Abstract
Background & Aims
Transforming growth factor-β1 (TGF-β1) plays important roles in chronic liver diseases, including metabolic dysfunction–associated steatotic liver disease (MASLD). MASLD involves various biological processes including dysfunctional cholesterol metabolism and contributes to progression to metabolic dysfunction–associated steatohepatitis and hepatocellular carcinoma. However, the reciprocal regulation of TGF-β1 signaling and cholesterol metabolism in MASLD is yet unknown.
Methods
Changes in transcription of genes associated with cholesterol metabolism were assessed by RNA sequencing of murine hepatocyte cell line (alpha mouse liver 12/AML12) and mouse primary hepatocytes treated with TGF-β1. Functional assays were performed on AML12 cells (untreated, TGF-β1 treated, or subjected to cholesterol enrichment [CE] or cholesterol depletion [CD]), and on mice injected with adenovirus-associated virus 8–control/TGF-β1.
Results
TGF-β1 inhibited messenger RNA expression of several cholesterol metabolism regulatory genes, including rate-limiting enzymes of cholesterol biosynthesis in AML12 cells, mouse primary hepatocytes, and adenovirus-associated virus–TGF-β1–treated mice. Total cholesterol levels and lipid droplet accumulation in AML12 cells and liver tissue also were reduced upon TGF-β1 treatment. Smad2/3 phosphorylation after 2 hours of TGF-β1 treatment persisted after CE or CD and was mildly increased after CD, whereas TGF-β1–mediated AKT phosphorylation (30 min) was inhibited by CE. Furthermore, CE protected AML12 cells from several effects mediated by 72 hours of incubation with TGF-β1, including epithelial–mesenchymal transition, actin polymerization, and apoptosis. CD mimicked the outcome of long-term TGF-β1 administration, an effect that was blocked by an inhibitor of the type I TGF-β receptor. In addition, the supernatant of CE- or CD-treated AML12 cells inhibited or promoted, respectively, the activation of LX-2 hepatic stellate cells.
Conclusions
TGF-β1 inhibits cholesterol metabolism whereas cholesterol attenuates TGF-β1 downstream effects in hepatocytes.
Keywords: Cholesterol Metabolism, TGF-β1, Hepatic Fibrosis, MASLD
Summary.
Transforming growth factor-β1 down-regulates the expression of multiple genes involved in cholesterol metabolism and thus reduces cholesterol levels in hepatocytes. Cholesterol depletion in hepatocytes causes epithelial–mesenchymal transition, actin polymerization, cell apoptosis, and signals for hepatic stellate cell activation in a transforming growth factor-β1–dependent manner, whereas cholesterol enrichment attenuates the related outcomes.
The multifactorial metabolic dysfunction-associated steatotic liver disease (MASLD) is the most common cause of chronic liver disease.1 It encompasses a spectrum of pathologic conditions ranging from simple steatosis, metabolic dysfunction–associated steatohepatitis (MASH), and fibrosis/cirrhosis, which can progress further to hepatocellular carcinoma and liver failure. At the early stage of MASLD development, hepatocytes accumulate various lipids (triglycerides, free fatty acids, and cholesterol). This induces steatosis and increases the vulnerability to injury caused by lipotoxicity, mitochondrial dysfunction, cellular stress, and inflammation, thought to be required to progress to MASH.2 In this process, defective hepatocyte regeneration for the replenishment of dead cells and the adverse factors, including inflammatory factors, dietary factors, and lipopolysaccharide, are the proposed hits that drive the progression of MASLD to fibrosis.3 Later on, these insults lead to the activation of hepatic stellate cells (HSCs), accumulating collagen and extracellular matrix production/deposition and portal hypertension, thus worsening liver fibrosis to cirrhosis and even hepatocellular carcinoma (HCC).4
Hepatic lipotoxicity implies exposure to, or accumulation of, certain lipid species within hepatic cells that may initiate MASLD.5 Potentially lipotoxic molecules include cholesterol,6,7 free fatty acids,5 and diacylglycerol.8 Fifty percent of total cholesterol biosynthesis in human beings occurs in the liver, where dysregulation of cholesterol homeostasis in MASLD leads to cholesterol accumulation.9 Excess dietary cholesterol has been shown consistently to cause the development of experimental MASH in different animal models, including high-fat diet, obesity, and hyperphagia.10, 11, 12 In addition, cholesterol is essential for membrane structure and function because it packs with phospholipids to form lipid microdomains, affects membrane fluidity and modulates membrane trafficking, host–pathogen interaction, and signal transduction,13 including transforming growth factor-β (TGF-β) signaling.14, 15, 16, 17, 18 TGF-β1 is a 25-kilodalton homodimeric peptide, playing significant roles in various biological processes, such as extracellular matrix formation, epithelial–mesenchymal transition (EMT), cell proliferation and differentiation, apoptosis, immune and inflammatory responses, and metabolic pathways.19, 20, 21, 22, 23 TGF-βs signal through binding to plasma membrane complexes of type II (TβR-II) and type I (TβR-I) TGF-β receptors.24, 25, 26 Apart from signaling to the canonical Smad2/3 pathway,25,27,28 and depending on the cell type and context, TGF-β also stimulates several non-Smad pathways,29, 30, 31, 32, 33 of which a prominent one in hepatocytes is the phosphoinositide 3-kinase/protein kinase B (AKT) pathway.17 Statin treatment, which inhibits cholesterol synthesis, is associated with significant protection from steatosis, inflammation, and fibrosis among patients with MASLD and/or MASH.34 Moreover, diets with excess cholesterol mitigate liver fibrosis through oxidative stress–induced HSC-specific apoptosis in mice.35 Cholesterol-lowering statins also were reported to promote basal pancreatic ductal adenocarcinoma through activation of TGF-β signaling, leading to EMT.36 However, how cholesterol affects TGF-β signaling in the progression of MASLD is yet unknown, and it is unclear whether and how TGF-β affects cholesterol regulation.
In this study, we investigated the crosstalk between TGF-β1 signaling and cholesterol levels by using RNA-sequencing (RNA-seq) analysis and functional assays in hepatocytes, hepatic stellate cells, and wild-type mice overexpressing TGF-β1.
Results
TGF-β1 Inhibits Cholesterol Metabolic Processes According to RNA-Seq Data Analysis
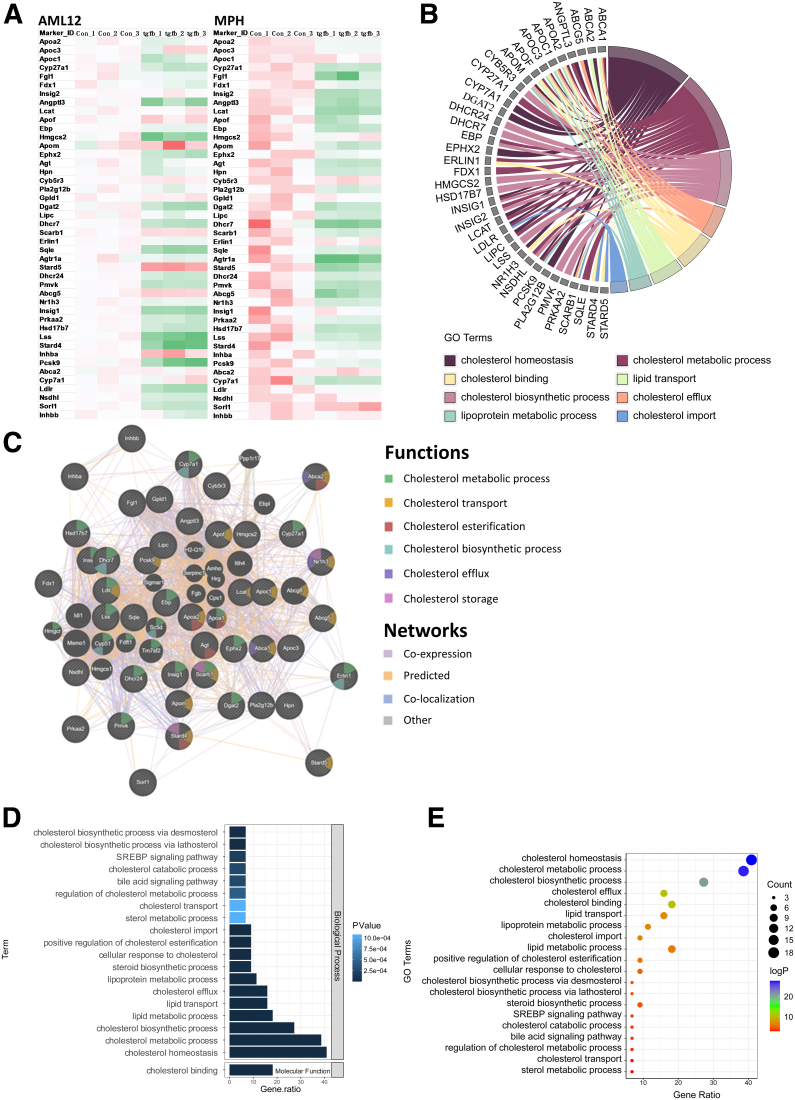
In MASLD, more cholesterol accumulates in the liver and the homeostasis of its metabolism becomes dysregulated in hepatocytes.37 Because TGF-β1 signaling is activated under these conditions,38 we investigated the effects of TGF-β1 on cholesterol metabolism in hepatocytes. Three control and TGF-β1 treatment groups of AML12 and mouse primary hepatocyte (MPH) cells were subjected to RNA-seq, and the genes involved in cholesterol metabolism were analyzed. TGF-β1 treatment for 24 hours significantly inhibited transcription of multiple cholesterol metabolism genes, as shown in the heatmaps of AML12 and MPH cells (Figure 1A). These include, among others, Apoa2, Apoc3, Cyp27a1, Fgl1, Insig2, Lcat, Hmgcs2, Aqle, Abcg5, Lss, and Cyp7a1. The chord diagram shows that these down-regulated genes are related to cholesterol homeostasis, biosynthesis, metabolic processes, efflux, import, cholesterol binding, lipid transport, and lipoprotein metabolism (Figure 1B), hence supporting the critical role of TGF-β1 in reducing cholesterol metabolism in hepatocytes. Furthermore, networking analysis shows that the majority of these genes display coexpression and colocalization (Figure 1C). Analysis of the signaling pathways shows that there also is notable down-regulation of genes involved in enhancing cholesterol esterification, bile acid signaling pathways, sterol regulatory element binding protein signaling pathway, and cellular response to cholesterol (Figure 1D and E).
Figure 1.
TGF-β1 inhibits cholesterol metabolism in hepatocytes. (A) Heatmap of the down-regulated genes involved in cholesterol metabolism upon 24-hour TGF-β1 treatment in AML12 cells and MPHs. number of independent experiments [n] = 3. (B) The chord diagram illustrates the main biological processes of the decreased genes. (C) Networking interpretation of the down-regulated genes. (D and E) Signaling pathway analysis of the inhibited cholesterol metabolic genes. GO, Gene Ontology.
TGF-β1 Inhibits Cholesterol Biosynthesis and Accumulation in Hepatocytes
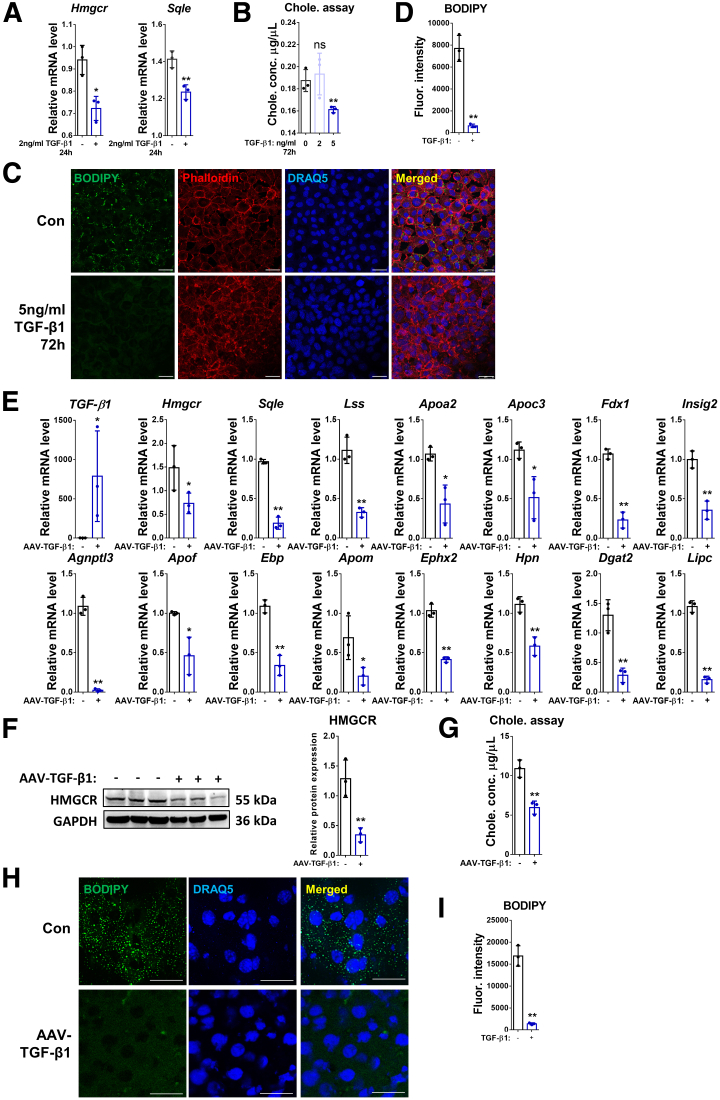
According to the RNA-seq analysis, TGF-β1 inhibits expression of multiple genes involved in cholesterol biosynthesis. This includes the 3 crucial players of the cholesterol biosynthetic pathway, 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgcr), squalene monooxygenase, and lanosterol synthase, which were verified by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) in AML12 cells (control vs TGF-β1–treated). This result shows that 24-hour incubation with TGF-β1 had an inhibitory effect on messenger RNA (mRNA) expression of Hmgcr and Sqle compared with the control group (Figure 2A). The cholesterol assay showed a reduced concentration of cholesterol in AML12 cells incubated with 5 ng/mL TGF-β1 for 72 hours, but not with 2 ng/mL TGF-β1 (Figure 2B). Treatment with 5 ng/mL TGF-β1 (72 h) also reduced lipid droplet accumulation in AML12 cells, identified by BODIPY (D-3922; Life Technologies, Waltham, MA) staining (Figure 2C and D). Next, we examined the effects of overexpressing TGF-β1 on cholesterol metabolism in the liver by injecting mice with adenovirus-associated virus (AAV)8–TGF-β1 (or AAV8–control). RT-qPCR showed that overexpression of TGF-β1 reduced, among others, the mRNA levels of Hmgcr, Sqle, Lss, Apoa2, Apoc3, Fdx1, Insig2, Agnptl3, Apof, and Ebp (Figure 2E). Western blot further confirmed that HMGCR, the rate-limiting enzyme of cholesterol biosynthesis, also is reduced at the protein level in the liver of AAV–TGF-β1–treated mice (Figure 2F). In addition, the level of total cholesterol in the liver tissue was decreased by TGF-β1 overexpression (Figure 2G). Accordingly, lipid droplet accumulation was decreased notably in the liver tissue of AAV8–TGF-β1–injected mice compared with the control group (Figure 2H and I).
Figure 2.
TGF-β1 inhibits cholesterol biosynthesis and accumulation in hepatocytes. (A) Relative mRNA levels of Hmgcr and Sqle were determined by RT-qPCR in AML12 cells treated with/without TGF-β1 for 24 hours. (B) Total cholesterol concentration was examined in AML12 cells treated (or not) with 2 or 5 ng/mL TGF-β1 for 72 hours. (C) Lipid droplet accumulation in AML12 cells was examined by BODIPY staining. Phalloidin was used for F-actin staining. (D) Quantification of F-actin staining. (E) Relative mRNA levels of TGF-β1, and genes involved in cholesterol metabolism as shown in the figure were determined by RT-qPCR in liver tissue of mice treated with AAV8–control or AAV8–TGF-β1. (F) Protein expression of HMGCR in the liver tissue of control or AAV8–TGF-β1 mice was determined by Western blot. (G) Total cholesterol concentration was examined in the liver tissue of mice treated with AAV8–control or AAV8–TGF-β1. (H) Lipid droplet accumulation in liver tissue of mice treated with AAV8–control or AAV8–TGF-β1 was examined by BODIPY staining. (I) Quantification of BODIPY staining. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, and NS = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. For immunofluorescence, DRAQ5 was used to stain the nuclei. Scale bars: 25 μm. Images were chosen representatively from 3 independent experiments. Quantification of protein expression or staining was performed using ImageJ (National Institutes of Health). Chole., cholesterol; Con, control; conc., concentration; Fluor., fluorescence.
Cholesterol Depletion and Enrichment Have Different Effects on TGF-β1–Mediated Smad2/3 and AKT Phosphorylation
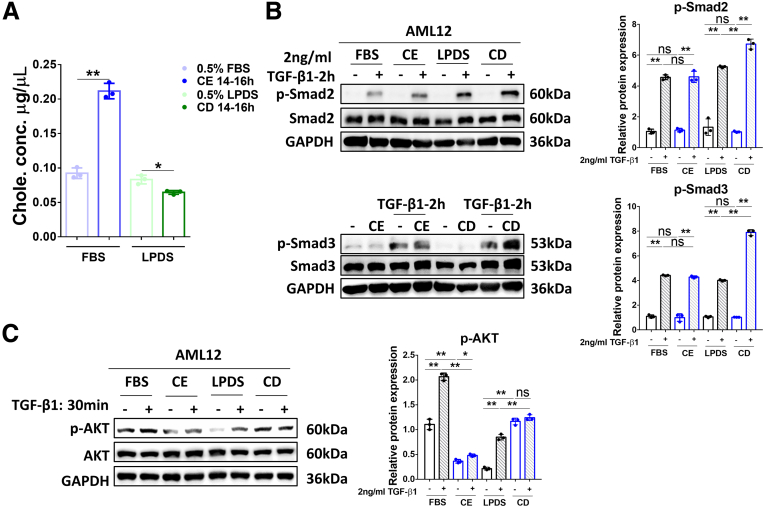
Because TGF-β1 inhibits cholesterol biosynthesis (Figures 1 and 2), we next tested the effects of cholesterol on TGF-β1 signaling. Incubation with cholesterol-methyl-β-cyclodextrin (MβCD) complex (5 mmol/L MβCD, 300 μg/mL cholesterol) or with 50 μmol/L lovastatin plus 50 μmol/L mevalonate (MVL) induced cholesterol enrichment (CE) or cholesterol depletion (CD), respectively, in AML12 cells, compared with the control groups (Figure 3A). To investigate the effect of CE and CD on TGF-β1 signaling, AML12 cells were subjected to CE or CD treatments (or untreated; control) for 14–16 hours, as described in Materials and Methods. Subsequently, the cells were serum-starved (0.5% fetal bovine serum [FBS] or lipoprotein-deficient serum [LPDS]) for 4 hours, before treatment with or without 2 ng/mL TGF-β1 for 2 hours (Smad2/3 signaling) or 30 minutes (AKT signaling). Western blot analysis for phosphorylated Smad2/3 (p-Smad2/3) and Smad2/3 showed that Smad2/3 phosphorylation in response to TGF-β1 (2 h) proceeded after either CE or CD treatments, with no effects for CE and some enhancement of p-Smad2/3 by CD (Figure 3B). On the other hand, CE treatment (but not CD) prevented TGF-β1–mediated (30 min) AKT phosphorylation at Ser473 (Figure 3C).
Figure 3.
Alteration of the cholesterol level in AML12 cells affect different TGF-β1–mediated signaling to Smad2/3 and to AKT. (A) Total cholesterol concentration was examined in AML12 cells treated with 5 mmol/L cholesterol–MβCD complex (CE) or 50 μmol/L lovastatin and 50 μmol/L MVL (CD), respectively, for 14–16 hours. (B) AML12 cells treated for CE, CD, or untreated (control) then were stimulated with 2 ng/mL TGF-β1 for 2 hours. The levels of p-Smad2 and total Smad2 (upper panels) or p-Smad3 and total Smad3 (lower panels) were determined by Western blot analysis. (C) Cells treated to alter their cholesterol levels as described earlier were incubated with 2 ng/mL TGF-β1 for 30 minutes and subjected to Western blot of p-AKT and total AKT. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, NS = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Quantification of protein expression was performed using ImageJ (National Institutes of Health). Chole., cholesterol; p-AKT, phosphorylated-protein kinase B.
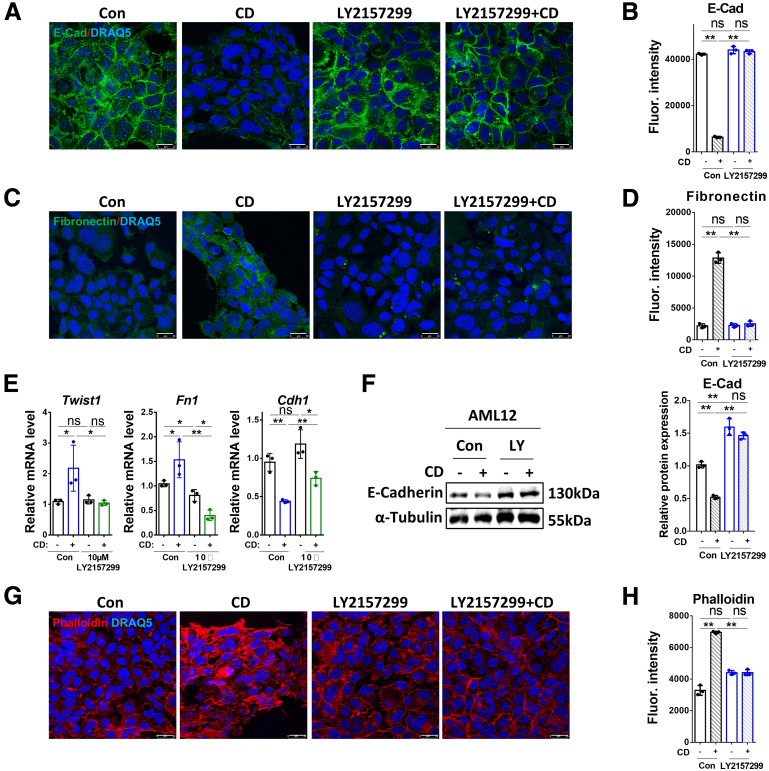
TGF-β1–Induced EMT and Stress Fiber Formation Are Inhibited by Cholesterol Enrichment and Promoted by Cholesterol Depletion
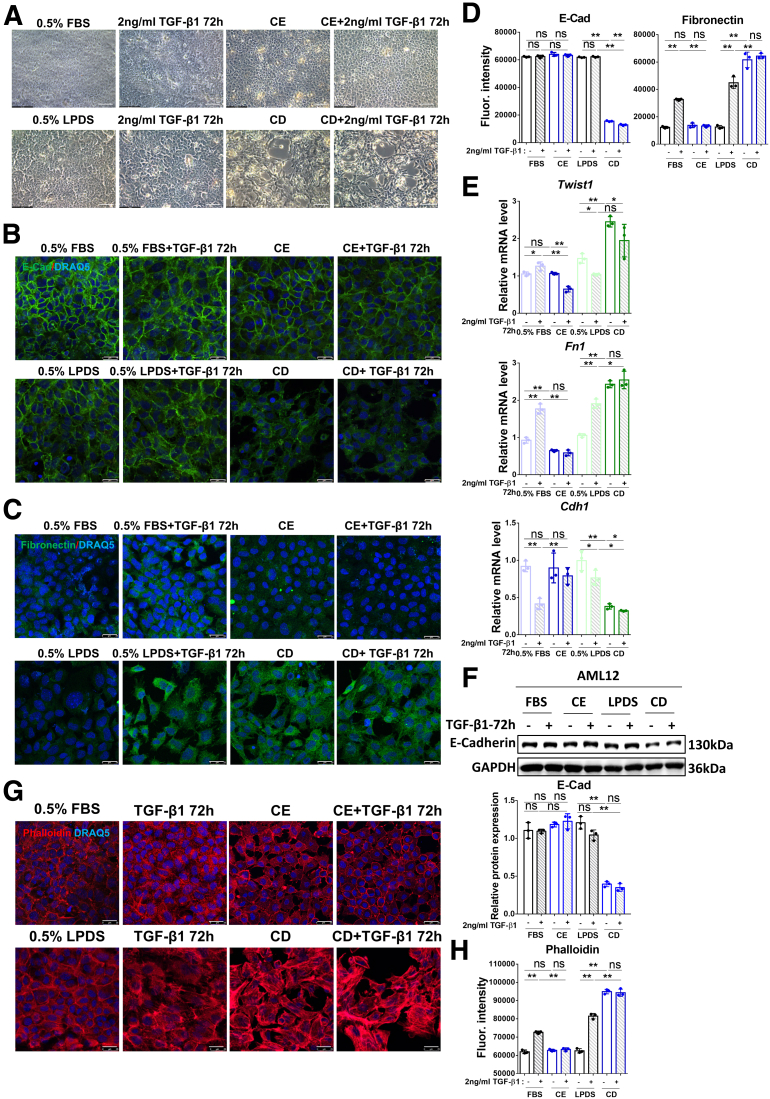
AML12 cells undergo morphologic changes after CE or CD treatments, which also affect morphologic changes induced by prolonged incubation (72 h) with TGF-β1 (Figure 4A). We therefore investigated the effects of TGF-β1 on EMT and actin polymerization in AML12 (untreated, or subjected to CE or CD). TGF-β1 treatment for 72 hours induced delocalization of E-cadherin from the cell membrane, and its membrane localization was re-established by CE treatment (Figure 4B). However, E-cadherin membrane localization was decreased further by CD treatment alone (Figure 4B). TGF-β1–induced fibronectin expression in the membrane was abrogated by CE, but enhanced further by CD, as shown by immunofluorescence (Figure 4C). The quantification of the immunofluorescence staining of E-cadherin and fibronectin are shown in Figure 4D. RT-qPCR confirmed the up-regulation of the mRNA levels of EMT markers Twist1 and Fn1, and the down-regulation of Cdh1 by TGF-β1, and the prevention of such TGF-β1–induced changes by CE; on the other hand, they could be phenocopied further by CD (Figure 4E). E-cadherin expression at the protein level was decreased in the CD incubation group, as identified by Western blot (Figure 4F). Although TGF-β1 inhibited Cdh1 mRNA expression (Figure 4E, third histogram), there was no obvious decrease in its protein level (Figure 4F). According to the immunofluorescence results (Figure 4B and D), TGF-β1 mainly disrupted E-cadherin membrane localization rather than its expression. In addition, actin polymerization induced by TGF-β1 was reduced to normal levels by CE, whereas CD dramatically promoted this outcome, as indicated by staining of F-actin with phalloidin (Figure 4G and H). These findings indicate that CE, which alters the balance between the stimulation of the Smad2/3 and AKT pathways, prevented TGF-β1–induced EMT and actin polymerization, perhaps because of its inhibitory effect on AKT signaling (Figure 3C). This did not occur in the case of CD treatment, in which AKT phosphorylation was not inhibited, whereas that of Smad2 may have been increased (Figure 3B). This different balance between the 2 pathways may be involved in the further aggravation by CD of TGF-β1–mediated outcomes in AML12 cells.
Figure 4.
CE inhibits TGF-β1–induced EMT and stress fiber formation whereas CD promotes these effects. (A) Brightfield images of the morphology of AML12 cells treated with CE/CD combined with/without 2 ng/mL TGF-β1 for 72 hours. Scale bars: 50 μm. (B–D) The expression and location of E-cadherin and fibronectin were determined by immunofluorescence in AML12 cells treated with CE/CD combined with/without 2 ng/mL TGF-β1 for 72 hours. (E) Relative mRNA levels of Twist, Fn1, and Cdh1, and (F) protein levels of E-cadherin were determined by RT-qPCR and Western blot, respectively, in AML12 cells with the conditioned treatment as shown in the figure. (G) Alexa Fluor 568 phalloidin staining showing actin polymerization in AML12 cells treated with CE/CD combined with/without 2 ng/mL TGF-β1 for 72 hours. (H) Quantification of phalloidin staining. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, NS = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. For immunofluorescence, DRAQ5 was used to stain the nuclei. Scale bars: 25 μm. Images were chosen representatively from 3 independent experiments. Quantification of protein expression or staining was performed using ImageJ (National Institutes of Health). E-cad, E-cadherin; Fluor., fluorescence.
Cholesterol Depletion Promotes EMT and Actin Polymerization in a TGF-β1–Dependent Manner
In view of the induction of EMT and actin polymerization by CD, we investigated whether these effects depend on the kinase activity of TβRI. AML12 cells were pretreated for 2 hour with 10 μmol/L of the TβR-I kinase inhibitor LY2157299, followed by 14-hour CD treatment. The AML12 cells then were incubated (or not) with LY2157299 for 72 hours. CD treatment induced delocalization of E-cadherin from the cell membrane, which was rescued by LY2157299, as shown by E-cadherin immunofluorescence (Figure 5A and B). Moreover, the induction of fibronectin by CD also was inhibited by LY2157299 (Figure 5C and D). These results also were evident in the effects of CD on the mRNA expression of EMT markers, which were abolished by LY2157299 (Figure 5E). A similar pattern was observed for the expression of E-cadherin, as measured by Western blot (Figure 5F). Furthermore, CD-mediated actin polymerization was reduced to normal levels by treatment with the inhibitor (Figure 5G and H). These results suggest that CD-induced EMT and actin polymerization depend on signaling by the TGF-β receptors.
Figure 5.
CD-mediated EMT and actin polymerization depend on TGF-β1. (A–D) The expression and location of E-cadherin and fibronectin were determined by immunofluorescence in AML12 cells treated with CD combined with/without 10 μmol/L LY2157299 for 72 hours. (E) Relative mRNA levels of Twist, Fn1, and Cdh1, and (F) protein levels of E-cadherin were determined by RT-qPCR and Western blot, respectively, in AML12 cells with the conditioned treatment as shown in the figure. (G) Alexa Fluor 568 phalloidin staining showing actin polymerization in AML12 cells treated with CD combined with/without 10 μmol/L LY2157299 for 72 hours. (H) Quantification of phalloidin staining. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, ns = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. For immunofluorescence, DRAQ5 was used to stain the nuclei. Scale bars: 25 μm. Images were chosen representatively from 3 independent experiments. Quantification of protein expression or staining was performed using ImageJ (National Institutes of Health). Con, control; E-cad, E-cadherin; Fluor., fluorescence; LY, LY2157299.
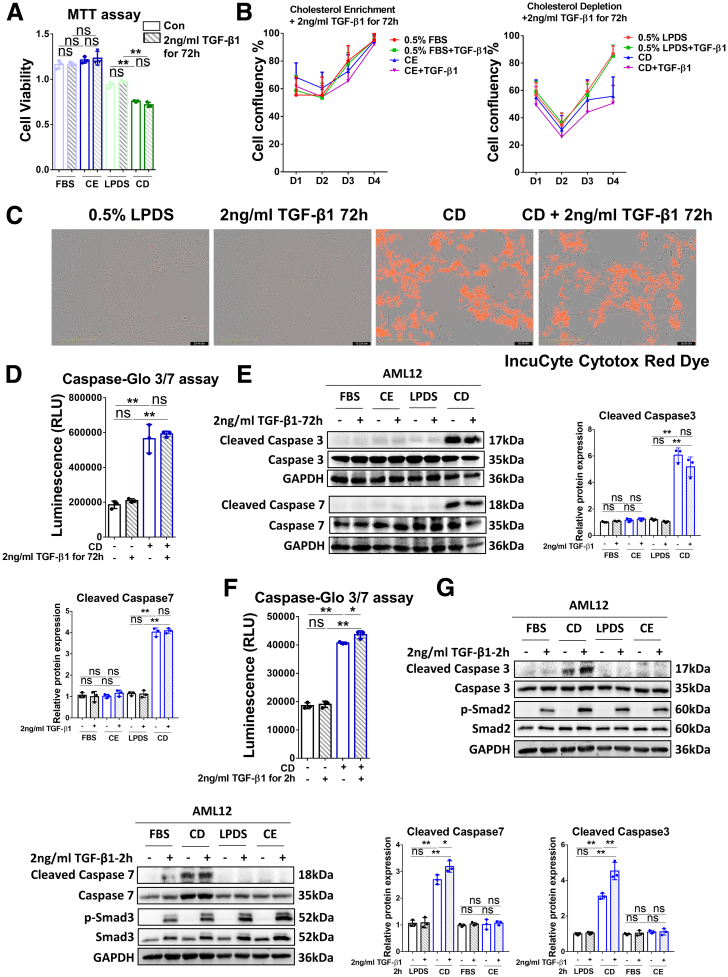
Cholesterol Depletion Induces Apoptosis Through the Caspase 3/7 Pathway
To investigate the effect of long-term TGF-β1 treatment in combination with CE or CD on cell viability, we performed a MTT assay. As shown in Figure 6A, this assay displayed that CD treatment significantly inhibited cell proliferation, whereas CE or TGF-β1 treatment alone did not have an obvious effect. Next, we conducted cell death measurements with IncuCyte Cytotox Red Dye. Cell number analysis confirmed that CD inhibited cell proliferation and markedly promoted cell death, whereas a 72-hour incubation with TGF-β1 alone or CE treatment without or with TGF-β1 did not have a significant effect on cell viability (Figure 6B and C). To further identify the mechanisms underlying CD-induced cell death, we performed the caspase-Glo 3/7 assay and examined expression of cleaved and total caspase 3/7 by Western blot. These studies showed that CD treatment enhanced the signals of cleaved caspase 3/7, whereas treatment with TGF-β1 for 72 hours alone did not affect the luminescence measured (Figure 6D). Western blot analysis further supported the notion that CD, but not CE, significantly increased cleaved caspase 3/7 (Figure 6E). Together, the MTT, IncuCyte Cytotox Red Dye, and caspase-Glo 3/7 assays suggest that the CD-mediated reduction in cell viability occurs through induction of apoptosis.
Figure 6.
CD induces cell apoptosis through the caspase 3/7 pathway. (A) The viability of AML12 cells treated with CE/CD ± TGF-β1 incubation for 72 hours was examined by MTT assay. (B) The confluency of AML12 cells subjected to the conditioned treatments indicated were analyzed by the IncuCyte machine. (C) Cell apoptosis was examined by IncuCyte Cytotox Red Dye in AML12 cells treated with/without CD ± TGF-β1 for 72 hours. Scale bars: 50 μm. (D) Cleaved caspase signals were tested by the caspase–Glo 3/7 assay kit in AML12 cells treated with/without CD ± TGF-β1 for 72 hours. (E) Protein levels of cleaved caspase 3/7 and caspase 3/7 were determined by Western blot in AML12 cells treated for CE/CD combined with/without 2 ng/mL TGF-β1 for 72 hours. (F) Cleaved caspase signals were tested by the caspase–Glo 3/7 assay kit in AML12 cells treated with/without CD ± TGF-β1 for 2 hours. (G) Protein levels of cleaved caspase 3/7, caspase 3/7, p-Smad2/3, and total Smad2/3 were determined by Western blot analysis in AML12 cells subjected (or not; FBS) to CE/CD, combined with/without 2 ng/mL TGF-β1 for 2 hours. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, ns = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Quantification of protein expression was performed using ImageJ (National Institutes of Health). Images were chosen representatively from 3 independent experiments. Con, control; RLU, relative light unit.
Our results suggest that prolonged treatment (72 h) with TGF-β1 does not suffice for induction of apoptosis. However, it was reported that TGF-β1 signaling induced apoptosis39 and could be enhanced by cholesterol depletion.36 We therefore tested the short-term (2 h) effect of TGF-β1 on cell apoptosis with/without CE or CD treatments. The caspase-Glo 3/7 assay showed that incubation with TGF-β1 for 2 hours alone did not have any effect on the luminescence, whereas CD treatment alone notably increased the caspase signals. Of note, CD treatment combined with 2 hours of TGF-β1 treatment increased the assays' readout substantially, as compared with the CD-alone group (Figure 6F). In addition, Western blot showed that a 2-hour incubation with TGF-β1 further enhanced cleaved caspase 3/7 and p-Smad2/3 levels, compared with the CD treatment group (Figure 6G). In conclusion, CD significantly promoted cell apoptosis, and short-term incubation (2 h) with TGF-β1 further enhanced this effect through the activation of caspase 3/7 pathways. These findings are in accord with the reported potentiation of TGF-β1 signaling by cholesterol depletion.36
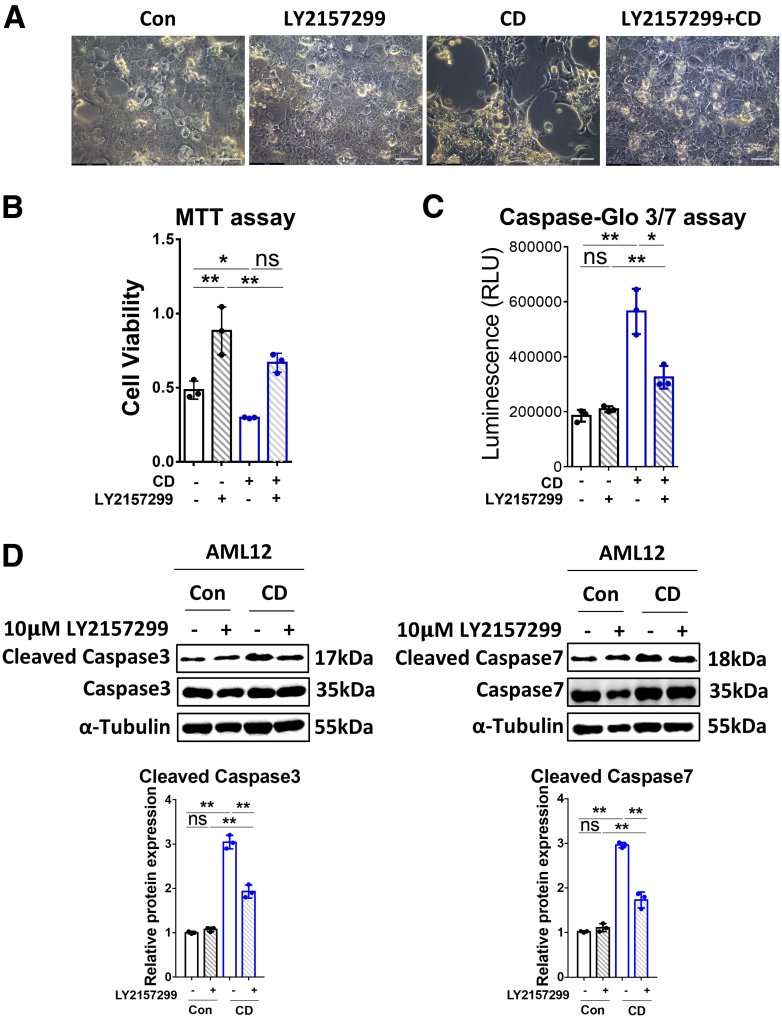
Cholesterol Depletion Promotes Cell Apoptosis Through TGF-β1 Signaling
To explore whether the induction of apoptosis by CD involves TGF-β1 signaling, we tested the effect of treating AML12 cells by LY2157299 (10 μmol/L, 2 h) before CD treatment (14 h), followed by incubation for up to 72 hours with the inhibitor kept in. As shown in Figure 7A, the cell numbers were reduced in the CD-treated group, an effect that was rescued by LY2157299. Quantification using the MTT assay showed that the TβR-I inhibitor promoted, whereas CD inhibited, cell proliferation compared with the control group (Figure 7B). Notably, LY2157299 retained cell viability despite CD treatment (Figure 7B). The caspase-Glo 3/7 assay (Figure 7C) and Western blot for the cleaved caspase 3/7 (Figure 7D) further confirmed that blocking TGF-β1 signaling partially inhibited CD-induced expression of cleaved caspase 3 and 7. Taken together, these findings suggest that CD-induced cell apoptosis is dependent on TGF-β1 signaling.
Figure 7.
CD promotes cell apoptosis through TGF-β1 signaling. (A) Brightfield images of AML12 cells treated (72 h) with CD alone, LY2157299 (10 μmol/L), or their combination. Untreated cells served as control. Scale bars: 50 μm. (B) The viability of AML12 cells treated for 72 hours with/without CD ± 10 μmol/L LY2157299 was examined by MTT assay. (C) Cleaved caspase signals were tested by the caspase–Glo 3/7 assay kit on AML12 cells subjected (or not; control) to CD treatment ± 10 μmol/L LY2157299 for 72 hours. (D) Western blot of cleaved and total caspase 3/7 in AML12 cells subjected to CD treatment with/without 10 μmol/L LY2157299 for 72 hours. For RT-qPCR, mouse Ppia was used as endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01, NS = P > .05. For Western blot, glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. Quantification of protein expression was performed using ImageJ (National Institutes of Health). Images were chosen representatively from 3 independent experiments. Con, control; RLU, relative light unit.
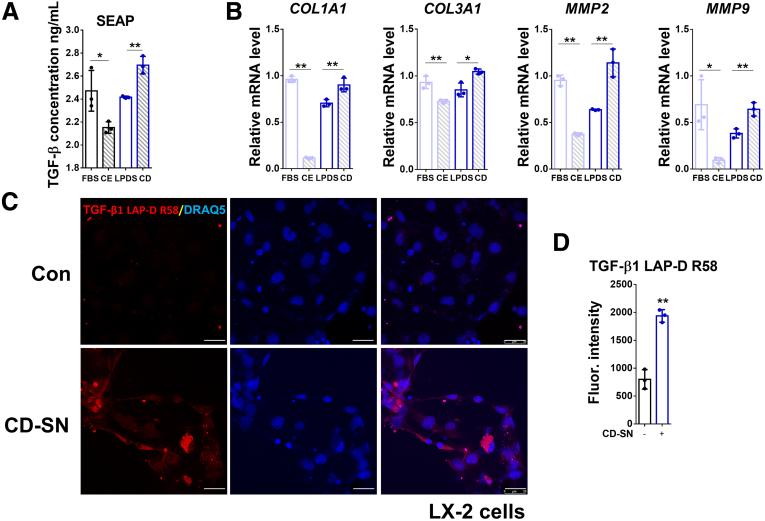
Supernatants From CE-Treated and CD-Treated AML12 Cells Have Opposing Effects on HSC Activation
Next, we investigated the effects of cholesterol levels on the crosstalk between hepatocytes (AML12) and immortalized hepatic stellate cells (LX-2). To do so, AML12 cells were subjected (or not; control) to CE or CD treatments, followed by 72 hours pf incubation without CE- or CD-inducing reagents. Supernatants from AML12 cells were collected, and LX-2 cells were incubated for 72 hours with these supernatants. Then, the conditioned LX-2 medium or the cells themselves were collected for further analysis by the secreted embryonic alkaline phosphatase (SEAP) reporter assay to quantify the amount of active TGF-β1 present in conditioned LX-2 supernatants, and the cells were subjected to RT-qPCR of HSC-specific hepatic fibrosis markers. The SEAP assay results indicated that the active TGF-β present in conditioned LX-2 supernatants was decreased in the CE group, whereas it was increased in the CD group (Figure 8A). Consistent with the changes observed for levels of active TGF-β1, HSC activation, or HSC-specific fibrosis markers, including COL1A1, COL3A1, MMP2, and MMP9, notably were inhibited in LX-2 cells incubated with supernatants from CE-treated AML12 cells. Conversely, these parameters were increased significantly in the groups of LX-2 cells incubated with supernatants of CD-treated AML12 cells (Figure 8B). Furthermore, we used an antibody recognizing the latent TGF-β1 latency-associated peptide (LAP) breakdown product (LAP-D R58) that is retained in the extracellular matrix upon latent TGF-β1 activation. Consistent with the SEAP assay result (Figure 8A), immunofluorescence staining showed an induction of LAP-D R58 in LX-2 cells treated with cholesterol-lowering agents (Figure 8C and D).
Figure 8.
Conditioned media from CE- and CD-treated AML12 cells have opposite effects on HSC activation. (A) Active TGF-β1 concentration in the culture medium of LX-2 cells treated with conditioned supernatant of AML12 cells was examined by SEAP activity assay. (B) Relative mRNA levels of COL1A1, COL3A1, MMP2, and MMP9 were determined by RT-qPCR in LX-2 cells treated with conditioned AML12 supernatants as shown in the figure. (C) Expression and location of TGF-β1 LAP-D (R58) were detected by immunofluorescent staining in LX-2 cells incubated with control or CD-treated supernatants of AML12 cells. (D) Quantification of TGF-β1 LAP-D (R58) staining. For RT-qPCR, human PPIA was used as the endogenous control. Bars represent means ± SD (n = 3). ∗P < .05, ∗∗P < .01. For immunofluorescence, DRAQ5 was used to stain the nuclei. Scale bars: 25 μm. Images were chosen representatively from 3 independent experiments. Quantification of staining was performed using ImageJ (National Institutes of Health). CD-SN, cholesterol depletion-treated cell supernatant; Con, control; Fluor., fluorescence.
These data suggest that CE-treated AML12 hepatocytes protect LX-2 HSCs from activation, whereas CD-treated AML12 cells activate stellate cells and promote subsequent fibrosis.
TGF-β1 Signaling and Cholesterol Metabolism in Patient Cohorts
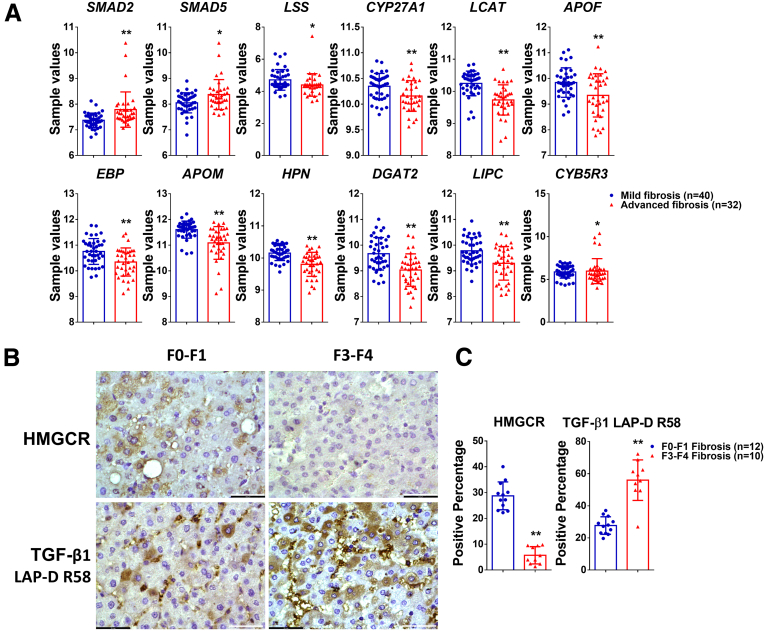
To verify the correlation of TGF-β signaling and cholesterol metabolism in patients suffering from chronic liver diseases, we extracted genes linked to TGF-β signaling and cholesterol metabolism from the Gene Expression Omnibus data set (GSE49541) that comprises mRNA expression data of liver tissue from nonalcoholic fatty liver disease patients with mild (F0–F1, n = 40) or advanced (F3–F4, n = 32) fibrosis. Comparison of transcript levels in these different patient sets shows an increase in SMAD2 and SMAD3 levels in advanced fibrosis whereas cholesterol metabolism–associated genes including LSS, CYP27A1, LCAT, and APOF are decreased, compared with patients with mild fibrosis (Figure 9A). These data are in accord with a scenario in which excessive TGF-β1 signaling induces a deregulation of cholesterol metabolism. Next, we performed immunohistochemistry staining of TGF-β1 LAP-D R58 and HMGCR in the liver tissue of MASLD patients with F0–F1 (n = 12) or F3–F4 (n = 10) fibrosis. As shown in Figure 9B and C, progression from F0–F1 to F3–F4 fibrosis correlates with increased LAP-D R58 staining, indicative of active TGF-β1 signaling, whereas HMGCR expression is reduced in hepatocytes from advanced vs early stage fibrosis. These data confirm, at the protein level, the tendencies observed at the level of gene transcripts in the GSE49541 data set.
Figure 9.
TGF-β1 signaling and cholesterol metabolism in patient cohorts. (A) mRNA expression of SMAD2, SMAD5, and cholesterol metabolism–related genes in liver tissue from patients with F0–F1 fibrosis or F3–F4 fibrosis, extracted from Gene Expression Omnibus data set GSE49541. (B and C) Staining and quantification of HMGCR and TGF-β1 LAP-D R58 in the liver tissue of MASLD patients with F0–F1 or F3–F4 fibrosis. Bars represent means ± SD. ∗P < .05, ∗∗P < .01. Scale bars: 43.5 μm. Images were chosen representatively from immunohistochemistry staining of patients with F1–F2 fibrosis (n = 12) or F3–F4 fibrosis (n = 10). Quantification of staining was performed using ImageJ (National Institutes of Health).
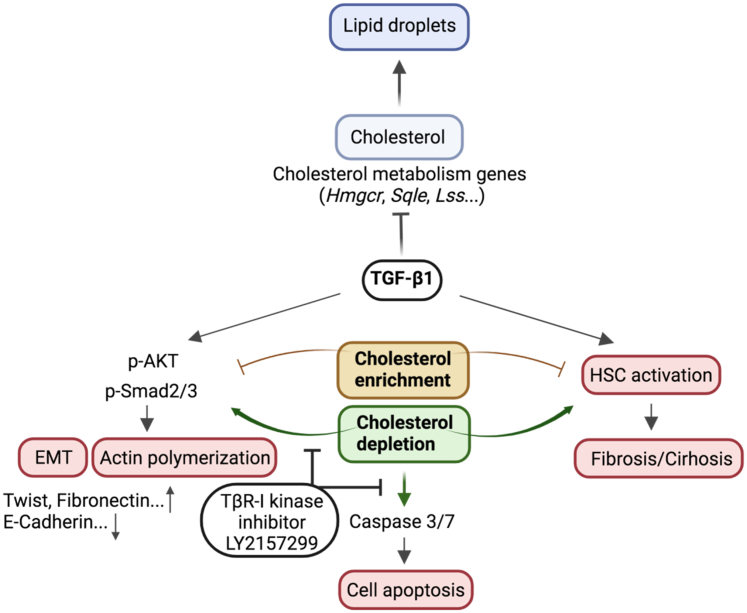
In conclusion, our data support the notion of reciprocal regulation of TGF-β1 and cholesterol in hepatocytes. TGF-β1 down-regulates the expression of multiple genes involved in cholesterol biosynthesis, associated metabolic processes, homeostasis, and transport, and thus may reduce cholesterol levels in this cell type. On the other hand, excess cholesterol attenuates TGF-β1 downstream effects in the liver, including EMT, actin polymerization, cell apoptosis, and HSC activation (Figure 10).
Figure 10.
Schematic representation of the crosstalk between the TGF-β1 pathway and cholesterol metabolism. TGF-β1 inhibits cholesterol biosynthesis and lipid droplet accumulation in hepatocytes. CE inhibits TGF-β1–induced EMT, actin polymerization, and HSC activation, whereas CD enhanced these effects in a TGF-β1–dependent manner. Furthermore, CD induces cell apoptosis through the caspase 3/7 pathway, which is reliant on TGF-β1 signaling. Figure was created with BioRender.com. p-AKT, phosphorylated-protein kinase B.
Discussion
TGF-β1 signaling and cholesterol level are 2 significant factors that affect the progression of MASLD.38 Yet, knowledge of the effects of TGF-β1 on cholesterol metabolism and the underlying mechanisms is incomplete. In this study, we found that TGF-β1 inhibits expression of multiple genes involved in cholesterol metabolism, including cholesterol homeostasis (Apoa2, Apoa1, Hpn, Fdft1, Angptl3), cholesterol biosynthesis (Hmgcs1, Sqle, Lss, Pmvk, Cyp5r3), cholesterol transport (Abca2, Abcg5, Ldlr, Stard5), cholesterol esterification (Soat2, Stard4, Lcat), and others. This goes along with reduced accumulation of lipid droplets because the excess cholesterol is esterified by acyl CoA:cholesterol acyltransferase (ACAT/SOAT) to cholesterol esters, and are either stored as a cholesterol reservoir in cytosolic lipid droplets or released as a major constituent of plasma lipoproteins.6,9,40 Further research is required to determine how TGF-β1 signaling affects the expression of these genes involved in cholesterol metabolism. It is possible that some transcription factors of TGF-β1 downstream pathways show common binding sites on the promoters of the aforementioned cholesterol metabolism–associated genes, thus inhibiting their transcription and expression. Interestingly, we discovered increased cholesterol levels in the plasma of AAV8–TGF-β1–injected mice despite the fact that TGF-β1 decreased cholesterol production and accumulation in the liver tissue of the same animals overexpressing TGF-β1. First, the half-life of plasma cholesterol that exceeds several days might serve as an explanation41; second, ABCA1, which exports cholesterol into the blood circulation, is induced strongly by TGF-β1 treatment42; third, the RNA-seq analysis has shown that the mRNA expression of Ldlr was reduced in both AML12 cells and MPHs upon TGF-β1 administration (Figure 1A), which is responsible for the uptake of cholesterol from blood to hepatocytes and its defects are correlated to hypercholesterolemia,43,44 indicating that blood cholesterol levels and local liver-specific cholesterol levels do not have to correlate. It also implies that more research is needed on cholesterol import and export in addition to its biosynthesis.
In this study, we also focused on investigating the roles of CE and CD in regulating TGF-β1 downstream biological effects (Figure 4, Figure 5, Figure 6, Figure 7, Figure 8). We found that in AML12 cells, CE inhibits TGF-β1–induced EMT and actin polymerization (Figure 4), cell apoptosis (Figure 6), and HSC activation/fibrosis (Figure 8). On the other hand, CD has the opposite effects (Figure 4, Figure 6, and 8), and its ability to promote these processes depends on TGF-β1 signaling, as indicated by its disruption by a TβR-I inhibitor, LY2157299 (Figures 5 and 7). These observations may be related to the report that Sirtuin (Sirt)1/Sirt6 play important roles in TGFβ/Smad pathway inhibition by deacetylation of Smad2 and Smad3.45,46 Moreover, forkhead box O3 (Foxo3) and Krüppel-like factor 6 (Klf6) can bind to the promoters of EMT genes, such as Snail and Twist, leading to an induction of their transcription,38 and cell division cycle 42 (Cdc42) forms a complex with neuronal wskott-aldrich syndrome protein (N-WASP) and actin related protein 2/3 (Arp2/3) to promote actin polymerization.47 The involvement of the earlier-described genes in cholesterol homeostasis has been suggested by several studies. Thus, increased Sirt1/Sirt6 were shown to decrease cholesterol level.48 In addition, it was found that Foxo3/Klf6 and Sirt6 are important for cholesterol homeostasis through controlling sterol regulatory element binding protein 2 expression, resulting in improvement of hypercholesterolemia in diet-induced or genetically obese mice.49 These studies imply that there may be a negative feedback regulation between cholesterol to Sirt1/6 and Foxo3/Klf6. As for the small guanosine triphosphatase Cdc42, it was shown to be required for the nickmann-pick C1-like 1 (NPC1L1) endocytic recycling compartment, a transporter of cholesterol uptake from intestine and bile, which also requires complex formation with the motor protein myosin Vb and actin filaments.47,50, 51, 52 CD could stimulate the formation of guanosine triphosphate-bound, active Cdc42 with increased activity for NPC1L1, resulting in the association of NPC1L1 with myosin Vb and actin, culminating in further NPC1L1 translocation to the plasma membrane by promoting actin polymerization (Figure 4G and H). In addition, this report shows caspase 3/7–dependent cell apoptosis induced by CD in AML12 cells; nevertheless, more research is required to understand the underlying mechanisms. We further show that hepatocytes subjected to CE protect HSCs from activation and then against subsequent fibrosis, whereas CD has the opposite consequences for HSC activation. These results are supported by a study in which similar effects were reported to occur after cholesterol enrichment, inducing oxidative stress and HSC cell death that mitigated liver fibrosis.35 Further investigation is needed to fully understand the interaction between CE/CD-treated hepatocytes and HSC activation.
Lipid droplet accumulation is associated with the progression of MASLD, in which changes in the accumulation of free fatty acids, triglycerides, cholesterol, and phospholipids occur at the early stages.53 Notably, the lipid alterations correlate with liver disease development. Thus, blood levels of fatty acids, triglycerides, ceramides, and bile acids from patients with MASLD all are increased, whereas phospholipids and cholesterol are depleted compared with healthy individuals not suffering from MASLD.54,55 Several studies have reported that the most important metabolic dysregulations are the high level of free fatty acids and triglycerides, both in the bloodstream and the liver tissue of patients with MASLD.56, 57, 58, 59 The function of cholesterol in MASLD development is less clear; 1 study found that cholesterol promotes MASLD through stabilizing TAZ (WW domain containing transcription regulator 1) in hepatocytes,60 whereas another study reported that cholesterol mitigates liver fibrosis in the MASH model via inducing HSC death.35 Other studies have reported that serum cholesterol levels and cholesterol synthesis are reduced significantly in HCC patients61 because high serum levels of cholesterol were said to suppress HCC tumorigenesis through the activation of natural killer cells.62 Cholesterol overload in hepatocytes leads to endoplasmic reticulum (ER) stress and mitochondrial dysfunction,63 but cholesterol also can promote hepatocyte proliferation.64 Rauchbach et al35 found that HSCs are more sensitive to cholesterol-induced apoptosis compared with hepatocytes. In our study, cholesterol enrichment by incubation with cholesterol–MβCD complex (300 μg/mL cholesterol) inhibited TGF-β1–mediated EMT and actin polymerization (Figure 4), did not affect AML12 cell proliferation or cell death (Figure 6), and protected HSCs from activation (Figure 8). Taken together, these findings imply that different cell types respond differently to altered cholesterol levels, probably related to the respective cells’ physiological state. Further studies are needed to explore this controversy. On the other hand, although several studies have reported that statin treatment benefits patients with MASLD, the statin effect is largely independent of its inhibitory effect on cholesterol synthesis. Rather, it appears to depend on the anti-inflammatory effects of the statin treatment and the improvement in endothelial function, which stabilize the atheromas in vessels, thus preventing heart attacks.65 Moreover, several studies have reported adverse effects of statin therapy, in which the most common events are associated with statin intolerance, including myopathy, myalgia, and gut dysbiosis.66,67 Our studies on AML12 cells68 have shown that statin-mediated CD modulates the balance between TGF-β signaling to Smad2/3 vs Akt. We found that the statin-induced CD of AML12 cells enhances EMT (Figures 4 and 5), apoptosis (Figures 6 and 7), and HSC activation (Figure 8) in a TGF-β–dependent manner. These effects are in accord with the report that cholesterol pathway inhibition induces TGF-β signaling to promote basal differentiation in pancreatic cancer,36 and raises the possibility that CD induced by statin treatment to patients with CLD, such as MASLD, may increase the risk of fibrosis progression, hepatocarcinoma formation, and metastasis.
In conclusion, in the liver, TGF-β1 signaling and cholesterol negatively regulate each other in an interdependent manner. In terms of therapeutic consequence, our findings suggest that the adverse effects encountered with cholesterol-lowering treatments always should be considered when treating MASLD patients with statins. This is of special relevance to TGF-β–targeted therapies.
Materials and Methods
Reagents and Antibodies
Lovastatin (cat. PHR1285; Sigma-Aldrich, Taufkirchen, Germany); MVL (cat. M4667; Sigma-Aldrich); MβCD soluble complex (cat. C4951; Sigma-Aldrich); TGF-β1 (cat. 100-21; PeproTech, Hamburg, Germany); LY2157299 (cat. HY-13226; MedChemExpress; Hoelzel Biotech, Cologne, Germany); MTT (cat. 11684795910; Sigma-Aldrich); IncuCyte Cytotox Red Dye (cat. 4632; Sartorius, Göttingen, Germany); BODIPY (D-3922; Life Technologies), caspase 3 antibody (cat. 9662; Cell Signaling Technology, Danvers, MA); cleaved caspase 3 antibody (cat. 9664; Cell Signaling Technology); caspase 7 antibody (cat. 9492; Cell Signaling Technology); cleaved caspase 7 antibody (cat. 8438; Cell Signaling Technology); p-Smad2 (Ser465/467) (cat. 3108; Cell Signaling Technology); Smad2 (cat. 5339; Cell Signaling Technology); p-Smad3 (cat. ab63403; Abcam); Smad3 (cat. 9523S; Cell Signaling Technology); phospho-Akt (Ser473) (cat. 4060; Cell Signaling Technology); Akt (cat. 9272; Cell Signaling Technology); E-cadherin antibody (cat. 3195; Cell Signaling Technology); HMGCR (cat. 271595; Santa Cruz Biotechnology, Santa Cruz, CA); caspase–Glo 3/7 Assay Systems (cat. 8090; Promega, Madison, WI); AlexaFluor 488 AffiniPure F(ab')2 Fragment Donkey Anti-Rabbit IgG (H+L) (cat. 711-546-152; Jackson ImmunoResearch, West Grove, PA); DRAQ5 (cat. 4084L; Cell Signaling Technology); glyceraldehyde-3-phosphate dehydrogenase antibody (cat. 32233; Santa Cruz Biotechnology); and Alexa Fluor 568 Phalloidin (cat. A12380; Invitrogen, Waltham, MA).
Human Samples
Fibrotic liver tissue was collected during surgery by the Department of General, Visceral, Vascular and Pediatric Surgery, Saarland University Medical Center (Homburg/Saar, Germany). The study protocol was approved by local ethics committees (143/21, 154/10, and 169/02). Written informed consent was obtained from the patients or their representatives.
Animal Experiments
C57BL/6 mice received a single tail vein injection of 2 × 1011 AAV8–control or AAV8–TGF-β1 for 7 days before collection of liver tissue and blood samples. AAV8–control and AAV8–TGF-β1 were generated by VectorBuilder (Neu-Isenburg, Germany). All experiments were conducted with 8-week-old male mice. Each group contained 3 mice. Animal experiments were performed in accordance with national guidelines for animal welfare and approved by the local animal care committee of the state of Baden-Württemberg, Germany (ethics approval 35-9185.81/G-172/15).
Mouse Primary Hepatocyte Isolation
C57BL/6 mice were used for primary hepatocyte isolation as previously described.69 After anesthesia with intraperitoneal injection of 10% ketamine hydrochloride (5 mg/100 mg body weight) and 2% xylazine hydrochloride (1 mg/100 mg body weight), the mouse liver was perfused sequentially with 50 mL perfusion buffer (Krebs–Henseleit buffer with 0.5 mmol/L EDTA; Sigma) and 50 mL collagenase A buffer (Krebs–Henseleit buffer with 0.1 mmol/L Ca2Cl and 0.4 mg/mL collagenase A; Sigma). Then, the perfused liver was removed and separated using tweezers to release hepatocytes. After pouring cells through a 70-mm mesh, the cell solution was centrifuged at 50 × g at 4°C for 5 minutes. To remove dead cells, hepatocytes were mixed with balanced Percoll solution (in Hank’s buffer), followed by centrifugation at 50 × g at 4°C for 10 minutes. The cell pellet was resuspended in culture medium for plating.
Cell Culture and Treatment
The AML12 murine hepatocyte cell line (cat. CRL-2254; American Type Culture Collection, Manassas, VA) was established from hepatocytes of a mouse transgenic for human TGF-α. They showed typical hepatocyte features such as peroxisomes and bile canalicular–like structure, and retained the capacity to express high levels of mRNA for hepatocyte functional genes.70 AML12 cells were grown at 37°C in 5% CO2 in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, penicillin, streptomycin, and L-glutamine as previously described.71 They were seeded initially for 12 hours either with 2 mL/well of DMEM supplemented with 10% FBS, or with 10% LPDS (all from Sigma-Aldrich). For CD, cells were treated with 50 μmol/L lovastatin and 50 μmol/L MVL in DMEM containing 10% LPDS; the addition of MVL at this concentration prevented excessive reduction of mevalonate, resulting in reduced cholesterol but maintaining normal farnesylation and geranylgeranylation.72 For CE, cells were treated in complete medium (with 10% FBS) with cholesterol–MβCD complex (5 mmol/L MβCD, 300 μg/mL cholesterol). After the first 14 hours of the earlier-described treatments, all groups were starved for 4–6 hours with the same aforementioned media, respectively, containing either 0.5% FBS or LPDS instead of the original 10% FBS. After starvation, the cells were stimulated with 2 ng/mL TGF-β1 in DMEM supplemented with either 0.5% FBS or 0.5% LPDS for 2 or 72 hours. After stimulation, the supernatants were collected for treatment of HSC LX-2 cells (see later). For RNA isolation (for RT-qPCR), the cells were lysed with TRIzol RNA Isolation Reagents (cat. 15596-018; Thermo Fisher Scientific, Waltham, MA). For protein analysis, they were lysed with RIPA buffer containing 20 mmol/L Tris–HCl (pH 7.2), 150 mmol/L NaCl, 2% (v/v) NP-40, 0.1% (w/v) sodium dodecyl sulfate (SDS), 0.5% (w/v) sodium deoxycholate, Complete mixture of proteinase inhibitors (Roche, Mannheim, Germany), and phosphatase inhibitor cocktail (Sigma-Aldrich), and collected for further protein analysis. For experiments on the effects of the supernatants of the AML12 cells on LX-2 HSCs, the latter were kept in DMEM supplemented with 1% L-glutamine, 1% penicillin/streptomycin, and 2% FBS, and starved for 4 hours with starvation medium before 72-hour treatment with AML12 supernatants.
The MFB-F11 cell line is a murine fibroblast cell line isolated from TGF-β knockout mice (Tgfb-/-), described by Tesseur et al,73 and were transfected with a Smad-binding element controlling a secreted alkaline phosphatase gene (SEAP). MFB-F11 cells were kept in high-glucose DMEM (11965092; Thermo Fisher Scientific) supplemented with 1% L-glutamine (25030081; Thermo Fisher Scientific), 1% P/S (15140122; Thermo Fisher Scientific), and 10% FBS (10439024; Thermo Fisher Scientific) without sodium pyruvate as an additive. MFB-F11 cells were seeded in 96-well plates (650160; Greiner Bio-One, Frickenhausen, Germany) at a density of 5 × 104 per well. After starvation for 2 hours, conditioned supernatant was added.
All cell lines were checked routinely for the absence of mycoplasma contamination via PCR.
RNA Isolation and qRT-PCR
RNA isolation was performed using the TRIzol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. A total of 500 ng RNA was used for complementary DNA (cDNA) synthesis using a commercially available cDNA synthesis kit (Thermo Fisher Scientific). A total of 20 μL of the mixture containing 5 μL cDNA (diluted 1:10), 4 μL Power SYBR Green Master Mix, and 10 μmol/L forward and reverse primers were used for real-time PCR in a StepOnePlus Real-Time PCR system (all from Applied Biosystems, Waltham, MA). The RT-qPCR amplification protocol comprised a polymerase activation step for 15 minutes at 95°C, a subsequent amplification step for 15 seconds at 95°C, 20 seconds at 60°C, and 20 seconds at 72°C for 40 cycles. A melting curve was established to validate specificity for each PCR analysis with the following: protocol 15 seconds at 95°C and 1 minute at 60°C, from 60°C to 95°C with +0.3°C every 15 seconds. Human peptidylprolyl isomerase A was used as a housekeeping gene for normalization of gene expression. Expressions were calculated with the ΔΔCt method described previously.74 Primer sequences were retrieved from the PrimerBank (Massachusetts General Hospital, Boston, MA) online resource and ordered from Eurofins Genomics (Eurofins Scientific, Luxembourg) (Table 1).
Table 1.
Primers for qRT-PCR
| Primer | Forward | Reverse |
|---|---|---|
| hCOL1A1 (human Collagen 1α1) | GAGGGCCAAGACGAAGACATC | CAGATCACGTCATCGCACAAC |
| hCOL3A1 (human Collagen 3α1) | TTGAAGGAGGATGTTCCCATCT | ACAGACACATATTTGGCATGGTT |
| hMMP-2 (human Matrix metalprotease-2) | TACAGGATCATTGGCTACACACC | GGTCACATCGCTCCAGACT |
| hMMP-9 (human Matrix metalprotease-9) | TGTACCGCTATGGTTACACTCG | GGCAGGGACAGTTGCTTCT |
| hPPIA (human Peptidylprolyl isomerase A) | AGGGTTCCTGCTTTCACAGA | CAGGACCCGTATGCTTTAGG |
| mCdh1 (mouse E-Cadherin) | CAGGTCTCCTCATGGCTTTGC | CTTCCGAAAAGAAGGCTGTCC |
| mFN1 (mouse Fibronectin) | GATGTCCGAACAGCTATTTACCA | CCTTGCGACTTCAGCCACT |
| mHmgcr (mouse 3-Hydroxy-3-methyl-glutaryl-coenzyme A reductase) | AGCTTGCCCGAATTGTATGTG | TCTGTTGTGAACCATGTGACTTC |
| mLss (mouse Lanosterol Synthase) | GTGTCTTGGCTGGGTGATAA | GACACCAACACTGACCCTATC |
| mPPIA (mouse Peptidylprolyl isomerase A) | GAGCTGTTTGCAGACAAAGTT | CCCTGGCACATGAATCCTGG |
| mSqle (mouse Squalene Epoxidase) | ATAAGAAATGCGGGGATGTCAC | ATATCCGAGAAGGCAGCGAAC |
| mTGF-β1 (mouse Transforming growth factor beta 1) | AGGGCTACCATGCCAACTTC | CCACGTAGTAGACGATGGGC |
| mTwist1 (mouse Twist basic helix-loop-helix transcription factor 1) | CAGCAAGATCCAGACGCTCAAG | ACACGGAGAAGGCGTAGCTGAG |
| mApoa2 (mouse Apolipoprotein A2) | GCAGACGGACCGGATATGC | GCTGCTCGTGTGTCTTCTCA |
| mApoc3 (mouse Apolipoprotein C3) | AGGCTACTGGAGCAAGTTTACT | ATAGCTGGAGTTGGTTGGTCC |
| mFdx1 (mouse Ferredoxin 1) | CAAGGGGAAAATTGGCGACTC | TTGGTCAGACAAACTTGGCAG |
| mInsig2 (mouse Insulin induced gene 2) | GGAGTCACCTCGGCCTAAAAA | CAAGTTCAACACTAATGCCAGGA |
| mAgnptl3 (mouse Angiopoietin like 3) | TCTACTGTGATACCCAATCAGGC | CATGTTTCGTTGAAGTCCTGTGA |
| mApof (mouse Apolipoprotein F) | ATAGCCTCCGACTCATCCTGA | TCTGCATCTGGTATCCCAACTT |
| mEbp (mouse EBP cholestenol delta-isomerase) | ATGACCACCAATACGGTCCC | GCCAACCAGGATATGCGAAGT |
| mApom (mouse Apolipoprotein M) | GTGGTACTTTATTGCGGGAGC | CCACTTTTCGTGCGGATGGTA |
| mEphx2 (mouse Epoxide hydrolase 2) | ACCACTCATGGATGAAAGCTACA | TCAGGTAGATTGGCTCCACAG |
| mHpn (mouse Hepsin) | GCGGTGTTTGACAAGACGGA | CACGCAAAAGAAGCCCGAT |
| mDgat2 (mouse Diacylglycerol O-acyltransferase 2) | GCGCTACTTCCGAGACTACTT | GGGCCTTATGCCAGGAAACT |
| mLipc (mouse Lipase C, hepatic type) | CTCAGCACCCGGAAACACT | CGCACTCACTATCTTCCAGATCC |
hPPIA, human peptidylprolyl isomerase A; Lss, lanosterol synthase; Sqle, squalene monooxygenase.
Western Blot
Cultured cells were dissolved in RIPA lysis buffer (1% Triton X-100 (Sigma-Aldrich), 50 mmol/L Tris [pH 7.5], 300 mmol/L NaCl, 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 1 mmol/L EDTA, and 0.1% SDS), supplemented with phosphatase inhibitors (Sigma-Aldrich). A DC Protein Assay (Bio-Rad, Hercules, CA) was performed according to the manufacturer's protocol to measure the sample protein concentration. Quantification was performed in a Tecan Infinite M200 (Tecan Group AG, Männedorf, Switzerland) microplate reader using the microplate reader's own protein assay protocol (absorbance, 595 nm). The Western blot sample protein concentration was calculated using a standard curve of absorbance plotted against predefined bovine serum albumin concentrations (0, 0.125, 0.25, 0.5, 1, 1.5, and 2 mg/mL). For each sample, 20 μg protein was separated by 10% SDS–polyacrylamide gel electrophoresis gels and were blotted onto a high-resolution nitrocellulose membrane (MERCK, Darmstadt, Germany). Membranes were blocked with 5% Albumin Bovine Fraction V (SERVA, Heidelberg, Germany) in TBST (Tris-buffered saline with 0.1% Tween 20 detergent) at room temperature for 1 hour. Subsequently, the membrane was incubated with primary antibodies overnight at 4°C. The next day, after washing with TBST 3 times, all membranes were incubated with horseradish peroxidase–linked anti-mouse or anti-rabbit secondary antibodies. Chemiluminescence was determined in a Fusion SL chemiluminescence reader (Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany) from membranes incubated in Western Lightening Plus-ECL solution (PerkinElmer, Waltham, MA).
Determination of Cholesterol Concentration
Total cholesterol concentration was measured using a cholesterol assay kit (K603-100; BioVision, Milpitas, CA). A total of 1 × 106 AML12 cells (untreated, or treated to reduce or enrich with cholesterol) were extracted with 200 μL chloroform:isopropanol:NP40 (7:11:0.1). After centrifugation for 10 minutes at 15,000 × g, the organic phase was transferred to a new tube, air-dried at 50°C to remove the chloroform, and vacuumed for 30 minutes to remove trace organic solvent. The dried lipids were dissolved with 200 μL cholesterol assay buffer and vortexed to homogeneity. A total of 1–50 μL of the extracted sample was used per assay and adjusted to a volume of 50 μL with cholesterol assay buffer. A total of 50 μL Reaction Mix containing 44 μL cholesterol assay buffer, 2 μL cholesterol probe, 2 μL cholesterol enzyme mix, and 2 μL cholesterol esterase were added to each sample for incubation at 37°C for 1 hour. Absorbance was measured at 570 nm using the Infinite 200 Spectrophotometer (Tecan Group AG).
MTT Assay
Proliferation measurements were performed using the MTT assay. Four thousand cells per well were seeded in quadruplicate in a 96-well plate. After attachment, cells were treated as described in the section Cell Culture and Treatment. At the end of all other experiments, the remaining cell culture medium was discarded, and 100 μL new culture medium and 10 μL MTT reagent were added to each well and incubated for 4 hours at 37°C to enable the formation of formazan crystals. After aspiration of the medium, 100 μL/well dimethyl sulfoxide were added, and the plate was incubated on a shaker at room temperature for 10 minutes to dissolve the formazan crystals. Absorbance was measured at 570 nm using the Infinite 200 Spectrophotometer.
Immunofluorescence Staining
AML12 cells or LX-2 HSCs were fixed with 4% paraformaldehyde (PFA) for 15 minutes at room temperature, washed 3 times with phosphate-buffered saline (PBS), and permeabilized and blocked with 1% bovine serum albumin and 0.5% Triton X-100 in PBS for 1 hour. The cells were incubated overnight at 4°C with the E-cadherin or fibronectin antibody (1:200 in PBS). After washing 3 times, the cells were incubated with the secondary antibody (AlexaFluor 488 AffiniPure F(ab')2 Fragment Donkey Anti-Rabbit IgG [H+L], 711-546-152, 1:200; Jackson ImmunoResearch, West Grove, PA) and the fluorescent DNA dye DRAQ5 (1:1000, 4084L; Cell Signaling Technology) in PBS for 1 hour at room temperature. After washing 3 times, the cells were mounted using the DakoCytomation fluorescent mounting medium (S3023; DAKO). Stained samples were analyzed with a TCS SP8 upright Confocal Microscope (Leica, Wetzlar, Germany).
Phalloidin Staining
AML12 cells were fixed, washed, and permeabilized as described earlier, and incubated with Alexa Fluor 568 phalloidin (1:250) and DRAQ5 (1:1000) in PBS for 1 hour at room temperature. After washing 3 times, the cells were mounted using Dakocytomation fluorescent mounting medium. Stained samples were analyzed by confocal microscopy as described previously.
Lipid Droplet Staining
AML12 cells or cryosections (4-μm thick) were washed briefly with PBS 3 times, followed by incubation with BODIPY (1:250) in PBS for 30 minutes at room temperature. After washing with PBS twice, the cells or sections were fixed with 4% PFA and stained with phalloidin and DRAQ5, and analyzed by confocal microscopy as described earlier.
Apoptosis Measurement by IncuCyte Cytotox Red Dye
Apoptosis was measured with IncuCyte Cytotox Red Dye. Four thousand AML12 cells were seeded per well in quadruplicate in a 96-well plate. After attachment, cells were treated as described in Cell Culture and Treatment. TGF-β1 was added first, followed by 100 μL 2x Cytotox Dye to each well containing 100 μL cell culture medium. Then, the plate was introduced into the IncuCyte chamber placed within a CO2 incubator; images were captured every 2 hours for 72 hours by the IncuCyte Live-Cell analysis system (Leica).
Caspase-Glo 3/7 Assay
Caspase 3/7 signals were measured using the caspase-Glo 3/7 assay. Four thousand cells per well were seeded in quadruplicate in 96-well plates. After attachment, cells were treated as described in Cell Culture and Treatment. At the end of all other experiments, the cell culture medium was discarded and 100 μL caspase-Glo 3/7 reagent were added to each well. Then, the plate was incubated on a shaker at 300–500 rpm for 30 seconds, followed by incubation at room temperature for 1 hour. All samples then were transferred to a white-walled, 96-well plate and their luminescence was measured using the Infinite 200 Spectrophotometer.
SEAP Assay
After the collection of conditioned medium from MFB-F11 TGF-β reporter cells after a 48-hour incubation, SEAP activity and TGF-β concentration were determined using the Great EscAPe SEAP (631738; TaKaRa Bio, Kusatsu, Japan) chemiluminescence kit to quantify SEAP activity (excitation wavelength, 360 nm; emission wavelength, 449 nm). Chemiluminescence was measured in a Tecan Infinite M200 microplate reader (signal integration time, 10,000 ms). TGF-β concentration was calculated from SEAP activity based on a standard curve from predefined TGF-β concentrations (0, 0.1, 0.25, 0.5, 1, 2, 5, 10, and 20 ng/mL).
RNA-Seq Microarray Analysis
Total RNA was isolated from AML12 cells or MPHs treated with TGF-β1 (or not; control) for 24 hours. Acceptable RNA quality was confirmed by capillary electrophoresis on an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA). Gene expression profiling was performed using arrays of the mouse MoGene 2.0 type. Biotinylated antisense cRNA then was prepared according to the Affymetrix standard labeling protocol with the GeneChip WT Plus Reagent Kit and the GeneChip Hybridization, Wash and Stain Kit. Afterward, the hybridization on the chip was performed on a GeneChip Hybridization oven 640. Dyeing took place in the GeneChip Fluidics Station 450. Thereafter, chips were scanned with the GeneChip Scanner 3000. All equipment used for RNA-seq microarray analysis was obtained from Affymetrix (Santa Clara, CA) where not otherwise specified.
Statistical Analysis
Statistical analyses were performed with GraphPad Prism version 6.0 software. The 2-tailed Student t test was used to compare 2 independent groups. One-way analysis of variance was adopted to test for statistical differences between the means of 2 groups. Variables were described by means and SD. Statistical significance was indicated as follows: ∗P < .05, ∗∗P < .01, NS > .05. Quantification of protein expression and imaging studies was performed using ImageJ software (National Institutes of Health).
All authors had access to the study data and have reviewed and approved the final version of this manuscript. Results will be made accessible to fellow researchers upon request to the corresponding author(s).
Acknowledgments
The authors acknowledge the support of the LIMa Live Cell Imaging at Microscopy Core Facility Platform Mannheim.
Sai Wang and Frederik Link contributed equally to this work.
CRediT Authorship Contributions
Sai Wang (Data curation: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead)
Frederik Link (Data curation: Supporting; Investigation: Lead; Methodology: Supporting; Writing – original draft: Lead)
Mei Han (Formal analysis: Supporting; Investigation: Supporting)
Roohi Chaudhary (Investigation: Supporting; Writing – review & editing: Supporting)
Anastasia Asimakopoulos (Investigation: Supporting; Writing – review & editing: Supporting)
Roman Liebe (Formal analysis: Supporting; Investigation: Supporting)
Ye Yao (Formal analysis: Supporting)
Seddik Hammad (Formal analysis: Supporting; Investigation: Supporting)
Anne Dropmann (Investigation: Supporting)
Marinela Krizanac (Investigation: Supporting)
Claudia Rubie (Investigation: Supporting)
Laura Kim Feiner (Investigation: Supporting)
Matthias Glanemann (Investigation: Supporting)
Matthias P. A. Ebert (Supervision: Supporting; Writing – review & editing: Supporting)
Ralf Weiskirchen (Conceptualization: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – review & editing: Equal)
Yoav I. Henis (Conceptualization: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – review & editing: Equal)
Marcelo Ehrlich (Conceptualization: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – review & editing: Equal)
Steven Dooley (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by German–Israeli collaborative project grants from the Deutsche Forschungsgemeinschaft (German Research Foundation), projects 437145127 and 452602471 (WE 2554/13-1 to R.W. and Y.I.H., and DO 373/19-1 to S.D. and M.E.), the BMBF (German Federal Ministry of Education and Research) Project LiSyM-Cancer grant PTJ-FKZ: 031L0257A, and the Stiftung fur Biomedizinische Alkoholforschung.
References
- 1.Targher G., Byrne C.D. Clinical review: nonalcoholic fatty liver disease: a novel cardiometabolic risk factor for type 2 diabetes and its complications. J Clin Endocrinol Metab. 2013;98:483–495. doi: 10.1210/jc.2012-3093. [DOI] [PubMed] [Google Scholar]
- 2.Su Q., Kumar V., Sud N., et al. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54–63. doi: 10.1016/j.addr.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Bertolani C., Marra F. The role of adipokines in liver fibrosis. Pathophysiology. 2008;15:91–101. doi: 10.1016/j.pathophys.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Silvente-Poirot S., Cancer Poirot M. Cholesterol and cancer, in the balance. Science. 2014;343:1445–1446. doi: 10.1126/science.1252787. [DOI] [PubMed] [Google Scholar]
- 6.Phillips M.C. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289:24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang T.Y., Chang C.C., Cheng D. Acyl-coenzyme A:cholesterol acyltransferase. Annu Rev Biochem. 1997;66:613–638. doi: 10.1146/annurev.biochem.66.1.613. [DOI] [PubMed] [Google Scholar]
- 8.Luo J., Jiang L., Yang H., et al. Routes and mechanisms of post-endosomal cholesterol trafficking: a story that never ends. Traffic. 2017;18:209–217. doi: 10.1111/tra.12471. [DOI] [PubMed] [Google Scholar]
- 9.Luo J., Yang H., Song B.L. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 10.Wouters K., van Bilsen M., van Gorp P.J., et al. Intrahepatic cholesterol influences progression, inhibition and reversal of non-alcoholic steatohepatitis in hyperlipidemic mice. FEBS Lett. 2010;584:1001–1005. doi: 10.1016/j.febslet.2010.01.046. [DOI] [PubMed] [Google Scholar]
- 11.Savard C., Tartaglione E.V., Kuver R., et al. Synergistic interaction of dietary cholesterol and dietary fat in inducing experimental steatohepatitis. Hepatology. 2013;57:81–92. doi: 10.1002/hep.25789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rooyen D.M., Larter C.Z., Haigh W.G., et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology. 2011;141:1393–1403. doi: 10.1053/j.gastro.2011.06.040. 1403 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sezgin E., Levental I., Mayor S., et al. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C.L., Huang S.S., Huang J.S. Cholesterol modulates cellular TGF-beta responsiveness by altering TGF-beta binding to TGF-beta receptors. J Cell Physiol. 2008;215:223–233. doi: 10.1002/jcp.21303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razani B., Zhang X.L., Bitzer M., et al. Caveolin-1 regulates transforming growth factor (TGF)-beta/SMAD signaling through an interaction with the TGF-beta type I receptor. J Biol Chem. 2001;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- 16.Zuo W., Chen Y.G. Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell. 2009;20:1020–1029. doi: 10.1091/mbc.E08-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer C., Godoy P., Bachmann A., et al. Distinct role of endocytosis for Smad and non-Smad TGF-beta signaling regulation in hepatocytes. J Hepatol. 2011;55:369–378. doi: 10.1016/j.jhep.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Shapira K.E., Ehrlich M., Henis Y.I. Cholesterol depletion enhances TGF-beta Smad signaling by increasing c-Jun expression through a PKR-dependent mechanism. Mol Biol Cell. 2018;29:2494–2507. doi: 10.1091/mbc.E18-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taipale J., Saharinen J., Keski-Oja J. Extracellular matrix-associated transforming growth factor-beta: role in cancer cell growth and invasion. Adv Cancer Res. 1998;75:87–134. doi: 10.1016/s0065-230x(08)60740-x. [DOI] [PubMed] [Google Scholar]
- 20.Markowitz S., Wang J., Myeroff L., et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 21.Heldin C.H., Vanlandewijck M., Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooley S., ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Fan W., Link F., et al. Transforming growth factor beta latency: a mechanism of cytokine storage and signalling regulation in liver homeostasis and disease. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2021.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 26.Ehrlich M., Gutman O., Knaus P., et al. Oligomeric interactions of TGF-beta and BMP receptors. FEBS Lett. 2012;586:1885–1896. doi: 10.1016/j.febslet.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 27.Moustakas A., Heldin C.H. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 28.Budi E.H., Duan D., Derynck R. Transforming growth factor-beta receptors and Smads: regulatory complexity and functional versatility. Trends Cell Biol. 2017;27:658–672. doi: 10.1016/j.tcb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Bakin A.V., Tomlinson A.K., Bhowmick N.A., et al. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 30.Moustakas A., Heldin C.H. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y.E. Non-Smad signaling pathways of the TGF-beta family. Cold Spring Harb Perspect Biol. 2017;9:a022129. doi: 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo K. Signaling cross talk between TGF-beta/Smad and other signaling pathways. Cold Spring Harb Perspect Biol. 2017;9:a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorrentino A., Thakur N., Grimsby S., et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 34.Dongiovanni P., Petta S., Mannisto V., et al. Statin use and non-alcoholic steatohepatitis in at risk individuals. J Hepatol. 2015;63:705–712. doi: 10.1016/j.jhep.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Rauchbach E., Zeigerman H., Abu-Halaka D., et al. Cholesterol induces oxidative stress, mitochondrial damage and death in hepatic stellate cells to mitigate liver fibrosis in mice model of NASH. Antioxidants (Basel) 2022;11:536. doi: 10.3390/antiox11030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabitova-Cornell L., Surumbayeva A., Peri S., et al. Cholesterol pathway inhibition induces TGF-beta signaling to promote basal differentiation in pancreatic cancer. Cancer Cell. 2020;38:567–583 e11. doi: 10.1016/j.ccell.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannou G.N. The role of cholesterol in the pathogenesis of NASH. Trends Endocrinol Metab. 2016;27:84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S., Duan Q., Wu R., et al. Pathophysiological communication between hepatocytes and non-parenchymal cells in liver injury from NAFLD to liver fibrosis. Adv Drug Deliv Rev. 2021;176 doi: 10.1016/j.addr.2021.113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozaki I., Hamajima H., Matsuhashi S., et al. Regulation of TGF-beta1-Induced pro-apoptotic signaling by growth factor receptors and extracellular matrix receptor integrins in the liver. Front Physiol. 2011;2:78. doi: 10.3389/fphys.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han M., Nwosu Z.C., Pioronska W., et al. Caveolin-1 impacts on TGF-beta regulation of metabolic gene signatures in hepatocytes. Front Physiol. 2019;10:1606. doi: 10.3389/fphys.2019.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eagar T.N., Turley D.M., Padilla J., et al. CTLA-4 regulates expansion and differentiation of Th1 cells following induction of peripheral T cell tolerance. J Immunol. 2004;172:7442–7450. doi: 10.4049/jimmunol.172.12.7442. [DOI] [PubMed] [Google Scholar]
- 42.Hu Y.W., Wang Q., Ma X., et al. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells. J Atheroscler Thromb. 2010;17:493–502. doi: 10.5551/jat.3152. [DOI] [PubMed] [Google Scholar]
- 43.Cuchel M., Bruckert E., Ginsberg H.N., et al. Homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J. 2014;35:2146–2157. doi: 10.1093/eurheartj/ehu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Defesche J.C., Gidding S.S., Harada-Shiba M., et al. Familial hypercholesterolaemia. Nat Rev Dis Primers. 2017;3 doi: 10.1038/nrdp.2017.93. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z.H., Zhang Y., Wang X., et al. SIRT1 activation attenuates cardiac fibrosis by endothelial-to-mesenchymal transition. Biomed Pharmacother. 2019;118 doi: 10.1016/j.biopha.2019.109227. [DOI] [PubMed] [Google Scholar]
- 46.Holian J., Qi W., Kelly D.J., et al. Role of Kruppel-like factor 6 in transforming growth factor-beta1-induced epithelial-mesenchymal transition of proximal tubule cells. Am J Physiol Renal Physiol. 2008;295:F1388–F1396. doi: 10.1152/ajprenal.00055.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie C., Li N., Chen Z.J., et al. The small GTPase Cdc42 interacts with Niemann-Pick C1-like 1 (NPC1L1) and controls its movement from endocytic recycling compartment to plasma membrane in a cholesterol-dependent manner. J Biol Chem. 2011;286:35933–35942. doi: 10.1074/jbc.M111.270199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammer S.S., Vieira C.P., McFarland D., et al. Fasting and fasting-mimicking treatment activate SIRT1/LXRalpha and alleviate diabetes-induced systemic and microvascular dysfunction. Diabetologia. 2021;64:1674–1689. doi: 10.1007/s00125-021-05431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao R., Xiong X., DePinho R.A., et al. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y.Y., Fu Z.Y., Wei J., et al. A LIMA1 variant promotes low plasma LDL cholesterol and decreases intestinal cholesterol absorption. Science. 2018;360:1087–1092. doi: 10.1126/science.aao6575. [DOI] [PubMed] [Google Scholar]
- 51.Ge L., Wang J., Qi W., et al. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 2008;7:508–519. doi: 10.1016/j.cmet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Chu B.B., Ge L., Xie C., et al. Requirement of myosin Vb.Rab11a.Rab11-FIP2 complex in cholesterol-regulated translocation of NPC1L1 to the cell surface. J Biol Chem. 2009;284:22481–22490. doi: 10.1074/jbc.M109.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul B., Lewinska M., Andersen J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayo R., Crespo J., Martinez-Arranz I., et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: results from discovery and validation cohorts. Hepatol Commun. 2018;2:807–820. doi: 10.1002/hep4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puri P., Daita K., Joyce A., et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology. 2018;67:534–548. doi: 10.1002/hep.29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ooi G.J., Meikle P.J., Huynh K., et al. Hepatic lipidomic remodeling in severe obesity manifests with steatosis and does not evolve with non-alcoholic steatohepatitis. J Hepatol. 2021;75:524–535. doi: 10.1016/j.jhep.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 57.Puri P., Baillie R.A., Wiest M.M., et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 58.Kotronen A., Velagapudi V.R., Yetukuri L., et al. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 2009;52:684–690. doi: 10.1007/s00125-009-1282-2. [DOI] [PubMed] [Google Scholar]
- 59.Oresic M., Hyotylainen T., Kotronen A., et al. Prediction of non-alcoholic fatty-liver disease and liver fat content by serum molecular lipids. Diabetologia. 2013;56:2266–2274. doi: 10.1007/s00125-013-2981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X., Cai B., Yang X., et al. Cholesterol stabilizes TAZ in hepatocytes to promote experimental non-alcoholic steatohepatitis. Cell Metab. 2020;31:969–986 e7. doi: 10.1016/j.cmet.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J.M., Skill N.J., Maluccio M.A. Evidence of aberrant lipid metabolism in hepatitis C and hepatocellular carcinoma. HPB (Oxford) 2010;12:625–636. doi: 10.1111/j.1477-2574.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin W.H., Yang Z.S., Li M., et al. High serum levels of cholesterol increase antitumor functions of nature killer cells and reduce growth of liver tumors in mice. Gastroenterology. 2020;158:1713–1727. doi: 10.1053/j.gastro.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 63.Horn C.L., Morales A.L., Savard C., et al. Role of cholesterol-associated steatohepatitis in the development of NASH. Hepatol Commun. 2022;6:12–35. doi: 10.1002/hep4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaminsky-Kolesnikov Y., Rauchbach E., Abu-Halaka D., et al. Cholesterol induces Nrf-2- and HIF-1alpha-dependent hepatocyte proliferation and liver regeneration to ameliorate bile acid toxicity in mouse models of NASH and fibrosis. Oxid Med Cell Longev. 2020;2020 doi: 10.1155/2020/5393761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Istvan E.S., Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 66.Joy T.R., Hegele R.A. Narrative review: statin-related myopathy. Ann Intern Med. 2009;150:858–868. doi: 10.7326/0003-4819-150-12-200906160-00009. [DOI] [PubMed] [Google Scholar]
- 67.Caparros-Martin J.A., Lareu R.R., Ramsay J.P., et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5:95. doi: 10.1186/s40168-017-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaudhary R., Goodman L.S., Wang S., et al. Cholesterol modulates type I-type II TGF-β receptor complexes and alters the balance between Smad and Akt signaling in hepatocytes. Commun Biol. 2024;7:8. doi: 10.1038/s42003-023-05654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S., Feng R., Wang S.S., et al. FOXA2 prevents hyperbilirubinaemia in acute liver failure by maintaining apical MRP2 expression. Gut. 2023;72:549–559. doi: 10.1136/gutjnl-2022-326987. [DOI] [PubMed] [Google Scholar]
- 70.Wu J.C., Merlino G., Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Roeyen C.R.C., Martin I.V., Drescher A., et al. Identification of platelet-derived growth factor C as a mediator of both renal fibrosis and hypertension. Kidney Int. 2019;95:1103–1119. doi: 10.1016/j.kint.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 72.Bonetti P.O., Lerman L.O., Napoli C., et al. Statin effects beyond lipid lowering--are they clinically relevant? Eur Heart J. 2003;24:225–248. doi: 10.1016/s0195-668x(02)00419-0. [DOI] [PubMed] [Google Scholar]
- 73.Tesseur I., Zou K., Berber E., et al. Highly sensitive and specific bioassay for measuring bioactive TGF-beta. BMC Cell Biol. 2006;7:15. doi: 10.1186/1471-2121-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]