels A and B, no protein added. (C) Silver-stained SDS-polyacrylamide gel of 2 fpu of Sp1 and 20 ng of p50. Sizes are indicated in kilodaltons.
The dissociation constants of Sp1 and NF-κB to consensus Sp1 and NF-κB binding sites are in the same order.
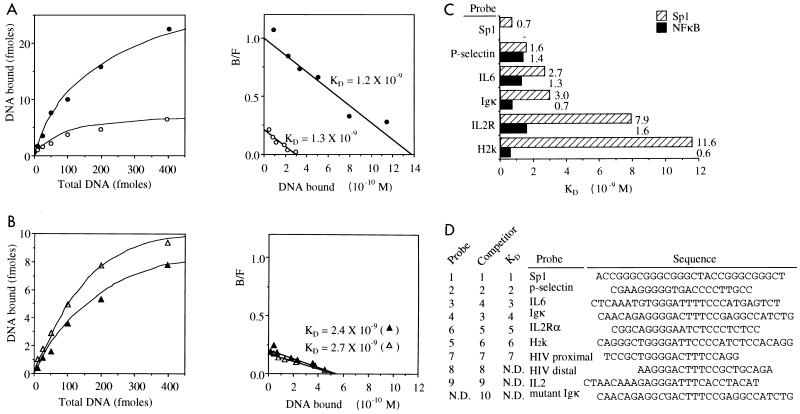
We next determined the dissociation constants for the Sp1- and p50-p65 interactions with NF-κB or Sp1 GC box binding sites by Scatchard analysis of quantitative EMSA using nuclear extracts of TNF-α-stimulated and unstimulated extracts under the same experimental conditions (Fig. 5A). The low-level nuclear NF-κB activity detected in unstimulated HeLa cells with the IL-6 probe and the elevated NF-κB activity after TNF-α stimulation revealed KDs in the nanomolar range of 1.2 × 10−9 and 1.3 × 10−9 M, respectively (Fig. 5A). Sp1 bound to the IL-6 probe with KDs of 2.4 × 10−9 and 2.7 × 10−9 M in extracts of stimulated and unstimulated cells, i.e., with only twofold-lower affinity than NF-κB. The KDs determined for Sp1 and NF-κB with the P-selectin probe were almost equal, 1.6 × 10−9 and 1.4 × 10−9 M, respectively (Fig. 5C). The consensus Sp1 GC box sequence revealed a dissociation constant of 7 × 10−10 M for Sp1. The affinity of Sp1 to its GC box has also been determined to values in the 10−10 M range by others (32). The order of the dissociation constants determined for the various sites matches very well with the relative affinities observed when the sites were used as probes (Fig. 2B) or as competitors (Fig. 2A) (see Fig. 5D for summary). Our data thus indicate that Sp1 binds with a significant affinity to a subset of NF-κB binding sites. This suggests that Sp1 should interact functionally with some NF-κB binding sites and that the different relative binding strengths expected for various NF-κB hetero- and homodimers may determine that some can compete with Sp1 whereas others cannot. An inspection of the DNA sequences of the NF-κB sites preferred by Sp1 did not reveal any obvious high similarity to Sp1 consensus sequences. Only the presence of the typical G3 or G4 stretch found in both halves of the palindromic NF-κB sites is similar to the G richness of Sp1 sites (Fig. 5D).
FIG. 5.
Determination of the dissociation constants for the interaction of Sp1 and NF-κB with Sp1 and NF-κB binding sites by quantitative EMSA and Scatchard analysis. (A and B) Binding curves (left) and Scatchard plots (right) for complex formation of p65-p50 (A) or Sp1 (B) with the IL-6 probe in nuclear extracts of nonstimulated and TNF-α (10 ng/ml, 30 min)-stimulated HeLa cells (open and closed symbols, respectively). B/F, bound/free. (C) Summary of the dissociation constants determined in HeLa cell nuclear extracts for the interaction of Sp1 or NF-κB with various NF-κB and Sp1 sites, as indicated. (D) Alignment of the Sp1 and NF-κB binding sites used in this study in order of relative affinity to Sp1. The left columns indicate similar orders of affinity as determined when we used them as probes, as competitors, or for KD determination. ND, not determined.
Sp1 transactivates homologous and heterologous promoters through NF-κB binding sites: functional interference between NF-κB and Sp1.
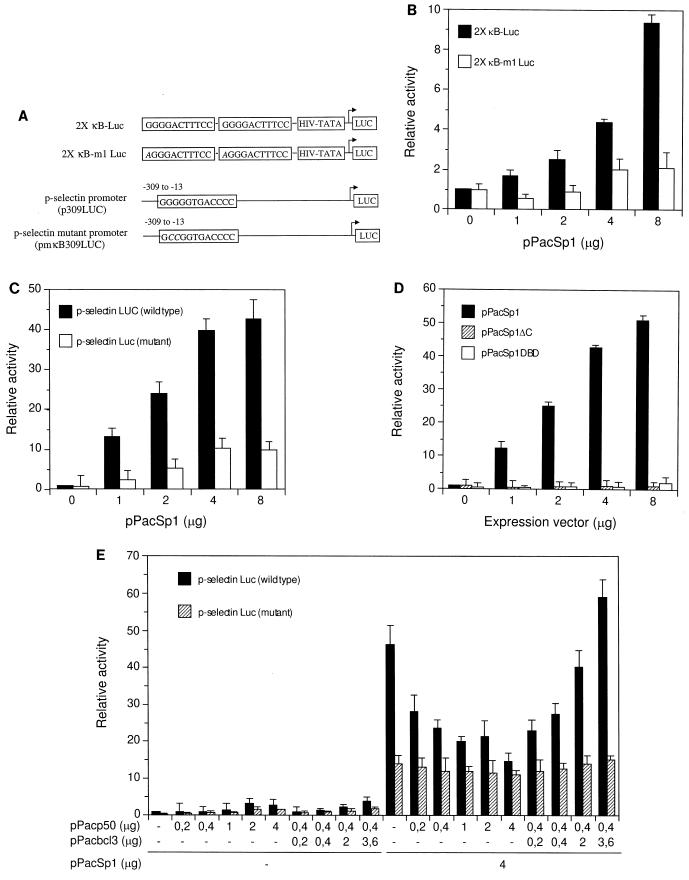
To assess a functional role for the interaction of Sp1 with NF-κB binding sites, we constructed luciferase reporter plasmids containing tandem Ig-κB sites in front of the HIV TATA box. These sites contained either the wild-type NF-κB binding site or a one-base mutation in the first residue of the G4 motif in the binding site (Fig. 6A). These constructs were cotransfected with increasing amounts of an Sp1 expression vector into Drosophila SL2 cells, which are devoid of endogenous Sp1 (9). Sp1 indeed strongly activated the promoter containing the wild-type κB site in both a dose-dependent and κB site-dependent manner (Fig. 6B); the mutant κB site, with only one base mutated in the G4 stretch, was only very weakly responsive. Under comparable conditions, the wild-type Ig-κB site conferred a fourfold-higher activation by transfected p50-p65 compared to Sp1 (data not shown). We furthermore tested the P-selectin promoter and a mutant thereof containing only two base changes in its NF-κB binding site (Fig. 6A). Sp1 potently activated transcription of the wild-type promoter up to 40-fold (Fig. 6C). In contrast, the mutant promoter was only weakly activated. Thus, Sp1 activates transcription through NF-κB binding sites in both homologous and heterologous promoter contexts. Transcriptional induction by Sp1 was dependent on its transactivation domains and on the DNA binding domain. Activation was lost when Sp1 mutant proteins which either lacked the transactivation domains (in pPacSp1DBD) or the DNA binding domain (in pPacSp1DC) were expressed (Fig. 6D).
FIG. 6.
Sp1 activates transcription through Igκ and P-selectin NF-κB binding sites. Functional interference with p50 and Bcl-3 was analyzed. (A) Schematic presentation of heterologous HIV-TATA luciferase reporter genes containing two tandem wild-type or mutant Igκ NF-κB binding sites and of the P-selectin luciferase promoter constructs with either the wild-type or mutated NF-κB binding site in the context of the complete promoter sequence (42). (B) Sp1 activates through Igκ NF-κB binding sites. Drosophila SL2 cells were transfected with reporter constructs containing wild-type (2× κB-Luc, 8 μg) or mutant (2× κB-m1 Luc, 8 μg) Igκ NF-κB binding sites with increasing amounts of pPacSp1 expression vector, as indicated. (C) Sp1 activates transcription through a P-selectin promoter NF-κB binding site. SL2 cells were transfected with the P-selectin promoter luciferase construct (8 μg) harboring a wild-type or mutated NF-κB binding site along with increasing amounts of Sp1 expression vector. (D) Activation through the P-selectin NF-κB site by Sp1 requires both DNA binding and transactivation domains of Sp1. Cell were transfected with P-selectin constructs as in panel C along with expression vectors encoding Sp1 (pPacSp1, amino acids 83 to 778) or Sp1 lacking the zinc finger region (pPacSp1ΔC, amino acids 83 to 611) or containing only the DNA binding domain of Sp1 (pPacSp1DBD, amino acids 612 to 778), as indicated. (E) Functional interference between Sp1, p50, and Bcl-3 at the P-selectin NF-κB binding site. The P-selectin wild-type or mutant promoter was transfected into SL2 cells with increasing amounts of p50 or constant amounts of p50 and increasing amounts of Bcl-3. In addition, cells were transfected without or with Sp1 expression vector, as indicated. Relative luciferase activity is shown in panels B to E as mean values with standard deviations of three to four independent experiments.
The P-selectin promoter NF-κB site preferentially binds homodimers of p52 or p50 and is superactivated by Bcl-3 (42). Since binding of Sp1 or p50 should be mutually exclusive and p50 in fact displaces Sp1 from this site (Fig. 4B), a functional interference between Sp1 and NF-κB–p50 is expected. To address this problem, P-selectin promoter constructs with a wild-type or mutated NF-κB site were transfected into Drosophila SL2 cells. Without transfected Sp1, transcription of both wild-type and mutant promoters was severely impaired and very poorly responded to p50 or to p50 and Bcl-3 (Fig. 6E, left half). With transfected Sp1, the P-selectin promoter was strongly activated, and this activation was gradually decreased to the level of the mutant promoter when increasing amounts of p50 were transfected (Fig. 6E). A strong activation could be retrieved by cotransfection of increasing amounts of Bcl-3 in a dose-dependent manner. In contrast, the mutant P-selectin promoter showed an elevated level of transcription in the presence of Sp1, presumably mediated by cryptic sites elsewhere in the promoter, but did not respond to p50 or to p50 and Bcl-3.
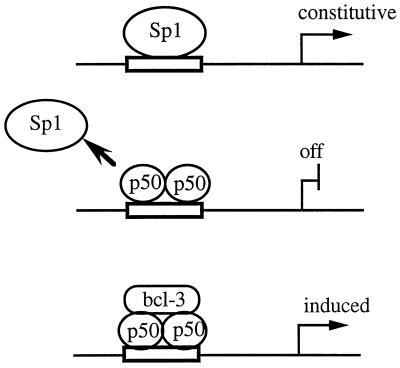
Thus, the P-selectin NF-κB binding site can confer a complex transcriptional regulation, depending on the relative affinities and abundance of the activators. In the absence of p50 homodimers it can be constitutively activated by Sp1, depending on the amount of Sp1. With excess p50 it is repressed, and in the presence of p50 and Bcl-3 it is strongly induced (Fig. 7).
FIG. 7.
Hypothetical scheme for the functional interaction of Sp1 with NF-κB sites. Sp1 occupies an NF-κB site and constitutively activates transcription in the absence of NF-κB (top). The exchange of Sp1 by NF-κB/Rel at certain sites (e.g., P-selectin) allows a switch from constitutive activation by Sp1 to repression by transcriptionally inactive p50 homodimers (center). The repressed gene can be induced depending on the availability of the Bcl-3 coactivator, which forms a ternary complex (bottom).
DISCUSSION
We have found a previously unrecognized mechanism of functional interference between NF-κB and Sp1. Cellular and recombinant Sp1 bind to a subset of NF-κB binding sites, including the elements in the IL-6 and P-selectin promoters, with a high affinity. For the most strongly interacting sites, a similar dissociation constant was determined as for a bona fide GC box Sp1 binding site. Several experimental procedures were used to characterize the interaction between Sp1 and NF-κB sites. We could place the relative affinities of Sp1 to eight different NF-κB sites in comparison to a GC box Sp1 site into the same order when comparing them either as probes or as competitors or when determining the dissociation constants. Cellular or recombinant NF-κB proteins competed with Sp1 for the binding site, implying overlapping base recognition of both factors. In transfected cells, Sp1 could stimulate transcription through NF-κB sites, and cotransfection of Sp1 with NF-κB–p50 indicated functional competition for the same site. Mutated NF-κB sites used in in vitro binding assays or in transfection experiments further supported the conclusion that Sp1 recognizes an overlapping motif in the NF-κB binding site. A one-base mutation in the Igκ site affecting a G4 motif at its third position abrogated DNA binding of both NF-κB and Sp1. Mutation in the first position strongly diminished transcriptional activation by Sp1. Similarly, mutation of the second or third guanine in a G5 motif in the P-selectin NF-κB site fully eliminated functional interaction with both Sp1 and NF-κB–p50. Despite the clearly overlapping features recognized by either Sp1 or NF-κB, it is unclear how Sp1 recognizes these sites. The augmenting number of binding sites identified for Sp1 has led to the deduction of increasingly degenerate consensus sequences (7, 8, 11, 23), but there is no perfect fit of these (e.g., [G/T][G/A]GG[C/A]G[G/T][G/A][G/A][C/T]) to any of the NF-κB sites preferentially bound by Sp1. In methylation interference experiments, we observed that methylation of all four G’s in the G4 motif of the Igκ site top strand blocked binding of purified Sp1 (data not shown). It is therefore possible that consecutive strings of G residues in NF-κB sites are part of a degenerate Sp1 site bound by one molecule. Alternatively, two Sp1 molecules may bind cooperatively to the NF-κB site with contacts to the strings of G residues on both sites of the palindrome.
The KDs for Sp1 bound to known Sp1 sites determined in other studies via quantitative footprinting assays or EMSAs were between 4.6 × 10−10 and 3.1 × 10−9 M (20, 32), in good agreement with our KD for a GC box Sp1 site (7 × 10−10 M) and close to the KDs that we measured for the Sp1 interaction with IL-6, P-selectin, or Igκ NF-κB binding sites (1.6 × 10−9 to 3.0 × 10−9 M). In contrast, the KDs reported for NF-κB vary greatly, from 6.5 × 10−10 M to the extremely high affinity of 3 × 10−13 M (15, 30, 34, 67). This wide range of affinities is largely due to the different experimental conditions chosen. In this study, we determined the relative dissociation constants of cellular Sp1 and NF-κB measured under the same experimental conditions, e.g., in the presence of the same competitor DNA and the same buffer components. The KDs which we determined for p50-p65 were between 6 × 10−10 M (H2K site) and 1.4 × 10−9 M (P-selectin site).
Recently, the crystal structure of p50 homodimers bound to a symmetrical NF-κB binding site has been determined (16, 38). The p50 homodimer wraps into the major groove, nearly enclosing the 10-bp binding site. Bases including all guanine residues in both half sites of the symmetrical binding site are contacted, and most of the binding site, except for a small part of the minor groove, is covered (16, 38). Sp1 is assumed to interact with guanine residues in the major groove, covering 9 consecutive residues, as has been derived from methylation protection and X-ray data (6, 43). The extended contacts made by Sp1 and NF-κB thus exclude simultaneous binding. This is in contrast to the synergistic interaction of NF-κB and the high-mobility-group protein HMG I(Y) at the virus-inducible enhancer element of the beta interferon gene (57). HMG I(Y) binds to a central region in the NF-κB site in the minor groove which is accessible in p50-DNA complexes (16, 38, 57).
NF-κB binding sites have been shown to be recognized by further transcription factors not belonging to the NF-κB/Rel family, including PRDII-BF1 (also called MBP-1 or HIV-EP1) and other highly related proteins (14, 37). PRDII-BF1 binds to NF-κB sites and contains several zinc finger structures, including the C2H2 type also found in Sp1. The functional role of PRDII-BF1 for the activity of NF-κB sites in vivo is unknown, but studies with chimeric PRDII-BF1 proteins suggest that it may act as a repressor (37). Furthermore, RBP-Jκ, also called RBP or CBF1, the mammalian homolog of Drosophila Suppressor of Hairless, binds to certain NF-κB sites and acts as a transcriptional repressor (26, 52). Sp1 is thus the first case of a non-Rel transcriptional activator that binds to a subset of NF-κB sites.
The recognition of subsets of NF-κB sites by Sp1 provides a number of possibilities for combinatorial regulation (Fig. 7). The interaction of these sites with Sp1 may raise the basal expression levels of NF-κB-dependent genes in the absence of activated NF-κB. Alternatively, it may provide a means to silence NF-κB target genes by competition for the binding site without functionally contributing to the basal expression level. In the latter case, Sp1 could compete selectively with specific NF-κB/Rel heteromers.
Although Sp1 is primarily regarded as a ubiquitous factor required in a variety of cell types for the activation of essential genes, both its expression levels and its activity are highly regulated. The expression levels of Sp1 vary strongly, up to 100-fold, in different tissues of the mouse as well as at different stages of development (49). A potential physiological importance of the high binding affinities determined here for Sp1 to certain NF-κB sites is therefore possible. In fact, single NF-κB knockout experiments, such as for p65 (RelA) or c-Rel, have demonstrated that basal expression levels of NF-κB target genes are not affected (5, 28). Although this may be largely due to compensation effects between the various NF-κB/Rel family members, the role of ubiquitous transcriptions factors such as Sp1 acting through certain NF-κB binding sites may provide an additional explanation.
ACKNOWLEDGMENTS
We thank Guntram Suske (IMT, Philipps University Marburg) for providing Sp1 expression vectors and for critical comments on the manuscript and Rodger P. McEver (University of Oklahoma Health Sciences Center) for the P-selectin constructs.
This work was in part supported by grant SFB 344 from the DFG to C.S.
REFERENCES
- 1.Augustin L B, Felsheim R F, Min B H, Fuchs S M, Fuchs J A, Loh H H. Genomic structure of the mouse delta opioid receptor gene. Biochem Biophys Res Commun. 1995;207:111–119. doi: 10.1006/bbrc.1995.1160. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 3.Ballard D W, Bohnlein E, Lowenthal J W, Wano Y, Franza B R, Greene W C. HTLV-I tax induces cellular proteins that activate the κB element in the IL-2 receptor α gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Baldwin A S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 5.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 6.Berg J M. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc Natl Acad Sci USA. 1992;89:11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 8.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 9.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 10.Courey A J, Holtzman D A, Jackson S P, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 11.Courey A J, Tjian R. Mechanism of transcriptional control as revealed by studies of human transcription factor Sp1. In: Yamamoto K R, McKnight S L, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 743–769. [Google Scholar]
- 12.Daniel S, Zhang S, DePaoli-Roach A A, Kim K H. Dephosphorylation of Sp1 by protein phosphatase 1 is involved in the glucose-mediated activation of the acetyl-CoA carboxylase gene. J Biol Chem. 1996;271:14692–14697. doi: 10.1074/jbc.271.25.14692. [DOI] [PubMed] [Google Scholar]
- 13.Demczuk S, Harbers M, Vennstrom B. Identification and analysis of all components of a gel retardation assay by combination with immunoblotting. Proc Natl Acad Sci USA. 1993;90:2574–2578. doi: 10.1073/pnas.90.7.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan C M, Maniatis T. A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev. 1990;4:29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- 15.Fujita T, Nolan G P, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh G, van Duyne G, Ghosh S, Sigler P B. Structure of NF-κB p50 homodimer bound to a κB site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 17.Grilli M, Chiu J J, Lenardo M J. NF-κB and Rel: participants in an multiform transcriptional regulatory system. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 18.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- 20.Harrington M A, Jones P A, Imagawa M, Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci USA. 1988;85:2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano F, Tanaka H, Makino Y, Okamoto K, Hiramoto M, Handa H, Makino I. Induction of the transcription factor AP-1 in cultured human colon adenocarcinoma cells following exposure to bile acids. Carcinogenesis. 1996;17:427–433. doi: 10.1093/carcin/17.3.427. [DOI] [PubMed] [Google Scholar]
- 22.Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 24.Jones P L, Kucera G, Gordon H, Boss J M. Cloning and characterization of the murine manganous superoxide dismutase-encoding gene. Gene. 1995;153:155–161. doi: 10.1016/0378-1119(94)00666-g. [DOI] [PubMed] [Google Scholar]
- 25.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 26.Kannabiran C, Zeng X, Vales L D. The mammalian transcriptional repressor RBP CBF1 regulates interleukin-6 gene expression. Mol Cell Biol. 1997;17:1–9. doi: 10.1128/mcb.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr L D, Ransone L J, Wamsley P, Schmitt M J, Boyer T G, Zhou Q, Berk A J, Verma I M. Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-κB. Nature. 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- 28.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation humoral immunity and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 29.Krappmann D, Wulczyn F G, Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 30.Kretzschmar M, Meisterernst M, Scheidereit C, Li G, Roeder R G. Transcriptional regulation of the HIV-1 promoter by NF-κB in vitro. Genes Dev. 1992;6:761–774. doi: 10.1101/gad.6.5.761. [DOI] [PubMed] [Google Scholar]
- 31.Kriwacki R V, Schultz S C, Steitz T M, Caradonna J P. Sequence-specific recognition of DNA by zinc-finger peptides derived from the transcription factor Sp1. Proc Natl Acad Sci USA. 1992;89:9759–9763. doi: 10.1073/pnas.89.20.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letovsky J, Dynan W S. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombardi L, Ciana P, Cappellini C, Trecca D, Guerrini L, Migliazza A, Maiolo A T, Neri A. Structural and functional characterization of the promoter regions of the NFKB2 gene. Nucleic Acids Res. 1995;23:2328–2336. doi: 10.1093/nar/23.12.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews J R, Kaszubska W, Turcatti G, Wells T N, Hay R T. Role of cysteine62 in DNA recognition by the P50 subunit of NF-κB. Nucleic Acids Res. 1993;21:1727–1734. doi: 10.1093/nar/21.8.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisterernst M, Gander I, Rogge L, Winnacker E-L. A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res. 1988;16:4419–4435. doi: 10.1093/nar/16.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moll T, Czyz M, Holzmuller H, Hofer-Warbinek R, Wagner E, Winkler H, Bach F H, Hofer E. Regulation of the tissue factor promoter in endothelial cells. Binding of NF-κB-, AP-1-, and Sp1-like transcription factors. J Biol Chem. 1995;270:3849–3857. doi: 10.1074/jbc.270.8.3849. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt C, Seeler J S, Nirula A, Shurland D L, Gaynor R B. Regulation of human immunodeficiency virus enhancer function by PRDII-BF1 and c-rel gene products. J Virol. 1992;66:244–250. doi: 10.1128/jvi.66.1.244-250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller C W, Rey F A, Sodeoka M, Verdine G L, Harrison S C. Structure of the NF-κB p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 39.Naumann M, Scheidereit C. Activation of NF-κB in vivo is regulated by multiple phosphorylations. EMBO J. 1994;13:4597–4607. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naumann M, Wulczyn F G, Scheidereit C. The NF-κB precursor p105 and the proto-oncogene product Bcl-3 are IκB molecules and control nuclear translocation of NF-κB. EMBO J. 1993;12:213–222. doi: 10.1002/j.1460-2075.1993.tb05647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neish A S, Williams A J, Palmer H J, Whitley M Z, Collins T. Functional analysis of the human vascular cell adhesion molecules 1 promoter. J Exp Med. 1992;176:1583–1593. doi: 10.1084/jem.176.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan J, McEver R P. Regulation of the human P-selectin promoter by Bcl-3 and specific homodimeric members of the NF-κB/Rel family. J Biol Chem. 1995;270:23077–23083. doi: 10.1074/jbc.270.39.23077. [DOI] [PubMed] [Google Scholar]
- 43.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 44.Pazin M J, Sheridan P L, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 45.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NFκB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domains of RelAp65 and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rokaeus A, Waschek J A. Primary sequence and functional analysis of the bovine galanin gene promoter in human neuroblastoma cells. DNA Cell Biol. 1994;8:845–855. doi: 10.1089/dna.1994.13.845. [DOI] [PubMed] [Google Scholar]
- 48.Roman D G, Toledano M B, Leonard W J. Sp1 represses IL-2 receptor alpha chain gene expression. New Biol. 1990;2:642–647. [PubMed] [Google Scholar]
- 49.Saffer J D, Jackson S P, Annarella M B. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–2199. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanceau J, Kaisho T, Hirano T, Wietzerbin J. Triggering of the human interleukin-6 gene by interferon-gamma and tumor necrosis factor-alpha in monocytic cells involves cooperation between interferon regulatory factor-1, NF-κB, and Sp1 transcription factors. J Biol Chem. 1995;270:27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- 51.Scheidereit C, Krappmann D, Wulczyn F G. Regulation of the NF-κB family of transcriptions factors by protein phosphorylation. In: Clemens M, editor. Protein phosphorylation and cell growth regulation. Amsterdam, The Netherlands: Harwood Academic Publishers; 1996. pp. 163–196. [Google Scholar]
- 52.Shirakata Y, Shuman J D, Coligan J E. Purification of a novel MHC class I element binding activity from thymus nuclear extracts reveals that thymic RBP-Jκ/CBF1 binds to NF-κB-like elements. J Immunol. 1996;156:4672–4679. [PubMed] [Google Scholar]
- 53.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 54.Sif S, Gilmore T D. Interaction of the v-Rel oncoprotein with cellular transcription factor Sp1. J Virol. 1994;68:7131–7138. doi: 10.1128/jvi.68.11.7131-7138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stade B G, Messer G, Riethmuller G, Johnson J P. Structural characteristics of the 5′ region of the human ICAM-1 gene. Immunobiology. 1990;182:79–87. doi: 10.1016/S0171-2985(11)80585-1. [DOI] [PubMed] [Google Scholar]
- 56.Sugimoto K, Tsuboi A, Miyatake S, Arai K, Arai N. Inducible and non-inducible factors co-operatively activate the GM-CSF promoter by interacting with two adjacent DNA motifs. Int Immunol. 1990;2:787–794. doi: 10.1093/intimm/2.8.787. [DOI] [PubMed] [Google Scholar]
- 57.Thanos D, Maniatis T. The high mobility group protein HMG IY is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 58.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 59.Ueberla K, Lu Y C, Chung E, Haseltine W A. The NF-κB p65 promoter. J AIDS Res. 1993;6:227–230. [PubMed] [Google Scholar]
- 60.Ueda A, Okuda K, Ohno S, Shirai A, Igarashi T, Matsunaga K, Fukushima J, Kawamoto S, Ishigatsubo Y, Okubo T. NF-κB and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J Immunol. 1994;153:2052–2063. [PubMed] [Google Scholar]
- 61.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 62.Viswanathan M, Yu M, Mendoza L, Yunis J J. Cloning and transcription factor-binding sites of the human c-rel proto-oncogene promoter. Gene. 1996;170:271–276. doi: 10.1016/0378-1119(95)00773-3. [DOI] [PubMed] [Google Scholar]
- 63.Vlach J, Garcia A, Jacque J-M, Rodriguez M S, Michelson S, Virelizier J-L. Induction of Sp1 phosphorylation and NF-κB-independent HIV promoter domain activity in T lymphocytes stimulated by okadaic acid. Virology. 1995;208:753–761. doi: 10.1006/viro.1995.1207. [DOI] [PubMed] [Google Scholar]
- 64.Wood L D, Farmer A A, Richmond A. HMGIY and Sp1 in addition to NF-κB regulate transcription of the MGSA/GROα gene. Nucleic Acids Res. 1995;23:4210–4219. doi: 10.1093/nar/23.20.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wulczyn F G, Krappmann D, Scheidereit C. The NF-κB and IκB gene families: mediators of immune response and inflammation. J Mol Med. 1996;74:749–769. doi: 10.1007/s001090050078. [DOI] [PubMed] [Google Scholar]
- 66.Yurochko A D, Kowalik T F, Huong S-M, Huang E-S. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zabel U, Schreck R, Baeuerle P A. DNA binding of purified transcription factor NF-κB. Affinity, specificity, Zn2+ dependence and differential half-site recognition. J Biol Chem. 1991;266:252–260. [PubMed] [Google Scholar]