Abstract
Histones are the fundamental components of the nucleosome. Physiologically relevant variation is introduced into this structure through chromatin remodeling, addition of covalent modifications, or replacement with specialized histone variants. The histone H3 family contains an evolutionary conserved variant, H3.3, which differs in sequence in only five amino acids from the canonical H3, H3.1, and was shown to play a role in the transcriptional activation of genes. Histone H3.3 contains a serine (S) to alanine (A) replacement at amino acid position 31 (S31). Here, we demonstrate by both MS and biochemical methods that this serine is phosphorylated (S31P) during mitosis in mammalian cells. In contrast to H3 S10 and H3 S28, which first become phosphorylated in prophase, H3.3 S31 phosphorylation is observed only in late prometaphase and metaphase and is absent in anaphase. Additionally, H3.3 S31P forms a speckled staining pattern on the metaphase plate, whereas H3 S10 and H3 S28 phosphorylation localizes to the outer regions of condensed DNA. Furthermore, in contrast to phosphorylated general H3, H3.3 S31P is localized in distinct chromosomal regions immediately adjacent to centromeres. These findings argue for a unique function for the phosphorylated isoform of H3.3 that is distinct from its suspected role in gene activation.
Keywords: mitosis, cell cycle, subtype modification
The building blocks of eukaryotic chromatin are nucleosomes that consist of 147 bp of DNA wrapped around histone octamers containing two copies each of the core histones H3, H4, H2A, and H2B (1). Covalent modifications of core histones, mostly occurring at the N termini, such as acetylation, methylation, and phosphorylation, can alter the conformation of nucleosomes and/or function as specific binding sites for enzymes that, in turn, alter chromatin structure (2-6). Alternatively, variation in chromatin can be introduced through the incorporation of variant histones into the nucleosome. For example, in mammals, in addition to the centromere-specific CENP-A variant, two nonallelic variants of the canonical histone H3 (H3.1) have been identified. One of these, H3.2, is closely related to H3.1 and only differs in a cysteine-serine substitution at amino acid position 96 and belongs to the family of S phase-dependent subtypes (7). In contrast, the second isoform, H3.3, is expressed throughout the cell cycle and differs from H3.1 in five amino acids. H3.3 contains a serine (S) at position 31, whereas an alanine (A) is found at this position in H3.1 and H3.2 (see Fig. 1 A). The other differences between H3.1/H3.2 and H3.3 are substitutions in the globular domain of the proteins and are crucial for their distinctive deposition during cell cycle (8). In Drosophila cells, H3.3 is found predominantly at active loci (8) and is enriched in covalent modifications associated with active chromatin, such as K4 and K79 methylation (9). Recently, H3.1 and H3.3 were shown to be deposited into chromatin by two distinct histone chaperone complexes, CAF-1 and HIRA, which are necessary to mediate DNA synthesis-dependent and -independent nucleosome assembly, respectively (10). Furthermore, Tagami and colleagues (10) showed that histones H3 and H4 might exist as dimeric units that are important intermediates in nucleosome formation: namely, that H3.1 and H3.3 do not assemble together in one nucleosome, but rather, pair as “homodimers.” Taken together, these observations suggest that nucleosomes within euchromatin are enriched in the histone H3.3 variant.
Fig. 1.
Specificity of the H3.3 S31P antibody. (A) Alignment of N-terminal tail of H3 of different species (Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melangolaster; Tt, Tetrahymena thermophila; Sc, Saccharomyces cerivisiae). S10, S28, and S31 are highlighted with gray boxes; amino acid positions are indicated at the bottom. S31, unique to H3.3, is indicated by an arrow. (B) The H3.3 S31P antibody recognizes only mitotic histones. Immunoblot with total acid-extracted histones from asynchronous growing (A) and nocodazole-arrested mitotic (M) HeLa cells. Only histones from mitotically arrested cells show a strong signal with the H3.3 S31P antibody. (C) Peptide competitions confirm specificity of the H3.3 S31P antibody. Peptides, as indicated, were incubated with the H3.3 S31P antibody before addition to a membrane with mitotic HeLa histones. In all cases, Ponceau staining of the membrane shows equal loading of histones.
Presented here is a demonstration of an H3.3 variant-specific posttranslational modification; we show that H3.3 is phosphorylated on S31 (H3.3 S31P) in vivo. H3.3 S31P is a mitosis-specific modification that differs from other mitotic marks, such as H3 S10P and H3 S28P, in both timing and localization. Whereas H3 S10 and H3 S28 phosphorylation first appear in prophase, persist until anaphase, and localize to outermost peripheral regions of the condensed DNA, H3.3 S31P is present only in late prometaphase and metaphase and forms a discrete speckled pattern over the dense region of the metaphase plate. Furthermore, like H3 S10P and H3 S28P, H3.3 S31P is excluded from centromeres. However, H3.3 S31P is enriched in distinct chromosomal areas immediately adjacent to centromeres. In contrast, H3 S10P and H3 S28P localize to the outer regions of chromosomes. Therefore, we provide evidence for a H3.3 variant-specific modification that is unique in its appearance in timing and localization during mitosis as compared with other general H3 mitotic marks. Our studies underscore an emerging theme, incorporation of histone variants into nucleosomes can alter the posttranlational modification “patterns” of the nucleosomes that incorporate them. Presumably, these changes, in turn, alter the DNA-templated processes in which these nucleosomes participate.
Materials and Methods
Cell Lines. HeLa and HEK293 cells were grown in Iscove's DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2.
Histone Isolation. Nuclei were isolated by hypotonic lysis in buffer containing 10 mM Tris·HCl (pH 8.0), 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.4 mM PMSF, and protease and phosphatase inhibitors. Pelleted nuclei were extracted by using 0.4 M sulfuric acid. The acid-soluble histones were precipitated with trichloroacetic acid and resuspended in water.
RP-HPLC. Histones from nocodazole-arrested HEK293 and HeLa cells were separated by RP-HPLC on a C8 column (220 × 4.6-mm Aquapore RP-300, PerkinElmer) by using a linear gradient of 35-60% solvent B (solvent A, 5% acetonitrile, 0.1% trifluoroacetic acid; solvent B, 90% acetonitrile, 0.1% trifluoroacetic acid) over 75 min at 1.0 ml/min on a Beckman Coulter System Gold 126 Pump Module and 166/168 Detector. The H3-containing fractions were dried under vacuum and stored at -80°C.
Immunoblotting (IB). Total acid-extracted or RP-HPLC-purified histone H3 fractions were separated on SDS/PAGE gel, transferred onto poly(vinylidene difluoride) membranes (Millipore), and stained with Ponceau S (Sigma) to ensure proper protein transfer. After incubation with primary antibodies that recognize specific histones or histone modifications (see Antibodies) and addition of a horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia), membranes were incubated with ECL-Plus substrate (Amersham Pharmacia), and proteins were detected by exposure to x-ray film (Amersham Pharmacia).
For peptide competition, the H3.3 S31P antibody was incubated for 1 h at 4°C with the following peptides (1 μg/ml) before its addition to the membrane: H3 unmodified, amino acids 19-35; H3 S10P, amino acids 1-20 with S10 phosphorylated; H3 S28P, amino acids 19-38 with S28 phosphorylated; H3.3 unmodified, amino acids 27-34; and H3.3 S31P, amino acids 27-34 with S31 phosphorylated. All peptides were synthesized and verified by MS at the Proteomics Resource Center of The Rockefeller University.
Immunofluorescence (IF) Microscopy. HeLa cells were grown on coverslips, washed, fixed in 1% formaldehyde-PBS solution, and permeabilized. After stepwise incubation with primary and then secondary fluorescent antibody, cells were stained with DAPI (Sigma) and mounted with Gel/Mount solution (Biomeda, Foster City, CA). For costaining with two primary rabbit antibodies the same procedure was performed twice with an additional blocking step in between. Stained cells were analyzed on an Axioskop 2 plus and Axioplan 2 microscope (Zeiss), and image processing was carried out with spot advanced (Diagnostic Instruments, Sterling Heights, MI), meta vue (Universal Imaging, Downingtown, PA), and photoshop (Adobe Systems, San Jose, CA) software packages.
Chromosome Spreads (CS). HeLa cells were arrested in mitosis by 16-h treatment with 200 ng/ml nocodazole (Sigma) and harvested by mitotic shake-off, and CS were prepared according to Perez-Burgos et al. (11). Stained chromosomes were analyzed on a Delta Vision Image Restoration 1 × 71 microscope (Olympus), and image processing was carried out with meta vue (Universal Imaging) and photoshop (Adobe Systems) software packages.
Antibodies. Antibodies were used in the following dilutions: H3.3 S31P (Abcam, Cambridge, MA), IB 1:1,000, IF and CS 1:100; H3 S10P (Upstate Biotechnology, Lake Placid, NY), IB 1:50,000, IF and CS 1:2,000; H3 S28P (Upstate Biotechnology), IB 1:5,000, IF and CS 1:1,100; CENP-A S7P (Upstate Biotechnology), IF 1:1,000; H3 (Upstate Biotechnology), IB 1:1,000; Aurora-B (Pharmingen), IF and CS 1:1,000; horseradish peroxidase-conjugated IgG (Amersham Pharmacia), IB 1:5,000; Rhodamine red-conjugated IgG (Jackson ImmunoResearch), IF and CS 1:1,000; Alexa Fluor 488-conjugated IgG (Invitrogen), IF and CS 1:1,000.
Results
H3.3 S31P Is a Mitosis-Specific Modification. In mammals, the canonical histones H3.1 and H3.2 differ in five and four amino acids, respectively, from the H3 variant H3.3. One of these sequence replacements is found at amino acid position 31, where H3.1 and H3.2 contain an alanine (A) and H3.3 possesses a serine (S) (Fig. 1 A, see arrow). H3.3 is evolutionary highly conserved and found in organisms from yeast to mammals, all containing S31 (S32 in Tetrahymena). This finding suggests that H3.3 is the common ancestor of histone H3 and that S31 might have a unique function. Because this residue is highly conserved, and because other serines in the N terminus (S10 and S28) have been shown to be phosphorylated during mitosis (9-11), we asked whether H3.3 S31 is phosphorylated and whether its phosphorylation might be cell cycle-dependent.
To address these questions, we used an antibody raised against a H3.3 S31P peptide (Abcam). Immunoblots with total acid-extracted histones from HeLa cells showed that H3.3 S31 was phosphorylated only in mitotic (nocodazole-arrested) and not in asynchronous growing cells (Fig. 1B). To validate the specificity of this antibody, we performed peptide competitions with a series of H3 synthetic peptides on immunoblots (Fig. 1C). Only the H3.3 S31P peptide fully competed away the H3.3 S31P antibody signal on mitotic HeLa histones, suggesting that the H3.3 S31P antibody specifically recognizes H3.3 phosphorylated on S31.
To determine whether the observed H3.3 S31P antibody staining of mitotic histones (Fig. 1B) was specific to H3.3, human histone H3 isoforms (H3.1, H.3.2, and H3.3) from HEK293 cells arrested in mitosis were fractionated by RP-HPLC (Fig. 2A). Histone H3 eluted in two distinct peaks under our RP-HPLC conditions (Fig. 2 A, peak 1 is indicated by 1, peak 2 is indicated by 2). Analysis of the HPLC fractions by MS showed that peak 1 contained a mixture of both H3.2 and H3.3, and peak 2 contained H3.1 (data not shown). Mass spectra recorded on the material eluting on the front side of peak 1 were assigned to H3.2, and spectra recorded from the late eluting shoulder were assigned to the isoform H3.3 (see arrow in Fig. 2 A and data not shown). Analysis of the H3.3 containing fractions by tandem MS showed that H3.3 S31 is phosphorylated in vivo and therefore confirmed our biochemical data showing that phosphorylation of H3.3 S31 is a unique mitotic mark (for details see Supporting Text and Fig. 5A, which are published as supporting information on the PNAS web site). S28 or S31 phosphorylation is easily detected on the H3.3 fragments (amino acids 27-40) by tandem MS. Interestingly, to date, no evidence has been obtained for a H3.3 fragment containing dual phosphorylated S28 and S31 (see Supporting Text and Fig. 5B). These data suggest that phosphorylation of S31 happens only in the absence of S28 phosphorylation, although we cannot exclude the existence of a rare population of dual phosphorylated peptides beyond our current MS detection limits. To further validate the purity of our H3 separation by RP-HPLC, all fractions from peak 1 and peak 2 were separated on SDS/PAGE gels, and proteins were visualized by Coomassie blue staining (data not shown).
Fig. 2.
S31 is phosphorylated in H3.3 during mitosis. (A) RP-HPLC profile of total histones from mitotic HEK293 cells shows two H3 peaks (1 and 2). Peak 1 contains H3.2 and H3.3 (shoulder, see arrow), whereas H3.1 is found only in peak 2. (B) IB analyses of RP-HPLC H3 fractions from mitotic HEK293 cells. Mitotic H3 containing fractions 65-75 were separated by SDS/PAGE gel and detected with antibodies against H3.3 S31P, H3 S10P, H3 S28P, and H3, as loading control in immunoblots. Phosphorylation of H3.3 at S31 is seen only in fractions containing H3.3 (fractions 68 and 69), but not in fractions containing H3.2 and H3.1. Antibodies against H3 S10P, H3 S28P, and general H3, as loading control, stained all mitotic H3-containing fractions. (C) Phosphatase treatment removes phospho-marks from mitotic histone samples. H3.2 and H3.3 containing mitotic fractions were separated by SDS/PAGE, blotted onto poly(vinylidene difluoride) membranes, and stained with antibodies against H3.3 S31P, H3 S10P, H3 S28P, and H3, as control. Treatment of mitotic H3.2 and H3.3 proteins with λ-phosphatase (Upstate Biotechnology) leads to essentially complete removal of all phospho marks assayed. Lane 1, H3 protein in water; lane 2, H3 protein with buffer and DTT, incubated for 30 min at 37°C; lane 3, the same as lane 2 with the addition of λ-phosphatase.
We then used the above H3-containing fractions for IB before probing the blots with antibodies against H3.3 S31P and H3 S10P/S28P. A general H3 antibody was used as a loading control (Fig. 2B). H3 S10P and H3 S28P antibodies stained all mitotic histone H3-containing fractions, regardless of the isoforms present. In contrast, the H3.3 S31P antibody recognized only H3 found in fractions that contained the H3.3 variant (fractions 68 and 69) and failed to react with fractions that contained H3.2 and H3.1. We also confirmed the specificity of the H3.3 S31P antibodies toward mitotic H3.3 by separating all three H3 species on 2D Triton acid-urea gels. Immunoblots with the H3.3 S31P antibody showed exclusive staining of the H3.3 spot and not spots for H3.1/H3.2 (data not shown).
Additionally, λ-phosphatase treatment of H3.2- and H3.3-containing fractions removed essentially all of the phospho-specific signal seen with H3.3 S31P and H3 S10P/S28P antibodies. This result underscores the dependency of the antibody on the presence of the phospho group in the context of the H3.3 sequence (Fig. 2C). Taken together, these data suggest that H3.3 is specifically phosphorylated at S31 during mitosis. Furthermore, we demonstrated the specificity of the H3.3 S31P antibody toward H3.3 phosphorylated at S31. These findings stimulated further research aimed to define the mitotic stages that exhibit phosphorylation on H3.3 S31 (see below).
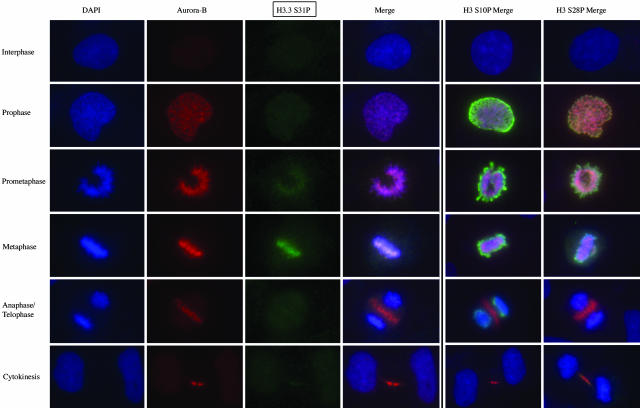
H3.3 S31P Is Restricted to Late Prometaphase and Metaphase Stages of Mitosis. In mammalian cells, site-specific phosphorylation of H3 S10 and H3 S28 is initiated during prophase, peaks during metaphase, diminishes during anaphase, and disappears during telophase (12-14). H3 S10P is also required for the initiation, but not maintenance, of mammalian chromosome condensation (15). Therefore, we conducted IF experiments to determine at what stage of mitosis H3.3 becomes phosphorylated on S31. To better distinguish between the different mitotic stages, we used an antibody against the so-called passenger protein Aurora-B kinase. Aurora-B first localizes to chromosomes in prophase, becomes concentrated at the centromere in prometaphase and metaphase, moves to the central spindle in anaphase, and eventually localizes to the midbody in cytokinesis (reviewed in ref. 16). Costaining of HeLa cells with H3.3 S31P, H3 S10P, and H3 S28P antibodies (green), Aurora-B kinase antibody (red), and DAPI to visualize DNA (blue) showed that phosphorylation of H3.3 S31 is initiated during late prometaphase and peaks in metaphase (Fig. 3 and see also Fig. 6A, which is published as supporting information on the PNAS web site). Phosphorylation of H3.3 S31 is not observed in interphase cells and is minimal in very early (prophase and early prometaphase) and late mitotic stages (anaphase and telophase). Therefore, the timing of H3.3 S31 phosphorylation differs from that observed for H3 S10P and H3 S28P, which are easily seen in prophase and anaphase stages (Fig. 3 Right).
Fig. 3.
H3.3 is phosphorylated at S31 during metaphase. Shown is IF microscopy of different mitotic stages of HeLa cells with antibodies against H3.3 S31P, H3 S10P, and H3 S28P (green) and all costained with Aurora-B antibody (red) and DAPI (blue). Cells in different mitotic stages are depicted from top to bottom and staining with antibodies is shown from left to right (see text for details). (Magnification: ×63.)
Localization of H3.3 S31P is also distinct from that observed for H3 S10P and H3 S28P. Strong staining of outermost, peripheral DNA regions was observed with the H3 S10P and H3 S28P antibodies (Figs. 3 and 6A). In contrast, the H3.3 S31P antibody showed speckled staining of the dense region of the metaphase plate and no enrichment in outer DNA regions. To better visualize this effect, we costained late prometaphase and metaphase HeLa cells with H3.3 S31P (green) and H3 S10P/S28P (red) antibodies and confirmed that these different phosphorylation “marks” localize to different regions in the mitotic chromatin (Fig. 6A). Additionally, costaining of HeLa cells with H3.3 S31P antibody (green), CENP-A S7P antibody (red), and DAPI (blue) confirmed that in prophase, when CENP-A becomes first phosphorylated on S7 by Aurora-B (17), H3.3 S31 is not phosphorylated (see Fig. 6B). We conclude that H3.3 S31P is a unique mitosis mark that is restricted to late prometaphase and metaphase stages of mitosis in HeLa cells.
H3.3 S31P Surrounds but Is Excluded from Centromeres. Because H3.3 S31P occurs at a very defined time frame during mitosis (late prometaphase and metaphase) and seems to be present in different chromosomal regions than H3 S10P/S28P, we investigated the exact localization of H3.3 S31 phosphorylation on metaphase chromosomes. Interestingly, IF of CS with H3.3 S31P and H3 S10P/S28P antibodies showed staining of distinct areas of metaphase chromosomes (Fig. 4). Consistent with previous research (16), we found H3 S10P and H3 S28P on the outer regions of chromosomes, excluded from centromeric and inner DNA-dense regions, and enriched in telomeric areas. In contrast, we observed H3.3 S31P on a distinct chromosomal location but not on the outer regions of chromosomes. To probe the location of H3.3 S31P further, we conducted costaining experiments on CS with antibodies to both H3.3 S31P (green) and Aurora-B kinase (red). The latter protein is known to localize to inner centromeres at metaphase (17). We found that H3.3 S31P and Aurora-B kinase occupy chromosomal regions in close proximity, but did not colocalize. Staining of CS from HeLa cells, expressing H3.3 fused to Flag/hemagglutinin peptide tags [gift from Y. Nakatani, Harvard Medical School, Boston (10)], with antibodies against hemagglutinin and H3.3 S31P showed that H3.3 localizes to the entire chromosome. In contrast, phosphorylation of S31 occurred only in distinct areas surrounding the centromeres (data not shown). These data suggest that only a subpopulation of H3.3 is phosphorylated at S31. We conclude that localization of H3.3 S31P on metaphase chromosomes is distinct from that of phosphorylated H3 S10/S28 and is restricted to regions immediately surrounding centromeres.
Fig. 4.
Phosphorylation of H3.3 S31 resides in a distinct chromosomal region immediately surrounding centromeric regions on metaphase chromosomes. CS of HeLa cells were stained with antibodies against H3.3 S31P, H3 S10P, and H3 S28P (green) and costained with an antibody against Aurora-B (red). DNA was visualized with DAPI (blue). H3.3 S31P antibody, in contrast to H3 S10P and H3 S28P antibodies, which stain outer regions of chromosomes, shows specific staining of chromosomal regions adjacent to centromeres. Higher magnification of one chromosome (star) is shown on the right of each staining panel. (Magnification ×100.)
Discussion
Historically, chromatin domains have been broadly classified into two forms: euchromatin, regions that are decondensed and thought to represent loci that are transcriptionally active, and heterochromatin, regions that are highly compacted and associated with silent DNA. Interestingly, some of these specialized domains in chromatin are enriched or diminished for specific histone variants, such as the H3 variant H3.3. For example, in Drosophila cells, H3.3 was found exclusively in active loci (8) and enriched in “active” modifications (9). However, we know of no other H3.3-specific modifications, i.e., marks that can distinguish it from its major H3 counterparts, H3.1 and H3.2. We show here that H3.3 is phosphorylated on the unique serine at amino acid position 31 during mitosis in vivo. In addition to CENP-AS7P (15), we have shown the existence of a H3 variant-specific covalent modification that cannot occur on the major members of the H3 family.
Histone phosphorylation occurs in many organisms and is associated with a wide range of biological functions, such as apoptosis (H2B S14P) (18), DNA repair (H2A.X S139P) (19), inducible gene activation (H3 S10P) (20), and mitosis (H3 S10P, H3 S28P, and T11P) (12, 14, 21). Whereas a mitosis-specific phosphorylation of H3 S10 and H3 S28 is necessary for the initiation of chromosome condensation in ciliates (22), a causal link is less clear in other organisms (23). Members of the Aurora AIR2-Ipl1 serine/threonine kinase family (Aurora-B in mammals) are responsible for many of these phosphorylation marks (24), and their action is counterbalanced by the activity of type 1 phosphatases (PP1), which remove the phosphate from H3 S10 in late anaphase and telophase (25). Although phosphorylation of H3.3 S31, H3 S10P, and H3 S28P occurs during mitosis, the initiation and progression of phosphorylation at H3.3 S31 differs from that of H3 S10/S28 in both time and space. Because of these differences, it is unlikely that Aurora-B kinase is responsible for H3.3 S31 phosphorylation. Additionally, comparison of sequences surrounding known histone targets of Aurora-B, H3 S10, H3 S28, and CENP-A S7 (17) suggests that this kinase has a preference for basic amino acids in positions -2 and -1 with regard to the phospho-acceptor site (see Fig. 1A). In position -2, all of the above sequences contain an arginine, which is likely important for Aurora-B target recognition and/or binding. In contrast, the -2 and the -1 positions relative to H3.3 S31 are alanine and proline. Moreover, Aurora-B, a member of the so-called passenger protein family (26), first localizes to chromosomes in prophase at the time H3 S10/S28 and CENP-A S7 phosphorylation is initiated, then becomes concentrated at the centromeres in prometaphase and metaphase, and eventually moves to the central spindle in anaphase (ref. 27 and reviewed in ref. 16). Because we did not observe overlapping staining of H3.3 S31 phosphorylation and Aurora-B on metaphase chromosomes (Fig. 4), it seems likely that a different mitotic kinase other than Aurora-B is responsible for phosphorylating H3.3 at S31.
Our studies report the existence of a unique H3.3 variant modification and add weight to emerging data, suggesting differences in biological function of H3.3 compared with H3.1 and H3.2 (6, 7). It is possible that, unlike H3 S10 and H3 S28 phosphorylation, phosphorylation of S31 is not necessary for chromosome condensation, especially if H3.3 S31P occurs only in the absence of S28P. Because of the late appearance of S31 phosphorylation in only a subpopulation of H3.3 in late prometaphase and metaphase and its distinct distribution around centromeres, it seems unlikely that this modification plays any pivotal role in the initial chromosome condensation event. What function then does H3.3 S31P serve? Speculative functions might include a role in the protection of euchromatin from the spreading of pericentric heterochromatin or a role in marking some H3.3 for replacement with canonical histone H3. Additionally, it is presently unknown whether a “transacting” binding protein exists that recognizes the phospho group on H3.3 S31, and if so, in what functional pathway it is involved. Because H3.3 S31 is conserved from yeast to human it will be of interest to determine whether S31P is also used during mitosis (or meiosis) in organisms other than human. Ongoing studies such as these may reveal insights into the function of the phosphorylation of S31 in H3.3 during mitosis in those organisms that use this mark.
Supplementary Material
Acknowledgments
We thank members of the C.D.A. and D.F.H. laboratories for insightful discussions and help; Emily Bernstein, Elizabeth Duncan, Christian Janzen, Joanna Wysocka, and Sean Taverna for critical review of the manuscript; and Alison North from The Rockefeller University Bio-Imaging Facility for advice and help with microscopy. This work was supported by National Institutes of Health Grants GM 40922 (to C.D.A.) and GM 37537 (to D.F.H.).
Author contributions: S.B.H. and C.D.A. designed research; S.B.H., B.A.G., M.K., S.P.B., and J.S. performed research; S.B.H., B.A.G., M.K., S.P.B., J.S., and D.F.H. analyzed data; S.B.H. and B.A.G. wrote the paper; and D.F.H. and C.D.A. corrected the manuscript and provided discussion.
Abbreviations: IB, immunoblotting; IF, immunofluorescence; CS, chromosome spreads.
References
- 1.van Holde, K. E. (1988) Chromatin (Springer, New York).
- 2.Wolffe, A. P. (1998) J. Exp. Zool. 282, 239-244. [PubMed] [Google Scholar]
- 3.Marmorstein, R. (2001) Nat. Rev. Mol. Cell Biol. 2, 422-432. [DOI] [PubMed] [Google Scholar]
- 4.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 5.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. [DOI] [PubMed] [Google Scholar]
- 6.Iizuka, M. & Smith, M. M. (2003) Curr. Opin. Genet. Dev. 13, 154-160. [DOI] [PubMed] [Google Scholar]
- 7.Franklin, S. G. & Zweidler, A. (1977) Nature 266, 273-275. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad, K. & Henikoff, S. (2002) Mol. Cell 9, 1191-1200. [DOI] [PubMed] [Google Scholar]
- 9.McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. (2004) Proc. Natl. Acad. Sci. USA 101, 1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. (2004) Cell 116, 51-61. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Burgos, L., Peters, A. H., Opravil, S., Kauer, M., Mechtler, K. & Jenuwein, T. (2004) Methods Enzymol. 376, 234-254. [DOI] [PubMed] [Google Scholar]
- 12.Gurley, L. R., D'Anna, J. A., Barham, S. S., Deaven, L. L. & Tobey, R. A. (1978) Eur. J. Biochem. 84, 1-15. [DOI] [PubMed] [Google Scholar]
- 13.Paulson, J. R. & Taylor, S. S. (1982) J. Biol. Chem. 257, 6064-6072. [PubMed] [Google Scholar]
- 14.Goto, H., Yasui, Y., Nigg, E. A. & Inagaki, M. (2002) Genes Cells 7, 11-17. [DOI] [PubMed] [Google Scholar]
- 15.Van Hooser, A., Goodrich, D. W., Allis, C. D., Brinkley, B. R. & Mancini, M. A. (1998) J. Cell Sci. 111, 3497-3506. [DOI] [PubMed] [Google Scholar]
- 16.Ducat, D. & Zheng, Y. (2004) Exp. Cell Res. 301, 60-67. [DOI] [PubMed] [Google Scholar]
- 17.Zeitlin, S. G., Barber, C. M., Allis, C. D. & Sullivan, K. F. (2001) J. Cell Sci. 114, 653-661. [DOI] [PubMed] [Google Scholar]
- 18.Cheung, W. L., Ajiro, K., Samejima, K., Kloc, M., Cheung, P., Mizzen, C. A., Beeser, A., Etkin, L. D., Chernoff, J., Earnshaw, W. C. & Allis, C. D. (2003) Cell 113, 507-517. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Capetillo, O., Lee, A., Nussenzweig, M. & Nussenzweig, A. (2004) DNA Repair (Amst) 3, 959-967. [DOI] [PubMed] [Google Scholar]
- 20.Clayton, A. L. & Mahadevan, L. C. (2003) FEBS Lett. 546, 51-58. [DOI] [PubMed] [Google Scholar]
- 21.Preuss, U., Landsberg, G. & Scheidtmann, K. H. (2003) Nucleic Acids Res. 31, 878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, Y., Mizzen, C. A., Cook, R. G., Gorovsky, M. A. & Allis, C. D. (1998) Proc. Natl. Acad. Sci. USA 95, 7480-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu, J. Y., Sun, Z. W., Li, X., Reuben, M., Tatchell, K., Bishop, D. K., Grushcow, J. M., Brame, C. J., Caldwell, J. A., Hunt, D. F., et al. (2000) Cell 102, 279-291. [DOI] [PubMed] [Google Scholar]
- 24.de la Barre, A. E., Gerson, V., Gout, S., Creaven, M., Allis, C. D. & Dimitrov, S. (2000) EMBO J. 19, 379-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murnion, M. E., Adams, R. R., Callister, D. M., Allis, C. D., Earnshaw, W. C. & Swedlow, J. R. (2001) J. Biol. Chem. 276, 26656-26665. [DOI] [PubMed] [Google Scholar]
- 26.Adams, R. R., Carmena, M. & Earnshaw, W. C. (2001) Trends Cell Biol. 11, 49-54. [DOI] [PubMed] [Google Scholar]
- 27.Crosio, C., Fimia, G. M., Loury, R., Kimura, M., Okano, Y., Zhou, H., Sen, S., Allis, C. D. & Sassone-Corsi, P. (2002) Mol. Cell. Biol. 22, 874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.