Abstract
A large body of evidence from viral systems has established that transcription factors play an important and direct role in activating viral DNA replication. Among the transcriptional activation domains that can stimulate viral DNA replication are acidic domains such as those derived from herpes simplex virus VP16 and the tumor suppressor p53. Here we show that acidic activation domains can also activate a cellular origin of replication in a chromosomal context. When tethered to the yeast ARS1 (autonomously replicating sequence 1) origin of replication, both VP16 and p53 activation domains can enhance origin function. In addition, the C-terminal acidic region of the yeast transcription factor ABF1, which normally activates the ARS1 origin, is sufficient for activating ARS1 function when tethered to the origin. Mutations at residues Trp-53 and Phe-54 of a 20-residue (41 to 60) activation region of p53 abolish the activation of both replication and transcription, suggesting that the same structural determinants may be employed to activate both processes in yeast. Furthermore, using a two-dimensional gel electrophoresis method, we demonstrate that the GAL4-p53 chimeric activator can activate initiation of chromosomal replication from an origin inserted at the native ARS1 locus. These findings strongly suggest functional conservation of the mechanisms used by the acidic activation domains to activate viral DNA replication in mammalian cells and chromosomal replication in yeast.
The eukaryotic origins of DNA replication characterized to date contain two functional elements: a core sequence that determines the site of initiation and a nearby auxiliary element that stimulates initiation efficiency (11, 16, 36, 55). Analogous to the TATA box of a transcription promoter, the core sequence of an origin of replication serves as the binding site for an initiator protein which in turn nucleates the assembly of a large initiation protein complex. The auxiliary elements of an origin usually contain binding sites for proteins that in other DNA contexts function as transcription factors. It has been well established that transcription factors play an important and direct role in viral DNA replication (15, 30, 64).
Most of our current understanding of transcription factors’ role in replication comes from studies of DNA tumor viruses (17, 33, 52, 63, 68). For example, the flanking auxiliary sequences of the simian virus 40 and polyomavirus origins are located adjacent to the core region that forms the binding site for the large T antigen, the viral initiator protein. The auxiliary sequences contain binding sites for several cellular transcription factors such as Sp1, AP1 and p53 for simian virus 40 and AP1 and PEA3 for polyomavirus. These cis elements act synergistically to increase the initiation frequency up to 1,000-fold (17, 25, 26). Heterologous transcription factors can also activate viral replication when tethered to the origins of replication. For example, factors such as NF-κB, VP16, E1A, bovine polyomavirus E2, and GAL4 can stimulate polyomavirus DNA replication (1, 24, 66). The functional promiscuity of these proteins in activation of replication is reminiscent of the similar behavior of these activators in transcriptional activation.
Transcription factors have also been implicated in activation of cellular DNA replication. In the case of Saccharomyces cerevisiae, a detailed mutational analysis of one origin, ARS1 (autonomously replicating sequence 1), has led to identification of two essential elements, A and B (10, 44). Element A contains an 11-bp consensus sequence that is conserved among all origins in S. cerevisiae. It is the binding site for the initiator protein called origin recognition complex (ORC) (2). The B element is composed of three functional sequences, B1, B2, and B3, which are collectively essential for origin function (44) and are conserved in another origin, ARS307 (50, 59). The B1 element is important for ORC binding and additional functions in replication initiation (51, 54). The function of the B2 element is unclear. B3 is the binding site for the ABF1 protein, which, in other contexts functions as a transcriptional activator or repressor protein (6, 8, 18, 19, 46). ABF1 binding sites have been found in several other ARSs; for example, the two ABF1 binding sites at ARS121 can function as far as 1 kb from the A element (19, 65). Like enhancers in viral replication, the function of the B3 element of ARS1 in plasmid replication can be replaced by binding sites for other yeast transcription factors, such as GAL4 and RAP1 (44). However, since a large number of the sequences that function as ARSs in plasmids do not serve as replicators in their native chromosomal contexts (46), it is not known whether heterologous transcription factors can substitute for the ABF1 function in initiation of replication from the chromosomal ARS1 locus.
Acidic domains are the most extensively studied type of activation domain in eukaryotic transcription factors (60). These activation regions, including those from yeast GAL4 and GCN4, mammalian p53, and herpes simplex virus VP16, contain a significant number of negatively charged residues. One salient feature of acidic activation domains is their ability to activate transcription in cells from a variety of eukaryotic species, including yeasts, plants, and mammals (48). This finding indicates that the mechanisms employed by acidic activators to stimulate transcription are highly conserved in the eukaryotic kingdom. By analogy to transcriptional activation, several acidic activation domains, such as those from VP16 and p53, activate viral replication when tethered to the viral origins of DNA replication (13, 29, 39). However, it is unclear whether the acidic activators that activate viral DNA replication can facilitate chromosomal DNA replication as well.
In this report, we analyze the abilities of several acidic activation domains to stimulate DNA replication in S. cerevisiae. We show that acidic activation domains, when tethered to the origin of replication, can enhance replication efficiency of the ARS1 origin in the native chromosomal locus as well in a circular plasmid. The data also define the C-terminal acidic region of the yeast transcription factor ABF1 as a domain important for activating normal ARS1 function. These results strongly suggest a functional conservation of the mechanism used by acidic activators to activate DNA replication in both yeast and higher eukaryotes.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strain used in this study, BP1ΔH, is derived from BP1 (gal4::HIS4 ura3-52 leu2-2,112 his4-519 ade1) (3). The modified strain contains a replacement of the HIS4 far-upstream sequence between nucleotides 68624 and 68985 of chromosome III (coordinates from the Stanford SC database) with the sequence GGAT.
To construct the GAL4-responsive ARS1 test plasmid (pARS1/-B23/G24), an oligonucleotide containing a wild-type GAL4 binding site (5′ CGGAGTACTGTCCTCCG 3′) was inserted at the polylinker region of the plasmid --B1A (where the first and second hyphens indicate mutated B3 and B2 boxes, respectively; the corresponding wild-type construct is B3B2B1A) described previously (44). The resulting plasmid contains a linker substitution at B2, a double-point mutation at B3, and a wild-type GAL4 binding site distal to the A element. There is a 37-bp linker sequence between the mutated B3 element and the GAL4 site. The control ARS1 plasmid used for the assays shown in Fig. 1 contains a mutant GAL4 binding site (5′CTTTAGCAAGGAAGGTG3′) inserted at the same polylinker sites as the wild-type GAL4 binding site.
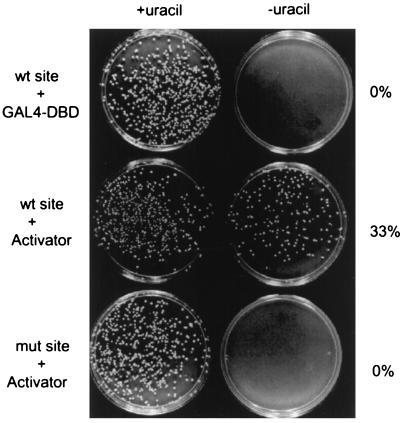
FIG. 1.
GAL4-derived transcription factors can activate ARS function in a site-dependent manner. ARS1 plasmids that carry either a wild-type (wt) (top and middle panels) or a mutant (mut) (bottom panel) GAL4 site were assayed for plasmid stability. The GAL4 DBD, either alone (top panel) or fused to a duplicate sequence from the activation domain of VP16 (middle and bottom panels), was expressed in the yeast cells. After nonselective growth for 30 h at 30°C, the cells were plated on medium with (left) or without (right) uracil. The ratio of the colony numbers on the two plates is indicative of the stability of the test plasmid. For induction of the GAL4 derivatives, a final concentration of 10 μM copper sulfate was used.
To construct the yeast strain that contained the GAL4-responsive ARS1 sequence at the ARS1 chromosomal locus, site-directed mutagenesis was used to replace the B3 element of the ARS1 sequence (5′ AATTTCGTCAAAAATGC 3′) with a GAL4 binding site (5′ CGGAGTACTGTCCTCCG 3′). In the same ARS1 sequence, the B1 and B2 elements and the sequence in between were also replaced by an Xho linker sequence. An Xba-EcoRI genomic fragment containing the modified ARS1 was then cloned into the corresponding restriction sites of the integration vector pJJ244 (32). The wild-type ARS1 sequence in the chromosome was replaced by the modified ARS1 by a “pop-in and pop-out” gene replacement method (34). The correct product of homologous recombination was verified by PCR and Southern blotting analysis.
The copper-inducible expression vector was derived from a pRS305-based expression vector described previously (57). The original GAL1 promoter was replaced with the CUP1 promoter region from nucleotides 212747 to 213179 of chromosome VIII (coordinates from the Stanford SC database) (7), and the POU domain was replaced with GAL4 amino acids 1 to 94 (the GAL4 DNA binding domain [DBD]) (58). All GAL4 fusion constructs were made by cloning the PCR fragments of various activation domains into the XbaI and BamHI sites immediately following the GAL4 DBD sequence. The β-galactosidase reporter construct contained two GAL4 binding sites (with the same sequence as shown above) in front of the HIS4 TATA sequence and the lacZ gene. The UAS of the HIS4 promoter (sequence between 68616 and 68453 of chromosome III) was deleted. The plasmid was integrated at the 3′ untranslated region of the LYS2 gene, between nucleotides 468962 and 468943 of chromosome II (coordinates from the Stanford SC database). pGAL4-2xVN8 contained DNA sequence that encoded two repeats of an eight-residue sequence from the VP16 activation domain (DFDLDMLG). The double-mutant derivatives of the GAL4-p53 fusion proteins contain either an L22Q/W23S or a W53Q/F54S mutation (9, 42).
Plasmid stability assay.
The assay was performed as described previously (44). The expression vectors and the ARS1 test plasmids were transformed separately into yeast cells by the standard lithium acetate method (34). After 30 h of growth in nonselective liquid medium, the cultures were diluted and equal numbers of cells were plated on selective (synthetic complete medium [SCM]-Leu-Ura) and nonselective (SCM-Leu) plates. Unless otherwise stated, all liquid media used contained 50 μM copper sulfate for induction of the GAL4-derived fusion proteins. The stability value for each GAL4 derivative is an average of data from at least three independent experiments, each using colonies from a separate transformation.
β-Galactosidase assay.
The transcription assay was performed and the specific activity was calculated according to a standard protocol (34). Each GAL4 derivative was tested in at least three independent experiments.
Immunoblotting.
Total cell lysates were prepared by the bead-beating method (34). The lysates were normalized for the total protein amounts and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After being transferred to a nitrocellulose membrane, the samples were probed with a monoclonal antibody against the hemagglutinin (HA) epitope (12CA5; Amersham). The immunoblots were developed with an ECL kit from Amersham.
2-D gel electrophoresis.
Genomic DNA was prepared and replication intermediates were analyzed exactly as described previously (41). Yeast cultures were grown in SCM-Leu with 100 μM copper sulfate, except for the nonselective medium used for the positive control shown in Fig. 4B. Cultures were harvested at an optical density at 600 nm of 1.0 to 1.2. A total of 500 ml of culture was used for each two-dimensional (2-D) sample.
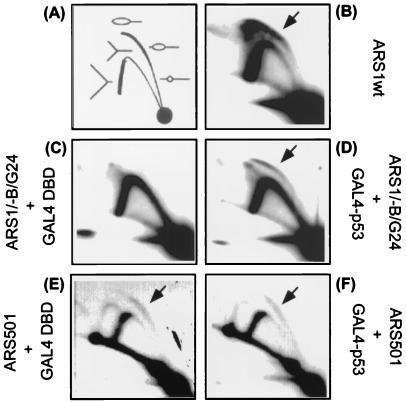
FIG. 4.
A GAL4-p53 chimeric activator stimulates replication initiated from the native chromosomal ARS1 locus. (A) Schematic diagram illustrating the patterns produced by bubble- and Y-shaped replication intermediates in the 2-D gel assay (20). See the text for more explanation. (B) Chromosomal replication intermediates from the parental wild-type ARS1 (ARS1wt) region. For the yeast cells used in panels C to F, the B elements at the ARS1 chromosomal locus were replaced with a GAL4 binding site. Either the GAL4 DBD alone (C and E) or GAL4-p53(41-60) (D and F) was expressed in these cells. A final concentration of 100 μM copper sulfate was used. Logarithmically growing cells were harvested. Yeast DNA was isolated, digested with restriction endonucleases, and enriched for replication intermediates. In panels C and D, the replication intermediates from a 5-kb NcoI-digested fragment containing the ARS1 region were identified by using a radioactive probe prepared from the same DNA fragment. The position of the bubble arc induced by the activator is indicated by arrows. As an internal control, the same membrane was stripped and reprobed with a radioactive fragment containing the ARS501 origin. Panels E and F show the replication intermediates from the ARS501 locus in the presence of GAL4 DBD alone and GAL4-p53(41-60), respectively.
RESULTS
Plasmid stability assay for studying activation of replication by transcriptional activation domains.
ARS1 in S. cerevisiae represents one of the best-characterized cellular origins in eukaryotes. The B3 element at ARS1 is the binding site for the yeast transcription factor ABF1 and can be functionally replaced by the GAL4 binding site (44). Characterization of ARS1 function largely has been done by using a plasmid stability assay (44). In this assay, yeast cells are transformed with a test plasmid containing the ARS of interest. Transformants are first grown in selective medium selecting for a marker on the plasmid, diluted into nonselective medium where the plasmid can be lost, and subsequently grown for approximately 14 generations. The percentage of yeast cells that retain the plasmid is then determined. With this assay, the wild-type ARS1 plasmid is retained in approximately 50% of yeast cells, whereas a crippled ARS1 plasmid with two mutant B elements is present in no more than 1 to 2% of yeast cells (44).
To assess the abilities of various transcriptional activation domains to stimulate ARS function, a GAL4 binding site was inserted next to a mutated B3 element. The B2 element of the same ARS1 sequence was also substituted with an 8-bp linker. Since this modified ARS1 sequence had both B2 and B3 elements inactivated, an ARS/CEN plasmid bearing the ARS1 sequence (pARS1/-B23/G24) displayed a very low stability in the absence of a GAL4-derived activator (0.6% ± 0.49%; Fig. 2C). In a separate set of constructs, transcriptional activation domains were fused to the GAL4 DBD. The N termini of the fusion proteins were also tagged with the HA epitope to facilitate their detection in yeast. The fusion gene was under the control of a copper-inducible promoter, and the construct used in this study was integrated into a chromosomal locus. Use of the inducible expression vectors alleviated the toxicity otherwise caused by high constitutive expression of some GAL4-derived activators (3). The integrated constructs also prevented possible functional interference in vivo between an expression plasmid and the test plasmid in the same yeast cells.
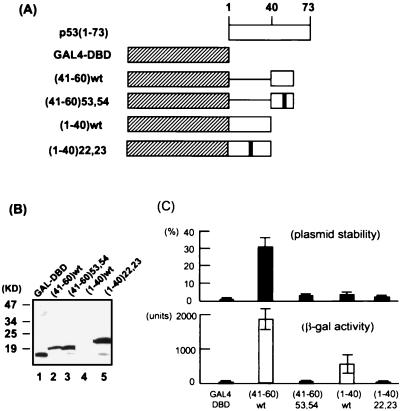
FIG. 2.
Activation of ARS function and transcription by various GAL4-p53 fusion proteins. (A) Schematic diagram of the GAL4-p53 fusion proteins. The double-mutant derivatives contain either an L22Q/W23S or a W53Q/F54S mutation. wt, wild type. (B) Expression of the GAL4 derivatives in yeast. The fusion proteins were detected by immunoblotting using a monoclonal antibody (12CA5) raised against the HA epitope that was tagged to the N terminus of each protein. A final concentration of 50 μM copper sulfate was used. (C) Comparison of the abilities of various activation domains in activation of plasmid stability (top) and transcription (bottom). The pARS1/-B23/G24 test plasmid was used for the stability assay. For the transcription assay, a GAL4-responsive β-galactosidase (β-gal) reporter construct was integrated in chromosome.
We first compared the GAL4 DBD alone with a GAL4 derivative (GAL4-2xVN8) fused to two repeats of an eight-residue sequence from the VP16 activation domain (DFDLDMLG). This chimeric activator was previously demonstrated to strongly activate yeast transcription (57). As shown in the top panel of Fig. 1, the ARS1 plasmid with a wild-type GAL4 binding site had 0% stability in the presence of the GAL4 DBD alone. In contrast, expression of the GAL4-derived activator resulted in a significant elevation of plasmid stability (33%; middle panel). This activation was dependent on GAL4 DNA binding, since the same activator had no effect on the stability of an ARS1 plasmid containing a mutant GAL4 binding site (bottom panel). This experiment confirmed and extended previous findings that the ABF1 site at ARS1 could be functionally substituted by binding sites for other yeast transcription factors. This assay also provided a convenient way to examine the potential of various transcriptional activation domains in activating ARS function.
A 20-residue peptide from the p53 activation domain can strongly activate replication and transcription.
The transcriptional activation domain of p53 has been mapped to the N-terminal 40 residues of the protein (62). However, sequences immediately following the region also contribute to p53 transcriptional activity (12). In fact, a recent study has identified a new activation domain between amino acids (aa) 40 and 83 (9). GAL4 fusions with either subdomain (aa 1 to 40 or 40 to 83) can activate transcription in both mammalian and yeast cells. We and others have shown previously that the N-terminal 73 residues of p53 activate viral DNA replication when fused to the GAL4 DBD (29, 39). To test whether the two subdomains of p53 can independently activate ARS function in yeast, the regions between residues 1 and 40 and residues 41 and 60 were fused separately to the GAL4 DBD (Fig. 3A), and the chimeric proteins were tested in both the plasmid stability assay and the β-galactosidase assay for the ability to enhance ARS1 plasmid stability and gene expression, respectively.
FIG. 3.
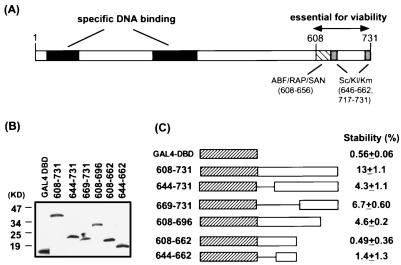
The C-terminal region of ABF1 is sufficient for activation of ARS function when fused to the GAL4 DBD. (A) Schematic diagram showing the important features of the ABF1 protein. Also indicated are the homologous regions among ABF1, RAP1, and SAN1 and those among the ABF1 proteins from S. cerevisiae (Sc), Kluyveromyces lactis (Kl), and Kluyveromyces marxianus (Km). (B) Expression of various GAL4-ABF1 fusion proteins in yeast. A final concentration of 50 μM copper sulfate was used. The proteins were detected by immunoblotting using the HA antibody. (C) Abilities of various GAL4 fusion proteins to activate the stability of the GAL4-responsive ARS1 plasmid (pARS1/-B23/G24). Cells were grown in nonselective medium at 30°C for 30 h before plating on nonselective and selective plates.
As shown in the upper panel of Fig. 2C, the GAL4 DBD alone supported a very low plasmid stability (0.67% ± 0.49%). In contrast, the GAL4-p53(41-60) fusion protein significantly stimulated the stability of the test plasmid (30% ± 7.8%). Activation was dependent on the wild-type GAL4 binding site engineered at the ARS1 origin (data not shown). In a parallel study, GAL4-p53(41-60) also strongly activated transcription from a GAL4-responsive β-galactosidase reporter gene (1,868 ± 305 U versus 12.4 ± 10.5 U by the GAL4 DBD alone; lower panel of Fig. 2C). It has been shown previously that mutations at residues Trp-53 and Phe-54 of p53 abolish the transcriptional activity of the second subdomain (9). When the same mutations were introduced into the GAL4-p53(41-60) fusion protein, they abrogated activation of both plasmid stability and transcription in yeast (3.3% ± 0.98% and 12.4 ± 5.7 U, respectively; Fig. 2C). Although we could not exclude the possibility that the double mutations changed the global structure of the 20-aa region of p53, the data were consistent with the notion that the same amino acid residues were used to activate both DNA replication and transcription. Taken together, these results strongly suggest that the p53 region between residues 41 and 60 contains a potent activation domain for yeast transcription and ARS function.
Given the potency of the first p53 subdomain in activating mammalian and yeast transcription (9, 62), it was of interest to determine whether it had a similar stimulatory effect on ARS function. Despite repeated efforts, we were unable to detect the GAL4-p53(1-40) protein from the yeast lysate in an immunoblotting assay (Fig. 2B, lane 4). However, expression of this protein did yield a significant increase in β-galactosidase activity in the transcription assay compared to the GAL4 DBD alone (456 ± 241 U versus 12 ± 10 U), whereas little stimulation could be observed in the plasmid stability assay (3.5% ± 1.5%; Fig. 2C). This result suggested that a low level of expression of the activator was sufficient for transcriptional activation but not for replication. Since the lack of activation of ARS function might be due to the relatively low sensitivity of the plasmid stability assay, it was not possible to determine whether the first subdomain of p53 had an inherent ability to activate replication. Mutations at residues 22 and 23 previously have been shown to abrogate the transcriptional activity of p53 (42). Strikingly, GAL4-p53(1-40) with mutations at these two positions was present in abundance in the yeast lysate (Fig. 2B, lane 5), which was a further indication that the wild-type GAL4-p53(1-40) might be growth inhibitory. However, the mutant protein did not stimulate either replication or transcription (2.1% ± 0.7% or 6.7 ± 1.9 U, respectively; Fig. 2C).
While the low protein level of GAL4-p53(1-40) in yeast complicated the study of its potential in activating ARS function, it is of interest that an almost identical fusion protein has been shown to undergo rapid degradation in mammalian cells (28). In the latter case, the oncoprotein Mdm2 binds the first 42 residues of p53 and promotes p53 degradation. Furthermore, mutations at residues 22 and 23 that disrupt Mdm2 binding also prevent Mdm2-dependent degradation (28, 37). The same mutations also dramatically increase the protein levels of the GAL4-p53(1-40) fusion protein in yeast, which raises an interesting possibility that this region of p53 can bind a yeast protein with a function similar to that of the Mdm2 protein in mammalian cells. Alternatively, strong heterologous activation domains that are toxic to yeast cell growth may be preferentially targeted by the protein degradation machinery.
The C-terminal region of ABF1 is important for activation of ARS function.
Having characterized heterologous activation domains of DNA replication, we next determined what type of protein domain normally activated initiation of replication at the ARS1 origin. Yeast ABF1 is a multifunctional protein that has been implicated in replication, transcriptional activation, and mating-type silencing (8). The full-length 731-residue protein can be divided into two regions (Fig. 3A): the first 530 residues are sufficient for specific DNA binding (27), and the remainder of the ABF1 protein is essential for cell viability (53). Although the function of the highly negatively charged C-terminal region is not well understood, it has been proposed to act as an acidic activation domain in stimulating both replication and transcription (22). To test this hypothesis, we fused the GAL4 DBD to the C-terminal 123 aa of ABF1. As shown in Fig. 3C, this chimeric protein (608-731) clearly enhanced the stability of the GAL4-responsive ARS1 test plasmid, indicating that the C-terminal portion of ABF1 was sufficient for stimulating ARS function when tethered to the origin. However, when tested in the transcriptional activation assay, the same fusion protein yielded only 55 ± 17 U of β-galactosidase activity, in comparison with 1,868 ± 305 U for GAL-p53(41-60) and 12 ± 10 U for the GAL4 DBD alone. Thus, despite the fact that the C-terminal domain of ABF1 has a much higher net negative charge than the p53 activation domain (residues 41 to 60), it is a much weaker transcriptional activator under the conditions used in these experiments.
In an attempt to determine which part of the ABF activation domain was required for stimulating ARS function, a series of deletion mutants was generated from either the N- or C-terminal end of the domain. All mutant proteins were expressed in yeast at levels similar to that of the wild-type protein (Fig. 3B). Deletion of the first 36 aa from the N terminus of the region (Fig. 3B, 644-731) reduced the ability of the GAL4-ABF1 fusion protein to activate ARS function, although the remaining portion still contained some residual activity. Removal of the last 35 residues from the C terminus (608-696) also resulted in a partial decrease in activity, and further truncation (608-662) completely abolished activity. Therefore, both the N (608-644) and C (662-731) termini of the ABF1 activation domain appeared to contribute to stimulation of plasmid stability. This is reminiscent of previous findings that many acidic activation domains contain multiple subdomains that collectively activate transcription (31, 43).
A GAL4-derived activator can stimulate initiation of DNA replication at the ARS1 chromosomal locus.
The plasmid stability assay has been valuable in dissecting the multiple genetic elements of the ARS1 sequence. All four cis elements that are involved in plasmid replication have been shown to contribute to initiation of DNA replication in their normal locations in the chromosome (44, 45). Plasmid stability, however, is affected both by the efficiency of its replication and by the efficiency of its segregation or nuclear retention. Thus, results from plasmid stability assays may not always reflect the contributions of cis-acting DNA sequences to the efficiency of initiation of DNA replication in the chromosome (46). When the ARS1 test plasmid was isolated from yeast cells at the completion of the plasmid stability assay and analyzed by DNA hybridization, there was a net increase in the total amount of plasmid DNA from the yeast population that expressed a GAL4-derived activator (data not shown). This finding supports the notion that replication efficiency of the test plasmid was indeed stimulated by the acidic activators.
To test whether GAL4-derived activators activate ARS1 replication in a chromosomal context, we used a 2-D gel electrophoresis technique to detect replication intermediates initiated from various derivatives of the chromosomal ARS1 (4). In this assay, chromosomal DNA from asynchronously growing cells was isolated and digested with a restriction endonuclease, and then the DNA fragments were separated by mass in the first dimension and subsequently by both mass and shape in the second dimension. Replication intermediates were separated from linear DNA fragments, forming arcs in the gel. Southern blotting analysis was then used to distinguish the following two types of replication intermediates: those origin-containing fragments with a characteristic bubble arc, and those that did not contain an origin but did contain a replication fork and migrated as a Y arc (Fig. 4A). A previous study has shown that simultaneous disruption of the three B elements at the ARS1 locus obliterates the bubble arc in the 2-D gel analysis, whereas removing only one or two functional B elements weakens but does not abolish the bubble arc (45). Since it is technically difficult to quantitate small differences in efficiency of origin firing by the 2-D gel method, we reasoned that a low baseline of replication in the absence of all three B elements might facilitate detection of any stimulatory effects by the GAL4-derived activators. Therefore, the three B elements at the ARS1 locus in chromosome IV were replaced with a single GAL4 binding site by homologous recombination.
The origin activity of the chromosomal ARS1 derivative was analyzed after digestion of the DNA to produce a 5-kb NcoI fragment that contained the ARS1 sequence. As shown in Fig. 4B, the yeast strain that contained the wild-type ARS1 gave rise to a strong bubble arc in addition to a Y arc. In keeping with previous findings (45), replacement of the B elements with the GAL4 site (-B1/-B2/-B3/G24) resulted in loss of the bubble arc, indicating loss of origin function in the absence of a GAL4 activator (Fig. 4C). The prominent Y arc detected in Fig. 4C is characteristic of fragments that were replicated exclusively by origins outside the fragment. Strikingly, expression of the GAL4-p53(41-60) activator in this strain resulted in a clear bubble arc (Fig. 4D), albeit weaker than that from the wild-type ARS1 sequence (Fig. 4B). The fact that origin function was only partially restored by the GAL4-derived activator was not unexpected, since the crippled ARS1 sequence lacked multiple B elements and GAL4-p53 might rescue function of only the B3 element. Indeed, the 2-D gel pattern shown in Fig. 4D was similar to that of the chromosomal ARS1 sequence with a single functional B element (45). As an internal control, the blot probed with the ARS1 fragment (Fig. 4C and D) was stripped and reprobed with a fragment encompassing the ARS501 region. In contrast to the ARS1 derivative, initiation of replication from ARS501 was not affected by the activator (Fig. 4E and F). Together with the stability assay, these data unequivocally establish that acidic activators can stimulate a cellular origin of replication in both the plasmid and chromosomal context.
DISCUSSION
Activation of DNA replication and transcription by acidic activation domains have many common characteristics. First, acidic activation domains are well known for their ability to activate transcription in a variety of eukaryotes (48). We and others have shown that the same group of activation domains can also activate chromosomal DNA replication in yeast as well as viral DNA replication in mammalian cells (this study and references 13, 29, and 39). Second, a small and well-defined peptide of VP16 and p53 activation domains is sufficient for stimulating both transcription and DNA replication. Third, the potency of an acidic activation domain in transcriptional activation in general correlates with that in activation of DNA replication (e.g., VP16 and p53 are stronger than ABF1). Finally, mutations in the acidic domains that abolish transcriptional activation also destroy their function in DNA replication. For instance, the GAL4-p53(41-60) fusion protein with mutations at residues Trp-53 and Phe-54 fails to activate yeast replication and transcription in our assays. In addition, it has been shown that mutations at the aromatic residue Phe-442 of the VP16 activation domain abrogate GAL4-VP16’s ability to activate transcription and polyomavirus DNA replication (14, 29). Taken together, these studies strongly suggest that some enhancers of replication and transcription may functionally overlap. Furthermore, there may be a conserved mechanism that the acidic activation domains utilize to activate DNA replication in yeast and higher eukaryotes.
Wiltshire et al. recently reported that a 50-aa C-terminal region of the ABF1 protein (aa 635 to 684) was sufficient to stimulate ARS121 function when tethered to the origin (67). This region partially overlaps one of the two regions that are shown in this study to activate ARS1 function. Unlike ARS1, activation of ARS121 by ABF1 cannot be replaced by other transcription factors (67), suggesting a special function of ABF1 at ARS121. Compared to the VP16 and p53 activation domains, the C-terminal region of ABF1 contains a relatively weak transcriptional activation domain. This finding is consistent with the previous observation that ABF1 activates transcription synergistically with other weak activators (5). Furthermore, a LexA-ABF1 fusion protein stimulates transcription only from a LexA-responsive promoter that contains other cis-regulatory elements (21). Therefore, transcriptional activation by ABF1 may require cooperation from other transcriptional activators that bind to the same promoter region. It is noteworthy that the 123-aa C-terminal region of ABF1 has a much higher net negative charge (−25) than the 20-aa region from the p53 activation domain (−7), yet the latter is a much stronger activator for both transcription and replication. It thus appears that the number of acidic residues in an activation domain may not be the primary determinant for the potency of activation of replication, just as previously shown for transcriptional activation (14, 42, 61). Rather, hydrophobic residues in the acidic activation domains may play a more critical role in activation of replication and transcription.
Intense work in the past few years has led to a better understanding of the mechanism(s) used by acidic activators to activate transcription. Acidic domains such as those of VP16 and p53 have been shown to contact several general factors in the transcriptional machinery in vitro, suggesting multiple pathways utilized by these domains to activate transcription (49). More recent studies have also demonstrated that acidic activators can counteract the nucleosomal repression of transcription and that the antirepression function may be mediated by several chromatin remodeling systems (23, 35, 47, 56). Compared with the studies of transcriptional activation, much less is known concerning the molecular basis for activation of DNA replication by acidic activators. The activation domains of VP16 and p53 interact with replication protein A (RPA), an essential component of the replication machinery (29, 39). Furthermore, mutations that disrupt the interaction between the acidic activation domains and RPA also abolish activation of DNA replication (29). In particular, Trp-53 and Phe-54 of p53, shown in this study to be critical for activation of replication, are important for p53 binding to RPA as well (38), suggesting recruitment of RPA to the origin as one potential mechanism for activation of replication by the acidic domains. However, other mechanisms for enhancing initiation of DNA replication are also possible. For example, in vitro biochemical studies have shown that the activation domains of VP16 and p53 can antagonize the nucleosomal repression of viral DNA replication (13, 40), thus raising the possibility that these transcription factors use similar strategies to overcome the nucleosomal repression in replication and transcription. Because the plasmid stability assay can offer powerful genetic screening possibilities in yeast, it should be feasible to examine more closely the proteins that interact with the activation domains to enhance DNA replication.
ACKNOWLEDGMENTS
We thank Leonard Guarente for the BP1 strain, Arnold Levine for the p53 mutant at residues 22 and 23, Yanfen Hu for help with plasmid construction, and Chun Liang for help with the 2-D gel analysis. R.L. is also grateful to Joyce Hamlin for helpful discussion and encouragement during the course of the work.
This work was supported in part by a Special Fellow Award from Leukemia Society of America, an ACS institutional research grant, and a start-up research fund from the University of Virginia (R.L.) and by funds from NIH grant RO1GM45436 to B.S.
REFERENCES
- 1.Baru M, Shlissel M, Manor H. The yeast GAL4 protein transactivates the polyomavirus origin of DNA replication in mouse cells. J Virol. 1991;65:3496–3503. doi: 10.1128/jvi.65.7.3496-3503.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 4.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 5.Buchman A R, Kornberg R D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990;10:887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchman A R, Lue N F, Kornberg R D. Connections between transcriptional activators, silencers, and telomers as revealed by functional anaylsis of a yeast DNA-binding protein. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butt T R, Sternberg E J, Gorman J A, Clark P, Hamer D, Rosenberg M, Crooke S T. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci USA. 1984;81:3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell J L, Newlon C S. Chromosomal DNA replication. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 41–146. [Google Scholar]
- 9.Candau R, Scolnick D M, Darpino P, Ying C Y, Halazonetis T D, Berger S L. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 10.Celniker S E, Sweder K, Spienc F, Bailey J E, Campbell J L. Deletion mutations affecting automonously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challberg M D, Kelly T J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- 12.Chang J, Kim D H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 13.Cheng L, Workman J L, Kingston R E, Kelly T J. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4-VP16. Proc Natl Acad Sci USA. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis M L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993;3:1161–1163. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 16.DePamphilis M L. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 17.DeVilliers J, Schaffner W, Tyndall C, Lupton S, Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984;312:242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- 18.Diffley J F X, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenberg S, Civalier C, Tye B-K. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc Natl Acad Sci USA. 1988;85:743–746. doi: 10.1073/pnas.85.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson B M, Brewer B J, Reynolds A E, Fangman W L. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- 21.Gonçalves P M, Maurer K, Amerongen G N, Bergkamp-Steffens K, Mager W H, Planta R J. C-terminal domains of general regulatory factors Abf1p and Rap1p in Saccharomyces cerevisiae display functional similarity. Mol Microbiol. 1996;19:535–543. doi: 10.1046/j.1365-2958.1996.404939.x. [DOI] [PubMed] [Google Scholar]
- 22.Gonçalves P M, Maurer K, Mager W H, Planta R J. Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res. 1992;20:2211–2215. doi: 10.1093/nar/20.9.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem. 1995;20:517–561. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 24.Guo Z-S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Z-S, Gutierrez C, Heine V, Sogo J M, DePamphilis M L. Origin auxiliary sequences can facilitate initiation of simian virus 40 DNA replication in vitro as they do in vivo. Mol Cell Biol. 1989;9:3593–3602. doi: 10.1128/mcb.9.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez L, Guo Z-S, Roberts J, DePamphilis M L. Simian virus 40 origin auxiliary sequences weakly facilitate T-antigen binding, but strongly facilitate DNA unwinding. Mol Cell Biol. 1990;10:1719–1728. doi: 10.1128/mcb.10.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halfter H, Kaverty B, Vandekerckhove J, Kiefer F, Gallwitz D. Sequence, expression and mutational analysis of BAF-1, a transcriptional activator and ARS-1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:4265–4272. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. The transcription protein VP16 and GAL4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 30.Heintz N H. Transcription factors and the control of DNA replication. Curr Opin Cell Biol. 1992;4:459–467. doi: 10.1016/0955-0674(92)90012-2. [DOI] [PubMed] [Google Scholar]
- 31.Hope I A, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 32.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 33.Jones K A, Kadonaga J T, Rosenfeld P J, Kelly T J, Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987;48:79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 35.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1992. [Google Scholar]
- 37.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 38.Leiter L M, Chen J, Marathe T, Tanaka M, Dutta A. Loss of transactivation and transrepression function, and not RPA binding, alters growth suppression by p53. Oncogene. 1996;12:2661–2668. [PubMed] [Google Scholar]
- 39.Li R, Botchan M R. The acidic transcription activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 40.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 43.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 44.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 45.Marahrens Y, Stillman B. Replicator dominance in a eukaryotic chromosome. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newlon C S, Theis J F. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 47.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcriptin by RNA polymerase II. Annu Rev Biochem. 1994;63:265–97. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 48.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 49.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 50.Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhode P R, Elsasser S, Campbell J L. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowley A, Cocker J H, Harwood J, Diffley J F X. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 56.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka M. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka M, Clouston W M, Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theis J F, Newlon C S. Domain B of ARS307 is modular and contributes to chromosomal replication origin function. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 61.Uesugi M, Nyanguile O, Lu H, Levine A, Verdine G L. Induced a helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 62.Unger T, Nau M M, Segal S, Minna I D. p53: a transdominant regulator of transcription whose function is ablated by mutations occurring in human cancer. EMBO J. 1992;11:1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van der Vliet P C. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 87–118. [Google Scholar]
- 65.Walker S S, Francesconi S C, Eisenberg S. A DNA replication enhancer in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wasylyk C, Schneikert J, Wasylyk B. Oncogene v-jun modulates DNA replication. Oncogene. 1990;5:1055–1058. [PubMed] [Google Scholar]
- 67.Wiltshire S, Raychaudhuri S, Eisenberg S. An Abf1p C-terminal region lacking transcriptional activation potential stimulates a yeast origin of replication. Nucleic Acids Res. 1997;25:4250–4256. doi: 10.1093/nar/25.21.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang L, Botchan M. Replication of bovine papillomavirus type 1 DNA initiates within an E2-responsive enhancer element. J Virol. 1990;64:5903–5911. doi: 10.1128/jvi.64.12.5903-5911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]